Submitted:
23 August 2023
Posted:
25 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
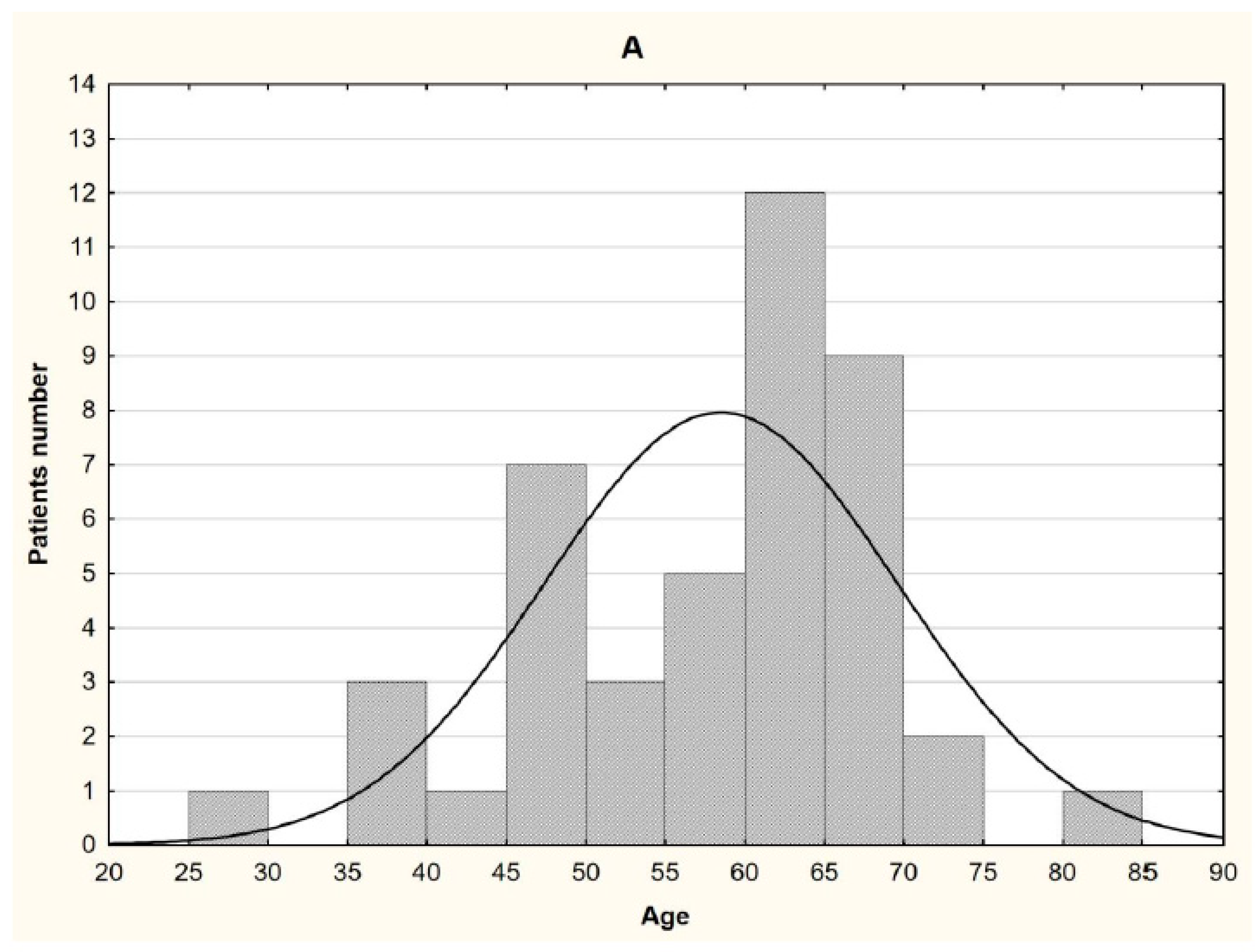
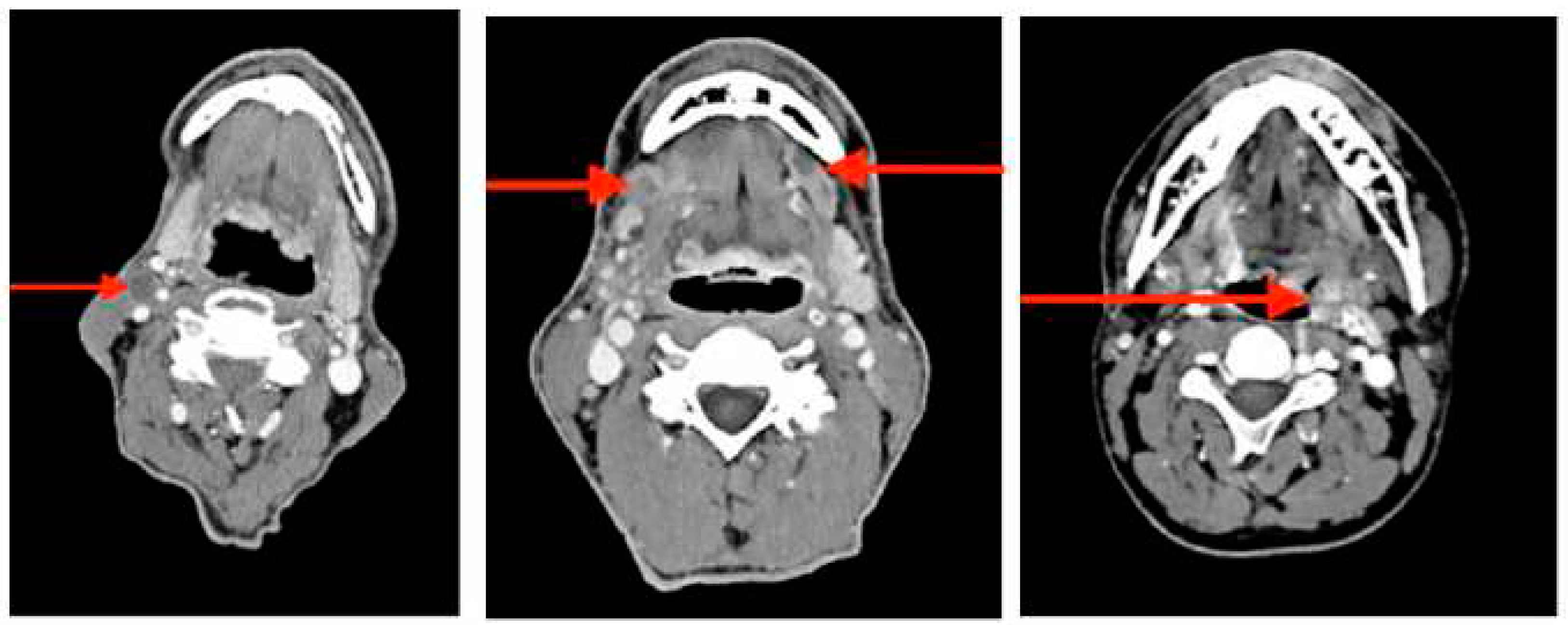
2.1. Patients characteristics
2.2. Sampling
2.3. DNA Extraction
2.4. DNA methylation analysis.
2.5. Statistical analysis
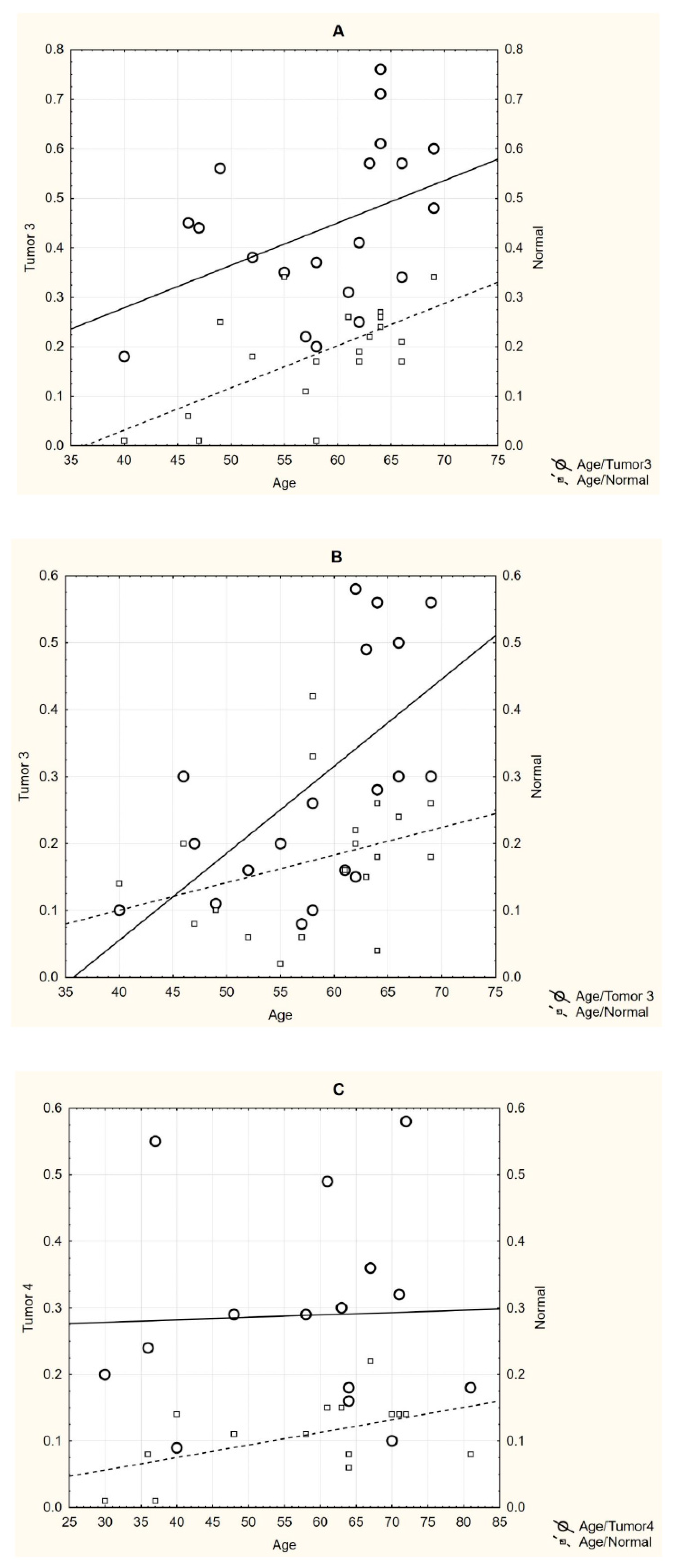
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Esteller, M.; Corn, P.G.; Baylin, S.B.; Herman, J.G. A gene hypermethylation profile of human cancer. Cancer Res. 2001, 61, 3225–3229. [Google Scholar] [PubMed]
- Gupta, S.; Kumar, P.; Maini, J.; Kaur, H. Epigenetic Biomarkers in Head and Neck Cancer. J. Cancer Genet. Biomarkers 2018, 1, 41–50. [Google Scholar] [CrossRef]
- Castilho, R.M.; Squarize, C.H.; Almeida, L.O. Epigenetic Modifications and Head and Neck Cancer: Implications for Tumor Progression and Resistance to Therapy. Int. J. Mol. Sci. 2017, 18, 1506. [Google Scholar] [CrossRef]
- Salameh, Y.; Bejaoui, Y.; El Hajj, N. DNA Methylation Biomarkers in Aging and Age-Related Diseases. Front. Genet. 2020, 11, 171. [Google Scholar] [CrossRef]
- Johansson, A.; Palli, D.; Masala, G.; Grioni, S.; Agnoli, C.; Tumino, R.; Giurdanella, M.C.; Fasanelli, F.; Sacerdote, C.; Panico, S.; et al. Epigenome-wide association study for lifetime estrogen exposure identifies an epigenetic signature associated with breast cancer risk. Clin. Epigenetics 2019, 11, 66. [Google Scholar] [CrossRef]
- Chen, L.; Ganz, P.A.; Sehl, M.E. DNA Methylation, Aging, and Cancer Risk: A Mini-Review. Front. Bioinform. 2022, 2. [Google Scholar] [CrossRef]
- Rahman, Q.B.; Iocca, O.; Kufta, K.; Shanti, R.M. Global Burden of Head and Neck Cancer. Oral Maxillofac. Surg. Clin. North Am. 2020, 32, 367–375. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Hashim, D.; Genden, E.; Posner, M.; Hashibe, M.; Boffetta, P. Head and neck cancer prevention: from primary prevention to impact of clinicians on reducing burden. Ann. Oncol. 2019, 30, 744–756. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef]
- Pfister, D.G.; Spencer, S.; Adelstein, D.; Adkins, D.; Anzai, Y.; Brizel, D.M.; Bruce, J.Y.; Busse, P.M.; Caudell, J.J.; Cmelak, A.J.; et al. Head and Neck Cancers, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2020, 18, 873–898. [Google Scholar] [CrossRef] [PubMed]
- Marcu, L.G.; Yeoh, E. A review of risk factors and genetic alterations in head and neck carcinogenesis and implications for current and future approaches to treatment. J. Cancer Res. Clin. Oncol. 2009, 135, 1303–1314. [Google Scholar] [CrossRef] [PubMed]
- WHO. int [Electronic resourse]: The world health report 2002: Reducing Risks, Promoting Healthy Life. – Available from: http://www.who.int/ whr/2002/en/whr02_en.pdf?ua=1. – Date of access: 20.06.2018. ISBN: 9241562072.
- WHO.int [Electronic resourse]: WHO global report on trends in tobacco smoking 2000–2025, 2015. – Available from: apps.who.int/iris/bitstre am/10665/156262/1/9789241564922_eng. – Date of access: 20.06.2018.
- Global Cancer Observatory. International Agency for Research on Cancer. World Health Organization. Available at: https://gco.iarc.fr/. Accessed on June 06, 2021.
- Demokan, S.; Dalay, N. Role of DNA methylation in head and neck cancer. Clin. Epigenetics 2011, 2, 123–150. [Google Scholar] [CrossRef] [PubMed]
- Maegawa, S.; Lu, Y.; Tahara, T.; Lee, J.T.; Madzo, J.; Liang, S.; Jelinek, J.; Colman, R.J.; Issa, J.-P.J. Caloric restriction delays age-related methylation drift. Nat. Commun. 2017, 8, 539. [Google Scholar] [CrossRef]
- Ovchinnikov, D.A.; Cooper, M.A.; Pandit, P.; Coman, W.B.; Cooper-White, J.J.; Keith, P.; Wolvetang, E.J.; Slowey, P.D.; Punyadeera, C. Tumor-suppressor Gene Promoter Hypermethylation in Saliva of Head and Neck Cancer Patients. Transl. Oncol. 2012, 5, 321–326. [Google Scholar] [CrossRef]
- Zhou, C.; Ye, M.; Ni, S.; Li, Q.; Ye, D.; Li, J.; Shen, Z.; Deng, H. DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics 2018, 13, 398–409. [Google Scholar] [CrossRef]
- Kurevlev, S.V.; Tskhovrebova, L.V.; Aghajanyan, A.V.; Fatkhudinov, T.K.; Gordon, K.B.; Azova, M.M. Methylation of the tumor associated genes in head and neck squamous cell carcinoma. Head Neck Tumors (HNT) 2023, 12, 61–70, (In Russ.). [Google Scholar] [CrossRef]
- Mete, O.; Wenig, B.M. Update from the 5th Edition of the World Health Organization Classification of Head and Neck Tumors: Overview of the 2022 WHO Classification of Head and Neck Neuroendocrine Neoplasms. Head Neck Pathol. 2022, 16, 123–142. [Google Scholar] [CrossRef]
- Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR, et al. (Eds.). AJCC Cancer Staging Manual. 2017. (8th edition). Springer International Publishing: American Joint Commission on Cancer.
- Krasnyi, A.M.; Kurevlev, S.V.; Sadekova, A.А.; Sefihanov, T.G.; Kometova, V.V.; Rodionov, V.V. Primary tumor gene methylation profile in patients with luminal HER2-negative breast cancer when metastasizing to regional lymph nodes. (Russian). Biomedicinskaya himia. 2021, 67, 88–94. [Google Scholar]
- Słomka, M.; Sobalska-Kwapis, M.; Wachulec, M.; Bartosz, G.; Strapagiel, D. High Resolution Melting (HRM) for High-Throughput Genotyping—Limitations and Caveats in Practical Case Studies. Int. J. Mol. Sci. 2017, 18, 2316. [Google Scholar] [CrossRef]
- Yamashita, K.; Hosoda, K.; Nishizawa, N.; Katoh, H.; Watanabe, M. Epigenetic biomarkers of promoter DNA methylation in the new era of cancer treatment. Cancer Sci. 2018, 109, 3695–3706. [Google Scholar] [CrossRef] [PubMed]
- Brait, M.; Ling, S.; Nagpal, J.K.; Chang, X.; Park, H.L.; Lee, J.; Okamura, J.; Yamashita, K.; Sidransky, D.; Kim, M.S. Cysteine Dioxygenase 1 Is a Tumor Suppressor Gene Silenced by Promoter Methylation in Multiple Human Cancers. PLOS ONE 2012, 7, e44951. [Google Scholar] [CrossRef] [PubMed]
- Minatani, N.; Waraya, M.; Yamashita, K.; Kikuchi, M.; Ushiku, H.; Kojo, K.; Ema, A.; Nishimiya, H.; Kosaka, Y.; Katoh, H.; et al. Prognostic Significance of Promoter DNA Hypermethylation of cysteine dioxygenase 1 (CDO1) Gene in Primary Breast Cancer. PLOS ONE 2016, 11, e0144862–e0144862. [Google Scholar] [CrossRef] [PubMed]
- Meller, S.; Zipfel, L.; Gevensleben, H.; Dietrich, J.; Ellinger, J.; Majores, M.; Stein, J.; Sailer, V.; Jung, M.; Kristiansen, G.; et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics 2016, 11, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.; Yamashita, K.; Ushiku, H.; Katoh, H.; Ishii, S.; Tanaka, T.; Yokoi, K.; Suzuki, M.; Ooizumi, Y.; Igarashi, K.; et al. The clinical significance of cysteine dioxygenase type 1 methylation in Barrett esophagus adenocarcinoma. Dis. Esophagus 2017, 30, 1–9. [Google Scholar] [CrossRef]
- Yang, T.-M.; Leu, S.-W.; Li, J.-M.; Hung, M.-S.; Lin, C.-H.; Lin, Y.-C.; Huang, T.-J.; Tsai, Y.-H.; Yang, C.-T. WIF-1 promoter region hypermethylation as an adjuvant diagnostic marker for non-small cell lung cancer-related malignant pleural effusions. J. Cancer Res. Clin. Oncol. 2008, 135, 919–924. [Google Scholar] [CrossRef]
- He, B.; You, L.; Uematsu, K.; Xu, Z.; Lee, A.Y.; Matsangou, M.; McCormick, F.; Jablons, D.M. A Monoclonal Antibody against Wnt-1 Induces Apoptosis in Human Cancer Cells. Neoplasia 2004, 6, 7–14. [Google Scholar] [CrossRef]
- Paluszczak, J.; Sarbak, J.; Kostrzewska-Poczekaj, M.; Kiwerska, K.; Jarmuż-Szymczak, M.; Grenman, R.; Mielcarek-Kuchta, D.; Baer-Dubowska, W. The negative regulators of Wnt pathway—DACH1, DKK1, and WIF1 are methylated in oral and oropharyngeal cancer and WIF1 methylation predicts shorter survival. Tumor Biol. 2015, 36, 2855–2861. [Google Scholar] [CrossRef]
- Pannone, G.B.P.; Santoro, A.; Franco, R.; Aquino, G.; Longo, F.; Botti, G. WNT pathway in oral cancer: epigenetic inactivation of WNT-inhibitors. Oncol Rep. 2010, 24, 1035–1041. [Google Scholar]
- Supic, G.; Kozomara, R.; Jovic, N.; Zeljic, K.; Magic, Z. Hypermethylation of RUNX3 but not WIF1 gene and its association with stage and nodal status of tongue cancers. Oral Dis. 2011, 17, 794–800. [Google Scholar] [CrossRef]
- Chan, S.L.; Cui, Y.; van Hasselt, A.; Li, H.; Srivastava, G.; Jin, H.; Ng, K.M.; Wang, Y.; Lee, K.Y.; Tsao, G.S.W.; et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Lab. Investig. 2007, 87, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Yeh, K.-T.; Chang, J.-G.; Lin, T.-H.; Wang, Y.-F.; Shih, M.-C.; Lin, C.-C. Correlation between protein expression and epigenetic and mutation changes of Wnt pathway-related genes in oral cancer. Int. J. Oncol. 2003, 23, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.E.; Davies, T.J.; Mc Auley, M.T. The role of DNA methylation in ageing and cancer. Proc. Nutr. Soc. 2018, 77, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Stefanski, C.D.; Prosperi, J.R. Wnt-Independent and Wnt-Dependent Effects of APC Loss on the Chemotherapeutic Response. Int. J. Mol. Sci. 2020, 21, 7844. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. Molecular genetics and targeted therapy of WNT-related human diseases (Review). Int. J. Mol. Med. 2017, 40, 587–606. [Google Scholar] [CrossRef]
- Strzelczyk, J.K.; Krakowczyk. ; Owczarek, A.J. Aberrant DNA methylation of the p16, APC, MGMT, TIMP3 and CDH1 gene promoters in tumours and the surgical margins of patients with oral cavity cancer. J. Cancer 2018, 9, 1896–1904. [Google Scholar] [CrossRef]
- Ali, J.; Sabiha, B.; Jan, H.U.; Haider, S.A.; Khan, A.A.; Ali, S.S. Genetic etiology of oral cancer. Oral Oncol. 2017, 70, 23–28. [Google Scholar] [CrossRef]
- Stucci, L.S.; Internò, V.; Tucci, M.; Perrone, M.; Mannavola, F.; Palmirotta, R.; Porta, C. The ATM Gene in Breast Cancer: Its Relevance in Clinical Practice. Genes 2021, 12, 727. [Google Scholar] [CrossRef]
- Marabelli, M.; Cheng, S.-C.; Parmigiani, G. Penetrance ofATMGene Mutations in Breast Cancer: A Meta-Analysis of Different Measures of Risk. Genet. Epidemiology 2016, 40, 425–431. [Google Scholar] [CrossRef]
- Ai, L.; Vo, Q.N.; Zuo, C.; Li, L.; Ling, W.; Suen, J.Y.; Hanna, E.; Brown, K.D.; Fan, C.-Y. Ataxia-Telangiectasia-Mutated ( ATM ) Gene in Head and Neck Squamous Cell Carcinoma: Promoter Hypermethylation with Clinical Correlation in 100 Cases. Cancer Epidemiology Biomarkers Prev. 2004, 13, 150–156. [Google Scholar] [CrossRef]
- Lindemann, A.; Takahashi, H.; Patel, A.; Osman, A.; Myers, J. Targeting the DNA Damage Response in OSCC with TP53 Mutations. J. Dent. Res. 2018, 97, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Takahashi, S.; Ouchi, K.; Otsuki, Y.; Wakayama, S.; Ishioka, C. Different impacts of TP53 mutations on cell cycle-related gene expression among cancer types. Sci. Rep. 2023, 13, 4868. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 Mutations in Human Cancers: Origins, Consequences, and Clinical Use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008–a001008. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C.; Khandelwal, A.R.; Wolf, G.T.; Rodrigo, J.P.; Mäkitie, A.A.; Saba, N.F.; Forastiere, A.A.; Bradford, C.R.; Ferlito, A. TP53 mutations in head and neck cancer. Mol. Carcinog. 2022, 61, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.C. p53 tumor suppressor gene: at the crossroads of molecular carcinogenesis, molecular epidemiology, and cancer risk assessment. Environ. Heal. Perspect. 1996, 104 (Suppl. 3), 435–439. [Google Scholar] [CrossRef]
- Golusinski, P.; Lamperska, K.; Pazdrowski, J.; Golusinski, W. Analiza występowania mutacji w obrębie genu TP53 u chorych na raka płaskonabłonkowego głowy i szyi [Analysis of mutations within the TP53 gene in patients with squamous cell carcinoma of the head and neck]. Otolaryngol. Polska 2011, 65, 114–121. [Google Scholar] [CrossRef]
- Di Fiore, R.; D'Anneo, A.; Tesoriere, G.; Vento, R. RB1 in cancer: Different mechanisms of RB1 inactivation and alterations of pRb pathway in tumorigenesis. J. Cell. Physiol. 2013, 228, 1676–1687. [Google Scholar] [CrossRef]
- Rusin, P.; Markiewicz, L.; Majsterek, I. Uwarunkowania genetyczne nowotworów głowy i szyi [Genetic predeterminations of head and neck cancer]. Postepy Hig Med Dosw (Online). Polish. 2008, 62, 490–501. [Google Scholar] [PubMed]
- Sabir, M.; Baig, R.M.; Ali, K.; Mahjabeen, I.; Saeed, M.; Kayani, M.A. Retinoblastoma (RB1) pocket domain mutations and promoter hyper-methylation in head and neck cancer. Cell. Oncol. 2014, 37, 203–213. [Google Scholar] [CrossRef]
| Symbol | Gene name | Location | Exon count | Gene ID | Transcripts | MIM | Gene type | Gene function | References |
|---|---|---|---|---|---|---|---|---|---|
|
APC GS; DP2; DP3; BTPS2; DESMD; DP2.5; PPP1R46 |
Adenomatous Polyposis Coli | 5q22.2 | 20 | 5624 | NM_000038.6 | 611731 | protein-coding | tumor suppressor | 37; 39 |
|
ATM AT1; ATA; ATC; ATD; ATE; ATDC; TEL1; TELO1 |
Ataxia-telangiectasia-mutated | 11q22.3 | 67 | 472 | NM_000051.4 | 607585 | protein-coding | cell cycle regulation. | 42-44 |
|
CDO1 (CDO-I) |
Cysteine dioxygenase type 1 | 5q22.3 | 9 | 1036 | NM_001323565.2 | 603943 | protein-coding | tumor suppressor | 26-28 |
|
TP53 (P53; BCC7; LFS1; BMFS5; TRP53) |
Tumor Protein P53 | 17p13.1 | 11 | 7157 | NM_000546.6. | 191170 | protein-coding | tumor suppressor | 45-50 |
|
RB1 (RB; pRb; OSRC; pp110; p105-Rb; PPP1R130; p110-RB1) |
Retinoblastoma 1 | 13q14.2 | 27 | 5925 | NM_000321.3... | 614041 | protein-coding | cell cycle regulator, tumor suppressor |
51-53 |
|
WIF1 (WIF-1) |
WNT inhibitory factor 1 | 12q14.3 | 10 | 11197 | NM_007191.5 | 605186 | protein-coding | tumor suppressor | 19; 30-36 |
| Patient ID | Tumor origin | ICD-10 | TNM classification |
Type of lesion | Histological glade | Gender | Age | Smoker |
|---|---|---|---|---|---|---|---|---|
| 1 | Tongue n=9 | C02.0 | T1N0M0 | Prim | SCC non-keratinizing | М | 50 | y |
| 2 | C02.1 | T2N0M0 | Prim | SCC non-keratinizing | F | 59 | n | |
| 3 | C02.1 | T3N0M0 | Rec | SCC keratinizing | F | 66 | y | |
| 4 | C02.1 | T3N0M0 | Rec | SCC non-keratinizing | F | 47 | n | |
| 5 | C02.1 | T3N0M0 | Prim | SCC keratinizing | М | 63 | n | |
| 6 | C02.1 | T3N0M0 | Prim | SCC keratinizing | М | 40 | n | |
| 7 | C02.1 | T3N0M0 | Prim | SCC keratinizing | M | 46 | n | |
| 8 | C02.1 | T3N1M0 | Prim | SCC non-keratinizing | М | 52 | n | |
| 9 | C02.1 | T4N2M0 | Prim | SCC keratinizing | М | 36 | n | |
| 10 | Oral cavity n=6 | C03.0 | T2N0M0 | Prim | SCC non-keratinizing | F | 59 | n |
| 11 | C03.0 | T3N0M0 | Prim | SCC keratinizing | M | 69 | y | |
| 12 | C03.0 | T4N0M0 | Prim | SCC keratinizing | М | 37 | y | |
| 13 | C03.1 | T4N0M0 | Prim | SCC non-keratinizing | М | 30 | n | |
| 14 | C03.1 | T4N0M0 | Prim | SCC non-keratinizing | F | 63 | n | |
| 15 | C03.1 | T4N0M0 | Prim | SCC non-keratinizing | F | 48 | n | |
| 16 | Floor of mouth n=3 | C04.1 | T2N0M0 | Prim | SCC keratinizing | F | 67 | y |
| 17 | C04.1 | T3N1M0 | Prim | SCC non-keratinizing | F | 66 | y | |
| 18 | C04.1 | T4N0M0 | Prim | SCC keratinizing | F | 64 | y | |
| 19 | Maxillary sinus n=6 | C31.0 | T3N0M0 | Prim | SCC non-keratinizing | M | 58 | y |
| 20 | C31.1 | T3N0M0 | Prim | SCC non-keratinizing | M | 55 | y | |
| 21 | C31.8 | T3N0M0 | Rec | SCC non-keratinizing | M | 64 | y | |
| 22 | C31.0 | T4N0M0 | Rec | SCC non-keratinizing | M | 41 | n | |
| 23 | С31.8 | T4aN0M0 | Prim | SCC non-keratinizing | M | 61 | n | |
| 24 | C31.0 | T4aN0M0 | Prim | SCC non-keratinizing | M | 67 | n | |
| 25 | Larynx n=20 | C32.1 | T1N2M0 | Prim | SCC non-keratinizing | М | 48 | n |
| 26 | C32.0 | T2N0M0 | Prim | SCC non-keratinizing | F | 55 | n | |
| 27 | С32.8 | T2N0M0 | Prim | SCC non-keratinizing | M | 70 | n | |
| 28 | С32.0 | T2N0M0 | Prim | SCC non-keratinizing | M | 69 | y | |
| 29 | C32.0 | T2N0M0 | Prim | SCC non-keratinizing | М | 58 | y | |
| 30 | C32.0 | T3N0M0 | Prim | SCC keratinizing | M | 64 | y | |
| 31 | C32.0 | T3N0M0 | Prim | SCC non-keratinizing | M | 62 | n | |
| 32 | C32.0 | T3N0M0 | Prim | SCC keratinizing | М | 62 | n | |
| 33 | C32.8 | T3N0M0 | Rec | SCC non-keratinizing | М | 64 | y | |
| 34 | C32.8 | T3N0M0 | Prim | SCC non-keratinizing | М | 69 | y | |
| 35 | C32.8 | T3N0M0 | Prim | SCC non-keratinizing | М | 58 | y | |
| 36 | C32.9 | T3N0M0 | Prim | SCC keratinizing | M | 61 | n | |
| 37 | C32.8 | T3N1M0 | Prim | SCC non-keratinizing | M | 49 | y | |
| 38 | C32.8 | T3N0M0 | Rec | SCC non-keratinizing | М | 69 | n | |
| 39 | C32.0 | T4N2M0 | Prim | SCC non-keratinizing | М | 64 | n | |
| 40 | C32.0 | T4N0M0 | Prim | SCC keratinizing | М | 71 | y | |
| 41 | C32.8 | T4aN0M0 | Prim | SCC keratinizing | M | 72 | y | |
| 42 | C32.8 | T4aN0M0 | Prim | SCC non-keratinizing | M | 58 | n | |
| 43 | С32.8 | T4aN2aM0 | Prim | SCC non-keratinizing | M | 70 | y | |
| 44 | C32.8 | T4aN2bM0 | Rec | SCC non-keratinizing | F | 81 | n |
| Gene | Forward primer sequence (5’ → 3’) |
Reverse primer sequence (5’ → 3’) |
Product size (bp*) |
|---|---|---|---|
| ATM | GTTGGTTATTGGTGGATATGG | TAATTCCAAAACCCAAACTCTTAAC | 696 |
| APC | GTTGGTTATTGGTGGATATGG | AACCTACAAAACCAAAAACCAACTA | 600 |
| CDO1 | GGGAGGATGA ATTTTATAGATTTG |
TAAACTTCCATA ATAACCTACACCTC |
396 |
| RB1 | GATAGGGATGAGGTTTATAGTTATTTATTA | AAAATCCTATCACCATTCTACAAAC | 770 |
| TP53 | GGATTATTTGTTTTTATTTGTTATGG | CAAAACTCCACTCCTCTACCTAAAC | 495 |
| WIF1 | GAGTGATGTT TTAGGGGT |
CCTCAACCA AAACTATTCC |
464 |
| Patients (n) Genes |
Sample | All (n=44) |
TNM ** | Type of lesion | SCC | Age | Smokers | ||||||
| before 50 years (n=10) | after 50 years (n=34) |
Yes | No | ||||||||||
| T2 (n=7) |
T3 (n=20) |
T4 (n=15) |
Prim (n=37) |
Rec (n=7) | non-keratinizing (n=30) | keratinizing (n=14) | (n=20) |
(n=24) |
|||||
| M ± m, Range | |||||||||||||
| APC | T | 0.36±0.17 (0.01÷0.54) |
0.31±0.14 (0.05÷0.47) |
0.32±0.18 (0.01÷0.54) |
0.31±0.16 (0.01÷0.52) |
0.30±0.18 (0.01÷0.54) |
0.36±0.16 (0.01÷0.47) |
0.36±0.14 (0.01÷0.54) |
0.24±0.20 (0.01÷0.52) |
0.32±0.17 (0.01÷0.50) |
0.31±0.17 (0.01÷0.54) |
0.31±0.16 (0.01÷0.50) |
0.31±0.20 (0.01÷0.54) |
| N | 0.28±0.18 (0.01÷0.56) |
0.22±0.18 (0.07÷0.48) |
0.31±0.18 (0.04÷0.56) |
0.24±0.18 (0.01÷0.56) |
0.27±0.19 (0.01÷0.56) |
0.34±0.14 (0.01÷0.46) |
0.32±0.17 (0.01÷0.56) |
0.21±0.19 (0.01÷0.56) |
0.27±0.18 (0.04÷0.50) |
0.28±0.19 (0.01÷0.56) |
0.27±0.17 (0.01÷0.50) |
0.29±0.20 (0.01÷0.56) |
|
| ATM | T | 0.35±0.18 (0.01÷0.54) |
0.31±0.20 (0.04÷0.54) |
0.34±0.19 (0.01÷0.54) |
0.37±0.18 (0.06÷0.54) |
0.34±0.19 (0.01÷0.54) |
0.38±0.19 (0.01÷0.53) |
0.39±0.18 (0.01÷0.54) |
0.28±0.18 (0.01÷0.53) |
0.30±0.23 (0.01÷0.53) |
0.37±0.17 (0.04÷0.54) |
0.33±0.19 (0.01÷0.54) |
0.37±0.18 (0.04÷0.54) |
| N | 0.30±0.19 (0.01÷0.57) |
0.27±0.22 (0.06÷0.57) |
0.30±0.19 (0.01÷0.56) |
0.29±0.17 (0.07÷0.49) |
0.31±0.19 (0.02÷0.57) |
0.27±0.21 (0.01÷0.49) |
0.36±0.18 (0.01÷0.57) |
0.20±0.18 (0.01÷0.55) |
0.29±0.19 (0.02÷0.50) |
0.30±0.19 (0.01÷0.57) |
0.30±0.18 (0.02÷0.57) |
0.30±0.20 (0.01÷0.56) |
|
| CDO1 | T | 0.41±0.16* (0.18÷0.76) |
0.30±0.11 (0.18÷0.48) |
0.44±0.16* (0.18÷0.76) |
0.42±0.16* (0.18÷0.71) |
0.38±0.15* (0.18÷0.71) |
0.51±0.18* (0.26÷0.76) |
0.43±0.15* (0.18÷0.71) |
0.36±0.16* (0.18÷0.66) |
0.38±0.15* (0.18÷0.66) |
0.41±0.16* (0.18÷0.76) |
0.44*±0.16 (0.18÷0.76) |
0.36±0.16 (0.18÷0.71) |
| N | 0.20±0.11 (0.01÷0.53) |
0.20±0.10 (0.08÷0.34) |
0.19±0.10 (0.01÷0.34) |
0.19±0.11 (0.01÷0.32) |
0.19±0.11 (0.01÷0.34) |
0.23±0.11 (0.01÷0.34) |
0.21±0.10 (0.01÷0.34) |
0.17±0.10 (0.01÷0.32) |
0.17±0.13 (0.01÷0.32) |
0.21±0.09 (0.01÷0.34) |
0.20±0.11 (0.01÷0.34) |
0.20±0.10 (0.01÷0.34) |
|
| TP53 | T | 0.10±0.06 (0.01÷0.20) |
0.08±0.05 (0.01÷0.18) |
0.09±0.06 (0.01÷0.20) |
0.11±0.06 (0.01÷0.20) |
0.10±0.06 (0.01÷0.20) |
0.10±0.08 (0.01÷0.20) |
0.10±0.06 (0.01÷0.18) |
0.10±0.04 (0.01÷0.17) |
0.10±0.06 (0.01÷0.20) |
0.10±0.06 (0.01÷0.20) |
0.10±0.07 (0.01÷0.20) |
0.10±0.05 (0.01÷0.18) |
| N | 0.09±0.07 (0.01÷0.21) |
0.08±0.07 (0.01÷0.19) |
0.09±0.06 (0.01÷0.20) |
0.09±0.07 (0.01÷0.21) |
0.09±0.07 (0.01÷0.21) |
0.07±0.05 (0.01÷0.16) |
0.08±0.07 (0.01÷0.21) |
0.09±0.06 (0.01÷0.21) |
0.15±0.04 (0.09÷0.21) |
0.07±0.06 (0.01÷0.20) |
0.10±0.07 (0.01÷0.20) |
0.07±0.06 (0.01÷0.21) |
|
| RB1 | T | 0.26±0.12 (0.01÷0.40) |
0.21±0.12 (0.01÷0.36) |
0.27±0.13 (0.01÷0.40) |
0.27±0.12 (0.01÷0.38) |
0.26±0.13 (0.01÷0.40) |
0.27±0.13 (0.01÷0.36) |
0.28±0.11 (0.01÷0.40) |
0.24±0.15 (0.01÷0.38) |
0.27±0.10 (0.06÷0.39) |
0.25±0.14 (0.01÷0.40) |
0.26±0.12 (0.01÷0.39) |
0.27±0.14 (0.01÷0.40) |
| N | 0.25±0.11 (0.01÷0.40) |
0.26±0.11 (0.01÷0.38) |
0.26±0.11 (0.01÷0.40) |
0.23±0.10 (0.01÷0.36) |
0.26±0.10 (0.01÷0.40) |
0.23±0.14 (0.01÷0.36) |
0.26±0.10 (0.01÷0.40) |
0.23±0.12 (0.01÷0.36) |
0.22±0.10 (0.01÷0.34) |
0.24±0.12 (0.01÷0.40) |
0.25±0.10 (0.01÷0.38) |
0.26±0.12 (0.01÷0.40 |
|
| WIF1 | T | 0.30±0.16* (0.08÷0.58) |
0.33±0.15 (0.14÷0.58) |
0.30±0.17 (0.08÷0.58) |
0.29±0.15* (0.09÷0.58) |
0.29±0.15* (0.08÷0.58) |
0.34±0.16* (0.09÷0.56) |
0.26±0.13 (0.08÷0.58) |
0.37±0.18* (0.08÷0.59) |
0.23±0.13* (0.09÷0.55) |
0.32±0.15 (0.09÷0.58) |
0.29*±0.16 (0.08÷0.58) |
0.32±0.16 (0.08÷0.58) |
| N | 0.16±0.08 (0.01÷0.42) |
0.21±0.05 (0.15÷0.30) |
0.18±0.10 (0.01÷0.42) |
0.11±0.05 (0.01÷0.22) |
0.16±0.07 (0.01÷0.42) |
0.18±0.08 (0.08÷0.26) |
0.16±0.09 (0.01÷0.42) |
0.15±0.08 (0.01÷0.30) |
0.10±0.06 (0.01÷0.20) |
0.18±0.08 (0.01÷0.20) |
0.14±0.06 (0.01÷0.26) |
0.18±0.11 (0.01÷0.42) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





