1. Introduction
The most common route for drug administration is oral (Stewart et al. 2016). Within the gastrointestinal system, duodenum and jejunum play the most crucial role in drug pharmacokinetics due to their large surface area and the abundance of transporters and metabolizing enzymes (Doherty und Pang 1997; Berggren et al. 2007). Therefore, appropriate 2D cell based models both mimicking the human intestinal epithelium in vitro and predicting the absorption of novel drug candidates are highly important. In drug discovery, the Caco-2 model has been widely used for ADME (absorption, distribution, metabolism, and excretion) purposes. This cell line is known to develop a well-differentiated polarized cell monolayer, form tight junctions (TJ) and microvilli structures in culture (Lea 2015). However, major drawbacks of the Caco-2 model are its adenocarcinoma origin and its considerably different gene and protein expression pattern of drug transporters (DT) and nuclear receptors (NRs) compared to jejunal tissue (Brück et al. 2017). Hence, Caco-2 cells are not fully representative of human small intestinal physiology.
The small intestinal epithelium is crucial for the absorption of nutrients and drugs and protects the gut against harmful xenobiotics by forming a tight barrier. Differentiated cell types mediate these functions: absorptive (enterocytes), enteroendocrine- (EECs), mucosecreting- (goblet cells, GCs) and Paneth cells (PCs) (Sancho et al. 2003), with enterocytes and GCs being most prevalent (Cheng und Leblond 1974; Potten 1998). In the apical membrane of enterocytes, several uptake (solute carrier-transporters; SLCs) and ATP-binding cassette (ABC) efflux transporters are located. SLCs facilitate drug absorption and are the most abundant transporters in the small intestine (Ménochet et al. 2022), while ABC transporters limit intestinal absorption and promote the excretion of endogenous substances (Hosomi et al. 2012). In addition, NRs function as modulators of drug metabolizing enzymes and DT expression (Urquhart et al. 2007).
In a previous study, we described a panel of ten 2D colorectal cancer cell lines with long lasting barrier integrity due to their basal activity and induction potential of Cytochrome P450 (CYP) 3A4. Of those lines, two candidates with higher CYP3A4 induction potential than Caco-2 cells were identified (Przybylla et al. 2022). The aim of the present study was to deepen this analysis by the assessment of intestinal cell differentiation and DT regulation at the transcriptional and protein level. Additionally, morphological and functional cell characteristics were visualized by immunofluorescent staining and scanning electron microscopy (SEM) as a measure of cellular differentiability.
2. Materials and Methods
2.1. Chemicals and reagents
Rifampin (RIF) was obtained from Carl Roth (Karlsruhe, Germany) and vitamin D3 (VD3) from Hycultec (Beutelsbach, Germany). Stock solutions of RIF (121.5 mM) and VD3 (1 mM) were prepared in DMSO and ethanol, respectively. The final concentration of DMSO was 0.1%.
2.2. Cell Culture
All ten HROC (Hansestadt Rostock, colorectal cancer) cell lines were obtained from the HROC collection (Mullins et al. 2019). Caco-2 cells were purchased from CLS (Eppelheim, Germany). Cell culture was performed as previously described (Przybylla et al. 2022).
For functional studies, cells were treated with 20 µM RIF and 100 nM VD3, respectively. Controls were treated with 0.1% DMSO or ethanol. Cells were harvested after incubation of 24 h, 48 h and 72 h.
For gene expression analysis and protein quantification, cells were seeded in 6-well plates (Sarstedt, Nümbrecht, Germany) at a density of 5 x 105 and were grown until reaching 60-70% confluence and ten days post-confluence, respectively. Cells were scraped and centrifuged at 4 °C at 1,200 rpm for 7 min, shock frozen with liquid nitrogen and stored at -20 °C for further analysis.
2.3. Gene Expression Analysis
Total RNA was isolated using Universal RNA Purification Kit (Roboklon, Berlin, Germany) by following the manufacturer’s protocol. RNA quantity was assessed by using NanoDrop 1000 spectrophotometer and ND-1000 spectrophotometer V3.7.1 software (Thermo Fisher Scientific, Waltham, USA). 1,000 ng RNA was reverse transcribed into cDNA by using reverse transcriptase (200 U/µl, Bioron, Römerberg, Germany). For quantative real time PCR (qRT-PCR) was performed using the Sibir Rox Hot Mastermix (Bioron) and 10 µM each of sense and antisense primers according to the manufacturer’s protocol. Samples were run on a ViiA7 Real-Time PCR system (Applied Biosystems, Thermo Fisher Scientific) using the following PCR conditions: 50 °C 2 min, 95 °C 10 min, followed by 35 cycles of: 95 °C 15 sec., 58 °C 1 min, 45 sec. 70 °C, melting curve: 95 °C 15 sec., 60 °C 15 sec., 95 °C 15 sec. Ct-values were normalized to GAPDH and relative mRNA expression levels were calculated by the 2-∆∆Ct method. For culture time-dependent gene expression analysis three normal epithelium biopsies from human small intestine (obtained from the BioBank Rostock) were pooled and included as reference.
Table 1.
Primers used for RT-PCR.
Table 1.
Primers used for RT-PCR.
| Gene name |
5’-3’ |
Sequences |
Product size (bp) |
| ABCB1 (P-gp) |
Forward
Reverse |
GCTGTCAAGGAAGCCAATGCCT
TGCAATGGCGATCCTCTGCTTC |
120 |
| ABCG2 (BCRP) |
Forward
Reverse |
GTTCTCAGCAGCTCTTCGGCTT
TCCTCCAGACACACCACGGATA |
145 |
| GAPDH |
Forward
Reverse |
GAAGGTGAAGGTCGGAGTC
GAAGATGGTGATGGGATTTC |
226 |
| SLC2A2 (GLUT-2) |
Forward
Reverse |
ATGTCAGTGGGACTTGTGCTGC
AACTCAGCCACCATGAACCAGG |
131 |
| SLCO2B1 (OATP2B1) |
Forward
Reverse |
TGGGCACAGAAAACACACCT
CGGCTGCCAAAATAGCTCAC |
265 |
| OCLN |
Forward
Reverse |
ATGGCAAAGTGAATGACAAGCGG
CTGTAACGAGGCTGCCTGAAGT |
124 |
| NR1I2 (PXR) |
Forward
Reverse |
GCTGTCCTACTGCTTGGAAGAC
CTGCATCAGCACATACTCCTCC |
124 |
| UGT1A6 |
Forward
Reverse |
GCAAAGCGCATGGAGACTAAGG
GGTCCTTGTGAAGGCTGGAGAG |
148 |
| NR1I1 (VDR) |
Forward
Reverse |
CGCATCATTGCCATACTGCTGG
CCACCATCATTCACACGAACTGG |
101 |
2.4. Immunocytochemistry
13 mm-diameter cover slips (Carl Roth) were sterilized with absolute ethanol and placed in 24-well plates (Sarstedt). Cells (104 cells per well) were seeded and grown until reaching confluence. All samples were fixed with 4% paraformaldehyde (Carl Roth) for 10 min at RT (room temperature); only for Zonula occludens-1 (ZO-1) staining fixation was done with 1:1 methanol/acetone for 20 min at -20 °C, respectively. After rinsing twice with PBS, samples were permeabilized with 0.5% Triton X-100 (AppliChem, Darmstadt, Germany) for 5 min at RT. Cells were washed and incubated with primary antibodies (diluted in PBS, given in
Table 2) for one hour at RT and then overnight at 4 °C, protected from light. Cells were washed and Hoechst 33342 (diluted 1:250 in PBS; Hycultec) was incubated for 5 min in the dark at RT. For imaging, the CytoViva® Enhanced Darkfield microscope system (CytoViva, Inc., Auburn, AL, USA) with a 60x oil objective and Ocular Software V2.0 (QImaging Ocular Software, Surrey, British Columbia, Canada) was used. Contrast adjustment and image overlay was performed by using Adobe Photoshop CS5 (Adobe Inc., San Jose, CA).
2.5. Protein Quantification by Liquid Chromatography–Mass Spectrometry/Mass Spectrometry (LC-MS/MS)
Protein abundance of PEPT1, OATP2B1, P-gp and BCRP and were determined using a validated LC-MS/MS-based targeted proteomics method (Gröer et al. 2013; Wenzel et al. 2021; Wenzel et al. 2022).
Cells were lysed and the membrane protein fraction was extracted using the ProteoExtract Native Membrane Protein Extraction kit (Merck KGaA, Darmstadt, Germany) according to the manufacturer’s protocol. The obtained membrane fraction was subjected to determination of the whole protein concentrations using the bicinchoninic acid assay (Thermo Fisher Scientific) and was adjusted to a maximum protein concentration of 2 mg/ml. Subsequently, 100 µL of each membrane fraction were mixed with 10 µL dithiothreitol (200 mM, Sigma-Aldrich, Taufkirchen, Germany), 40 µL ammonium bicarbonate buffer (50 mM, pH 7.8, Sigma-Aldrich), and 10 µL ProteaseMAX™ (1%, m/v, Promega, Mannheim, Germany) and incubated for 20 min at 60 °C (denaturation). After cooling down, 10 µL iodoacetamide (400 mM, Sigma-Aldrich) were added and the samples were incubated in a darkened water quench for 15 min at 37 °C (alkylation). For protein digestion, 10 µL trypsin (trypsin/protein ratio: 1/40, Promega) was added and the samples were incubated in a water quench for 16 h at 37 °C. Digestion was stopped by addition of 20 µL formic acid (10% v/v, Sigma-Aldrich). All samples were stored at -80 °C until further processing. The samples were centrifuged one more time for 15 min at 16,000 g and 4 °C. Finally, 50 µL of the digested membrane fraction were mixed with 50 µL isotope-labeled internal standard (IS) peptide mix (10 nmol/L of each IS). For preparation of the calibration curves, digested human serum albumin (2 mg/ml) was used as blank matrix and spiked with reference peptides to reach 0.1 - 25 nmol/L and with IS (final concentration, 5 nmol/L of each peptides). The following proteospecific peptides were used: P-gp, AGAVAEEVLAAIR; BCRP, SSLLDVLAAR; OATP2B1, SSPAVEQQLLVSGPGK und PEPT1, TLPVFPK. All peptides (purity > 97-99%) were from Thermo Fisher Scientific.
All sample preparation and digestion steps were performed using Protein LoBind tubes (Eppendorf, Hamburg, Germany). Protein quantification was conducted on a 5500 QTRAP triple quadrupole mass spectrometer (AB Sciex, Darmstadt, Germany) coupled to an Agilent Technologies 1260 Infinity system (Agilent Technologies, Berlin, Germany).
For each peptide, 3-4 mass transitions were monitored; the final protein abundance data (picomoles/mg protein) were calculated by normalization to the total protein content of the isolated membrane fraction.
2.6. Scanning Electron Microscopy
SEM was used to examine cell morphology and the extent of villous structures present in confluent HROC and Caco-2 cell monolayers. Cells were seeded on sterilized 13 mm-diameter glass cover slips (Plano #7013, Wetzlar, Germany) at a density of 104 cells per well in 24-well plates (Sarstedt). When reaching confluence, cells were washed with PBS containing Ca2+ and Mg2+ on the cover slips and then were fixed with a solution containing 2% glutaraldehyde and 1% paraformaldehyde in 0.1 M sodium phosphate buffer at pH 7.3 and stored at 4 °C. Further processing for SEM started with two washes in 0,1 M PBS followed by an ascending series of ethanol in distilled water (30% for 2 min, 50% for 5 min, 70%, 90% and pure ethanol 15 min each) prior to drying using CO2 in an Emitech K850 critical point dryer (Emitech /Quorum Technologies Ltd., East Sussex, UK). Surface conductivity was established by sputter coating the specimens with a gold layer of approximately 10-15 nm thickness in a Leica SCD 500 sputter coater (Leica Microsystems, Wetzlar, Germany). Cell surfaces were examined in a field emission scanning electron microscope (Merlin VP Compact, Carl Zeiss, Oberkochen, Germany) using an acceleration voltage of 5 kV at a working distance of 5-6 mm with a high efficiency secondary electron detector for digital image acquisition (image size of 1024x756 pixels).
2.7. Data Analysis
For gene expression analysis, QuantStudio™ Real-time PCR software 1.3 (Thermo Fisher Scientific) was used. Statistical analysis between treated and untreated groups and time to confluent growth on alterations of gene expression and protein levels, the paired two tailed Student’s t-test was used. Spearman’s rank correlation was computed for gene expression data and protein levels. GraphPad Prism software 9.0.0 (GraphPad Software, Inc., San Diego, USA) was used for calculation.
3. Results
3.1. Cell Morphology and -Differentiation
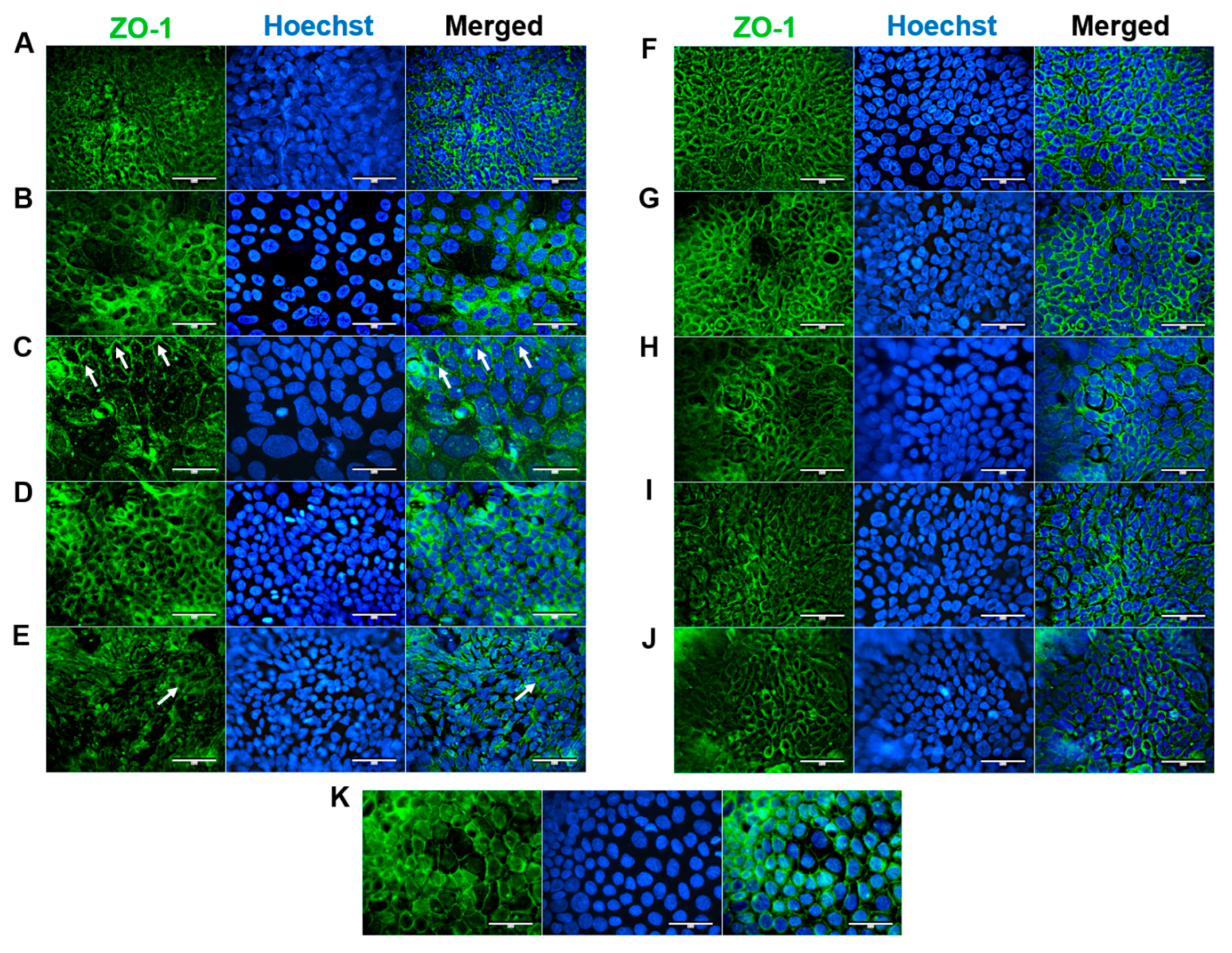
ZO-1 is essential for proper apical surface assembly and is typically localized at discrete sides of cell-cell contacts and within the cytoplasm (Howarth et al. 1992). ZO-1 was expressed in the apical junctions of all confluent HROC cell monolayers (shown in
Figure 1). This corresponds well with stable TEER values reported in our previous study (Przybylla et al. 2022). In HROC43, HROC80 T1 M1, HROC159 T2 M4, HROC183 T0 M2 and HROC217 T1 M2, ZO-1 was well organized and showed a consistent spiderweb-like staining pattern as found in Caco-2 cells (
Figure 1, K). A thin thread-like pattern was found in HROC32 (
Figure 1, A). ZO-1 expression was lower in HROC60, HROC126, HROC239 T0 M1 and HROC383 cells (
Figure 1, C, E, I and J, respectively). However, higher fluorescent signals in particular areas were found for HROC60.
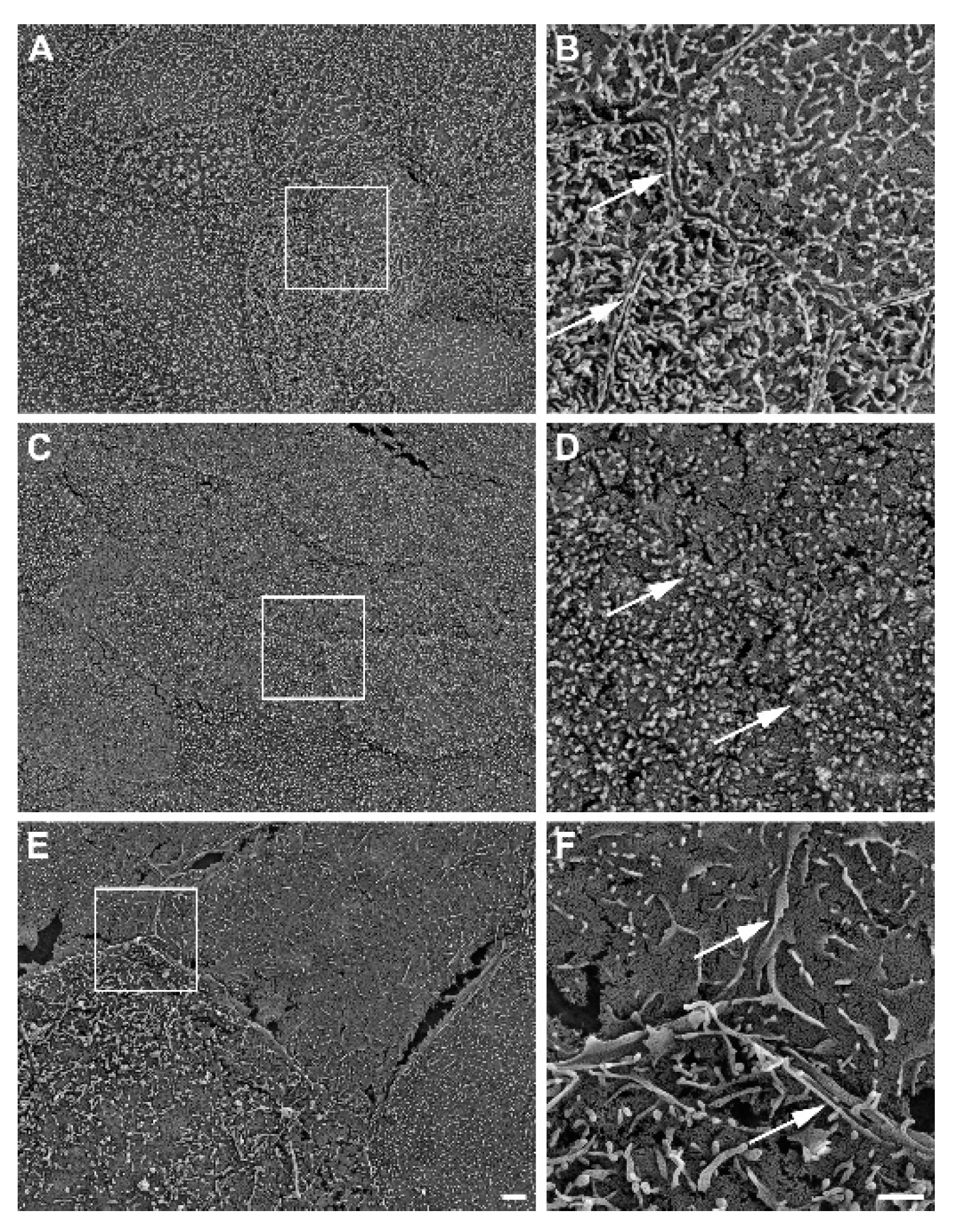
Further, SEM revealed well-developed cell-cell boundaries evident by protruding membrane ridges and alterations in villi decoration at the apical side of confluent HROC monolayers, indicating tight cell apposition. Representative photographs and resolution details are shown for HROC60, HROC159 T2 M4 and Caco-2 cells as a reference in
Figure 2.
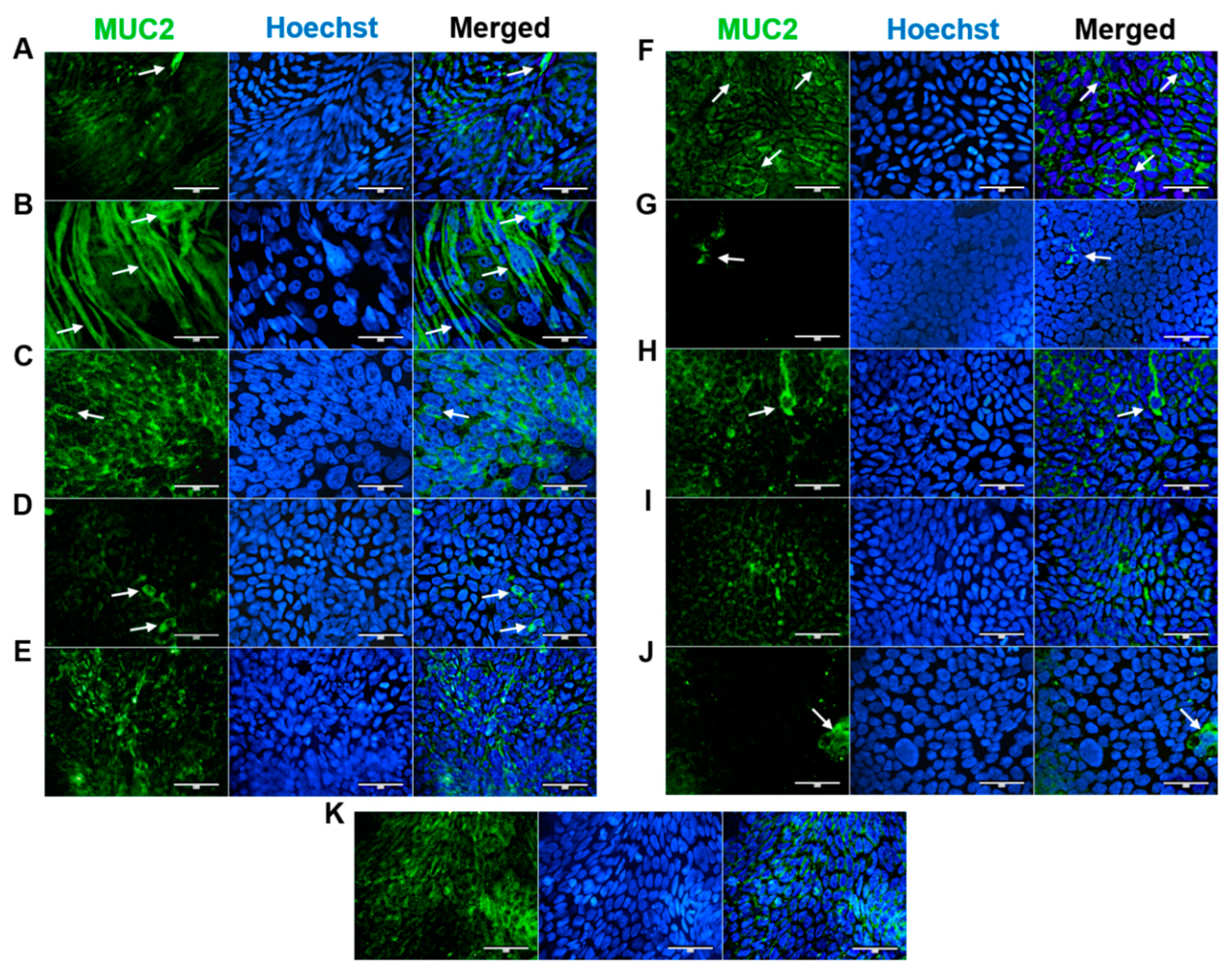
Intestinal mucus producing GCs lubricate luminal contents and are essential in the maintenance of intestinal homeostasis (Herath et al. 2020). Mucin 2 producing cells were clearly identified by a perinuclear staining patterns in HROC32, HROC43, HROC60, HROC80 T1 M1, HROC159 T2 M4, HROC183 T0 M2, HROC217 T1 M2 and HROC383 cells (
Figure 3). In HROC43, a strongly increased cytoplasmic secretion-like staining pattern was detected (
Figure 3, B). Strong homogenous fluorescence patterns were also observed for HROC60 and HROC159 T2 M4. HROC60 was characterized by fine fluorescent droplets and structures lateral to nuclei whereas signals were slightly lower in HROC159 T2 M4 with prominently arranged droplets in cytoplasmatic regions (
Figure 3, C and F). Signals were lower in HROC32, HROC80 T1 M1, HROC183 T0 M2 and HROC217 T1 M2. However, a scattered perinuclear fluorescence was observed in these lines with tail-like structures in HROC32 and HROC217 T1 M2 (
Figure 3, A and H). Unspecific fluorescent patterns were observed in HROC126, HROC239 T0 M1 and Caco-2 with no single cells detected (
Figure 3, E, I and K).
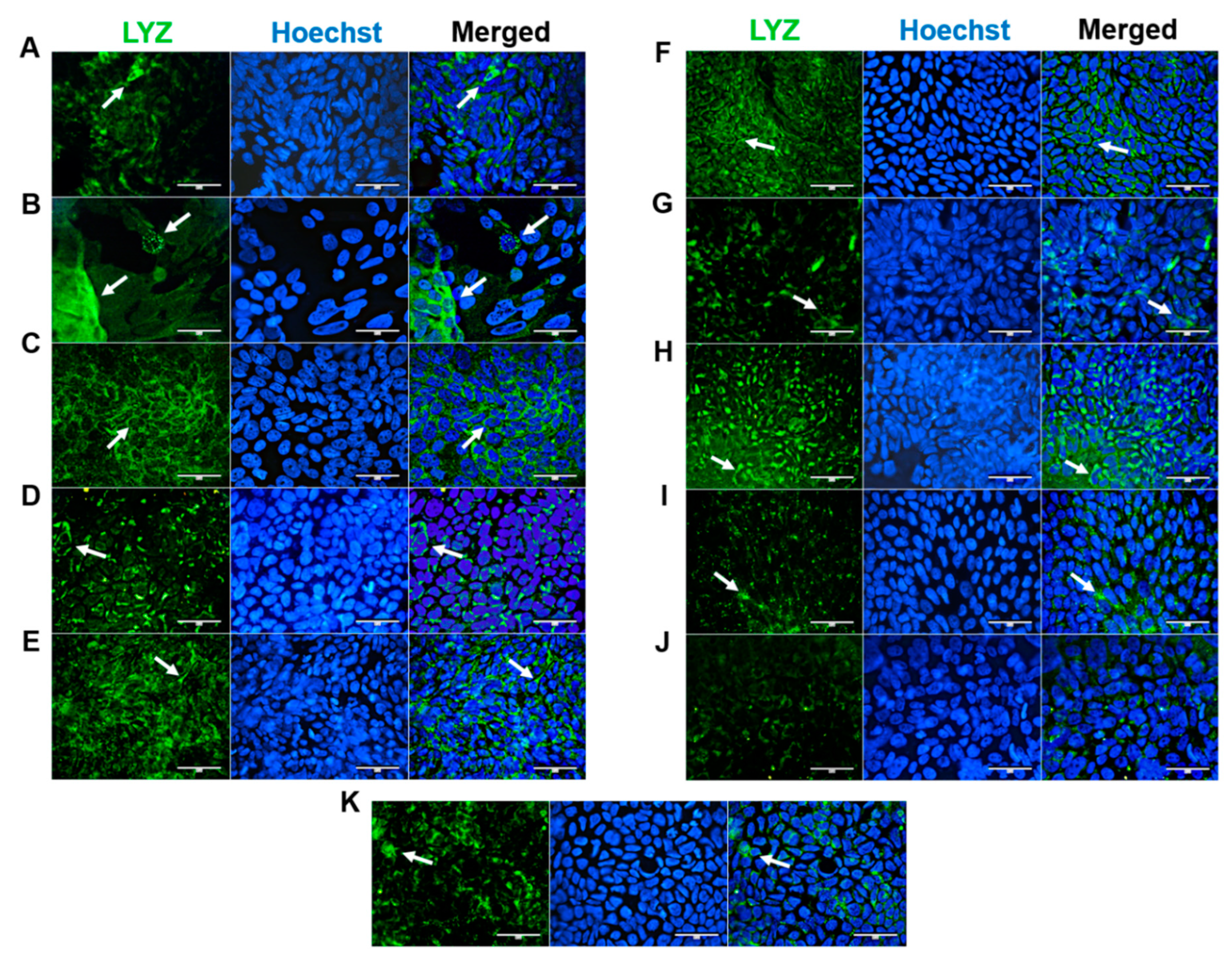
PCs secrete antimicrobial peptides and immunomodulating proteins. They are essential in modulating the microbiome and maintaining intestinal homeostasis (Lueschow und McElroy 2020). Immunostaining with the PC marker lysozyme led to high cytoplasmatic fluorescence in HROC43, characterized by fine droplets in nuclear and perinuclear regions (
Figure 4, B). Homogenous perinuclear fluorescent staining patterns were found for HROC60, HROC126 and HROC159 T2 M4, characterized by finely granular cytoplasmatic fluorescence in HROC60 (
Figure 4, C, E and F). HROC80 T1 M1 und HROC217 T1 M2 demonstrated perinuclear fluorescence patterns with single-side nuclei fitting structures (
Figure 4, D and H). Lower basic signals were observed for HROC32, HROC183 T0 M2 und Caco-2 (
Figure 4, A, G and K). However, partially specific perinuclear fluorescent patterns became evident in these lines. In contrast, a low fluorescence intensity with coarse-dropped structures in perinuclear and nuclear regions was found for HROC239 T0 M1 (
Figure 4, I). Fluorescence intensity was also less pronounced in HROC383, characterized by a weak perinuclear staining pattern (
Figure 4, J).
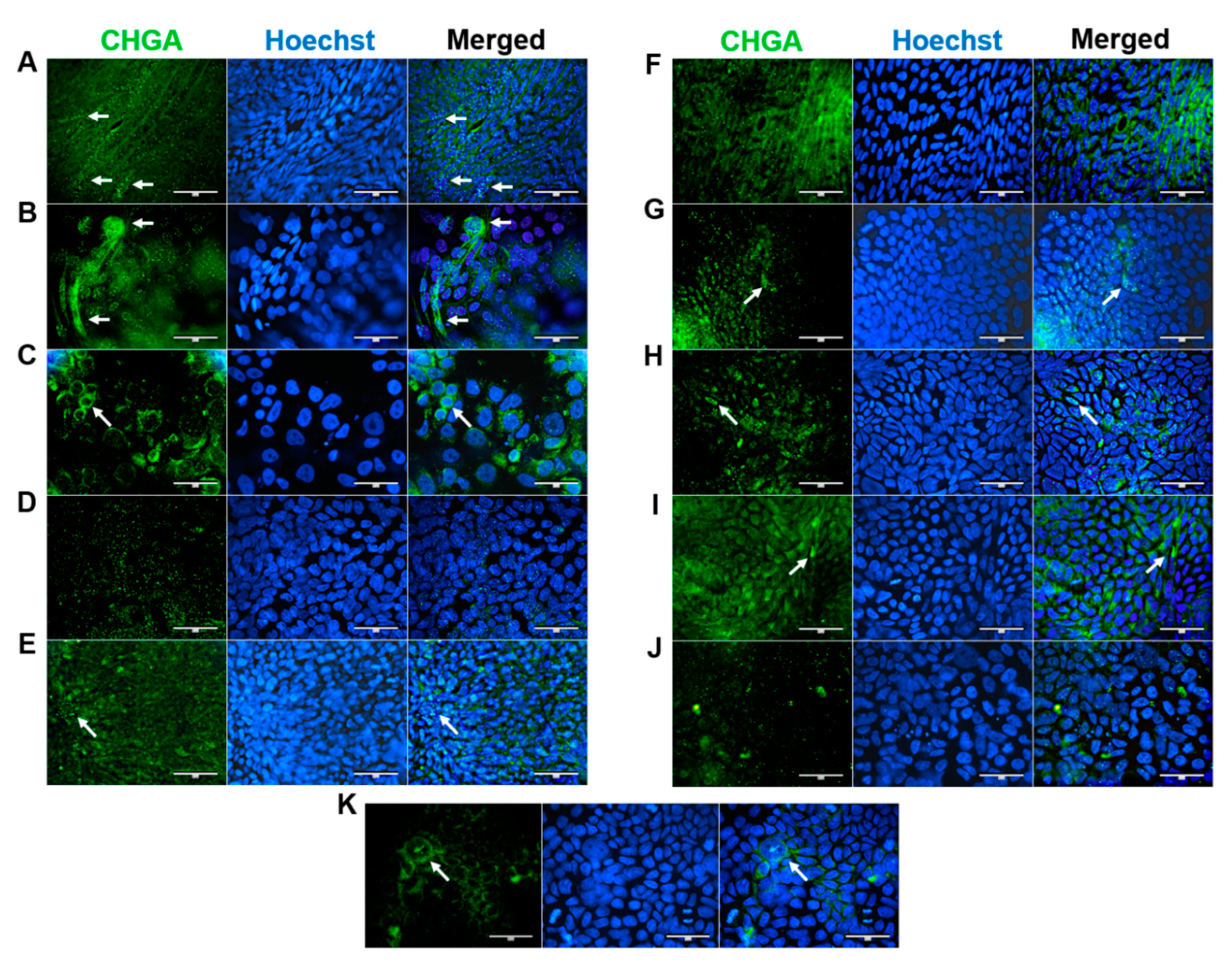
EECs secrete a variety of hormones and regulate digestion, appetite, gut motility and metabolism (Rehfeld 2004). Immunofluorescent staining of the EEC marker chromogranin A revealed finely granular fluorescence in all lines with single cells detected in HROC32, HROC43, HROC60, HROC126, HROC183 T0 M2, HROC217 T1 M2, HROC239 T0 M1 and Caco-2 cells (
Figure 5). Strongest signals were observed for HROC43 and HROC60. Fluorescence pattern of HROC43 was characterized by strong granular stainings in regions with pronounced perinuclear and nuclear fluorescence (
Figure 5, B). HROC60 showed a more homogenous and intense perinuclear fluorescence pattern with less fluorescent granular (
Figure 5, C). HROC183 T0 M2 showed perinuclear, HROC217 T1 M2 occasionally nuclear fluorescence in regions with pronounced granular staining (
Figure 5, G and H). HROC32, HROC126 und HROC159 T2 M4 were characterized by a homogenous cytoplasmatic basic fluorescence that was partly depicted as a perinuclear fluorescence pattern in HROC32 und HROC126 (
Figure 5, A, E and F). In Caco-2, fluorescence intensity was low with perinuclear fluorescence in regions with pronounced granular patterns (
Figure 5, K). Signals were also low in HROC80 T1 M1; however, granular fluorescence became evident in a high number of nuclei (
Figure 5, D). HROC239 T0 M1 was characterized by a prominent homogenous cytoplasmatic basic fluorescence, partially shown as arrowhead-like structures lateral to nuclei (
Figure 5, I). Lowest signals with a weak pronounced granular pattern and perinuclear fluorescence were observed for HROC383 (
Figure 5, J).
In sum, cell differentiation to specific cell types was strongly pronounced in HROC32, HROC43, HROC60, HROC80 T1 M1, HROC159 T2 M4, HROC183 T0 M2 and HROC217 T1 M2 cells. In contrast, cell differentiation was restricted in HROC126, HROC239 T0 M1, HROC383 and Caco-2 cells.
3.2. Detection of Intestinal Brush Border
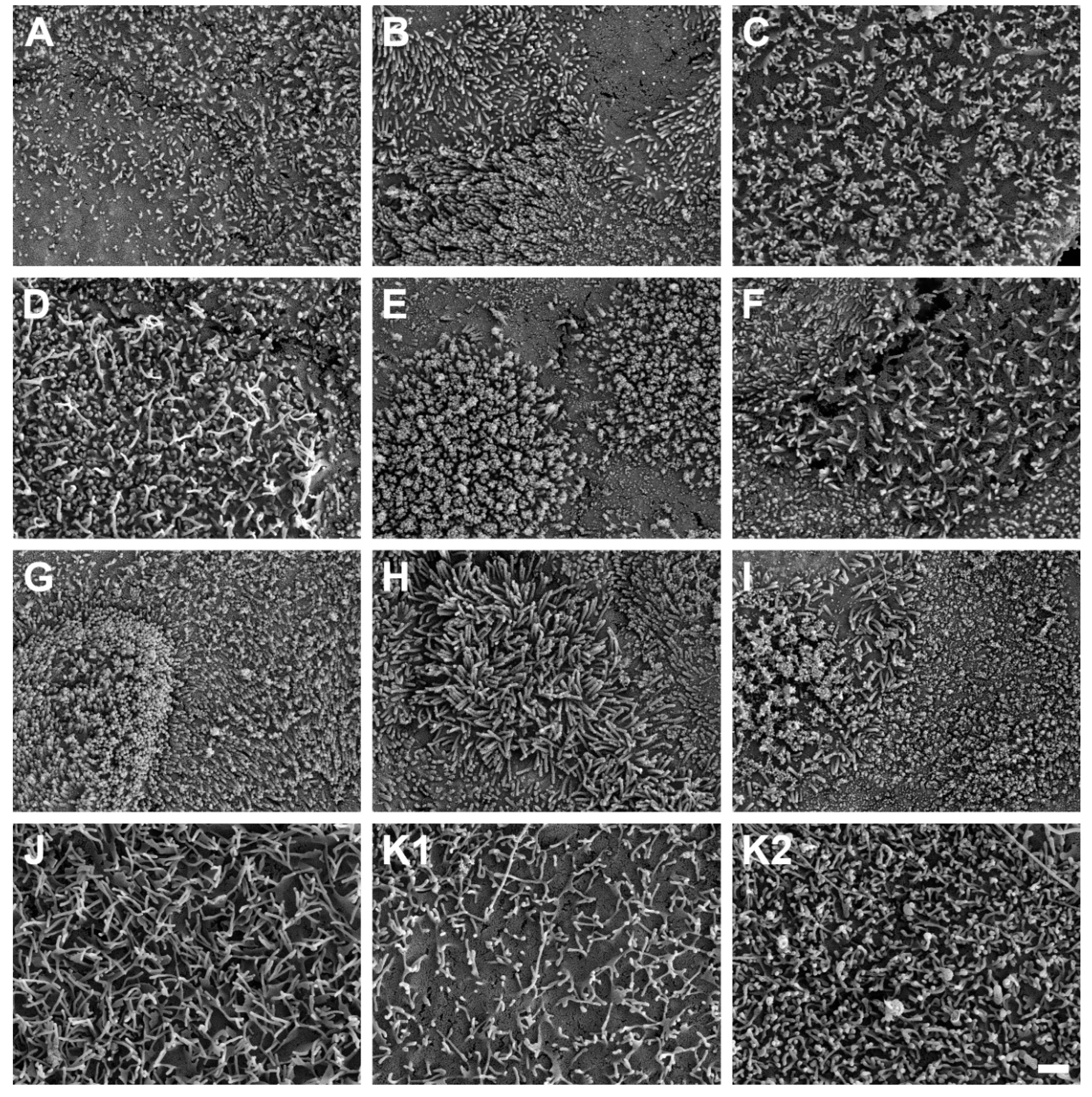
SEM micrographs demonstrated the presence of microvilli on the apical cell surface of all ten HROC cell lines analyzed after growth to cell monolayers (
Figure 6). Microvilli structures and surface decoration differ with respect to density and length of microvilli among the ten HROC cell lines examined (
Figure 6 A-J) and the Caco-2 cells analyzed for reference comparison (
Figure 6, K). For example, longer villous structures with higher densities compared to Caco-2 cells were observed in HROC183 T0 M2 and HROC217 T1 M2 cells (
Figure 6, G and H). Microvilli morphology and decoration in HROC80 T1 M1 cells were similar to Caco-2 cells (
Figure 6, D) whereas an altered density and length of microvilli compared to Caco-2 was observed in HROC60 cells (
Figure 6, C). Variations in the decoration with microvilli structures were also found among individual cells of the same cell line (e.g.
Figure 6, E), indicating that the extent of microvilli development in addition might be influenced by cell state and maturity. To allow for direct comparison, the HROC cells are depicted with fully developed microvilli decoration (
Figure 6, A-J) next to the Caco-2 reference cells for which both low and high cell surface microvilli densities are shown (
Figure 6, K1 and K2). In conclusion, all ten HROC cell lines show microvilli structures which mirror the phenotype of primary human small intestinal epithelial cell monolayers (Speer et al. 2019).
3.3. Culture Time-Dependent Gene Expression
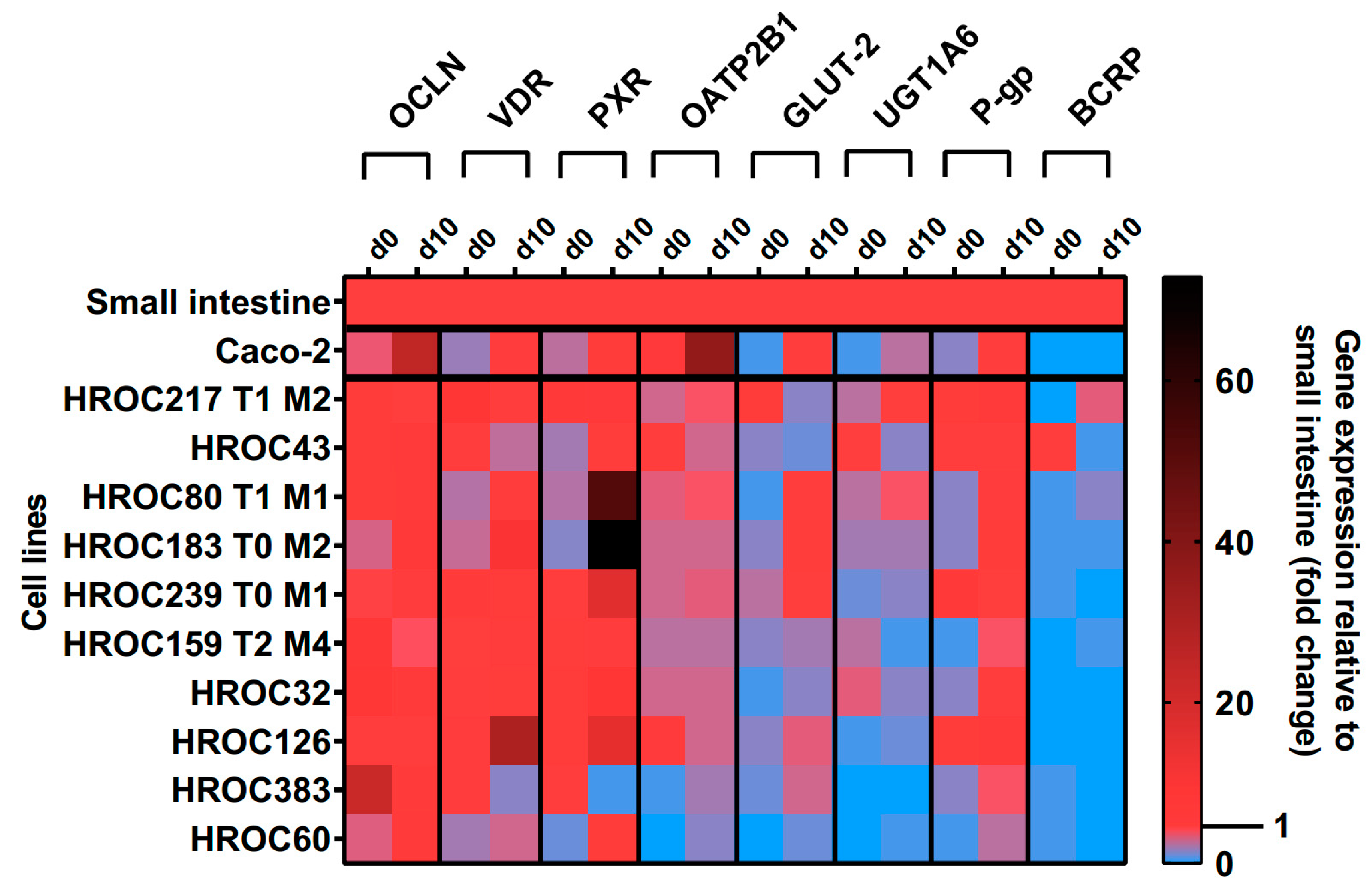
The effects of ten days post-confluence (d10) state on intestine-specific gene expression are shown in
Figure 7. A confluence-dependent increase of OCLN, VDR, PXR, OATP2B1, GLUT-2, UGT1A6 and P-gp mRNAs was most pronounced in HROC60, HROC80 T1 M1 and HROC183 T0 M2. Genes were stably expressed in HROC217 T1 M2 and, with the exception of GLUT-2, confluence-dependent changes were also observed for this line. Lowest effects were observed for HROC60 and HROC383 with expression levels staying distinctly below small intestinal expression. In HROC43, VDR, OATP2B1, UGT1A6 and BCRP mRNAs were higher expressed in the control group than at post-confluence state. UGT1A6 was barely expressed in HROC126 and absent in HROC383. A high up-regulation of PXR mRNA was observed in HROC183 T0 M2 (73-fold) and HROC80 T1 M1 (52-fold) at d10 post-confluence. OCLN and OATP2B1 were overexpressed in Caco-2 cells at post-confluence state (25- and 36- fold change). HROC32, HROC126 and Caco-2 lacked sufficient BCRP expression.
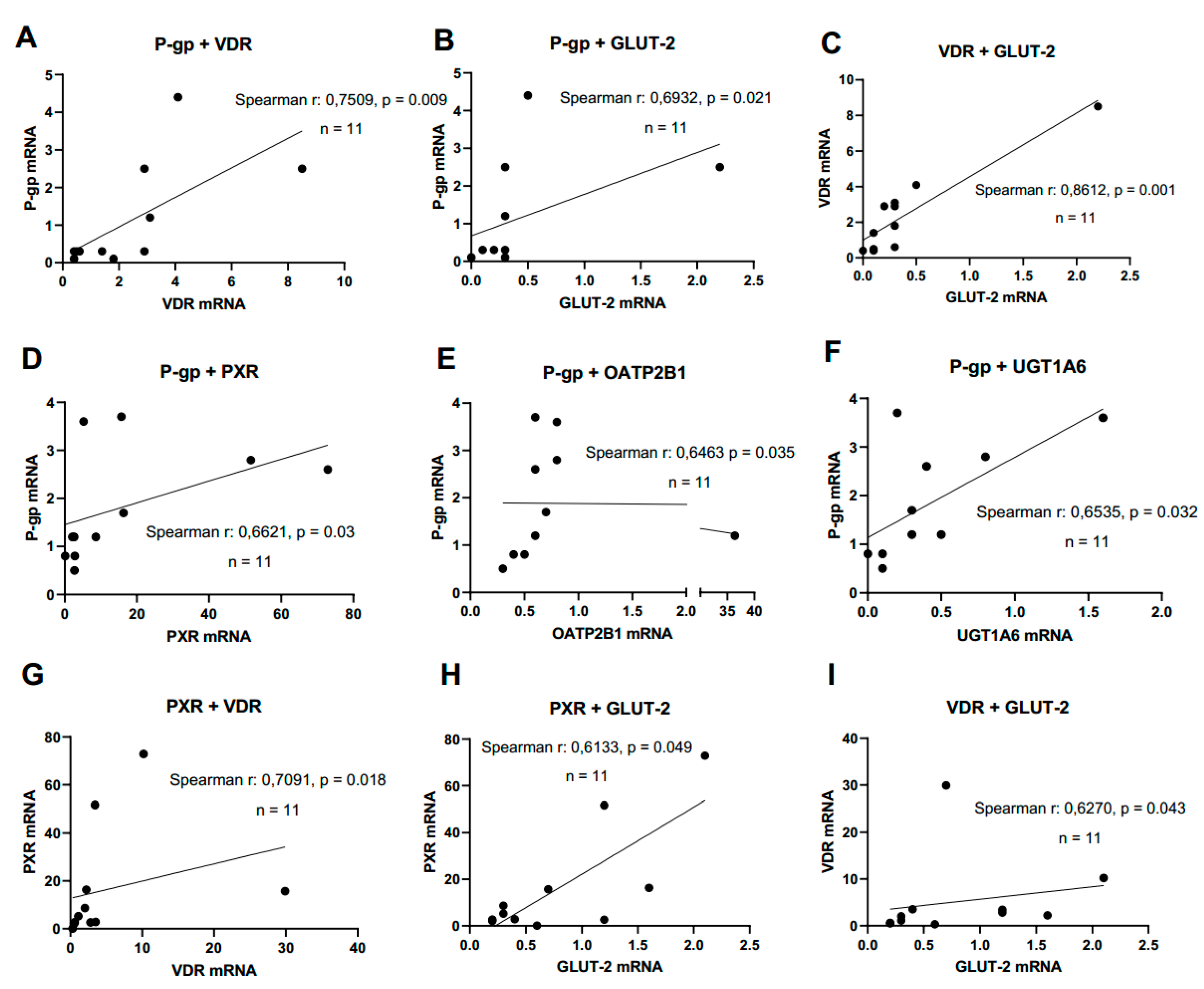
PXR mRNA expression significantly correlated with VDR mRNA (r = 0.709, p = 0.018) and GLUT-2 mRNA (r = 0.613, p = 0.049, shown in
Figure 8). Expression of P-gp mRNA correlated with mRNAs of PXR (r = 0.662, p = 0.03), OATP2B1 (r = 0.646 , p = 0.035) and UGT1A6 (r = 0.654, p = 0.032). Further, a significant correlation was found for VDR and GLUT-2 mRNAs (r = 0.627, p = 0.043) at d10 post-confluence. In sum, significant correlations between the expression of selected transporters and NRs were more abundant with an increase of culture time.
3.4. Culture Time-Dependent Protein Levels
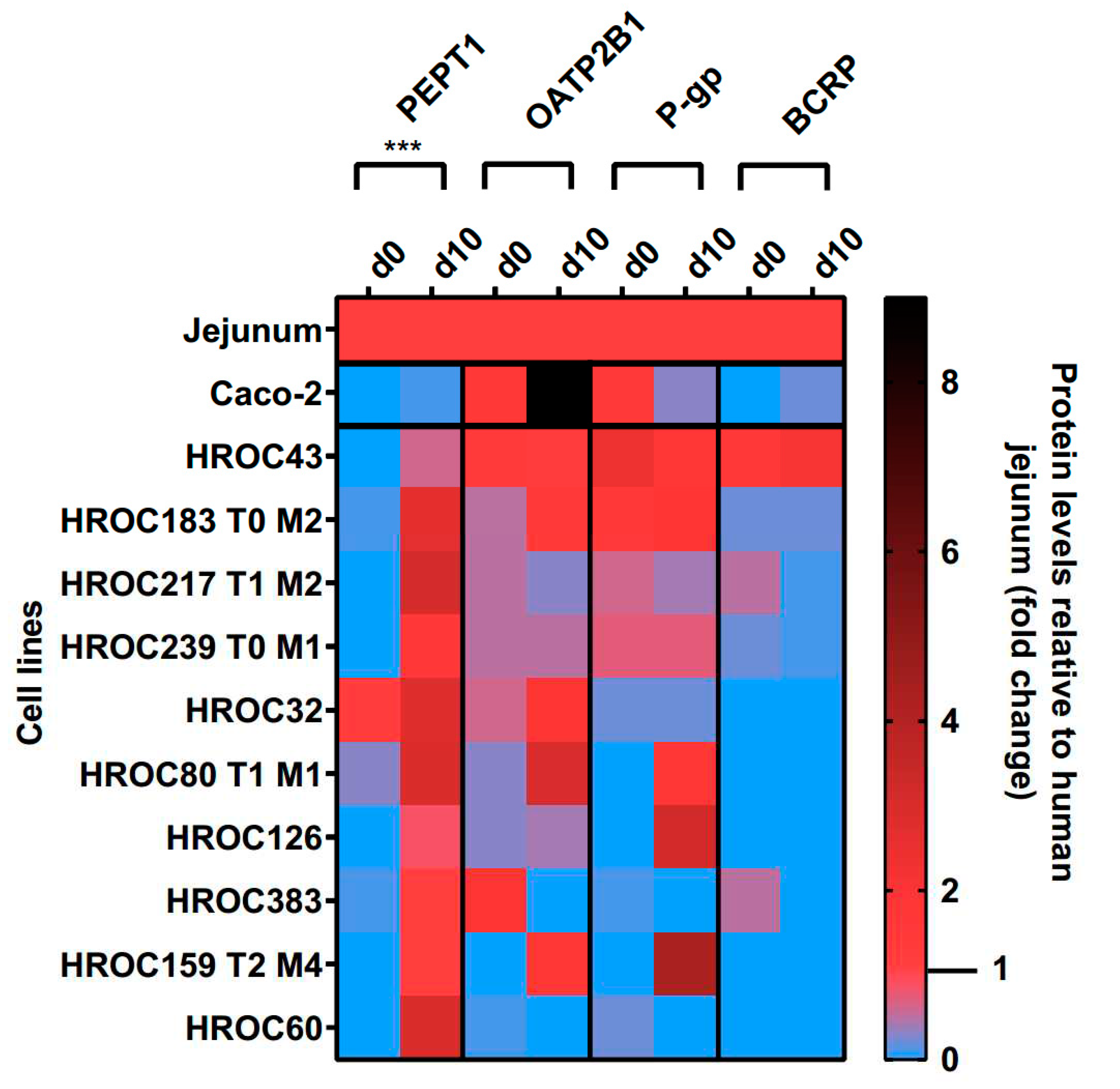
Protein levels of PEPT1, OATP2B1, P-gp and BCRP in the basal and d10 post-confluence state are shown in
Figure 9. A confluence-dependent increase of PEPT1 protein was observed in all lines and strongly correlated with an increase of culture time (p = 0.0004). Referred to human jejunal tissue, higher protein levels of PEPT1 were detected in HROC217 T1 M2 (3.1-fold), HROC60 (2.9-fold), HROC80 T1 M1 (2.9-fold) and HROC32 (2.8-fold), but were lowest in Caco-2 (0.1-fold). PEPT1 protein levels most comparable with human jejunum were found in HROC159 T2 M4 (1-fold), HROC383 (1-fold), HROC126 (0.8-fold) and HROC43 (0.6-fold) at d10 post-confluence state.
OATP2B1, P-gp and BCRP protein levels varied between both groups and were not significantly increased with prolonged confluence. Higher OATP2B1 protein levels compared to jejunum were detected in Caco-2 (9-fold), HROC80 T1 M1 (3-fold), HROC32 (1.9-fold) and HROC159 T2 M4 (1.8-fold) while protein levels most similar to jejunum were found for HROC43 (1.2-fold), HROC183 T0 M2 (1.4-fold) and HROC239 T0 M1 (0.5-fold) at d10 post-confluence state.
P-gp protein was higher abundant in HROC159 T2 M4 (4.2-fold), HROC126 (3.2-fold), HROC183 T0 M2 (1.9-fold) and HROC80 T1 M1 (1.8-fold) compared to levels at basal state with protein levels most similar to jejunum in HROC239 T0 M1 (0.7-fold), HROC217 T1 M2 (0.4-fold), HROC43 (1.8-fold) and HROC80 T1 M1 (1.8-fold). Lowest protein levels were detected in Caco-2 (0.3-fold) and HROC32 (0.2-fold). No expression was found for HROC60 and HROC383 at d10 post-confluence state.
Protein abundance of BCRP was highest in HROC43 (2.1-fold), HROC183 T0 M2 (0.2-fold) and Caco-2 (0.2-fold) and was absent in the remaining lines at d10 post-confluence. However, BCRP protein levels were closer to jejunal protein levels at basal state in HROC217 T1 M2 (0.5-fold), HROC383 (0.5-fold) and HROC239 T0 M1 (0.2-fold) compared to post-confluence.
In sum, protein levels of PEPT1, OATP2B1, P-gp and BCRP were higher in HROC43, HROC183 T0 M2, HROC217 T1 M2 and HROC239 T0 M1 compared to Caco-2 with protein levels closest to human jejunal tissue in HROC43. In total, HROC43 was the single most cell line demonstrating sufficient human jejunal-like protein levels of PEPT1, OATP2B1, P-gp and BCRP. CYP3A4 and PXR protein were absent in all lines.
3.5. Regulation of Intestinal Drug Transporter Expression
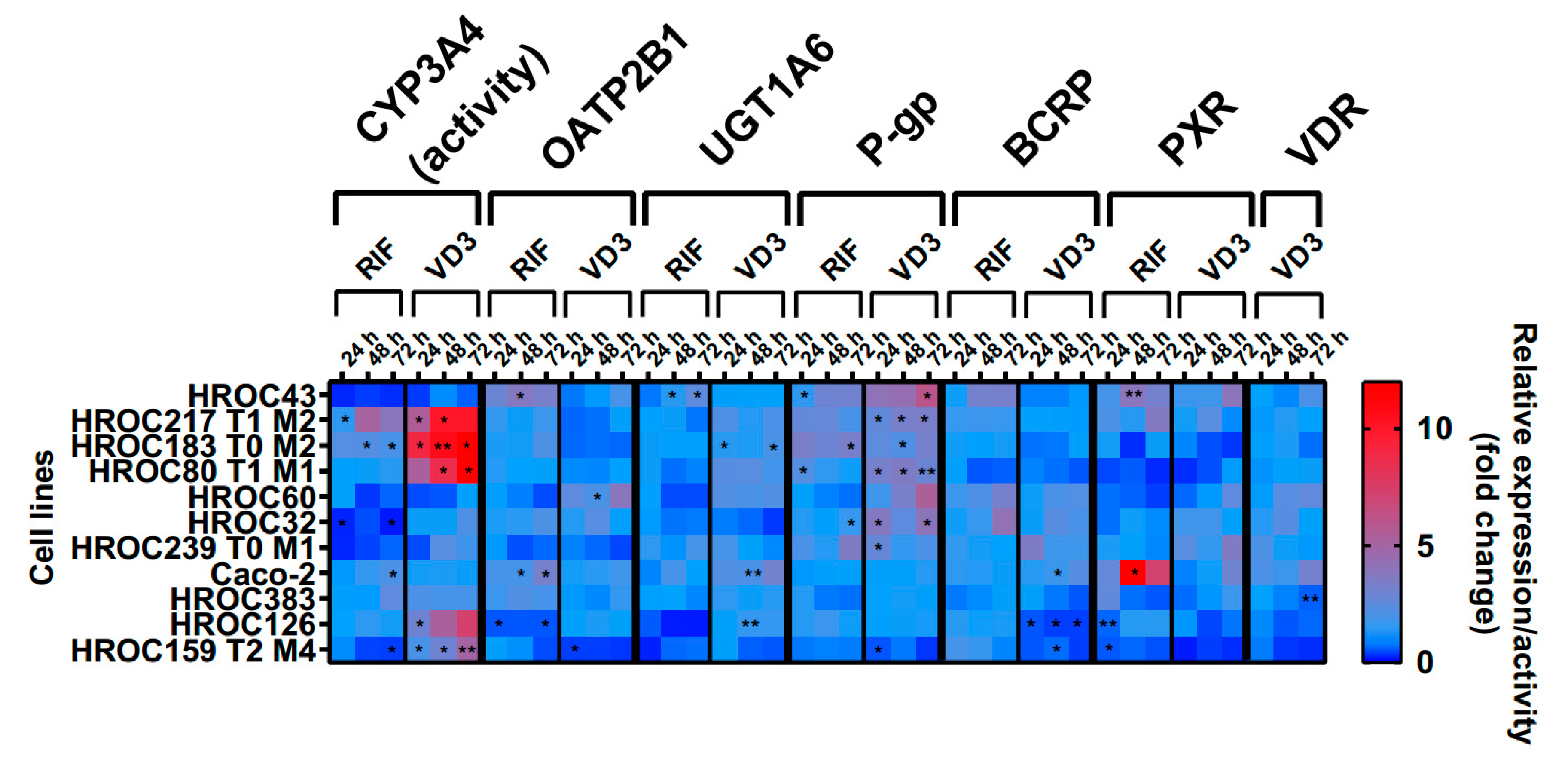
The effects of RIF and VD3 treatment on transporter expression are shown in
Figure 10. These data were combined with CYP3A4 activity levels from a recently published data set (Przybylla et al. 2022). In summary, drug treatment led to a high variability of drug response within all HROC lines. There was a stronger regulation of efflux transporters P-gp and BCRP in six HROC lines compared to Caco-2 cells. VD3 treatment led to higher increase of P-gp mRNA in seven HROC lines compared to Caco-2 cells (1.4-fold change at 72 h). It was highest in HROC43 (6.5-fold change at 72 h) and in HROC60 (3.8-fold change at 72 h). BCRP mRNA levels were also higher upregulated compared to Caco-2 (1.4-fold change at 48 h) in five HROC lines when treated with RIF and were highest in HROC32 (4.2-fold change at 72 h), HROC43 (3.3-fold change at 72 h), and HROC60 (3.2-fold change at 72 h). VDR-mediated induction of BCRP mRNA was higher in HROC239 T0 M1 (3.5-fold change at 24 h), HROC32 (2.3-fold change at 48 h), and HROC60 (2.2-fold change at 72 h) compared to Caco-2 (1.7-fold change at 48 h).
RIF and VD3 treatment led to higher regulation of CYP3A4 activity and P-gp mRNA in HROC183 T0 M2 and HROC217 T1 M2 compared to Caco-2. HROC60 showed comparable VDR-mediated induction of PXR-, UGT1A6-, and stronger P-gp- and OATP2B1- expression than Caco-2. Highest PXR-mediated regulation of CYP3A4 activity as well as OATP2B1-, UGT1A6-, P-gp-, and BCRP-expression was found in HROC43. Multiple DT were downregulated in HROC126 and HROC159 T2 M4. In HROC159 T2 M4, RIF treatment led to downregulation of CYP3A4 activity, as well as UGT1A6-, P-gp-, and PXR expression. Whereas VD3 treatment increased CYP3A4 activity but decreased OATP2B1-, UGT1A6-, P-gp-, BCRP-, PXR-, and VDR expression.
VDR mRNA was increased in HROC32 (2.4-fold change at 48 h), HROC43 (2.1-fold change at 72 h), HROC60 (2.6-fold change at 72 h), and HROC239 T0 M1 (2.1-fold change at 24 h) with VD3 at levels comparable to Caco-2 (3.2-fold change at 72 h). VD3-mediated induction of PXR mRNA was highest in HROC43 (3.9-fold change at 72 h) and was further observed for HROC239 T0 M1 (3.5-fold change at 72 h), Caco-2 (3-fold change at 72 h), HROC217 T1 M2 (2.3-fold change at 48 h), and HROC383 (1.7-fold change at 72 h).
In HROC383 cells, the only significant change was a decrease in VDR mRNA after VD3 treatment (0.6-fold change at 72 h). HROC383 cells appeared to be low sensitive to RIF and VD3 and were found to be comparable with Caco-2. However, a strong increase of PXR mRNA with RIF was observed in Caco-2 (16.6-fold change at 48 h). Overall, HROC43, HROC60, HROC80 T1 M1, HROC183 T0 M2 and HROC217 T1 M2 showed a clearly stronger response in gene regulation subsequent to RIF and VDR treatment than Caco-2.
4. Discussion
The aim of this study was to identify novel human intestinal in vitro models for studying cell differentiation and drug transport. Therefore, ten cell lines were first identified out of the broad HROC collection and were subsequently characterized concerning their potential to differentiate into intestinal epithelial cells (IECs) and to express intestinal proteins and DT with and without addition of regulating drugs – all in comparison to Caco-2 as the classical model. Our data show functional tight junction complexes and defined membrane border ridges as well as the formation of microvilli structures on the apical surface of all cell lines analyzed. This is indicative of enterocytic cell differentiation, intact cell-cell contacts and epithelial barrier integrity. Judged from the nuclear as well as cytoplasmic ZO-1 staining patterns within the lines, TJ complexes and thus intestinal barrier function were omnipresent. Since low ZO-1 expression is reported in poorly differentiated cell lines (Sommers et al. 1994), the weak signals in HROC126 and HROC383 suggest a lower grade of differentiation of these lines, which might also explain the limited number of relevant IECs detected in HROC126 and HROC383 monolayers.
We identified three lines, HROC43, HROC183 T0 M2 and HROC217 T1 M2, which closely resembled in vivo jejunal properties and would thus be very promising intestinal epithelium in vitro models. As monolayer models, these lines differentiated into all relevant IEC types and expressed a variety of DT, thereby closely resembling normal human intestinal epithelium (Sancho et al. 2003). Notably, compared to Caco-2, also a higher regulation of CYP3A4 activity and P-gp mRNA by RIF and VD3 was observed. Especially BCRP was highly expressed in these three HROC lines adding to the range of intestine-specific DT analyzable in these novel in vitro models.
Another interesting finding of our study was the identification of small intestinal-like gene expression in the rectal cancer cell line HROC239 T0 M1. There, also stable OATP2B1 mRNA and protein expression was observed, which is a little unexpected since OATP2B1 was not detectable in human rectal tissue (Schulte und Ho 2019). Still, HROC239 T0 M1 might be a useful in vitro model for studying rectal drug administration.
Our study outlined several drawbacks of the Caco-2 model. First, GC properties were lacking in the Caco-2 model (e.g. low mucin) confirming the findings of others (van Klinken et al. 1996). Lack of GCs can prompt an overestimation of drug permeability (Lea 2015). In contrast, HROC43 showed a distinct mucin expression, indicating GC-like properties of this cell line. Second, much higher OATP2B1 mRNA and protein levels were detected in long-term Caco-2 cultures compared to intestinal epithelium, again consistent with other studies (Brück et al. 2017; Pshezhetsky et al. 2007). Third, and as recently reported, Caco-2 cells showed low PXR-and VDR-mediated regulation of multiple DT and CYP3A4 activity (Przybylla et al. 2022).
To date, a large number of cell lines derived from human colon carcinomas were successfully established. They vary widely concerning differentiation states, proliferation and metabolic characteristics. However, most of them do not differentiate under standard culture conditions (Simon-Assmann et al. 2007).
In the present study, we clearly expanded the list of well-differentiating lines by identifying three HROC lines able to differentiate into IEC subtypes even under standard culture conditions; the latter might come handy for many researchers in the field. These lines are immortal but low-passaged, limiting culture-induced genetic alterations and sub-clonal varieties as described for Caco-2 (Ohura et al. 2016). Further, Caco-2 cells require about three weeks to complete cell differentiation (Antunes et al. 2013), limiting the usefulness for high throughput screening of new drug candidates (Cai et al. 2014). To overcome the latter issue, efforts to shorten this time period have repeatedly been undertaken (Chong et al. 1997; Sevin et al. 2013). HROC43 and HROC217 T1 M2 exhibited intestinal marker expression within a clearly shorter culture period and may thus be perfectly suited for high-throughput culture protocols.
Our study has three main limitations. First, we focused on drug transport and regulation mechanisms. Therefore, applications of the models included for nutrient absorption or further functional studies were not tested. Second, the number of transporters quantified on the protein level is smaller than the number of genes included in the expression analyses. Because measuring protein contents by LC-MS/MS is technically challenging and prone to variations, we focused on the detection of PEPT1, OATP2B1, P-gp and BCRP. Third, influence of RIF and VD3 on the protein levels of transporters was not analyzed. However, the findings of our study are new and are adequate to characterize the similarity of several HROC lines to human jejunum cell features. Thus, our data provide potentially useful information for new approaches and further investigations of transporters in selected lines.
In summary, we characterized a larger panel of novel human intestinal 2D cell models which differ in somatic mutational profiles (Przybylla et al. 2022), drug sensitivity (data not shown), degree of cell differentiation as measured by cell type marker expression, as well as individual regulation pattern of drug metabolizing enzymes and DT. They represent thus a novel and potentially important tool set for functional cell type studies, for ADME purposes, and for developing target-drug therapies; the latter both on the academic translational research as well as on more industrial high-throughput screening level.
Table 3.
Summary of HROC cell models.
Table 3.
Summary of HROC cell models.
| Cell line-specific advantages, limitations and applications for ADME purposes |
| Cell line |
Advantages |
Limitations |
Applications |
| HROC32 |
Presence of GCs, PCs, EECs, villi formation, protein abundance of PEPT1, OATP2B1 and P-gp, high basal CYP3A4 activity, high efflux effects
|
Lack of BCRP protein and mRNA, lower levels of GLUT-2 mRNA compared to small intestine, colonic origin |
VDR-mediated first-pass metabolism, cell differentiation, cell interaction, drug and nutrient absorption, HTS |
| HROC43 |
Well-developed TJs, closely resembles in vivo jejunal properties, presence of GCs, PCs, EECs, villi formation, high basal CYP3A4 activity, protein abundance of PEPT1, OATP2B1, P-gp, and BCRP, PXR-/VDR-mediated regulation of CYP3A4 activity + DT mRNA |
Lower levels of GLUT-2 mRNA compared to small intestine, colonic origin |
First-pass metabolism, drug- and nutrient absorption, permeability, host-microbe interaction, tissue regeneration, HTS |
| HROC60 |
Well-developed TJs, presence of GCs and EECs, villi formation, LYZ expressed, protein abundance of PEPT1,
high TEER, stable long-term culture, high VDR-mediated regulation of DT |
Limited degree of differentiation, poor abundance of CYP3A4, DT and tight junctions, limited reflection of the in vivo situation, colonic origin |
Gut-on-a-chip, tissue regeneration, drug and nutrient absorption, HTS |
| HROC80 T1 M1 |
Well-developed TJs, presence of GCs, villi formation, PXR-/VDR-mediated regulation of CYP3A4 + DT |
Lack of BCRP protein, limited degree of differentiation (lack of EECs and LYZ), poor PXR-mediated drug response, colonic origin |
First-pass metabolism, drug- and nutrient absorption, HTS |
| HROC126 |
EECs present, villi formation, VDR-mediated induction of CYP3A4 activity |
Limited degree of differentiation (lack of GCs and LYZ), rectal cancer origin, poor PXR-mediated regulation of DT, VDR overexpressed |
VDR-mediated first-pass metabolism, Rectal drug administration, drug and nutrient absorption, HTS |
| HROC159 T2 M4 |
Well-developed TJs, GCs present, LYZ expressed, villi formation, high basal CYP3A4 activity, appropriate VDR-mediated regulation of CYP3A4 activity |
Poor gene expression and protein levels of multiple DT, colonic origin |
First-pass metabolism, drug- and nutrient absorption, tissue regeneration, HTS |
| HROC183 T0 M2 |
Well-developed TJs, closely resembles in vivo jejunal properties, presence of GCs and EECs, LYZ expressed, villi formation, VDR-mediated induction of CYP3A4 activity, appropriate PXR-/VDR-mediated regulation of CYP3A4 activity and P-gp mRNA |
Low basal CYP3A4 activity, colonic origin |
PXR-mediated first-pass metabolism, drug- and nutrient absorption, HTS |
| HROC217 T1 M2 |
Well-developed TJs, closely resembles in vivo jejunal properties, presence of GCs and EECs, LYZ expressed, villi formation, protein abundance of PEPT1, OATP2B1, P-gp and BCRP, appropriate PXR-/VDR-mediated regulation of CYP3A4 activity and P-gp mRNA |
Lower protein levels of OATP2B1, P-gp and BCRP compared to jejunum, colonic origin |
PXR-/VDR-mediated first-pass metabolism, Drug and nutrient absorption, HTS |
| HROC239 T0 M1 |
Closely resembles in vivo jejunal properties, presence of EECs, villi formation, protein abundance of PEPT1, OATP2B1, P-gp and BCRP |
Rectal cancer origin, PXR overexpressed, low UGT1A6 mRNA expression, lack of GCs and PCs, colonic origin, lower protein levels of OATP2B1, P-gp and BCRP compared to jejunum |
Rectal drug administration, drug and nutrient absorption, HTS |
| HROC383 |
Presence of GCs and villi formation, protein abundance of PEPT1 and OATP2B1 |
Limited degree of differentiation, poor PXR- and VDR-mediated drug response, poor abundance of OATP2B1, UGT1A6 not expressed, colonic origin |
Drug and nutrient absorption, HTS |
Funding statement
This project was funded by the Monika Kutzner foundation. The DFG funded the CytoViva® Enhanced Darkfield Microscope (project number: 433722450).
Acknowledgments
We thank Karoline Schulz (Electron Microscopy Center) for expert technical support with SEM imaging and sample preparation and for help with figure editing.
Abbreviations
| ABC |
ATP-binding cassette; |
| ADME |
administration distribution metabolism excretion; |
| CYP3A4 |
Cytochrome P450 (CYP) 3A4; |
| GCs |
goblet cells; |
| DT |
drug transporter; |
| EECs |
enteroendocrine cells; |
| HROC |
Hansestadt Rostock CRC; |
| IECs |
intestinal epithelial cells; |
| LC-MS/MS |
liquid chromatography–mass spectrometry/mass spectrometry; |
| NRs |
nuclear receptors; |
| PCs |
Paneth cells; |
| PXR |
pregnane X receptor; |
| RIF |
rifampicin; |
| SLC |
solute carrier transporters; |
| TJ |
tight junction; |
| VD3 |
vitamin D3 |
| ZO-1 |
zonula occludens-1 |
References
- Antunes, Filipa; Andrade, Fernanda; Araújo, Francisca; Ferreira, Domingos; Sarmento, Bruno (2013): Establishment of a triple co-culture in vitro cell models to study intestinal absorption of peptide drugs. In: European journal of pharmaceutics and biopharmaceutics : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V 83 (3), S. 427–435. [CrossRef]
- Berggren, Sofia; Gall, Christine; Wollnitz, Nadine; Ekelund, Mats; Karlbom, Urban; Hoogstraate, Janet et al. (2007): Gene and protein expression of P-glycoprotein, MRP1, MRP2, and CYP3A4 in the small and large human intestine. In: Molecular pharmaceutics 4 (2), S. 252–257. [CrossRef]
- Brück, S.; Strohmeier, J.; Busch, D.; Drozdzik, M.; Oswald, S. (2017): Caco-2 cells - expression, regulation and function of drug transporters compared with human jejunal tissue. In: Biopharmaceutics & drug disposition 38 (2), S. 115–126. [CrossRef]
- Cai, Yike; Xu, Chenshu; Chen, Peiyi; Hu, Jinqing; Hu, Rong; Huang, Min; Bi, Huichang (2014): Development, validation, and application of a novel 7-day Caco-2 cell culture system. In: Journal of pharmacological and toxicological methods 70 (2), S. 175–181. [CrossRef]
- Cheng, H.; Leblond, C. P. (1974): Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. In: The American journal of anatomy 141 (4), S. 537–561. [CrossRef]
- Chong, S.; Dando, S. A.; Morrison, R. A. (1997): Evaluation of Biocoat intestinal epithelium differentiation environment (3-day cultured Caco-2 cells) as an absorption screening model with improved productivity. In: Pharmaceutical research 14 (12), S. 1835–1837. [CrossRef]
- Doherty, M. M.; Pang, K. S. (1997): First-pass effect: significance of the intestine for absorption and metabolism. In: Drug and chemical toxicology 20 (4), S. 329–344. [CrossRef]
- Gröer, C.; Brück, S.; Lai, Y.; Paulick, A.; Busemann, A.; Heidecke, C. D. et al. (2013): LC-MS/MS-based quantification of clinically relevant intestinal uptake and efflux transporter proteins. In: Journal of pharmaceutical and biomedical analysis 85, S. 253–261. [CrossRef]
- Herath, Madushani; Hosie, Suzanne; Bornstein, Joel C.; Franks, Ashley E.; Hill-Yardin, Elisa L. (2020): The Role of the Gastrointestinal Mucus System in Intestinal Homeostasis: Implications for Neurological Disorders. In: Frontiers in cellular and infection microbiology 10, S. 248. [CrossRef]
- Hosomi, Atsushi; Nakanishi, Takeo; Fujita, Takuya; Tamai, Ikumi (2012): Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. In: PloS one 7 (2), e30456. [CrossRef]
- Howarth, A. G.; Hughes, M. R.; Stevenson, B. R. (1992): Detection of the tight junction-associated protein ZO-1 in astrocytes and other nonepithelial cell types. In: The American journal of physiology 262 (2 Pt 1), C461-9. [CrossRef]
- Lea, Tor (2015): The Impact of Food Bioactives on Health: in vitro and ex vivo models. Caco-2 Cell Line. Hg. v. Kitty Verhoeckx, Paul Cotter, Iván López-Expósito, Charlotte Kleiveland, Tor Lea, Alan Mackie, et al. Cham (CH).
- Lueschow, Shiloh R.; McElroy, Steven J. (2020): The Paneth Cell: The Curator and Defender of the Immature Small Intestine. In: Frontiers in immunology 11, S. 587. [CrossRef]
- Ménochet, Karelle; Chanteux, Hugues; Henshall, Jamie; Nicolas, Jean-Marie; Wright, Sara; Asperen, Judith; Ungell, Anna-Lena (2022): Intestinal Epithelium and Drug Transporters. In: Edmund S. Kostewicz, Maria Vertzoni, Heather A. E. Benson und Michael S. Roberts (Hg.): Oral Drug Delivery for Modified Release Formulations, Bd. 10: Wiley, S. 39–64.
- Mullins, Christina S.; Micheel, Bianca; Matschos, Stephanie; Leuchter, Matthias; Bürtin, Florian; Krohn, Mathias et al. (2019): Integrated Biobanking and Tumor Model Establishment of Human Colorectal Carcinoma Provides Excellent Tools for Preclinical Research. In: Cancers 11 (10). [CrossRef]
- Ohura, Kayoko; Nishiyama, Hikaru; Saco, Saori; Kurokawa, Keisuke; Imai, Teruko (2016): Establishment and Characterization of a Novel Caco-2 Subclone with a Similar Low Expression Level of Human Carboxylesterase 1 to Human Small Intestine. In: Drug metabolism and disposition: the biological fate of chemicals 44 (12), S. 1890–1898. [CrossRef]
- Potten, C. S. (1998): Stem cells in gastrointestinal epithelium: numbers, characteristics and death. In: Philosophical transactions of the Royal Society of London. Series B, Biological sciences 353 (1370), S. 821–830. [CrossRef]
- Przybylla, Randy; Mullins, Christina Susanne; Krohn, Mathias; Oswald, Stefan; Linnebacher, Michael (2022): Establishment and Characterization of Novel Human Intestinal In Vitro Models for Absorption and First-Pass Metabolism Studies. In: International journal of molecular sciences 23 (17). [CrossRef]
- Pshezhetsky, Alexey V.; Fedjaev, Michael; Ashmarina, Lyudmila; Mazur, Alexander; Budman, Lorne; Sinnett, Daniel et al. (2007): Subcellular proteomics of cell differentiation: quantitative analysis of the plasma membrane proteome of Caco-2 cells. In: Proteomics 7 (13), S. 2201–2215. [CrossRef]
- Rehfeld, J. F. (2004): A centenary of gastrointestinal endocrinology. In: Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme 36 (11-12), S. 735–741. [CrossRef]
- Sancho, Elena; Batlle, Eduard; Clevers, Hans (2003): Live and let die in the intestinal epithelium. In: Current opinion in cell biology 15 (6), S. 763–770. [CrossRef]
- Schulte, Rachael R.; Ho, Richard H. (2019): Organic Anion Transporting Polypeptides: Emerging Roles in Cancer Pharmacology. In: Molecular pharmacology 95 (5), S. 490–506. [CrossRef]
- Sevin, E.; Dehouck, L.; Fabulas-da Costa, A.; Cecchelli, R.; Dehouck, M. P.; Lundquist, S.; Culot, M. (2013): Accelerated Caco-2 cell permeability model for drug discovery. In: Journal of pharmacological and toxicological methods 68 (3), S. 334–339. [CrossRef]
- Simon-Assmann, P.; Turck, N.; Sidhoum-Jenny, M.; Gradwohl, G.; Kedinger, M. (2007): In vitro models of intestinal epithelial cell differentiation. In: Cell biology and toxicology 23 (4), S. 241–256. [CrossRef]
- Sommers, C. L.; Byers, S. W.; Thompson, E. W.; Torri, J. A.; Gelmann, E. P. (1994): Differentiation state and invasiveness of human breast cancer cell lines. In: Breast cancer research and treatment 31 (2-3), S. 325–335. [CrossRef]
- Speer, Jennifer E.; Gunasekara, Dulan B.; Wang, Yuli; Fallon, John K.; Attayek, Peter J.; Smith, Philip C. et al. (2019): Molecular transport through primary human small intestinal monolayers by culture on a collagen scaffold with a gradient of chemical cross-linking. In: Journal of biological engineering 13, S. 36. [CrossRef]
- tewart, Katie D.; Johnston, Joseph A.; Matza, Louis S.; Curtis, Sarah E.; Havel, Henry A.; Sweetana, Stephanie A.; Gelhorn, Heather L. (2016): Preference for pharmaceutical formulation and treatment process attributes. In: Patient preference and adherence 10, S. 1385–1399. [CrossRef]
- Urquhart, Bradley L.; Tirona, Rommel G.; Kim, Richard B. (2007): Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. In: Journal of clinical pharmacology 47 (5), S. 566–578. [CrossRef]
- van Klinken, B. J.; Oussoren, E.; Weenink, J. J.; Strous, G. J.; Büller, H. A.; Dekker, J.; Einerhand, A. W. (1996): The human intestinal cell lines Caco-2 and LS174T as models to study cell-type specific mucin expression. In: Glycoconjugate journal 13 (5), S. 757–768. [CrossRef]
- Wenzel, Christoph; Drozdzik, Marek; Oswald, Stefan (2021): Mass spectrometry-based targeted proteomics method for the quantification of clinically relevant drug metabolizing enzymes in human specimens. In: Journal of chromatography. B, Analytical technologies in the biomedical and life sciences 1180, S. 122891. [CrossRef]
- Wenzel, Christoph; Gödtke, Lisa; Reichstein, Anne; Keiser, Markus; Busch, Diana; Drozdzik, Marek; Oswald, Stefan (2022): Gene Expression and Protein Abundance of Nuclear Receptors in Human Intestine and Liver: A New Application for Mass Spectrometry-Based Targeted Proteomics. In: Molecules (Basel, Switzerland) 27 (14). [CrossRef]
Figure 1.
Immunofluorescence analysis of ZO-1 protein localization. (A) HROC32, (B) HROC43, (C) HROC60, (D) HROC80 T1 M1, (E) HROC126, (F) HROC159 T2 M4, (G) HROC183 T0 M2, (H) HROC217 T1 M2, (I) HROC239 T0 M1, (J) HROC383, K) Caco-2. Cells were grown until reaching confluence, fixed in 1:1 methanol/acetone, permeabilized with 0.5% Triton X-100 and stained for localization of ZO-1 by direct immunofluorescence and for nuclear DNA by Hoechst staining. Bars = 50 µm.
Figure 1.
Immunofluorescence analysis of ZO-1 protein localization. (A) HROC32, (B) HROC43, (C) HROC60, (D) HROC80 T1 M1, (E) HROC126, (F) HROC159 T2 M4, (G) HROC183 T0 M2, (H) HROC217 T1 M2, (I) HROC239 T0 M1, (J) HROC383, K) Caco-2. Cells were grown until reaching confluence, fixed in 1:1 methanol/acetone, permeabilized with 0.5% Triton X-100 and stained for localization of ZO-1 by direct immunofluorescence and for nuclear DNA by Hoechst staining. Bars = 50 µm.
Figure 2.
Representative SEM photographs of stratified HROC60 (A, B) and HROC159 T2 M4 (C, D) epithelial monolayers in comparison with Caco-2 cells (E, F). High magnification images from the insert regions demonstrate the close apposition and sealing of membranes at the cell-cell boundaries (arrows). Scale bars are 2 µm for overview images and 1 µm for inserts.
Figure 2.
Representative SEM photographs of stratified HROC60 (A, B) and HROC159 T2 M4 (C, D) epithelial monolayers in comparison with Caco-2 cells (E, F). High magnification images from the insert regions demonstrate the close apposition and sealing of membranes at the cell-cell boundaries (arrows). Scale bars are 2 µm for overview images and 1 µm for inserts.
Figure 3.
Immunofluorescence analysis of MUC-2 protein localization. (A) HROC32, (B) HROC43, (C) HROC60, (D) HROC80 T1 M1, (E) HROC126, (F) HROC159 T2 M4, (G) HROC183 T0 M2, (H) HROC217 T1 M2, (I) HROC239 T0 M1, (J) HROC383 and K) Caco-2. Cells were grown until reaching confluence, fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and stained for localization of MUC-2 by direct immunofluorescence and for nuclear DNA by Hoechst staining. Bars = 50 µm.
Figure 3.
Immunofluorescence analysis of MUC-2 protein localization. (A) HROC32, (B) HROC43, (C) HROC60, (D) HROC80 T1 M1, (E) HROC126, (F) HROC159 T2 M4, (G) HROC183 T0 M2, (H) HROC217 T1 M2, (I) HROC239 T0 M1, (J) HROC383 and K) Caco-2. Cells were grown until reaching confluence, fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and stained for localization of MUC-2 by direct immunofluorescence and for nuclear DNA by Hoechst staining. Bars = 50 µm.
Figure 4.
Immunofluorescence analysis of LYZ protein localization. (A) HROC32, (B) HROC43, (C) HROC60, (D) HROC80 T1 M1, (E) HROC126, (F) HROC159 T2 M4, (G) HROC183 T0 M2, (H) HROC217 T1 M2, (I) HROC239 T0 M1, (J) HROC383 and K) Caco-2. Cells were grown until reaching confluence, fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and stained for localization of LYZ by direct immunofluorescence and for nuclear DNA by Hoechst staining. Bars = 50 µm.
Figure 4.
Immunofluorescence analysis of LYZ protein localization. (A) HROC32, (B) HROC43, (C) HROC60, (D) HROC80 T1 M1, (E) HROC126, (F) HROC159 T2 M4, (G) HROC183 T0 M2, (H) HROC217 T1 M2, (I) HROC239 T0 M1, (J) HROC383 and K) Caco-2. Cells were grown until reaching confluence, fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and stained for localization of LYZ by direct immunofluorescence and for nuclear DNA by Hoechst staining. Bars = 50 µm.
Figure 5.
Immunofluorescence analysis of CHGA protein localization. (A) HROC32, (B) HROC43, (C) HROC60, (D) HROC80 T1 M1, (E) HROC126, (F) HROC159 T2 M4, (G) HROC183 T0 M2, (H) HROC217 T1 M2, (I) HROC239 T0 M1, (J) HROC383 and K) Caco-2. Cells were grown until reaching confluence, fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and stained for localization of CHGA by direct immunofluorescence and for nuclear DNA by Hoechst staining. Bars = 50 µm.
Figure 5.
Immunofluorescence analysis of CHGA protein localization. (A) HROC32, (B) HROC43, (C) HROC60, (D) HROC80 T1 M1, (E) HROC126, (F) HROC159 T2 M4, (G) HROC183 T0 M2, (H) HROC217 T1 M2, (I) HROC239 T0 M1, (J) HROC383 and K) Caco-2. Cells were grown until reaching confluence, fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and stained for localization of CHGA by direct immunofluorescence and for nuclear DNA by Hoechst staining. Bars = 50 µm.
Figure 6.
Scanning electron microscope photographs of microvilli formations in HROC- and Caco-2 cells after reaching confluence. Cells with a dense coverage of microvilli are depicted (A) HROC32, (B) HROC43, (C) HROC60, (D) HROC80 T1 M1, (E) HROC126, (F) HROC159 T2 M4, (G) HROC183 T0 M2, (H) HROC217 T1 M2, (I) HROC239 T0 M1, and (J) HROC383. Note that microvilli decoration my vary among neighboring cells and thus for the Caco-2 reference low (K1) an high decoration (K2) with microvilli is depicted for reference (K) Caco-2. Scale bar is 1 µm.
Figure 6.
Scanning electron microscope photographs of microvilli formations in HROC- and Caco-2 cells after reaching confluence. Cells with a dense coverage of microvilli are depicted (A) HROC32, (B) HROC43, (C) HROC60, (D) HROC80 T1 M1, (E) HROC126, (F) HROC159 T2 M4, (G) HROC183 T0 M2, (H) HROC217 T1 M2, (I) HROC239 T0 M1, and (J) HROC383. Note that microvilli decoration my vary among neighboring cells and thus for the Caco-2 reference low (K1) an high decoration (K2) with microvilli is depicted for reference (K) Caco-2. Scale bar is 1 µm.
Figure 7.
Time-dependent gene expression of selected genes in HROC- and Caco-2 cells. Cells were cultured on 6-well plates and then grown until reaching confluence. Fold changes of GAPDH-normalized measurements between control (d0) and ten day post-confluence growth (d10) were calculated by performing the ∆∆Ct method. Reference tissue was set at 1.0.
Figure 7.
Time-dependent gene expression of selected genes in HROC- and Caco-2 cells. Cells were cultured on 6-well plates and then grown until reaching confluence. Fold changes of GAPDH-normalized measurements between control (d0) and ten day post-confluence growth (d10) were calculated by performing the ∆∆Ct method. Reference tissue was set at 1.0.
Figure 8.
Gene expression correlation analyses. Graphs showing Spearman rank correlation analyses of transporter and nuclear receptor expression levels in HROC- and Caco-2 cells at control state (A-C) and ten day post-confluence growth (D-I). Spearman rank correlation coefficient (r) and p-values were computed for each analyses. Data are shown as mean from three independent datasets.
Figure 8.
Gene expression correlation analyses. Graphs showing Spearman rank correlation analyses of transporter and nuclear receptor expression levels in HROC- and Caco-2 cells at control state (A-C) and ten day post-confluence growth (D-I). Spearman rank correlation coefficient (r) and p-values were computed for each analyses. Data are shown as mean from three independent datasets.
Figure 9.
Time-dependent protein expression levels of drug transporters in HROC- and Caco-2 cells. Cells were cultured on 6-well plates and then grown until reaching confluence. Protein levels detected by LC-MS/MS were calculated as fold changes relative to human jejunal tissue. Reference tissue was set at 1.0. ***p < 0.001 by paired Student’s t-test.
Figure 9.
Time-dependent protein expression levels of drug transporters in HROC- and Caco-2 cells. Cells were cultured on 6-well plates and then grown until reaching confluence. Protein levels detected by LC-MS/MS were calculated as fold changes relative to human jejunal tissue. Reference tissue was set at 1.0. ***p < 0.001 by paired Student’s t-test.
Figure 10.
Effects of rifampicin (RIF) and 1a,25-dihydroxyvitamin D3 (VD3) on mRNA expression in HROC- and Caco-2 cells. Cells were cultured on 6-well plates and then treated with vehicle (0.1% DMSO or 0.1% ethanol) or inducers (20 µM RIF or 100 nM VD3) for 72 hours. Data are presented as mean of three independent experiments. Fold changes of GAPDH-normalized measurements between control and inducer treatment were calculated by performing the 2-∆∆Ct method. Untreated control was set at 1.0. *p < 0.01, **p < 0.005 by paired Student’s t-test.
Figure 10.
Effects of rifampicin (RIF) and 1a,25-dihydroxyvitamin D3 (VD3) on mRNA expression in HROC- and Caco-2 cells. Cells were cultured on 6-well plates and then treated with vehicle (0.1% DMSO or 0.1% ethanol) or inducers (20 µM RIF or 100 nM VD3) for 72 hours. Data are presented as mean of three independent experiments. Fold changes of GAPDH-normalized measurements between control and inducer treatment were calculated by performing the 2-∆∆Ct method. Untreated control was set at 1.0. *p < 0.01, **p < 0.005 by paired Student’s t-test.
Table 2.
Antibodies used.
Table 2.
Antibodies used.
| Antibody |
Host |
Company |
Dilution |
Catalogue number |
| anti Chr-A |
mouse |
Santa Cruz |
1:50 |
sc-393941 |
| anti Lysozyme C |
mouse |
Santa Cruz |
1:50 |
sc-518012 |
| anti Mucin 2 |
mouse |
Santa Cruz |
1:50 |
sc-515032 |
| anti ZO-1 |
rat |
Santa Cruz |
1:50 |
sc-33725 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).