Submitted:
23 August 2023
Posted:
25 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Inflammatory Bowel Disease
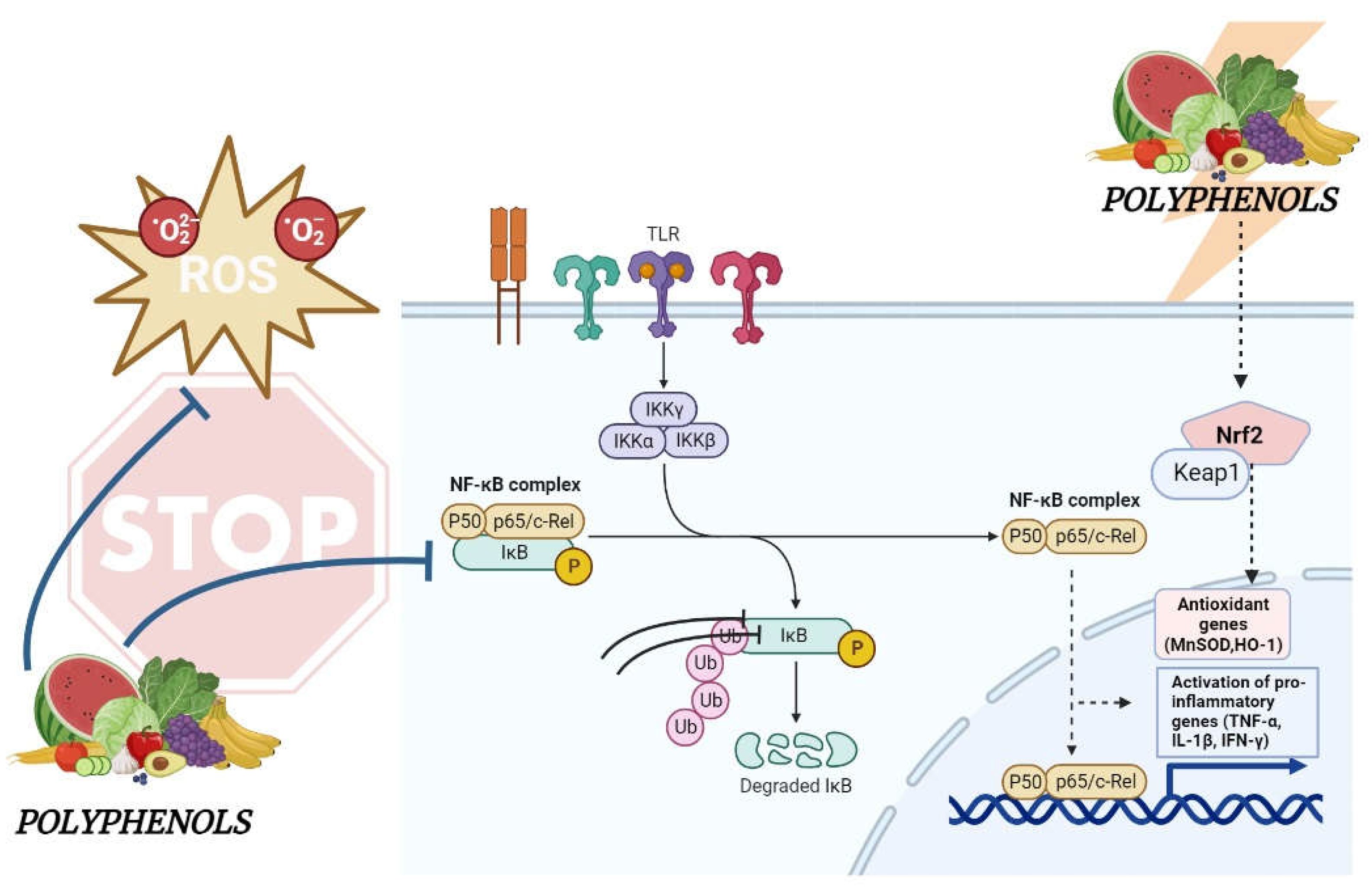
3. Potential Beneficial Effects of Dietary Polyphenols in IBD
4. Molecular Mechanisms Underlying the Effects of Dietary Polyphenols in IBD
5. Foodomics Approach to Investigate the Relationship between Food and Health
6. Genomics Approach
7. Transcriptomics Approach
8. Proteomics Approach
9. Metabolomic Approach
10. Trimethylamine-N-Oxide: A Novel Metabolomics-Based Biomarker of Inflammation and IBD Diagnosis
11. Gastrointestinal Protective Effects Mediated by Polyphenols: Foodomics-Based Approaches
12. Low Bioavailability and Bioaccessibility, Appropriate Doses and Side Effects of Polyphenols
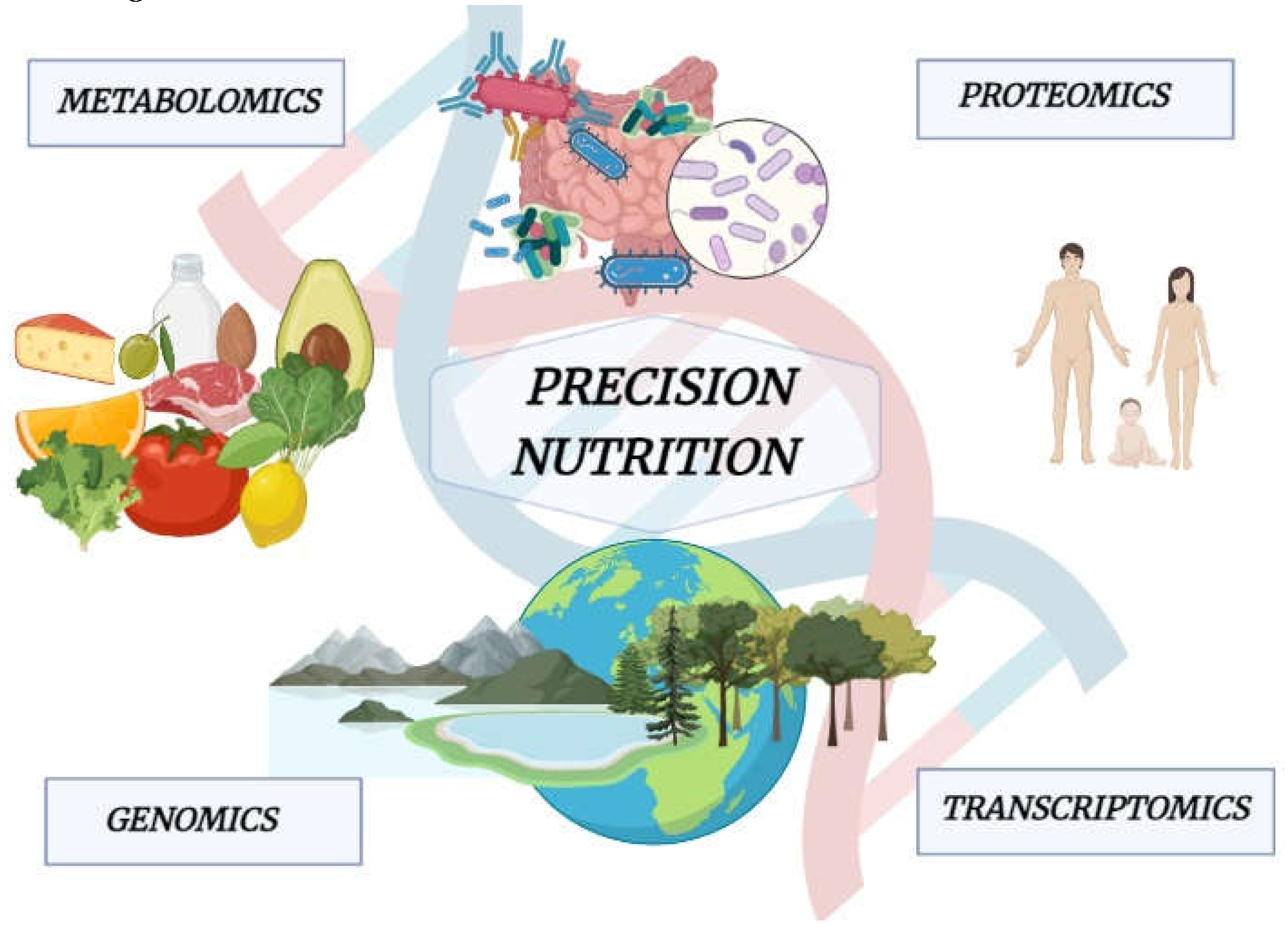
13. Precision Nutrition and IBD Prevention and Treatment
14. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Caruso, R.; Warner, N.; Inohara, N.; Núñez, G. NOD1 and NOD2: Signaling, Host Defense, and Inflammatory Disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, M. Leaky gut: Mechanisms, measurement and clinical implications in humans. Gut 2019, 68, 1516–1526. [Google Scholar] [CrossRef]
- Bibbò, S.; Ianiro, G.; Giorgio, V.; Scaldaferri, F.; Masucci, L.; Gasbarrini, A.; Cammarota, G. The role of diet on gut microbiota composition. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4742–4749. [Google Scholar] [PubMed]
- Zhang, Y.; Zhang, J.; Duan, L. The role of microbiota-mitochondria crosstalk in pathogenesis and therapy of intestinal diseases. Pharmacol. Res. 2022, 186, 106530. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. , Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef]
- Hughes, R.L.; Holscher, H.D. Fueling Gut Microbes: A Review of the Interaction between Diet, Exercise, and the Gut Microbiota in Athletes. Adv. Nutr. Int. Rev. J. 2021, 12, 2190–2215. [Google Scholar] [CrossRef]
- Ding, J.; Ouyang, R.; Zheng, S.; Wang, Y.; Huang, Y.; Ma, X.; Zou, Y.; Chen, R.; Zhuo, Z.; Li, Z.; et al. Effect of Breastmilk Microbiota and Sialylated Oligosaccharides on the Colonization of Infant Gut Microbial Community and Fecal Metabolome. Metabolites 2022, 12, 1136. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; Khalili, H.; Konijeti, G.G.; Higuchi, L.M.; de Silva, P.; Korzenik, J.R.; Fuchs, C.S.; Willett, W.C.; Richter, J.M.; Chan, A.T. , A prospective study of long-term intake of dietary fiber and risk of Crohn's disease and ulcerative colitis. Gastroenterology 2013, 145, 970–977. [Google Scholar] [CrossRef]
- Fernando, M.R.; Saxena, A.; Reyes, J.-L.; McKay, D.M. Butyrate enhances antibacterial effects while suppressing other features of alternative activation in IL-4-induced macrophages. Am. J. Physiol. Liver Physiol. 2016, 310, G822–G831. [Google Scholar] [CrossRef] [PubMed]
- Albenberg, L.G.; Wu, G.D. Diet and the Intestinal Microbiome: Associations, Functions, and Implications for Health and Disease. Gastroenterology 2014, 146, 1564–1572. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. , Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef]
- Wlodarska, M.; Luo, C.; Kolde, R.; D’hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Sonnenburg, J.L. Eating For Two: How Metabolism Establishes Interspecies Interactions in the Gut. Cell Host Microbe 2011, 10, 336–347. [Google Scholar] [CrossRef]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef]
- Hosomi, K.; Kiyono, H.; Kunisawa, J. Fatty acid metabolism in the host and commensal bacteria for the control of intestinal immune responses and diseases. Gut Microbes 2019, 11, 276–284. [Google Scholar] [CrossRef]
- Hartel, J.C.; Merz, N.; Grösch, S. How sphingolipids affect T cells in the resolution of inflammation. Front. Pharmacol. 2022, 13, 1002915. [Google Scholar] [CrossRef]
- Duncan, S.H.; Louis, P.; Thomson, J.M.; Flint, H.J. The role of pH in determining the species composition of the human colonic microbiota. Environ. Microbiol. 2009, 11, 2112–2122. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, N.; Gasbarrini, A.; Ponziani, F.R. Intestinal Barrier in Human Health and Disease. Int. J. Environ. Res. Public Health 2021, 18, 12836. [Google Scholar] [CrossRef] [PubMed]
- Tamburini, B.; La Manna, M.P.; La Barbera, L.; Mohammadnezhad, L.; Badami, G.D.; Azgomi, M.S.; Dieli, F.; Caccamo, N. Immunity and Nutrition: The Right Balance in Inflammatory Bowel Disease. Cells 2022, 11, 455. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, F.; Ferrante, A.; Guggino, G.; Triolo, G. The role of the gastrointestinal tract in the pathogenesis of rheumatic diseases. Best Pr. Res. Clin. Rheumatol. 2016, 30, 889–900. [Google Scholar] [CrossRef]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Yu, Y.R.; Rodriguez, J.R. Clinical presentation of Crohn’s, ulcerative colitis, and indeterminate colitis: Symptoms, extraintestinal manifestations, and disease phenotypes. Semin. Pediatr. Surg. 2017, 26, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Gastroenterology, H.N.A.S.F.P.; America, C.F.O.; Bousvaros, A.; A Antonioli, D.; Colletti, R.B.; Dubinsky, M.C.; Glickman, J.N.; Gold, B.D.; Griffiths, A.M.; Jevon, G.P.; et al. Differentiating Ulcerative Colitis from Crohn Disease in Children and Young Adults: Report of a Working Group of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the Crohn's and Colitis Foundation of America. J. Pediatr. Gastroenterol. Nutr. 2007, 44, 653–674. [Google Scholar] [CrossRef]
- Segal, J.P.; LeBlanc, J.-F.; Hart, A.L. Ulcerative colitis: An update. Clin. Med. 2021, 21, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Hibi, T.; Ogata, H. Novel pathophysiological concepts of inflammatory bowel disease. J. Gastroenterol. 2006, 41, 10–16. [Google Scholar] [CrossRef]
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. , Ulcerative colitis. Nat. Rev. Dis. Primers 2020, 6, 74. [Google Scholar] [CrossRef]
- Petronis, A.; Petroniene, R. Epigenetics of inflammatory bowel disease. Gut 2000, 47, 302–306. [Google Scholar] [CrossRef]
- Ji, Y.; Yang, Y.; Sun, S.; Dai, Z.; Ren, F.; Wu, Z. Insights into diet-associated oxidative pathomechanisms in inflammatory bowel disease and protective effects of functional amino acids. Nutr. Rev. 2022, 81, 95–113. [Google Scholar] [CrossRef]
- Emanuele, S.; D’anneo, A.; Calvaruso, G.; Cernigliaro, C.; Giuliano, M.; Lauricella, M. The Double-Edged Sword Profile of Redox Signaling: Oxidative Events As Molecular Switches in the Balance between Cell Physiology and Cancer. Chem. Res. Toxicol. 2018, 31, 201–210. [Google Scholar] [CrossRef]
- Dodson, M.; Castro-Portuguez, R.; Zhang, D.D. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019, 23, 101107. [Google Scholar] [CrossRef]
- Li, H.; Christman, L.M.; Li, R.; Gu, L. Synergic interactions between polyphenols and gut microbiota in mitigating inflammatory bowel diseases. Food Funct. 2020, 11, 4878–4891. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.A.; Hennet, T. Mechanisms and consequences of intestinal dysbiosis. Cell. Mol. Life Sci. 2017, 74, 2959–2977. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Kamada, N. Host-microbial Cross-talk in Inflammatory Bowel Disease. Immune Netw. 2017, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Khorsand, B.; Aghdaei, H.A.; Nazemalhosseini-Mojarad, E.; Nadalian, B.; Nadalian, B.; Houri, H. Overrepresentation of Enterobacteriaceae and Escherichia coli is the major gut microbiome signature in Crohn’s disease and ulcerative colitis; a comprehensive metagenomic analysis of IBDMDB datasets. Front. Cell. Infect. Microbiol. 2022, 12, 1015890. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, M.; Zirnbauer, R.; Schlager, S.; Mertens, D.; Gasche, N.; Sladek, B.; Herbold, C.; Bochkareva, O.; Emelianenko, V.; Vogelsang, H.; et al. coli are associated with disease activity in ulcerative colitis. Gut Microbes 2022, 14, 2143218. [Google Scholar] [CrossRef] [PubMed]
- Chervy, M.; Sivignon, A.; Dambrine, F.; Buisson, A.; Sauvanet, P.; Godfraind, C.; Allez, M.; Le Bourhis, L.; The Remind Group; Barnich, N. ; et al. Epigenetic master regulators HDAC1 and HDAC5 control pathobiont Enterobacteria colonization in ileal mucosa of Crohn’s disease patients. Gut Microbes 2022, 14, 2127444. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Gu, Y.; Jiang, T.; Che, H. Inhibition effect of PPAR-gamma signaling on mast cell-mediated allergic inflammation through down-regulation of PAK1/ NF-kappaB activation. Int. Immunopharmacol. 2022, 108, 108692. [Google Scholar] [CrossRef]
- Wang, L.; Hu, Y.; Song, B.; Xiong, Y.; Wang, J.; Chen, D. Targeting JAK/STAT signaling pathways in treatment of inflammatory bowel disease. Inflamm. Res. 2021, 70, 753–764. [Google Scholar] [CrossRef]
- Tsopmejio, I.S.N.; Yuan, J.; Diao, Z.; Fan, W.; Wei, J.; Zhao, C.; Li, Y.; Song, H. Auricularia polytricha and Flammulina velutipes reduce liver injury in DSS-induced Inflammatory Bowel Disease by improving inflammation, oxidative stress, and apoptosis through the regulation of TLR4/NF-κB signaling pathways. J. Nutr. Biochem. 2023, 111, 109190. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Wang, S.; Li, J. Treatment of Inflammatory Bowel Disease: A Comprehensive Review. Front. Med. 2021, 8, 2681. [Google Scholar] [CrossRef] [PubMed]
- Vulliemoz, M.; Brand, S.; Juillerat, P.; Mottet, C.; Ben-Horin, S.; Michetti, P. TNF-Alpha Blockers in Inflammatory Bowel Diseases: Practical Recommendations and a User’s Guide: An Update. Digestion 2020, 101, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Piodi, L.P.; Poloni, A.; Ulivieri, F.M. , Managing osteoporosis in ulcerative colitis: Something new? World J. Gastroenterol. 2014, 20, 14087–14098. [Google Scholar] [CrossRef] [PubMed]
- Park, K.T.; Bass, D. , Inflammatory bowel disease-attributable costs and cost-effective strategies in the United States: A review. Inflamm. Bowel. Dis. 2011, 17, 1603–1609. [Google Scholar] [CrossRef]
- Tsao, R. Chemistry and Biochemistry of Dietary Polyphenols. Nutrients 2010, 2, 1231–1246. [Google Scholar] [CrossRef]
- Rudrapal, M.; Khairnar, S.J.; Khan, J.; Bin Dukhyil, A.; Ansari, M.A.; Alomary, M.N.; Alshabrmi, F.M.; Palai, S.; Deb, P.K.; Devi, R. Dietary Polyphenols and Their Role in Oxidative Stress-Induced Human Diseases: Insights Into Protective Effects, Antioxidant Potentials and Mechanism(s) of Action. Front. Pharmacol. 2022, 13, 806470. [Google Scholar] [CrossRef]
- Niwano, Y.; Kohzaki, H.; Shirato, M.; Shishido, S.; Nakamura, K. Metabolic Fate of Orally Ingested Proanthocyanidins through the Digestive Tract. Antioxidants 2022, 12, 17. [Google Scholar] [CrossRef]
- Grosso, G.; Godos, J.; Lamuela-Raventos, R. ; Ray, S.; Micek, A.; Pajak, A.; Sciacca, S.; D'Orazio, N.; Del Rio, D.; Galvano, F. A comprehensive meta-analysis on dietary flavonoid and lignan intake and cancer risk: Level of evidence and limitations. Mol. Nutr. Food Res. 2016, 61. [Google Scholar] [CrossRef]
- De Blasio, A.; D’anneo, A.; Lauricella, M.; Emanuele, S.; Giuliano, M.; Pratelli, G.; Calvaruso, G.; Carlisi, D. The Beneficial Effects of Essential Oils in Anti-Obesity Treatment. Int. J. Mol. Sci. 2021, 22, 11832. [Google Scholar] [CrossRef] [PubMed]
- Pratelli, G.; Di Liberto, D.; Carlisi, D.; Emanuele, S.; Giuliano, M.; Notaro, A.; De Blasio, A.; Calvaruso, G.; D’anneo, A.; Lauricella, M. Hypertrophy and ER Stress Induced by Palmitate Are Counteracted by Mango Peel and Seed Extracts in 3T3-L1 Adipocytes. Int. J. Mol. Sci. 2023, 24, 5419. [Google Scholar] [CrossRef] [PubMed]
- Pratelli, G.; Carlisi, D.; D’anneo, A.; Maggio, A.; Emanuele, S.; Piccionello, A.P.; Giuliano, M.; De Blasio, A.; Calvaruso, G.; Lauricella, M. Bio-Waste Products of Mangifera indica L. Reduce Adipogenesis and Exert Antioxidant Effects on 3T3-L1 Cells. Antioxidants 2022, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- Rienks, J.; Barbaresko, J.; Oluwagbemigun, K.; Schmid, M.; Nöthlings, U. Polyphenol exposure and risk of type 2 diabetes: Dose-response meta-analyses and systematic review of prospective cohort studies. Am. J. Clin. Nutr. 2018, 108, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Grosso, G.; Stepaniak, U.; Micek, A.; Stefler, D.; Bobak, M.; Pająk, A. Dietary polyphenols are inversely associated with metabolic syndrome in Polish adults of the HAPIEE study. Eur. J. Nutr. 2016, 56, 1409–1420. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zamora-Ros, R.; Chan, S.; Cross, A.J.; Ward, H.; Jakszyn, P.; Luben, R.; Opstelten, J.L.; Oldenburg, B.; Hallmans, G.; et al. Dietary Polyphenols in the Aetiology of Crohn's Disease and Ulcerative Colitis-A Multicenter European Prospective Cohort Study (EPIC). Inflamm. Bowel. Dis. 2017, 23, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Larussa, T.; Oliverio, M.; Suraci, E.; Greco, M.; Placida, R.; Gervasi, S.; Marasco, R.; Imeneo, M.; Paolino, D.; Tucci, L.; et al. Oleuropein Decreases Cyclooxygenase-2 and Interleukin-17 Expression and Attenuates Inflammatory Damage in Colonic Samples from Ulcerative Colitis Patients. Nutrients 2017, 9, 391. [Google Scholar] [CrossRef]
- Vezza, T.; Algieri, F.; Rodríguez-Nogales, A.; Garrido-Mesa, J.; Utrilla, M.P.; Talhaoui, N.; Gómez-Caravaca, A.M.; Segura-Carretero, A.; Rodríguez-Cabezas, M.E.; Monteleone, G.; et al. Immunomodulatory properties of Olea europaea leaf extract in intestinal inflammation. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Daniel, K.; Vitetta, L.; Singh, M.A.F. Effects of olives and their constituents on the expression of ulcerative colitis: A systematic review of randomised controlled trials. Br. J. Nutr. 2021, 127, 1153–1171. [Google Scholar] [CrossRef]
- Singla, V.; Pratap Mouli, V.; Garg, S.K.; Rai, T.; Choudhury, B.N.; Verma, P.; Deb, R.; Tiwari, V.; Rohatgi, S.; Dhingra, R.; et al. Induction with NCB-02 (curcumin) enema for mild-to-moderate distal ulcerative colitis - a randomized, placebo-controlled, pilot study. J. Crohns Colitis 2014, 8, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.; Salomon, N.; Wu, J.C.; Kopylov, U.; Lahat, A.; Har-Noy, O.; Ching, J.Y.; Cheong, P.K.; Avidan, B.; Gamus, D.; et al. Curcumin in Combination With Mesalamine Induces Remission in Patients With Mild-to-Moderate Ulcerative Colitis in a Randomized Controlled Trial. Clin. Gastroenterol. Hepatol. 2015, 13, 1444–1449. [Google Scholar] [CrossRef] [PubMed]
- Kedia, S.; Bhatia, V.; Thareja, S.; Garg, S.; Mouli, V.P.; Bopanna, S.; Tiwari, V.; Makharia, G.; Ahuja, V. Low dose oral curcumin is not effective in induction of remission in mild to moderate ulcerative colitis: Results from a randomized double blind placebo controlled trial. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 147–154. [Google Scholar] [CrossRef]
- Masoodi, M.; Mahdiabadi, M.A.; Mokhtare, M.; Agah, S.; Kashani, A.H.F.; Rezadoost, A.M.; Sabzikarian, M.; Talebi, A.; Sahebkar, A. The efficacy of curcuminoids in improvement of ulcerative colitis symptoms and patients’ self-reported well-being: A randomized double-blind controlled trial. J. Cell. Biochem. 2018, 119, 9552–9559. [Google Scholar] [CrossRef]
- Sadeghi, N.; Mansoori, A.; Shayesteh, A.; Hashemi, S.J. The effect of curcumin supplementation on clinical outcomes and inflammatory markers in patients with ulcerative colitis. Phytotherapy Res. 2019, 34, 1123–1133. [Google Scholar] [CrossRef]
- Samsami-Kor, M.; Daryani, N.E.; Asl, P.R.; Hekmatdoost, A. Anti-Inflammatory Effects of Resveratrol in Patients with Ulcerative Colitis: A Randomized, Double-Blind, Placebo-controlled Pilot Study. Arch. Med Res. 2015, 46, 280–285. [Google Scholar] [CrossRef]
- Samsamikor, M.; Daryani, N.E.; Asl, P.R.; Hekmatdoost, A. Resveratrol Supplementation and Oxidative/Anti-Oxidative Status in Patients with Ulcerative Colitis: A Randomized, Double-Blind, Placebo-controlled Pilot Study. Arch. Med Res. 2016, 47, 304–309. [Google Scholar] [CrossRef]
- Rastegarpanah, M.; Malekzadeh, R.; Vahedi, H.; Mohammadi, M.; Elahi, E.; Chaharmahali, M.; Safarnavadeh, T.; Abdollahi, M. A randomized, double blinded, placebo-controlled clinical trial of silymarin in ulcerative colitis. Chin. J. Integr. Med. 2012, 21, 902–906. [Google Scholar] [CrossRef]
- Hanai, H.; Iida, T.; Takeuchi, K.; Watanabe, F.; Maruyama, Y.; Andoh, A.; Tsujikawa, T.; Fujiyama, Y.; Mitsuyama, K.; Sata, M.; et al. Curcumin Maintenance Therapy for Ulcerative Colitis: Randomized, Multicenter, Double-Blind, Placebo-Controlled Trial. Clin. Gastroenterol. Hepatol. 2006, 4, 1502–1506. [Google Scholar] [CrossRef]
- Morvaridi, M.; Jafarirad, S.; Seyedian, S.S.; Alavinejad, P.; Cheraghian, B. The effects of extra virgin olive oil and canola oil on inflammatory markers and gastrointestinal symptoms in patients with ulcerative colitis. Eur. J. Clin. Nutr. 2020, 74, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Venancio, V.P.; Fang, C.; Dupont, A.W.; Talcott, S.T.; Mertens-Talcott, S.U. Mango (Mangifera indica L.) polyphenols reduce IL-8, GRO, and GM-SCF plasma levels and increase Lactobacillus species in a pilot study in patients with inflammatory bowel disease. Nutr. Res. 2020, 75, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Peng, P.; Ding, N.; Jia, W.; Huang, C.; Tang, Y. Oxidative Stress, Inflammation, Gut Dysbiosis: What Can Polyphenols Do in Inflammatory Bowel Disease? Antioxidants 2023, 12, 967. [Google Scholar] [CrossRef] [PubMed]
- De Ferrars, R.M.; Czank, C.; Zhang, Q.; Botting, N.P.; Kroon, P.; Cassidy, A.; Kay, C. The pharmacokinetics of anthocyanins and their metabolites in humans. J. Cereb. Blood Flow Metab. 2014, 171, 3268–3282. [Google Scholar] [CrossRef]
- Crescenti, A.; Caimari, A.; Alcaide-Hidalgo, J.M.; Marine-Casado, R.; Valls, R.M.; Companys, J.; Salamanca, P.; Calderon-Perez, L.; Pla-Paga, L.; Pedret, A.; et al. Hesperidin Bioavailability Is Increased by the Presence of 2S-Diastereoisomer and Micronization-A Randomized, Crossover and Double-Blind Clinical Trial. Nutrients 2022, 14, 2481. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Liu, Y.; Kang, L.; Ye, H.; Zang, J.; Wang, J.; Han, D. Bifidobacterium animalis Promotes the Growth of Weaning Piglets by Improving Intestinal Development, Enhancing Antioxidant Capacity, and Modulating Gut Microbiota. Appl. Environ. Microbiol. 2022, 88, e0129622. [Google Scholar] [CrossRef]
- Fan, F.-Y.; Sang, L.-X.; Jiang, M. Catechins and Their Therapeutic Benefits to Inflammatory Bowel Disease. Molecules 2017, 22, 484. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, C. ; Abdullah; Tian, W. ; Qiu, Z.; Song, M.; Cao, Y.; Xiao, J., Hydroxytyrosol Alleviates Dextran Sulfate Sodium-Induced Colitis by Modulating Inflammatory Responses, Intestinal Barrier, and Microbiome. J. Agric. Food Chem. 2022, 70, 2241–2252. [Google Scholar] [PubMed]
- Astorga, J.; Gasaly, N.; Dubois-Camacho, K.; De la Fuente, M.; Landskron, G.; Faber, K.N.; Urra, F.A.; Hermoso, M.A. The role of cholesterol and mitochondrial bioenergetics in activation of the inflammasome in IBD. Front. Immunol. 2022, 13, 1028953. [Google Scholar] [CrossRef]
- Hegazy, S.K.; El-Bedewy, M.M. , Effect of probiotics on pro-inflammatory cytokines and NF-kappaB activation in ulcerative colitis. World J. Gastroenterol. 2010, 16, 4145–4451. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.D.; Goodlad, J.R.; Limb, G.A.; Powell, J.J.; Thompson, R.P.; Punchard, N.A. , Activation of nuclear factor kappa B in Crohn's disease. Inflamm. Res. 1998, 47, 440–445. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Hamalainen, M.; Nieminen, R.; Vuorela, P.; Heinonen, M.; Moilanen, E. , Anti-inflammatory effects of flavonoids: Genistein, kaempferol, quercetin, and daidzein inhibit STAT-1 and NF-kappaB activations, whereas flavone, isorhamnetin, naringenin, and pelargonidin inhibit only NF-kappaB activation along with their inhibitory effect on iNOS expression and NO production in activated macrophages. Mediat. Inflamm. 2007, 2007, 45673. [Google Scholar]
- Cho, D.-I.; Koo, N.-Y.; Chung, W.J.; Kim, T.-S.; Ryu, S.Y.; Im, S.Y.; Kim, K.-M. Effects of resveratrol-related hydroxystilbenes on the nitric oxide production in macrophage cells: Structural requirements and mechanism of action. Life Sci. 2002, 71, 2071–2082. [Google Scholar] [CrossRef]
- Cho, M.K.; Park, J.W.; Jang, Y.P.; Kim, Y.C.; Kim, S.G. , Potent inhibition of lipopolysaccharide-inducible nitric oxide synthase expression by dibenzylbutyrolactone lignans through inhibition of I-kappaBalpha phosphorylation and of p65 nuclear translocation in macrophages. Int. Immunopharmacol. 2002, 2, 105–116. [Google Scholar] [CrossRef]
- Kennedy, M.; Wilson, L.; Szabo, C.; Salzman, A.L. 5-aminosalicylic acid inhibits iNOS transcription in human intestinal epithelial cells. Int. J. Mol. Med. 1999, 4, 437–480. [Google Scholar] [CrossRef]
- Comalada, M.; Ballester, I.; Bailón, E.; Sierra, S.; Xaus, J.; Gálvez, J.; de Medina, F.S.; Zarzuelo, A. Inhibition of pro-inflammatory markers in primary bone marrow-derived mouse macrophages by naturally occurring flavonoids: Analysis of the structure–activity relationship. Biochem. Pharmacol. 2006, 72, 1010–1021. [Google Scholar] [CrossRef]
- Nicholas, C.; Batra, S.; Vargo, M.A.; Voss, O.H.; Gavrilin, M.A.; Wewers, M.D.; Guttridge, D.C.; Grotewold, E.; Doseff, A.I. , Apigenin blocks lipopolysaccharide-induced lethality in vivo and proinflammatory cytokines expression by inactivating NF-kappaB through the suppression of p65 phosphorylation. J. Immunol. 2007, 179, 7121–7127. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.K.; Aggarwal, R.S.; Sethi, G.; Aggarwal, B.B.; Ramesh, G.T. , Morin (3,5,7,2',4'-Pentahydroxyflavone) abolishes nuclear factor-kappaB activation induced by various carcinogens and inflammatory stimuli, leading to suppression of nuclear factor-kappaB-regulated gene expression and up-regulation of apoptosis. Clin. Cancer Res. 2007, 13, 2290–2297. [Google Scholar] [CrossRef]
- Sung, B.; Pandey, M.K.; Aggarwal, B.B. Fisetin, an inhibitor of cyclin-dependent kinase 6, down-regulates nuclear factor-kappaB-regulated cell proliferation, antiapoptotic and metastatic gene products through the suppression of TAK-1 and receptor-interacting protein-regulated IkappaBalpha kinase activation. Mol. Pharmacol. 2007, 71, 1703–1714. [Google Scholar]
- Kunnumakkara, A.B.; Nair, A.S.; Ahn, K.S.; Pandey, M.K.; Yi, Z.; Liu, M.; Aggarwal, B.B. Gossypin, a pentahydroxy glucosyl flavone, inhibits the transforming growth factor beta-activated kinase-1-mediated NF-kappaB activation pathway, leading to potentiation of apoptosis, suppression of invasion, and abrogation of osteoclastogenesis. Blood 2007, 109, 5112–5121. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Peng, W.H.; Tsai, K.D.; Hsu, S.L. Luteolin suppresses inflammation-associated gene expression by blocking NF-kappaB and AP-1 activation pathway in mouse alveolar macrophages. Life Sci. 2007, 81, 1602–1614. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Zhang, J.F.; Guo, B.S.; He, Q.; Wang, B.Y.; He, B.; Wang, C.Q. , Resveratrol inhibits macrophage expression of EMMPRIN by activating PPARgamma. Vascul Pharmacol. 2007, 46, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jung, H.S.; Giang, P.M.; Jin, X.; Lee, S.; Son, P.T.; Lee, D.; Hong, Y.S.; Lee, K.; Lee, J.J. , Blockade of nuclear factor-kappaB signaling pathway and anti-inflammatory activity of cardamomin, a chalcone analog from Alpinia conchigera. J. Pharmacol. Exp. Ther. 2006, 316, 271–278. [Google Scholar] [CrossRef]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-Like Receptor 2 Pathway Establishes Colonization by a Commensal of the Human Microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef]
- Brown, M.; Hughes, K.R.; Moossavi, S.; Robins, A.; Mahida, Y.R. Toll-like receptor expression in crypt epithelial cells, putative stem cells and intestinal myofibroblasts isolated from controls and patients with inflammatory bowel disease. Clin. Exp. Immunol. 2014, 178, 28–39. [Google Scholar] [CrossRef]
- Youn, H.S.; Lee, J.Y.; Saitoh, S.I.; Miyake, K.; Kang, K.W.; Choi, Y.J.; Hwang, D.H. , Suppression of MyD88- and TRIF-dependent signaling pathways of Toll-like receptor by (-)-epigallocatechin-3-gallate, a polyphenol component of green tea. Biochem. Pharmacol. 2006, 72, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Kim, S.Y.; Kim, Y.S.; Lee, W.-H.; Hwang, D.H.; Lee, J.Y. Suppression of the TRIF-dependent signaling pathway of Toll-like receptors by luteolin. Biochem. Pharmacol. 2009, 77, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Youn, H.S.; Lee, J.Y.; Fitzgerald, K.A.; Young, H.A.; Akira, S.; Hwang, D.H. Specific Inhibition of MyD88-Independent Signaling Pathways of TLR3 and TLR4 by Resveratrol: Molecular Targets Are TBK1 and RIP1 in TRIF Complex. J. Immunol. 2005, 175, 3339–3346. [Google Scholar] [CrossRef]
- Schulze-Osthoff, K.; Ferrari, D.; Riehemann, K.; Wesselborg, S. , Regulation of NF-kappa B activation by MAP kinase cascades. Immunobiology 1997, 198, 35–49. [Google Scholar] [CrossRef]
- Huang, S.; Wu, C.; Yen, G. Effects of flavonoids on the expression of the pro-inflammatory response in human monocytes induced by ligation of the receptor for AGEs. Mol. Nutr. Food Res. 2006, 50, 1129–1139. [Google Scholar] [CrossRef]
- Sahu, B.D.; Kumar, J.M.; Sistla, R. , Fisetin, a dietary flavonoid, ameliorates experimental colitis in mice: Relevance of NF-kappaB signaling. J. Nutr. Biochem. 2016, 28, 171–82. [Google Scholar] [CrossRef]
- Jiang, M.; Wu, Y.L.; Li, X.; Zhang, Y.; Xia, K.L.; Cui, B.W.; Lian, L.H.; Nan, J.X. , Oligomeric proanthocyanidin derived from grape seeds inhibited NF-kappaB signaling in activated HSC: Involvement of JNK/ERK MAPK and PI3K/Akt pathways. Biomed. Pharmacother. 2017, 93, 674–680. [Google Scholar] [CrossRef]
- Cho, S.Y.; Park, S.J.; Kwon, M.J.; Jeong, T.S.; Bok, S.H.; Choi, W.Y.; Jeong, W.I.; Ryu, S.Y.; Do, S.H.; Lee, C.S.; et al. Quercetin suppresses proinflammatory cytokines production through MAP kinases andNF-kappaB pathway in lipopolysaccharide-stimulated macrophage. Mol. Cell Biochem. 2003, 243, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Jan, R.-L.; Kuo, C.-H.; Chu, Y.-T.; Wang, W.-L.; Lee, M.-S.; Chen, H.-N.; Hung, C.-H. Natural Flavone Kaempferol Suppresses Chemokines Expression in Human Monocyte THP-1 Cells through MAPK Pathways. J. Food Sci. 2010, 75, H254–H259. [Google Scholar] [CrossRef]
- Xagorari, A.; Roussos, C.; Papapetropoulos, A. Inhibition of LPS-stimulated pathways in macrophages by the flavonoid luteolin. Br. J. Pharmacol. 2002, 136, 1058–1064. [Google Scholar] [CrossRef]
- Masuelli, L.; Marzocchella, L.; Quaranta, A.; Palumbo, C.; Pompa, G.; Izzi, V.; Canini, A.; Modesti, A.; Galvano, F.; Bei, R. Apigenin induces apoptosis and impairs head and neck carcinomas EGFR/ErbB2 signaling. Front. Biosci. 2011, 16, 1060–8. [Google Scholar] [CrossRef]
- Bae, J.-Y.; Choi, J.-S.; Choi, Y.-J.; Shin, S.-Y.; Kang, S.-W.; Han, S.J.; Kang, Y.-H. (−)Epigallocatechin gallate hampers collagen destruction and collagenase activation in ultraviolet-B-irradiated human dermal fibroblasts: Involvement of mitogen-activated protein kinase. Food Chem. Toxicol. 2008, 46, 1298–1307. [Google Scholar] [CrossRef]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef]
- Qin, S.; Hou, D.X. , Multiple regulations of Keap1/Nrf2 system by dietary phytochemicals. Mol Nutr Food Res 2016, 60, 1731–1755. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.F.; Revel, J.S.; Maier, C.S.; Rada, P.; Pardo, V.; Mobasher, M.A.; Martínez, I.G.; Ruiz, L.; González-Rodríguez. ; Sanchez-Ramos, C.; et al. Mitochondria-Centric Review of Polyphenol Bioactivity in Cancer Models. Antioxidants Redox Signal. 2018, 29, 1589–1611. [Google Scholar] [CrossRef]
- Kode, A.; Rajendrasozhan, S.; Caito, S.; Yang, S.-R.; Megson, I.L.; Rahman, I. Resveratrol induces glutathione synthesis by activation of Nrf2 and protects against cigarette smoke-mediated oxidative stress in human lung epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008, 294, L478–L488. [Google Scholar] [CrossRef]
- Tanigawa, S.; Fujii, M.; Hou, D.-X. Action of Nrf2 and Keap1 in ARE-mediated NQO1 expression by quercetin. Free. Radic. Biol. Med. 2007, 42, 1690–1703. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Chen, J.; Tanigawa, S.; Hou, D.-X. Gene expression profiling and pathway network analysis of hepatic metabolic enzymes targeted by baicalein. J. Ethnopharmacol. 2012, 140, 131–140. [Google Scholar] [CrossRef]
- Qin, S.; Deng, F.; Wu, W.; Jiang, L.; Yamashiro, T.; Yano, S.; Hou, D.X. , Baicalein modulates Nrf2/Keap1 system in both Keap1-dependent and Keap1-independent mechanisms. Arch. Biochem. Biophys. 2014, 559, 53–61. [Google Scholar] [CrossRef]
- Dietz, B.M.; Kang, Y.-H.; Liu, G.; Eggler, A.L.; Yao, P.; Chadwick, L.R.; Pauli, G.F.; Farnsworth, N.R.; Mesecar, A.D.; van Breemen, R.B.; et al. Xanthohumol Isolated from Humulus lupulus Inhibits Menadione-Induced DNA Damage through Induction of Quinone Reductase. Chem. Res. Toxicol. 2005, 18, 1296–1305. [Google Scholar] [CrossRef]
- Luo, Y.; Eggler, A.L.; Liu, D.; Liu, G.; Mesecar, A.D.; van Breemen, R.B. Sites of alkylation of human Keap1 by natural chemoprevention agents. J. Am. Soc. Mass Spectrom. 2007, 18, 2226–2232. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-C.; Nguyen, T.; Pickett, C.B. Phosphorylation of Nrf2 at Ser-40 by Protein Kinase C Regulates Antioxidant Response Element-mediated Transcription. J. Biol. Chem. 2002, 277, 42769–42774. [Google Scholar] [CrossRef]
- Nguyen, T.; Sherratt, P.J.; Huang, H.-C.; Yang, C.S.; Pickett, C.B. Increased Protein Stability as a Mechanism That Enhances Nrf2-mediated Transcriptional Activation of the Antioxidant Response Element. J. Biol. Chem. 2003, 278, 4536–4541. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Yang, C.S.; Pickett, C.B. The pathways and molecular mechanisms regulating Nrf2 activation in response to chemical stress. Free. Radic. Biol. Med. 2004. [Google Scholar] [CrossRef]
- Yao, P.; Nussler, A.; Liu, L.; Hao, L.; Song, F.; Schirmeier, A.; Nussler, N. Quercetin protects human hepatocytes from ethanol-derived oxidative stress by inducing heme oxygenase-1 via the MAPK/Nrf2 pathways. J. Hepatol. 2007, 47, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Jang, J.-H.; Li, M.-H.; Surh, Y.-J. Resveratrol upregulates heme oxygenase-1 expression via activation of NF-E2-related factor 2 in PC12 cells. Biochem. Biophys. Res. Commun. 2005, 331, 993–1000. [Google Scholar] [CrossRef]
- Yang, C.-M.; Huang, S.-M.; Liu, C.-L.; Hu, M.-L. Apo-8′-lycopenal Induces Expression of HO-1 and NQO-1 via the ERK/p38-Nrf2-ARE Pathway in Human HepG2 Cells. J. Agric. Food Chem. 2012, 60, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-W.; Wu, M.-J.; Liu, I.Y.-C.; Su, J.-D.; Yen, J.-H. Neurotrophic and Cytoprotective Action of Luteolin in PC12 Cells through ERK-Dependent Induction of Nrf2-Driven HO-1 Expression. J. Agric. Food Chem. 2010, 58, 4477–4486. [Google Scholar] [CrossRef]
- Rodríguez-Ramiro, I.; Ramos, S.; Bravo, L.; Goya, L.; Martín, M. . Procyanidin B2 induces Nrf2 translocation and glutathione S-transferase P1 expression via ERKs and p38-MAPK pathways and protect human colonic cells against oxidative stress. Eur. J. Nutr. 2011, 51, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.P.; Choi, J.H.; Choi, J.M.; Chung, Y.C.; Jeong, H.G. Protective mechanisms of anthocyanins from purple sweet potato against tert-butyl hydroperoxide-induced hepatotoxicity. Food Chem. Toxicol. 2011, 49, 2081–2089. [Google Scholar] [CrossRef]
- Chen, M.-C.; Ye, Y.-Y.; Ji, G.; Liu, J.-W. Hesperidin Upregulates Heme Oxygenase-1 To Attenuate Hydrogen Peroxide-Induced Cell Damage in Hepatic L02 Cells. J. Agric. Food Chem. 2010, 58, 3330–3335. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Hsu, M.; Hsieh, C.; Lin, J.; Lai, P.; Wung, B. Upregulation of heme oxygenase-1 by Epigallocatechin-3-gallate via the phosphatidylinositol 3-kinase/Akt and ERK pathways. Life Sci. 2006, 78, 2889–2897. [Google Scholar] [CrossRef] [PubMed]
- Granado-Serrano, A.B.; Martín, M.A.; Haegeman, G.; Goya, L.; Bravo, L.; Ramos, S. Epicatechin induces NF-κB, activator protein-1 (AP-1) and nuclear transcription factor erythroid 2p45-related factor-2 (Nrf2) via phosphatidylinositol-3-kinase/protein kinase B (PI3K/AKT) and extracellular regulated kinase (ERK) signalling in HepG2 cells. Br. J. Nutr. 2009, 103, 168–179. [Google Scholar] [CrossRef]
- Pullikotil, P.; Chen, H.; Muniyappa, R.; Greenberg, C.C.; Yang, S.; Reiter, C.E.; Lee, J.-W.; Chung, J.H.; Quon, M.J. Epigallocatechin gallate induces expression of heme oxygenase-1 in endothelial cells via p38 MAPK and Nrf-2 that suppresses proinflammatory actions of TNF-α. J. Nutr. Biochem. 2011, 23, 1134–1145. [Google Scholar] [CrossRef] [PubMed]
- Bak, M.-J.; Jun, M.; Jeong, W.-S. Procyanidins from Wild Grape (Vitis amurensis) Seeds Regulate ARE-Mediated Enzyme Expression via Nrf2 Coupled with p38 and PI3K/Akt Pathway in HepG2 Cells. Int. J. Mol. Sci. 2012, 13, 801–818. [Google Scholar] [CrossRef]
- Gao, S.S.; Choi, B.-M.; Chen, X.Y.; Zhu, R.Z.; Kim, Y.; So, H.; Park, R.; Sung, M.; Kim, B.-R. Kaempferol Suppresses Cisplatin-Induced Apoptosis Via Inductions of Heme Oxygenase-1 and Glutamate-Cysteine Ligase Catalytic Subunit in HEI-OC1 cells. Pharm. Res. 2009, 27, 235–245. [Google Scholar] [CrossRef]
- Jeong, G.-S.; Lee, D.-S.; Li, B.; Lee, H.-J.; Kim, E.-C.; Kim, Y.-C. Effects of sappanchalcone on the cytoprotection and anti-inflammation via heme oxygenase-1 in human pulp and periodontal ligament cells. Eur. J. Pharmacol. 2010, 644, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Sivandzade, F.; Prasad, S.; Bhalerao, A.; Cucullo, L. NRF2 and NF-қB interplay in cerebrovascular and neurodegenerative disorders: Molecular mechanisms and possible therapeutic approaches. Redox Biol. 2019, 21, 101059. [Google Scholar] [CrossRef]
- Wu, S.; Liao, X.; Zhu, Z.; Huang, R.; Chen, M.; Huang, A.; Zhang, J.; Wu, Q.; Wang, J.; Ding, Y. , Antioxidant and anti-inflammation effects of dietary phytochemicals: The Nrf2/NF-kappaB signalling pathway and upstream factors of Nrf2. Phytochemistry 2022, 204, 113429. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, F.; Bordoni, A. Foodomics: A new comprehensive approach to food and nutrition. Genes Nutr. 2012, 8, 1–4. [Google Scholar] [CrossRef] [PubMed]
- LeVatte, M.; Keshteli, A.H.; Zarei, P.; Wishart, D.S. Applications of Metabolomics to Precision Nutrition. Lifestyle Genom. 2021, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Braconi, D.; Bernardini, G.; Millucci, L.; Santucci, A. Foodomics for human health: Current status and perspectives. Expert Rev. Proteom. 2017, 15, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Rodríguez, G.; Montero, L.; Herrero, M.; Cifuentes, A.; Castro-Puyana, M. Capillary electromigration methods for food analysis and Foodomics: Advances and applications in the period March 2021 to March 2023. ELECTROPHORESIS 2023. [Google Scholar] [CrossRef] [PubMed]
- German, J.B.; Zivkovic, A.M.; Dallas, D.C.; Smilowitz, J.T. , Nutrigenomics and personalized diets: What will they mean for food? Annu. Rev. Food Sci. Technol. 2011, 2, 97–123. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R. , Next-generation sequencing platforms. Annu. Rev. Anal. Chem. 2013, 6, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Roh, S.W.; Abell, G.C.; Kim, K.-H.; Nam, Y.-D.; Bae, J.-W. Comparing microarrays and next-generation sequencing technologies for microbial ecology research. Trends Biotechnol. 2010, 28, 291–299. [Google Scholar] [CrossRef]
- Berlin, K.; Koren, S.; Chin, C.-S.; Drake, J.P.; Landolin, J.M.; Phillippy, A.M. , Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 2015, 33, 623–630. [Google Scholar] [CrossRef]
- Pushkarev, D.; Neff, N.F.; Quake, S.R. , Single-molecule sequencing of an individual human genome. Nat. Biotechnol. 2009, 27, 847–850. [Google Scholar] [CrossRef]
- Tuberosa, R.; Frascaroli, E.; Salvi, S. Leveraging plant genomics for better and healthier food. Curr. Opin. Food Sci. 2017, 16, 100–105. [Google Scholar] [CrossRef]
- Bashiardes, S.; Godneva, A.; Elinav, E.; Segal, E. Towards utilization of the human genome and microbiome for personalized nutrition. Curr. Opin. Biotechnol. 2018, 51, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Mattar, R.; de Campos Mazo, D.F.; Carrilho, F.J. , Lactose intolerance: Diagnosis, genetic, and clinical factors. Clin. Exp. Gastroenterol. 2012, 5, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, S.H. Precision (Personalized) Nutrition: Understanding Metabolic Heterogeneity. Annu. Rev. Food Sci. Technol. 2020, 11, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Bordoni, L.; Gabbianelli, R. Primers on nutrigenetics and nutri(epi)genomics: Origins and development of precision nutrition. Biochimie 2019, 160, 156–171. [Google Scholar] [CrossRef]
- Valdés, A.; Cifuentes, A.; León, C. Foodomics evaluation of bioactive compounds in foods. TrAC Trends Anal. Chem. 2017, 96, 2–13. [Google Scholar] [CrossRef]
- Soni, K.A.; Nannapaneni, R.; Tasara, T. , The contribution of transcriptomic and proteomic analysis in elucidating stress adaptation responses of Listeria monocytogenes. Foodborne Pathog. Dis. 2011, 8, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Malone, J.H.; Oliver, B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011, 9, 34–34. [Google Scholar] [CrossRef]
- Lancova, K.; Dip, R.; Antignac, J.-P.; Le Bizec, B.; Elliott, C.T.; Naegeli, H. , Detection of hazardous food contaminants by transcriptomics fingerprinting. TrAC Trends Anal. Chem. 2011, 30, 181–191. [Google Scholar] [CrossRef]
- Valdés, A.; Ibáñez, C.; Simó, C.; García-Cañas, V. Recent transcriptomics advances and emerging applications in food science. TrAC Trends Anal. Chem. 2013, 52, 142–154. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. , RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef]
- Wu, Q.; Yuan, H.; Zhang, L.; Zhang, Y. Recent advances on multidimensional liquid chromatography–mass spectrometry for proteomics: From qualitative to quantitative analysis—A review. Anal. Chim. Acta 2012, 731, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chandramouli, K.; Qian, P.-Y. Proteomics: Challenges, Techniques and Possibilities to Overcome Biological Sample Complexity. Hum. Genom. Proteom. 2009, 1, 239204. [Google Scholar] [CrossRef] [PubMed]
- Ferranti, P. The future of analytical chemistry in foodomics. Curr. Opin. Food Sci. 2018, 22, 102–108. [Google Scholar] [CrossRef]
- Tholey, A.; Becker, A. , Top-down proteomics for the analysis of proteolytic events-Methods, applications and perspectives. Mol. Cell Res. 2017, 1864, 2191–2199. [Google Scholar] [CrossRef]
- D’alessandro, A.; Zolla, L. We Are What We Eat: Food Safety and Proteomics. J. Proteome Res. 2011, 11, 26–36. [Google Scholar] [CrossRef]
- Almeida, A.; Bassols, A.; Bendixen, E.; Bhide, M.; Ceciliani, F.; Cristobal, S.; Eckersall, P.D.; Hollung, K.; Lisacek, F.; Mazzucchelli, G.; et al. Animal board invited review: Advances in proteomics for animal and food sciences. Animal 2015, 9, 1–17. [Google Scholar] [CrossRef]
- Puiggròs, F.; Solà, R.; Bladé, C.; Salvadó, M.-J.; Arola, L. Nutritional biomarkers and foodomic methodologies for qualitative and quantitative analysis of bioactive ingredients in dietary intervention studies. J. Chromatogr. A 2011, 1218, 7399–7414. [Google Scholar] [CrossRef]
- Xu, Y.-J.; Wu, X. Foodomics in microbiological investigations. Curr. Opin. Food Sci. 2015, 4, 51–55. [Google Scholar] [CrossRef]
- Gómez, E.; Muñoz, M.; Simó, C.; Ibáñez, C.; Carrocera, S.; Martín-González, D.; Cifuentes, A. Corrigendum to "Non-invasive metabolomics for improved determination of embryonic sex markers in chemically defined culture medium" [J. Chromatogr. A 1474 (2016) 138-144]. J. Chromatogr. A 2017, 1526, 167. [Google Scholar] [CrossRef]
- Gowda, G.A.N.; Djukovic, D. Overview of Mass Spectrometry-Based Metabolomics: Opportunities and Challenges. In Mass Spectrometry in Metabolomics; 2014; Volume 1198, pp. 3–12. [CrossRef]
- Laghi, L.; Picone, G.; Capozzi, F. Nuclear magnetic resonance for foodomics beyond food analysis. TrAC Trends Anal. Chem. 2014, 59, 93–102. [Google Scholar] [CrossRef]
- Emwas, A.H. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015, 1277, 161–193. [Google Scholar] [PubMed]
- Liu, Y.-R.; Xiao, B.-K.; Yang, J.-Y.; Guo, C.-H.; Shen, S.-J.; Tang, Z.-S.; Dong, J.-X.; Huang, R.-Q. , 1H-NMR and HPLC–MS/MS-based global/targeted metabolomic evaluation of Hypericum perforatum L. intervention for menopause. J. Funct. Foods 2015, 17, 722–741. [Google Scholar] [CrossRef]
- Kohler, I.; Giera, M. Recent advances in liquid-phase separations for clinical metabolomics. J. Sep. Sci. 2016, 40, 93–108. [Google Scholar] [CrossRef]
- Castro-Puyana, M.; Herrero, L.; González, M.; Gómara, B. Rapid and simultaneous determination of polychlorinated biphenyls and their main metabolites (hydroxylated and methyl sulfonyl) by gas chromatography coupled to mass spectrometry: Comparison of different ionisation modes. Anal. Chim. Acta 2013, 787, 148–154. [Google Scholar] [CrossRef]
- Jalandra, R.; Dalal, N.; Yadav, A.K.; Verma, D.; Sharma, M.; Singh, R.; Khosla, A.; Kumar, A.; Solanki, P.R. Emerging role of trimethylamine-N-oxide (TMAO) in colorectal cancer. Appl. Microbiol. Biotechnol. 2021, 105, 7651–7660. [Google Scholar] [CrossRef] [PubMed]
- Craciun, S.; Balskus, E.P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. USA 2012, 109, 21307–21312. [Google Scholar] [CrossRef]
- Campo, A.M.-D.; Bodea, S.; Hamer, H.A.; Marks, J.A.; Haiser, H.J.; Turnbaugh, P.J.; Balskus, E.P. Characterization and Detection of a Widely Distributed Gene Cluster That Predicts Anaerobic Choline Utilization by Human Gut Bacteria. Mbio 2015, 6, e00042–15. [Google Scholar] [CrossRef]
- Krueger, S.K.; Siddens, L.K.; Henderson, M.C.; Andreasen, E.A.; Tanguay, R.L.; Pereira, C.B.; Cabacungan, E.T.; Hines, R.N.; Ardlie, K.G.; Williams, D.E. Haplotype and functional analysis of four flavin-containing monooxygenase isoform 2 (FMO2) polymorphisms in Hispanics. Pharmacogenetics Genom. 2005, 15, 245–256. [Google Scholar] [CrossRef]
- Cashman, J.R.; Akerman, B.R.; Forrest, S.M.; Treacy, E.P. Population-specific polymorphisms of the human FMO3 gene: Significance for detoxication. Drug Metab. Dispos. 2000, 28, 169–173. [Google Scholar] [PubMed]
- Treacy, E.P.; Akerman, B.R.; Chow, L.M.L.; Youil, R.; Lin, C.B.; Bruce, A.G.; Knight, M.; Danks, D.M.; Cashman, J.R.; Forrest, S.M. Mutations of the flavin-containing monooxygenase gene (FMO3) cause trimethylaminuria, a defect in detoxication. Hum. Mol. Genet. 1998, 7, 839–845. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.W.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Suez, J.; Elinav, E. , You are what you eat: Diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol 2019, 16, 35–56. [Google Scholar] [CrossRef]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef]
- Wang, H.; Rong, X.; Zhao, G.; Zhou, Y.; Xiao, Y.; Ma, D.; Jin, X.; Wu, Y.; Yan, Y.; Yang, H.; et al. The microbial metabolite trimethylamine N-oxide promotes antitumor immunity in triple-negative breast cancer. Cell Metab. 2022, 34, 581–594. [Google Scholar] [CrossRef]
- Wu, K.; Yuan, Y.; Yu, H.; Dai, X.; Wang, S.; Sun, Z.; Wang, F.; Fei, H.; Lin, Q.; Jiang, H.; et al. The gut microbial metabolite trimethylamine N-oxide aggravates GVHD by inducing M1 macrophage polarization in mice. Blood 2020, 136, 501–515. [Google Scholar] [CrossRef]
- Chen, M.L.; Zhu, X.H.; Ran, L.; Lang, H.D.; Yi, L.; Mi, M.T. Trimethylamine-N-Oxide Induces Vascular Inflammation by Activating the NLRP3 Inflammasome Through the SIRT3-SOD2-mtROS Signaling Pathway. J Am Heart Assoc 2017, 6, e006347. [Google Scholar] [CrossRef]
- Mirji, G.; Worth, A.; Bhat, S.A.; El Sayed, M.; Kannan, T.; Goldman, A.R.; Tang, H.Y.; Liu, Q.; Auslander, N.; Dang, C.V.; et al. The microbiome-derived metabolite TMAO drives immune activation and boosts responses to immune checkpoint blockade in pancreatic cancer. Sci Immunol 2022, 7, eabn0704. [Google Scholar] [CrossRef]
- Roberts, A.B.; Gu, X.; Buffa, J.A.; Hurd, A.G.; Wang, Z.; Zhu, W.; Gupta, N.; Skye, S.M.; Cody, D.B.; Levison, B.S.; et al. Development of a gut microbe–targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 2018, 24, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M.; et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Denman, S.E.; Morrison, M.; Yu, Z.; Dore, J.; Leclerc, M.; McSweeney, C.S. , Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis 2010, 16, 2034–2042. [Google Scholar] [CrossRef]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef]
- Patel, N.; Alkhouri, N.; Eng, K.; Cikach, F.; Mahajan, L.; Yan, C.; Grove, D.; Rome, E.S.; Lopez, R.; Dweik, R.A. Metabolomic analysis of breath volatile organic compounds reveals unique breathprints in children with inflammatory bowel disease: A pilot study. Aliment. Pharmacol. Ther. 2014, 40, 498–507. [Google Scholar] [CrossRef]
- Hendrickson, B.A.; Gokhale, R.; Cho, J.H. Clinical Aspects and Pathophysiology of Inflammatory Bowel Disease. Clin. Microbiol. Rev. 2002, 15, 79–94. [Google Scholar] [CrossRef]
- Wilson, A.; Teft, W.A.; Morse, B.L.; Choi, Y.H.; Woolsey, S.; DeGorter, M.K.; Hegele, R.A.; Tirona, R.G.; Kim, R.B. , Trimethylamine-N-oxide: A Novel Biomarker for the Identification of Inflammatory Bowel Disease. Dig Dis Sci 2015, 60, 3620–3230. [Google Scholar] [CrossRef] [PubMed]
- Puiggròs, F.; Solà, R.; Bladé, C.; Salvadó, M.-J.; Arola, L. Nutritional biomarkers and foodomic methodologies for qualitative and quantitative analysis of bioactive ingredients in dietary intervention studies. J. Chromatogr. A 2011, 1218, 7399–7414. [Google Scholar] [CrossRef] [PubMed]
- García-Cañas, V.; Simó, C.; León, C.; Cifuentes, A. Advances in Nutrigenomics research: Novel and future analytical approaches to investigate the biological activity of natural compounds and food functions. J. Pharm. Biomed. Anal. 2010, 51, 290–304. [Google Scholar] [CrossRef]
- Valdés, A.; García-Cañas, V.; Rocamora-Reverte, L.; Gómez-Martínez. ; Ferragut, J.A.; Cifuentes, A. Effect of rosemary polyphenols on human colon cancer cells: Transcriptomic profiling and functional enrichment analysis. Genes Nutr. 2012, 8, 43–60. [Google Scholar] [CrossRef]
- Dolara, P.; Luceri, C.; De Filippo, C.; Femia, A.P.; Giovannelli, L.; Caderni, G.; Cecchini, C.; Silvi, S.; Orpianesi, C.; Cresci, A. Red wine polyphenols influence carcinogenesis, intestinal microflora, oxidative damage and gene expression profiles of colonic mucosa in F344 rats. Mutat. Res. Mol. Mech. Mutagen. 2005, 591, 237–246. [Google Scholar] [CrossRef]
- Wang, J.; Tang, L.; Zhou, H.; Zhou, J.; Glenn, T.C.; Shen, C.-L.; Wang, J.-S. Long-term treatment with green tea polyphenols modifies the gut microbiome of female sprague-dawley rats. J. Nutr. Biochem. 2018, 56, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Shan, S.; Zhang, C.; Shi, J.; Li, H.; Li, Z. Inhibitory Effects of Bound Polyphenol from Foxtail Millet Bran on Colitis-Associated Carcinogenesis by the Restoration of Gut Microbiota in a Mice Model. ACS Appl. Mater. Interfaces 2020, 68, 3506–3517. [Google Scholar] [CrossRef]
- Ruiz, L.; Hidalgo, C.; Blanco-Míguez, A.; Lourenço, A.; Sánchez, B.; Margolles, A. Tackling probiotic and gut microbiota functionality through proteomics. J. Proteom. 2016, 147, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Valdés, A.; Artemenko, K.A.; Bergquist, J.; García-Cañas, V.; Cifuentes, A. Comprehensive Proteomic Study of the Antiproliferative Activity of a Polyphenol-Enriched Rosemary Extract on Colon Cancer Cells Using Nanoliquid Chromatography–Orbitrap MS/MS. J. Proteome Res. 2016, 15, 1971–1985. [Google Scholar] [CrossRef] [PubMed]
- Barnett, M.P.; Cooney, J.M.; Dommels, Y.E.; Nones, K.; Brewster, D.T.; Park, Z.; Butts, C.A.; McNabb, W.C.; Laing, W.A.; Roy, N.C. , Modulation of colonic inflammation in Mdr1a(-/-) mice by green tea polyphenols and their effects on the colon transcriptome and proteome. J Nutr Biochem 2013, 24, 1678–1690. [Google Scholar] [CrossRef]
- Li, Z.; Xie, J.; Tian, X.; Li, K.; Hou, A.; Wang, Y. Proteomic changes in EHEC O157:H7 under catechin intervention. Microb. Pathog. 2018, 123, 9–17. [Google Scholar] [CrossRef]
- Pimentel, G.; Burton, K.J.; von Ah, U.; Bütikofer, U.; Pralong, F.P.; Vionnet, N.; Portmann, R.; Vergères, G. Metabolic Footprinting of Fermented Milk Consumption in Serum of Healthy Men. J. Nutr. 2018, 148, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Arroyo, S.; Gomez-Martinez, A.; Rocamora-Reverte, L.; Quirantes-Pine, R.; Segura-Carretero, A.; Fernandez-Gutierrez, A.; Ferragut, J.A. , Application of nanoLC-ESI-TOF-MS for the metabolomic analysis of phenolic compounds from extra-virgin olive oil in treated colon-cancer cells. J Pharm Biomed Anal 2012, 63, 128–34. [Google Scholar] [CrossRef]
- Li, Q.; Liang, X.; Guo, N.; Hu, L.; E. , M.P.; Wu, Y.; Xue, X.; Wu, L.; Wang, K. Protective effects of Bee pollen extract on the Caco-2 intestinal barrier dysfunctions induced by dextran sulfate sodium. BioMedicine 2019, 117, 109200. [Google Scholar] [CrossRef]
- Dihal, A.A.; van der Woude, H.; Hendriksen, P.J.M.; Charif, H.; Dekker, L.J.; Ijsselstijn, L.; de Boer, V.C.J.; Alink, G.M.; Burgers, P.C.; Rietjens, I.M.C.M.; et al. Transcriptome and proteome profiling of colon mucosa from quercetin fed F344 rats point to tumor preventive mechanisms, increased mitochondrial fatty acid degradation and decreased glycolysis. Proteomics 2008, 8, 45–61. [Google Scholar] [CrossRef]
- Di Nunzio, M.; Picone, G.; Pasini, F.; Caboni, M.F.; Gianotti, A.; Bordoni, A.; Capozzi, F. Olive oil industry by-products. Effects of a polyphenol-rich extract on the metabolome and response to inflammation in cultured intestinal cell. Food Res. Int. 2018, 113, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Seidel, D.V.; Azcárate-Peril, M.A.; Chapkin, R.S.; Turner, N.D. Shaping functional gut microbiota using dietary bioactives to reduce colon cancer risk. Semin. Cancer Biol. 2017, 46, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Ibañez, C.; Valdés, A.; Garcia-Cañas, V.; Simo, C.; Celebier, M.; Rocamora-Reverte, L.; Gómez-Martínez, Á.; Herrero, M.; Castro-Puyana, M.; Segura-Carretero, A.; et al. Global Foodomics strategy to investigate the health benefits of dietary constituents. J. Chromatogr. A 2012, 1248, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Mayta-Apaza, A.C.; Pottgen, E.; De Bodt, J.; Papp, N.; Marasini, D.; Howard, L.; Abranko, L.; Van de Wiele, T.; Lee, S.-O.; Carbonero, F. Impact of tart cherries polyphenols on the human gut microbiota and phenolic metabolites in vitro and in vivo. J. Nutr. Biochem. 2018, 59, 160–172. [Google Scholar] [CrossRef]
- Gomez-Lopez, I.; Lobo-Rodrigo, G.; Portillo, M.P.; Cano, M.P. Characterization, Stability, and Bioaccessibility of Betalain and Phenolic Compounds from Opuntia stricta var. Dillenii Fruits and Products of Their Industrialization. Foods 2021, 10, 1593. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, C.; Colombo, F.; Biella, S.; Stockley, C.; Restani, P. Polyphenols and Human Health: The Role of Bioavailability. Nutrients 2021, 13, 273. [Google Scholar] [CrossRef] [PubMed]
- Samba-Mondonga, M.; Constante, M.; Fragoso, G.; Calvé, A.; Santos, M.M. Curcumin induces mild anemia in a DSS-induced colitis mouse model maintained on an iron-sufficient diet. PLoS ONE 2019, 14, e0208677. [Google Scholar] [CrossRef]
- Inoue, H.; Akiyama, S.; Maeda-Yamamoto, M.; Nesumi, A.; Tanaka, T.; Murakami, A. High-dose green tea polyphenols induce nephrotoxicity in dextran sulfate sodium-induced colitis mice by down-regulation of antioxidant enzymes and heat-shock protein expressions. Cell Stress Chaperon 2011, 16, 653–662. [Google Scholar] [CrossRef]
- Kobayashi, H.; Murata, M.; Kawanishi, S.; Oikawa, S. Polyphenols with Anti-Amyloid beta Aggregation Show Potential Risk of Toxicity Via Pro-Oxidant Properties. Int J Mol Sci 2020, 21, 3561. [Google Scholar] [CrossRef]
- Inoue, H.; Maeda-Yamamoto, M.; Nesumi, A.; Tanaka, T.; Murakami, A. Low and Medium but Not High Doses of Green Tea Polyphenols Ameliorated Dextran Sodium Sulfate-Induced Hepatotoxicity and Nephrotoxicity. Biosci. Biotechnol. Biochem. 2013, 77, 1223–1228. [Google Scholar] [CrossRef]
- Gandhi, H.; Rathore, C.; Dua, K.; Vihal, S.; Tambuwala, M.M.; Negi, P. Efficacy of resveratrol encapsulated microsponges delivered by pectin based matrix tablets in rats with acetic acid-induced ulcerative colitis. Drug Dev. Ind. Pharm. 2020, 46, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Pérez, A.A.; Rodriguez-Nogales, A.; Ortiz-Cullera, V.; Algieri, F.; Garrido-Mesa, J.; Zorrilla, P.; Rodriguez-Cabezas, M.E.; Garrido-Mesa, N.; Utrilla, M.P.; De Matteis, L.; et al. Silk fibroin nanoparticles constitute a vector for controlled release of resveratrol in an experimental model of inflammatory bowel disease in rats. Int. J. Nanomed. 2014, 9, 4507–4520. [Google Scholar] [CrossRef]
- Pujara, N.; Wong, K.Y.; Qu, Z.; Wang, R.; Moniruzzaman, M.; Rewatkar, P.; Kumeria, T.; Ross, B.P.; McGuckin, M.; Popat, A. , Oral Delivery of beta-Lactoglobulin-Nanosphere-Encapsulated Resveratrol Alleviates Inflammation in Winnie Mice with Spontaneous Ulcerative Colitis. Mol Pharm 2021, 18, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Huguet-Casquero, A.; Xu, Y.; Gainza, E.; Pedraz, J.L.; Beloqui, A. Oral delivery of oleuropein-loaded lipid nanocarriers alleviates inflammation and oxidative stress in acute colitis. Int. J. Pharm. 2020, 586, 119515. [Google Scholar] [CrossRef] [PubMed]
- Marinho, S.; Illanes, M.; Ávila-Román, J.; Motilva, V.; Talero, E. Anti-Inflammatory Effects of Rosmarinic Acid-Loaded Nanovesicles in Acute Colitis through Modulation of NLRP3 Inflammasome. Biomolecules 2021, 11, 162. [Google Scholar] [CrossRef]
- Nguyen, T.-H.T.; Trinh, N.-T.; Tran, H.N.; Tran, H.T.; Le, P.Q.; Ngo, D.-N.; Tran-Van, H.; Van Vo, T.; Vong, L.B.; Nagasaki, Y. Improving silymarin oral bioavailability using silica-installed redox nanoparticle to suppress inflammatory bowel disease. J. Control. Release 2020, 331, 515–524. [Google Scholar] [CrossRef]
- Arafat, E.A.; Marzouk, R.E.; Mostafa, S.A.; Hamed, W.H.E. Identification of the Molecular Basis of Nanocurcumin-Induced Telocyte Preservation within the Colon of Ulcerative Colitis Rat Model. Mediat. Inflamm. 2021, 2021, 1–13. [Google Scholar] [CrossRef]
- Sébédio, J.-L. Metabolomics, Nutrition, and Potential Biomarkers of Food Quality, Intake, and Health Status. 2017, 82, 83–116. [CrossRef]
- Bush, C.L.; Blumberg, J.B.; El-Sohemy, A.; Minich, D.M.; Ordovás, J.M.; Reed, D.G.; Behm, V.A.Y. Toward the Definition of Personalized Nutrition: A Proposal by The American Nutrition Association. J. Am. Coll. Nutr. 2019, 39, 5–15. [Google Scholar] [CrossRef]
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised nutrition and health. BMJ 2018, 361, bmj–k2173. [Google Scholar] [CrossRef]
- De Toro-Martín, J.; Arsenault, B.J.; Després, J.-P.; Vohl, M.-C. Precision Nutrition: A Review of Personalized Nutritional Approaches for the Prevention and Management of Metabolic Syndrome. Nutrients 2017, 9, 913. [Google Scholar] [CrossRef]
- Sales, N.M.R.; Pelegrini, P.B.; Goersch, M.C. Nutrigenomics: Definitions and Advances of This New Science. J. Nutr. Metab. 2014, 2014, 1–6. [Google Scholar] [CrossRef]
- Vázquez-Fresno, R.; Llorach, R.; Urpi-Sarda, M.; Lupianez-Barbero, A.; Estruch, R.; Corella, D.; Fitó, M.; Arós, F.; Ruiz-Canela, M.; Salas-Salvadó, J.; et al. Metabolomic Pattern Analysis after Mediterranean Diet Intervention in a Nondiabetic Population: A 1- and 3-Year Follow-up in the PREDIMED Study. J. Proteome Res. 2014, 14, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Rezzi, S.; Ramadan, Z.; Fay, L.B.; Kochhar, S. Nutritional Metabonomics: Applications and Perspectives. J. Proteome Res. 2007, 6, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Gerasimidis, K.; Nikolaou, C.K.; Edwards, C.A.; McGrogan, P. , Serial fecal calprotectin changes in children with Crohn's disease on treatment with exclusive enteral nutrition: Associations with disease activity, treatment response, and prediction of a clinical relapse. J Clin Gastroenterol 2011, 45, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Cameron, F.L.; Gerasimidis, K.; Papangelou, A.; Missiou, D.; Garrick, V.; Cardigan, T.; Buchanan, E.; Barclay, A.R.; McGrogan, P.; Russell, R.K. , Clinical progress in the two years following a course of exclusive enteral nutrition in 109 paediatric patients with Crohn's disease. Aliment Pharmacol Ther 2013, 37, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ney, M.; Eslamparast, T.; Vandermeer, B.; Ismond, K.P.; Kroeker, K.; Halloran, B.; Raman, M.; Tandon, P. Systematic review of nutrition screening and assessment in inflammatory bowel disease. World J. Gastroenterol. 2019, 25, 3823–3837. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





