1. Introduction
The CLAVATA3/EMBRYO SURROUNDING REGION (CLE) peptides are a group of hormones that play a crucial role in plant development and growth. In shoot apical meristem (SAM),
CLV3 is expressed and regulates the size of the SAM by down-regulating expression of the
WUSCHEL (
WUS) gene encoding a homeodomain transcription factor. Other
CLE genes, including
CLE1,
CLE4,
CLE6, and
CLE7 can complement the enlarged meristem of the
clv3 mutants when they are expressed under the control of the
CLV3 promoter [
1,
2]. At the same time, the
CLE1,
CLE3,
CLE4, and
CLE7 genes are induced in roots under nitrogen-deficient conditions and regulate root growth and branching in response to nitrogen availability [
3].
CLE genes have been identified in various plant species, including
Arabidopsis thaliana, rice, and maize. Previously we have identified
CLE genes in potato
Solanum tuberosum L. (
StCLE) and found that expression level of several
CLE genes is regulated by nitrogen supply [
4]. Nitrogen is a macronutrient essential for plant growth and development, and it is often a limiting factor in crop production. Adequate nitrogen supply is necessary for the formation of healthy and high-yielding potato tubers. During the early stages of tuber development, nitrogen plays a role in the formation of stolons, which are stem-like structures that develop from the lower nodes of the potato plant. As tuber development progresses, nitrogen is required for the accumulation of starch and other nutrients in the tubers. Nitrogen deficiency during this stage can result in reduced tuber size and lower yield. However, excessive nitrogen supply can also have negative effects on tuber development. Excessive nitrogen levels can cause an overabundance of vegetative growth, which can have a negative impact on tuber development, ultimately resulting in lower yields and lower quality tubers. Therefore, it is important to maintain a balanced nitrogen supply throughout the growing season to ensure optimal tuber development in potato plants.
We previously found that the StCLE4 was induced in potato roots under N-rich growth conditions and suggested that StCLE4 could be a negative regulator of tuberization. In this study, we show that the StCLE4 promoter drives GUS reporter activity in the roots on N-rich media. Plants with overexpression of StCLE4 demonstrated a wus-like phenotype with arrested growth of the shoot apical meristem (SAM). Overexpression of StCLE4 also resulted in stimulation of root growth and non-swelling stolons that were converted into branches. The transcriptomic studies of StCLE4 overexpression plants revealed repression of the IT1 gene, a regulator of potato tuber initiation. Overall, overexpression phenotype and RNA-Seq expression profiles suggest roles of StCLE4 in regulation of shoot, stolon, and root growth.
2. Results
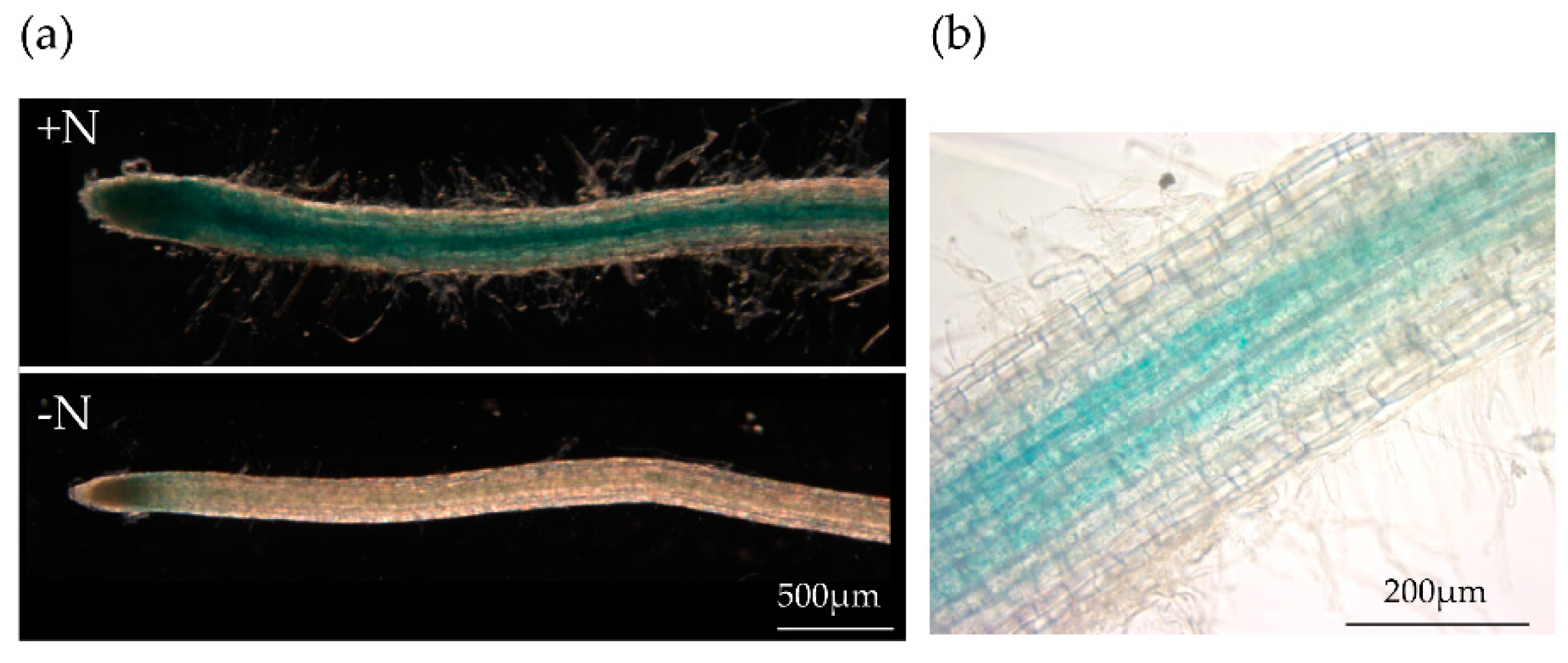
3.1. Promoter Activity of StCLE4 Gene in Potato Roots
The StCLE4 gene belongs to the same clade of CLEs as the nitrate-regulated CLE genes of A. thaliana (AtCLE1, -3, -4, and -7) [
4] that are induced upon N deprivation and suppress the initiation of lateral roots [
3]. Previously, we evaluated the expression of the StCLE4 gene in the roots of potato exposed to different N-availability conditions using RT-qPCR. Opposite to A. thaliana CLE genes, the StCLE4 gene was positively regulated by nitrogen supply [
4]. In this study to determine the expression pattern of StCLE4 in potato roots, we generated promCLE4:GUS fusion constructs using 2963 bp of the genomic region upstream of StCLE4. The StCLE4 promoter drove GUS activity in the vascular tissues of the root (
Figure 1b), and the reporter gene was expressed most strongly on N-rich media (
Figure 1a), confirming that this gene is activated under high nitrogen conditions.
3.2. Effects of Overexpressing StCLE4 and StCLE4G6T Genes on Root, Shoot and Stolon Growth
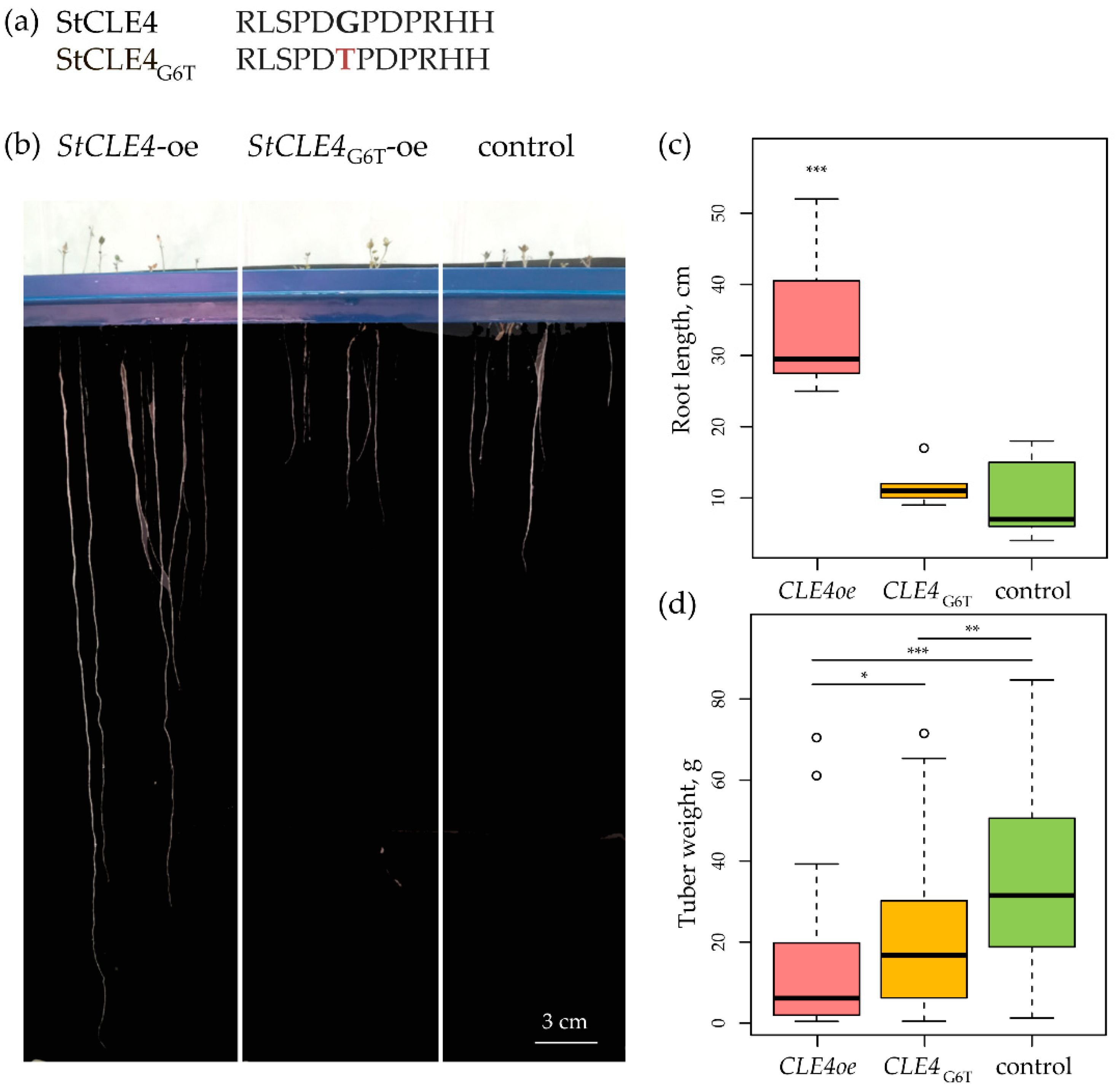
To further investigate the functions of StCLE4, we used antagonistic peptide technology [
5]. Specifically, we created an antagonistic StCLE4 construct by mutating a codon encoding glycine in the CLE motif-coding region (which is known to play a crucial role in the biological activity of CLE) to threonine-coding codon (G6T) (
Figure 2a). Vectors carrying the StCLE4 and StCLE4
G6T genes under the constitutive 35S promoter were obtained. Overexpression of StCLE4 resulted in stimulation of root growth in aeroponic nutrient medium without nitrogen (
Figure 2bc) and reduction of tuber weight (
Figure 2c). At the same time, we did not detect any alterations in growth of StCLE4
G6T overexpressing potato plants, except lower tuber weight (
Figure 2,
Figure S2).
Overexpression of StCLE4 also demonstrated phenotype similar to the phenotypes described for the overexpression of CLV3, CLE2, -3, -4, -5, -6, -7, -9, -10, -11, and -13 genes in A. thaliana [
6]. The observed phenotypes were characterized by arrest of leaf development from the SAM and subsequent recovery via the activation of axillary buds (
Figure 3a), which is very similar to wus loss-of-function mutants [
7]. In contrast to Arabidopsis plants overexpressing CLV3, CLE9, -10, -11, and -13, all potato plants overexpressing StCLE4 resumed growth after the initial SAM arrest (
Figure 3a). Also non-swelling stolons that converted into aerial shoot branches were observed on plants overexpressing StCLE4 (
Figure 3b,
Figure S2).
3.3. RNA-Sequencing of Transgenic Potato Leaves and Roots with StCLE4 and StCLE4G6T Overexpression
To study the effect of gene overexpression on the potato transcriptome, we performed RNA sequencing analysis of leaves and roots of transgenic potato plants overexpressing the StCLE4 and StCLE4
G6T genes. RNA was isolated from both nitrogen-rich and nitrogen-free media grown plants. The obtained RNA-seq data were analyzed using bioinformatics tools to identify differentially expressed genes (DEGs) between transgenic and control plants. Gene ontology analysis was performed to identify the biological processes affected by StCLE4 and StCLE4
G6T overexpression. The results showed that the overexpression of the StCLE4 and StCLE4
G6T genes altered the expression of genes involved in regulation of shoot development, response to gibberellin, leaf development, auxin homeostasis and other processes (
Table S2). Based on our RNA-Seq data we supposed that long-root phenotype can be caused by altering expression of auxin response genes, including SMALL AUXIN-UP RNAs (SAURs), Glycoside hydrolase 3 (GH3) and auxin response factor (
Table S2). Among DEGs in StCLE4 overexpressing leaves, the Identity of Tuber 1 (StIT1) gene was found. StIT1 encodes a TCP transcription factor that determines tuber identity and interacts with the tuberization signal SP6A. The stolons of it1 knockout mutants were converted into aerial shoot branches, instead of swelling [
8]. Based on our RNA-Seq analysis and the observed effects of StCLE4 overexpression it could be speculated that the StCLE4 peptide may initiate a signaling pathway that results in the inhibition of the StIT1 gene expression.
3.4. Yeast one-hybrid assay of interaction between StNLPs and the promoters of the StCLE4, StBEL5, StSP6A, and StIT1 genes
We next performed yeast one-hybrid (Y1H) assay to identify putative regulators of StCLE4. Previously we found, that crucial regulator of the photoperiod-dependent tuberization StBEL5 did not bind to DNA sequence corresponding to 500 bp upstream of the start codon of the StCLE4 gene [
4]. In this study we performed analysis of direct binding of key nitrate regulators NLPs to the promoter regions of StCLE4. NIN-like proteins (NLPs) are a family of transcription factors that play important roles in plant growth and development, as well as in plant-microbe interactions. The name "NIN-like" comes from the fact that these proteins are structurally similar to the Nodule Inception (NIN) protein, which is involved in the formation of root nodules in legume plants. One of the key functions of NLPs is the regulation of nitrate signaling and metabolism [
9,
10]. In Arabidopsis, the AtNLP6 and AtNLP7 proteins are crucial in regulating the expression of genes that respond to nitrate [
9,
10,
11]. Using NLP protein sequences from A. thaliana, Medicago truncatula, and Solanum lucopersicum as queries, three StNLP genes homologues to AtNLP6 and AtNLP7 were identified (StNLP3.1 (Soltu.DM.08G004830.1), StNLP3.2 (Soltu.DM.08G004830.2) and StNLP5 (Soltu.DM.08G029670.1)) (
Figure S3). Among two variants of StNLP3 gene, StNLP3.1 was selected for further investigation due to its higher expression levels in compared to StNLP3.2 (
Table S2) (means of transcripts per million (TPM) values are 1722 and 80, respectively). The StNLP5 gene is also highly expressed in all samples (mean of TPM = 2037).
NLPs are known to bind to nitrate-response elements (NREs) and activate the expression of nitrate-regulated genes in response to high nitrate levels [
11]. We searched for the NREs in the promoters of the StCLE4 gene, and in promoters of major regulators of tuber development (StBEL5, StSP6A, and StIT1 genes) using the motifs described in the studies by Nishida et al. [
12] and Laffont et al. [
13] In the promoters of the StIT1, StCLE4, and StBEL5 genes we found one, two and three putative NLP-binding sites, respectively (
Figure 4a,
Table S3). Next, we checked whether these putative NREs motifs could be bound by StNLP3.1 and StNLP5 using yeast one-hybrid assay. We found that StNLP3.1 and StNLP5 bound to the promoter of the StIT1 gene whereas no interaction was detected for the StCLE4 and StBEL5 promoters (
Figure 4b).
3. Discussion
Nitrogen is a crucial nutrient for the growth and development of potato plants, including tuber development. Previously, we found that the
StCLE4 gene was positively regulated by nitrogen supply and suggested that StCLE4 could be a negative regulator of tuberization. In this study we found that the
StCLE4 overexpressing plants demonstrated
wus-like shoots with arrested growth of the SAM and subsequent recovery of shoot growth via the activation of axillary buds. This phenotype was similar to that described for overexpression of
CLV3,
CLE2,
-3,
-4,
-5,
-6,
-7,
-9,
-10,
-11, and
-13 genes in
A. thaliana [
6]. Like Arabidopsis plants with
AtCLE1-7 overexpression, potato plants overexpressing the
StCLE4 gene resumed growth after the initial SAM arrest. Furthermore, the StCLE4 is close to these AtCLE proteins on phylogenetic tree [
4]. Besides this,
StCLE4 overexpression promoted root growth, and similar effect was also observed for
AtCLE2,
AtCLE4-7 overexpression [
6]. However, opposite to
A. thaliana CLE1,
-3,
-4, and
-7 genes, the
StCLE4 gene was positively regulated by nitrogen supply, like the
AtCLE2 and
AtCLE6 genes [
3]. At the same time
AtCLE2 and
AtCLE6 promoters drive specific GUS staining in the cells at the junction between the primary root and the lateral roots [
14], whereas
StCLE4 reporter expression was detected in the vascular tissues of the root. Therefore, StCLE4 is closely related to Arabidopsis AtCLE1-7 peptides according to phylogenetic analysis and functional studies; however, regulation of
StCLE4 activity in potato is different from that described for the
AtCLE1-7 genes.
Overexpression of
StCLE4 not only affected shoot apex and root growth but also promoted stolon conversion into branches. We performed of RNA-Seq analysis of leaves and roots of transgenic and control plants. At the same time downregulation of the
Identity of Tuber 1 (
StIT1) gene was found in the leaves of the
StCLE4 overexpressing plants grown in N-deficient media. IT1 is a transcription factor that directly interacts with mobile tuberigen, SP6A, and regulates tuber identity [
8]. The stolons of
it1 knockout mutants were converted into branches, instead of swelling [
8]. Opposite to
it1 mutants,
StCLE4 overexpressing plants produced tubers, but tuber weight of these tubers was lower than controls. Therefore, we suppose that StCLE4 promotes growth of stolons as shoots, similar to milder
it1 phenotype, and that
StCLE4 overexpressing phenotype could be explained by downregulation of
IT1, that in turn regulates stolon swelling.
The NLPs transcription factors can respond to nitrate signaling, regulate gene expression, and participate in N absorption and utilization. Previously, it was shown that NLP can regulate the expression of the
CLE genes [
13,
15,
16], and therefore we assumed that StNLP3 and StNLP5 can bind to the promoter region of
StCLE4. However, we did not detect this interaction in yeast one-hybrid system. Moreover, previously, we also did not find interaction of photoperiod-dependent tuberization factor StBEL5 and the promoter region of the
StCLE4 gene [
4], and therefore, we still do not know which transcription factor regulates the
StCLE4 gene expression. However, we found that StNLP3 and StNLP5 bound to the promoter of
StIT1. Previously, it was shown that N plays an important role in regulating tuberization [
17], and based on our data, we speculate that the StCLE4 peptide, as well as the StNLP3 and StNLP5 transcription factors, may act on N-depended tuberization pathway and regulate the expression of
StIT1, however, further investigations are necessary to study the mechanisms of such regulation in more detail. Further research on the mechanisms of
CLE genes regulation and their interactions with nitrogen signaling pathways will be crucial for understanding how plants respond to changes in nitrogen availability and for developing strategies to improve crop productivity.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Tubers of the
Solanum tuberosum cultivar Desirée was obtained from the stock of N. I. Vavilov All-Russian Institute of Plant Genetic Resources (Saint-Petersburg, Russia). The plants were propagated by two-node stem cuttings and were cultivated
in vitro on a Murashige and Skoog (MS) medium containing 0.8% (w/v) agar and 1% sucrose under long-day conditions (16 h light:8 h dark) at 22 °C. N-depleted or N-rich medium was prepared as described in [
18]. For investigation of effects of
StCLE4 and
StCLE4G6T overexpression on root growth, aeroponic nutrient medium without nitrate was used [
19]. Seven days old stem cuttings were transferred into aeroponic system in a controlled greenhouse (16 h light:8 h dark and 22 °C) and were grown for four weeks. The root length was measured using the ImageJ [
20] software, and a Tukey's test was used to compare the means of three groups. Stem cuttings were also transferred to the pots (9x9x9 cm) with vermiculite and grown in growth chamber for three months (16 h light:8 h dark and 22°C first two months; 8 h light:16 h dark and 22 °C for induction of tuber development). Twice a month plants were supplied with ¼ MS media without sucrose. Tubers, obtained from plants growing in the vermiculite, were planted in the field (Leningrad Region, Russia) in June 2023 and were grown for 2.5 months. Tuber weight of plants grown in the field was measured, and a Tukey's test was used to compare the means of three groups.
4.2. Constructs
The primers used for PCR are listed in
Table S2. Polymerase chain reaction (PCR) with Phusion High-Fidelity DNA Polymerase (Thermo Scientific, USA) was used to amplify coding sequences or predicted promoter fragments of genes from whole plant DNA of
Solanum tuberosum L. cv Desiree. PCR fragments were extracted from an agarose gel using a Cleanup Mini Kit (Evrogen, Russia) and cloned into the entry vector pDONR207. Then, coding sequences were cloned into the vector pMDC32 [
21], under the CaMV 35S promoter, via Gateway technology, according to the manufacturer’s instructions (Invitrogen, USA). Promoter sequences for GUS or GFP reporter analysis were cloned into the vector pBGWFS7 [
22] or pHm43GW (Invitrogen, USA). For yeast one-hybrid assay, the promoter fragments of the
StCLE4,
StSP6A, and
StIT1 genes (
Table S1) were cloned into the pHISLEU2GW vector (kindly provided by Dr. Rogers from Cardiff University). Coding sequences of
StNLP3.1 and
StNLP5 were introduced into pDEST22 (Invitrogen, USA). The resulting plasmids were transformed into Top10
Escherichia coli Competent Cells. Plasmid DNA was isolated using Plasmid Miniprep Kit (Evrogen, Russia). Each plasmid was checked by sequencing. The resulting binary vectors were transferred to the
Agrobacterium rhizogenes strain Arqua or
Agrobacterium tumefaciens strain AGL1. Agrobacterium cultures were grown overnight on a shaking incubator at 28-30°C in Luria-Bertani supplemented with antibiotics.
4.3. Transformation of potato
For hairy potato root transformation with the promStCLE4::GUS construct, stems were wounded and inoculated with the A. rhizogenes and then co-cultivated on Murashige and Skoog (MS) media with 10 g/L sucrose for 2 days. Transgenic hairy roots were grown in MS medium supplemented with cefotaxime (500 mg/L) to prevent Agrobacterium overgrowth.
Transgenic
S. tuberosum plants containing prom35S::StCLE4, prom35S::CLE4G6T constructs were generated by
Agrobacterium tumefaciens-mediated transformation mainly as described previously [
23,
24,
25]. The wounded leaf explants were incubated with agrobacteria in liquid MS medium with 16 g/L glucose for 2 days. Then the explants were moved to solidified MS medium (8 g/L agar) that induces callus growth (16 g/L glucose; 500mg/L cefotaxime; 3 mg/L hygromycin; 5 mg/L NAA; 0.1 mg/L BAP) and were incubated for 7–8 d under LD at 27°C. The explants were later transferred to a shoot regeneration MS medium (16 g/L glucose; 500mg/L cefotaxime; 3 mg/L hygromycin; 1 mg/L BAP; 0.1 mg/L GA) and were repeatedly transferred to fresh medium with the same composition every 10 days until shoots developed on the calli. The resulting 3 cm shoots were then transferred to MS medium (10 g/L sucrose; 500mg/L cefotaxime) for rooting.
4.4. Histochemical Assays
β-Glucuronidase (GUS) staining was performed as described [
26]. Incubation periods ranged from 2 to 16 h following vacuum infiltration. Roots were imaged using a ZEISS SteREO Discovery.V12 microscope.
4.5. RNA Isolation and RNA-Seq
For RNA-Seq analyses, total RNA was extracted from samples using the RNeasy Plant Mini Kit (Qiagen, USA). Enrichment of mRNA was performed using NEBNext® Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, USA). The libraries were prepared using NEBNext® Ultra™ II Directional (New England Biolabs, USA). For sequencing HiSeq PE Rapid Cluster Kit v2 and HiSeq Rapid SBS Kit v2 was used. Manufacturer’s instructions were used for all procedures. The libraries were sequenced on the Illumina HiSeq 2500 sequencing platform.
Reads quality control was performed with FASTQC v. 0.11.9 (
http://www.bioinformatics.babraham.ac.uk/ projects/fastqc/). FastQC reports were aggregated by MultiQC v. 1.14 [
27]. The trimmomatic v. 0.39 program [
28] was used to trim reads from technical artifacts. HISAT2 v. 2.2.1 [
29] was used as alignment program for mapping reads to a doubled monoploid potato DM 1-3 516 R44 genome v. 6.1. BAM files were sorted by samtools v. 1.13. Quantitation of RNA-Seq data was performed by StringTie v. 2.2.1 [
30]. The
StCLE and
StCEP (C-terminally encoded peptide) [
31] genes that have not been annotated previously as coding sequences were manually included in annotation. Differential gene expression analysis was performed using DeSeq2 v. 1.40.2 [
32] in R.
2.6. Yeast One-Hybrid Assay
The 2.5-kb sequences upstream of start-codon were used for the promoter analysis. Putative NRE motifs were identified using sequences revised in several recent studies [
12,
13] and MAST algorithms available on the MEME suite server (version 5.1.0,
http://alternate.meme-suite.org/tools/meme). Transformation of
Saccharomyces cerevisiae strain Y2HGold (Clontech) was performed as described in [
33]. A yeast one-hybrid assay was performed as described in [
34].
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org. Figure S1: The phenotype of
StCLE4G6T overexpressing plants that were grown in growth chamber; Figure S2: The phenotype of
StCLE4 (
StCLE4-oe) and
StCLE4G6T (
StCLE4G6T-oe) overexpressing plants, and control plants that were grown in the field; Figure S3: Phylogenetic tree of
Arabidopsis thaliana,
Medicago truncatula,
Solanum lycopersicum,
Oryza sativa,
Lotus japonicas, and
Solanum tuberosum NLP protein families; Table S1: Differentially expressed genes (DEGs) in leaves and roots from nitrogen-rich media grown control and transgenic plants; Table S2: List of primers; Table S3: Potential NRE sites in promoter regions of
StCLE4,
StBEL5,
StIT1, which were used for yeast-one-hybrid analyses.
Author Contributions
Conceptualization, L.L. and M.G.; methodology, M.G.; validation, M.G.; investigation, M.G.; resources, L.L. and M.G.; writing, M.G.; funding acquisition, M.G. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the Russian Science Foundation (grant no. 22-76-00022).
Data Availability Statement
All sequence reads of the RNA-Seq analysis have been deposited at the National Center for Biotechnology Information database under BioProject accession number PRJNA1007918.
Acknowledgments
We thank Dr. Maria Lebedeva (Department of Genetics and Biotechnology, Saint Petersburg State University) for discussions and critical reading of the manuscript. We are grateful to Alexey Masharsky (Centre for Molecular and Cell Technologies of Saint Petersburg State University Research Park) for library preparation and RNA sequencing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ni, J.; Clark, S. E. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant physiology. 2006, 140 (2), 726–733. [CrossRef]
- Meng, L.; Ruth, K. C.; Fletcher, J. C.; Feldman, L. The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity. Molecular plant. 2010, 3(4), 760–772. [CrossRef]
- Araya, T.; Von Wirén, N.; Takahashi, H. CLE peptides regulate lateral root development in response to nitrogen nutritional status of plants. Plant Signal. Behav. 2014, 9, e29302-1-3. [CrossRef]
- Gancheva, M.; Dodueva, I.; Lebedeva, M.; Lutova, L. CLAVATA3/EMBRYO SURROUNDING REGION (CLE) Gene Family in Potato (Solanum tuberosum L.): Identification and Expression Analysis. Agronomy 2021, 11, 984. [CrossRef]
- Song, X. F.; Guo, P.; Ren, S. C.; Xu, T. T.; Liu, C. M. Antagonistic peptide technology for functional dissection of CLV3/ESR genes in Arabidopsis. Plant physiology. 2013, 161(3), 1076–1085. [CrossRef]
- Strabala, T. J.; O'donnell, P. J.; Smit, A. M.; Ampomah-Dwamena, C.; Martin, E. J.; Netzler, N.; Nieuwenhuizen, N. J.; Quinn, B. D.; Foote, H. C.; Hudson, K. R. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant physiology. 2006, 140(4), 1331–1344. [CrossRef]
- Laux, T.; Mayer, K. F.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996, 122(1), 87–96. [CrossRef]
- Tang, D.; Jia, Y.; Zhang, J.; Li, H.; Cheng, L.; Wang, P.; Bao, Z.; Liu, Z.; Feng, S.; Zhu, X.; Li, D.; Zhu, G.; Wang, H.; Zhou, Y.; Zhou, Y.; Bryan, G. J.; Buell, C. R.; Zhang, C.; Huang, S. Genome evolution and diversity of wild and cultivated potatoes. Nature. 2022, 606(7914), 535–541. [CrossRef]
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nature communications. 2013, 4, 1713. [CrossRef]
- Castaings, L.; Camargo, A.; Pocholle, D.; Gaudon, V.; Texier, Y.; Boutet-Mercey, S.; Taconnat, L.; Renou, J. P.; Daniel-Vedele, F.; Fernandez, E.; Meyer, C.; Krapp, A. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. The Plant journal: for cell and molecular biology. 2009, 57(3), 426–435. [CrossRef]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nature communications. 2013, 4, 1617. [CrossRef]
- Nishida, H.; Nosaki, S.; Suzuki, T.; Ito, M.; Miyakawa, T.; Nomoto, M.; Tada, Y.; Miura, K.; Tanokura, M.; Kawaguchi, M.; Suzaki, T. Different DNA-binding specificities of NLP and NIN transcription factors underlie nitrate-induced control of root nodulation. The Plant cell. 2021, 33(7), 2340–2359. [CrossRef]
- Laffont, C.; Ivanovici, A.; Gautrat, P.; Brault, M.; Djordjevic, M.A.; Frugier, F. The NIN Transcription Factor Coordinates CEP and CLE Signaling Peptides That Regulate Nodulation Antagonistically. Nat. Commun. 2020, 11, 3167. [CrossRef]
- Jun, J.; Fiume, E.; Roeder, A. H.; Meng, L.; Sharma, V. K.; Osmont, K. S.; Baker, C.; Ha, C. M.; Meyerowitz, E. M.; Feldman, L. J.; Fletcher, J. C. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant physiology. 2010, 154(4), 1721–1736. [CrossRef]
- Nishida, H.; Tanaka, S.; Handa, Y.; Ito, M.; Sakamoto, Y.; Matsunaga, S.; Betsuyaku, S.; Miura, K.; Soyano, T.; Kawaguchi, M.; Suzaki, T. A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat Commun. 2018, 9, 499. [CrossRef]
- Soyano, T.; Hirakawa, H.; Sato, S.; Hayashi, M.; Kawaguchi, M. Nodule Inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proceedings of the National Academy of Sciences of the United States of America. 2014, 111(40), 14607–14612. [CrossRef]
- Ewing, E.E. The Role of Hormones In Potato (Solanum Tuberosum L.) Tuberization. In: Davies, P.J. (eds) Plant Hormones. Springer, Dordrecht. 1995. [CrossRef]
- Ohkubo, Y.; Tanaka, M.; Tabata, R.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 2017, 3, 17029. [CrossRef]
- Barker D.G.; Pfaff, T.; Moreau, D.; Groves, E.; Ruffel, S.; Lepetit, M.; Whitehand, S.; Maillet, F.; Nair, R.M.; Journet, E.–P. Growing M. truncatula: choice of substrates and growth conditions. In: Mathesius. U; Journet, E.–P.; Sumner, L.W. eds. The Medicago truncatula handbook. 2006. http://www.noble.org/MedicagoHandbook/.
- Schneider, C. A.; Rasband, W. S.; Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012, 9(7), 671–675. [CrossRef]
- Curtis, M. D., Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant physiology. 2003, 133(2), 462–469. [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in plant science. 2002, 7(5), 193–195. [CrossRef]
- Beaujean, A.; Sangwan, R.S.; Lecardonnel, A.; Sangwan-Norreel B.S. Agrobacterium-mediated transformation of three economically important potato cultivars using sliced internodal explants: An efficient protocol of transformation. J Exp Bot. 1998, 49(326):1589–95. [CrossRef]
- Banerjee A.K.; Prat, S.; Hannapel, D.J. Efficient production of transgenic potato (S. tuberosum L. ssp. andigena) plants via Agrobacterium tumefaciens mediated transformation. Plant Sci. 2006, 170, 732–738. [CrossRef]
- Kolachevskaya, O. O.; Alekseeva, V. V.; Sergeeva, L. I.; Rukavtsova, E. B.; Getman, I. A.; Vreugdenhil, D.; Buryanov, Y. I.; Romanov, G. A. Expression of auxin synthesis gene tms1 under control of tuber-specific promoter enhances potato tuberization in vitro. Journal of integrative plant biology. 2015, 57(9), 734–744. [CrossRef]
- Ilina, E. L.; Logachov, A. A.; Laplaze, L.; Demchenko, N. P.; Pawlowski, K.; Demchenko, K. N. Composite Cucurbita pepo plants with transgenic roots as a tool to study root development. Annals of Botany. 2012, 110(2): 479–489. [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [CrossRef]
- Love, M.I; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014, 15, 550. [CrossRef]
- Rutkovskaya, E.A.; Gancheva, M.S.; Lebedeva, M.A.; Lutova, L.A. Identification and Expression Analysis of CEP Genes in Potato. Russ J Genet. 2022, 58, 751–755. [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [CrossRef]
- Davies, S.E.W. Transcription factor interactions at the promoter of the Arabidopsis circadian clock gene LHY. PhD thesis, University of Warwick, England, 2013.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).