1. Introduction
Extended-spectrum cephalosporin resistance is a major threat worldwide, as the cephalosporins are often used as first-line antimicrobial agents for treating infections caused by Gram-negative bacteria [
1,
2]. Cephalosporin-resistant bacteria produce enzymes called extended-spectrum beta-lactamases (ESBLs). ESBL enzymes hydrolyze antibiotics, including penicillins and cephalosporins, making these drugs ineffective in treating infections [
3].
ESBL-producing
Enterobacteriaceae are rising a major public problem in healthcare settings and the community. In 2017, there were an estimated 197,400 cases among hospitalized patients and 9,100 estimated deaths in the United States [
1]. Among these
Enterobacteriaceae,
E. coli is one of the primary pathogens of antimicrobial-resistant clinical infections [
4]. The emergence of
E. coli strains resistant to extended-spectrum cephalosporins was observed after the 2000s and continuously reported at present.
To understand the genetic diversity of E. coli, multilocus sequence typing has been most frequently used [
5]. The prevalence of ESBL-producing
E. coli, particularly sequencing type (ST) 131, has been reported as the predominant sequence type [
6]. Following a study from Europe, the incidence of ESBL-producing
E. coli ST131 has been reported to be 20% in four European hospitals [
7]. Also,
E. coli ST131 isolates have been reported in Korea. ST131 is the predominant clone among ESBL-producing isolates in community and healthcare settings in Korea [
8]. Much of the literature focuses on an occurrence of clonal related antimicrobial-resistant bacteria is expected to contribute to understanding of transmission their pathway. Therefore, it is necessary to analyze the clonal diversity of the antimicrobial resistant strains.
The Korea Disease Control and Prevention Agency (KDCA) has collected and tested pathogenic E. coli isolates from patients with diarrhea to monitor antimicrobial resistance profiles. Between 2009 and 2018, a total of 187 third-generation cephalosporin-resistant pathogenic E. coli isolates were confirmed and the clonal complex (CC) 10 was the predominant clones. The objective of this study was to characterize ESBL-producing pathogenic E. coli CC10 strains obtained from diarrheal patients in recent decades, to improve the understanding of CC10 distribution in Korea and the world.
2. Results
2.1. Serotyping
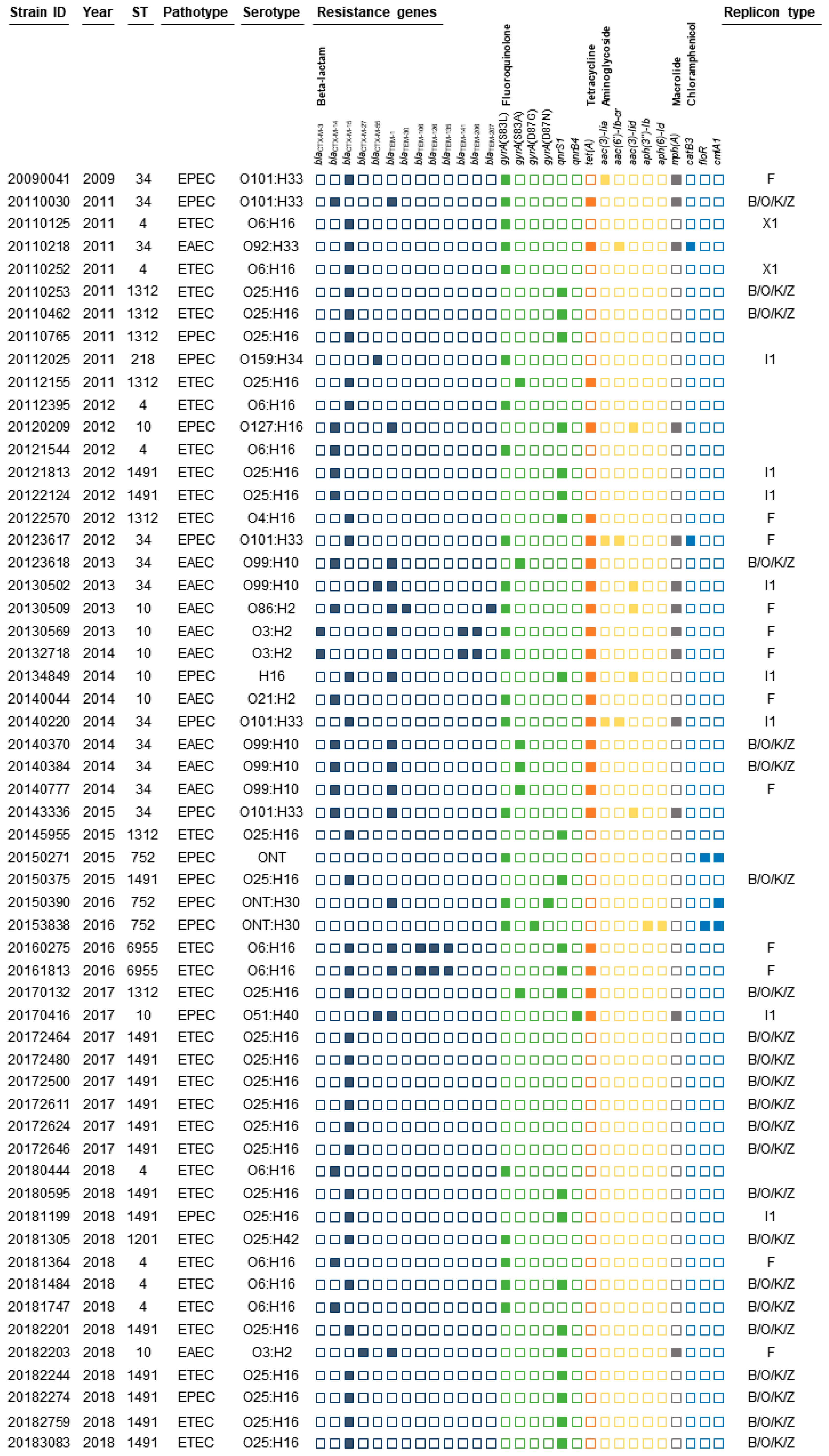
The 57 pathogenic
E. coli CC10 isolates were obtained from 16 regions in the Republic of Korea. The strains belonged to 12 O serogroups and expressed 8 different H antigens. The O serotypes were O25 (n=22), O6 (n=10), O101 (n=5), O99 (n=5), and O3 (n=3). The more common H serotypes were H16 (n=35), H33 (n=6), H2 (n=5), H10 (n=5) and H30 (n=3). The most prevalent serotype was O25:H16 (38.9%, 21/57), followed by O6:H16 (19.6%, 10/57), O99:H10 (9.8%, 5/57) and O101:H33 (9.8%, 5/57) (
Figure 1). In
silico FimH typing revealed 11 types of FimH (
Figure 3.2). Of the total CC10, 16 (31.4%), 9 (17.7%), 8 (15.7%), 5 (9.8%), 4 (7.8%), and 1(1.9%) were positive for FimH198, FimH54, FimH23, FimH30, FimH24, and FimH1194, respectively.
2.2. Distribution of genomic determinants of antimicrobial resistance
A total of 34 antimicrobial resistance genes/mutations were detected, involving six classes of antimicrobial agents, including beta-lactam (18 genes), fluoroquinolone (two genes and four mutations), tetracycline (one gene), aminoglycoside (five genes), macrolide (one gene) and chloramphenicol (three genes) (
Figure 1).
Beta-lactam resistance genes were detected in all CC10 isolates; blaCTX-M-15 and blaCTX-M-14 were most prevalent and were found in 26 (51%) and 15 (30%) isolates, respectively. Also, 17 strains carried blaTEM genes. Fluoroquinolone resistance genes/mutations were present in all isolates, of which quinolone resistance-determining regions (QRDRs) of gyrA genes mutation and plasmid-mediated quinolone resistance (PMQR) gene (qnrS1 and qnrB4) were found in 32 (62.7%) and 23 (45.1%) isolates, respectively. Mutations in the gyrA gene were observed at codons 83 and 87, producing the single-residue substitutions S83L, S83A, D87G and D87N. Multiple fluoroquinolone resistance associated mutations were detected in two isolates, specifically, double mutations in gyrA (S83L with D87G or D87N). Tetracycline resistance was identified in 22 (43.2%) isolates and macrolide resistance was confirmed in 13 (25.5%) isolates of genotypes tet(A) and mph(A), respectively. Aminoglycoside resistance genes were harbored by 10 isolates (19.6%), including aac(3)-Iia, aac(6')-Ib-cr, aac(3)-Iid, aph(3'')-Ib and aph(6)-Id. These isolates carried one or two resistance genes. Chloramphenicol resistance genes were detected in 5 (9.8%) isolates that were catB3, floR and cmlA1.
2.3. Transmission of bla gene
Characterization of blaCTX-M plasmids was performed to better understand the horizontal transfer of blaCTX-M using conjugation. CC10 strains capable of horizontal transfer through conjugation were identified as 82.5% (45/57) of the total. Twenty-three isolates carried an IncB/O/K/Z plasmid, twelve isolates carried an IncF-type plasmid, eight isolates carried an IncI1 plasmid, and two carried an IncX1 plasmid.
2.4. Analysis of the regions surrounding blaCTX-M genes
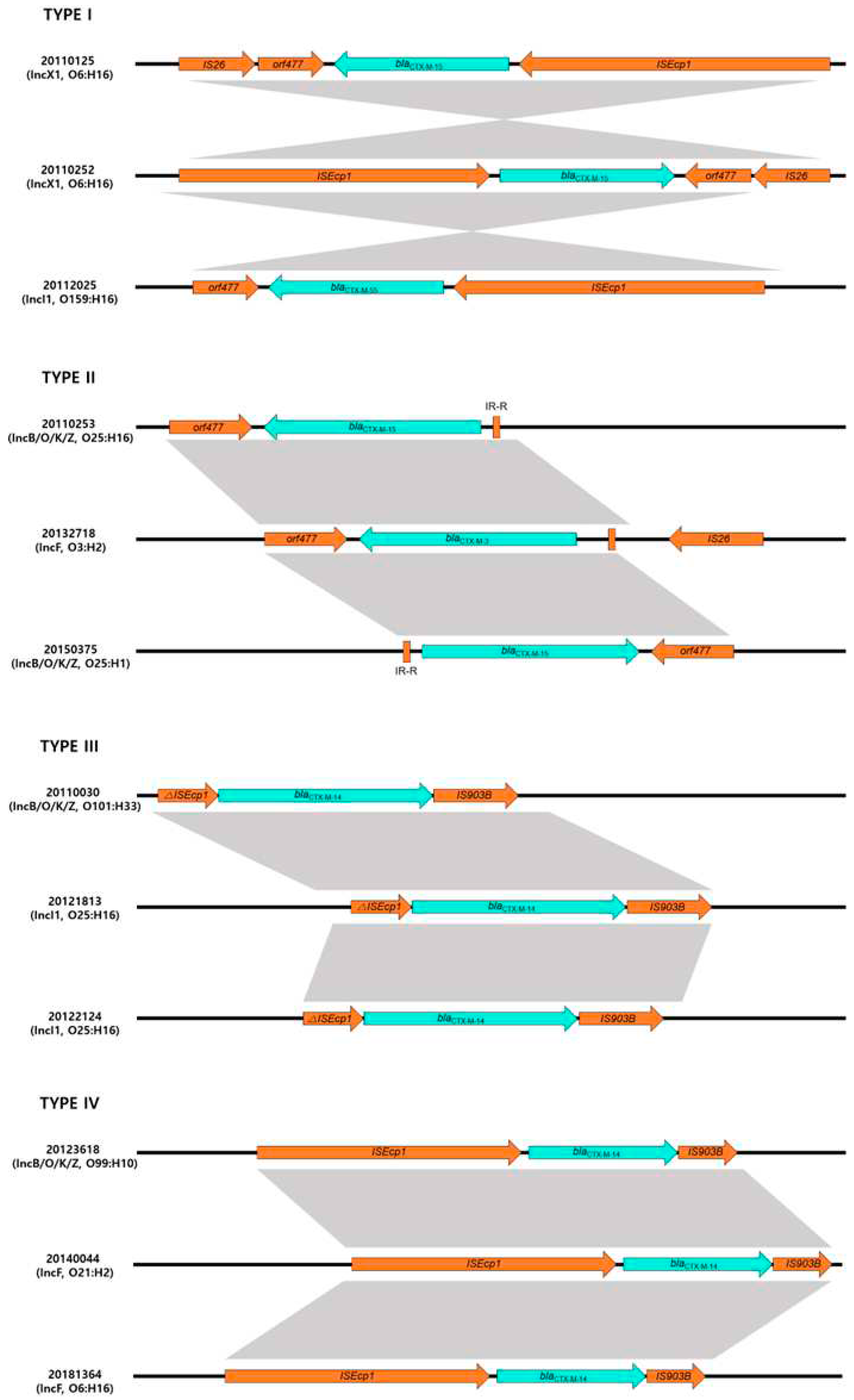
Four different structures [type I (10 isolates), type II (23 isolates), type III (3 Isolates) and type IV (9 isolates)] were identified regarding the genetic elements of
blaCTX-M (
Figure 2).
Type I genetic structure was found in 10 isolates producing blaCTX-M with ISEcp1-blaCTX-M-orf477 genetic structures. Type II genetic structure was most common and identified in 23 isolates. Analysis of the region flanking blaCTX-M revealed an orf477 downstream sequence with a spacer region between the inverted repeat (IR) sequences of ISEcp1 upstream. These genetic structures belong to the CTX-M-I group, such as blaCTX-M-3, -15 and -55 possessing isolates. However, Type III genetic structure was identified in three isolates that had a different genetic element flanking blaCTX-M, ISEcp1 upstream and downstream of the IS903 (IR-ISEcp1-blaCTX-M-IS903). Three isolates have type IV genetic structure, with ISEcp1-blaCTX-M-IS903 as a transposable element.
2.5. Whole-genome SNPs-based phylogeny of CC10
Phylogenetic analysis was performed with 265 genomes of
E. coli strains belonging to the international CC10, a whole-genome SNP phylogeny was generated using
E. coli K12-MG1655 as a reference (
Figure 3). To better understand the global population structure of
E. coli CC10, we have identified a genome alignment in which 14,260 SNPs were identified. Phylogeny analysis of 57 clinical pathogenic
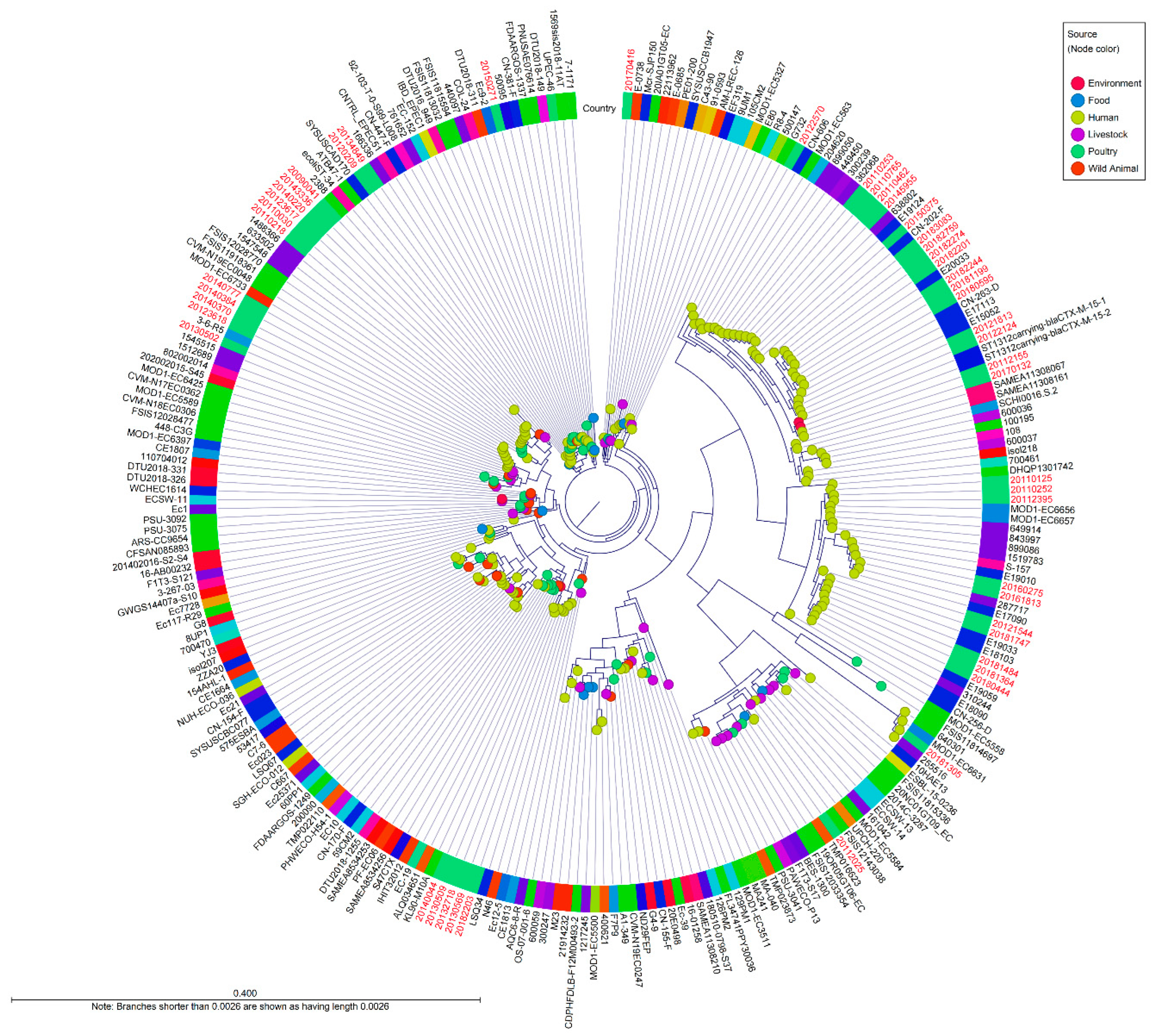
E. coli isolates in this study identified several distinct lineages, which comprised ST4, ST10, ST34, ST752, ST1312, and ST1491. There was no observed substantial clustering related to location or time of sampling during this study period.
Figure 3.
Whole genome SNP-derived phylogenetic tree of the CC10 isolates in a global context. The tree includes 265 international E. coli CC10 sequences, including the reference sequence of E. coli K-12 MG1655. The diagram depicts a phylogenetic tree with a genome alignment of 14,260 SNPs for 265 globally CC10 strains, including all publicly available isolates from Enterobase. The node color represents the source of isolations, the colored ring around the tree indicated the country of isolates.
Figure 3.
Whole genome SNP-derived phylogenetic tree of the CC10 isolates in a global context. The tree includes 265 international E. coli CC10 sequences, including the reference sequence of E. coli K-12 MG1655. The diagram depicts a phylogenetic tree with a genome alignment of 14,260 SNPs for 265 globally CC10 strains, including all publicly available isolates from Enterobase. The node color represents the source of isolations, the colored ring around the tree indicated the country of isolates.
There was observed substantial clustering related to CC10 isolated in the Republic of Korea from 27 reference strains. ST1312carrying-
blaCTX-M-15-1 and ST1312carrying-
blaCTX-M-15-2, isolated from river in Sweden in 2013, were closely related ranging from 100 to 109 SNP differences within this study strains (20112155 and 20170132) [
9]. These isolates had the O25:H16 serotype and common resistance genes, including
sul1,
dfrA14,
tet(A) and
blaCTX-M-15.
Five isolates (20121544, 20180444, 20181364, 20181484, 20181747) from this study were similar to isolates collected from patients in China between 2017 and 2019 (6 to 91 SNP differences); they also harbored
blaCTX-M-15 or
blaCTX-M-14 and had mutations in quinolone resistance-determining regions (E18090, E18013, E19033, E19059, E15052, E20033, E17113, E19010, E17090) [
10].
Five isolates (20123618, 20130502, 20140370, 20140384, 20140777) from this study were related to isolates 1512689, 1545515, 3-6-R5 from patients in the United Kingdom and Australia, with SNP differences ranging from 39 to 195. One isolate (287717) from the United Kingdom in 2016 was genetically similar to two isolates from this study (20160275, 20161813), with the same genetic determinants, plasmids, and serotype, with SNP differences ranging from 33 and 46 [
11]. Two clinical isolates from China in 2017 (CN-202-F, CN-263-D) carried same antimicrobial resistance genes, mutations, and plasmids as present in similar human isolates (20182244, 20180595, 20181199, 20182201, 20182274, 20183083, 20182759) in this study (54 to 58 SNP differences).
3. Discussion
ESBL-producing pathogenic E. coli sequence types are extremely genetically diverse in the past decade. During this period, CC10 was the most prominent types, comprising 57 isolates with 9 different STs. In this study, compared the CC10 of ESBL-producing clinical E. coli isolates derived from human in order to describe their characteristics.
All strains in association with ESBL showed high multidrug-resistant (MDR) occurrence. Although resistance rate was to tetracycline, followed by nalidixic acid, azithromycin, ciprofloxacin and trimethoprim/sulfamethoxazole. Most MDR in
E. coli associated with ESBLs has become a serious problem in public health because of dissemination of ESBL genes. It has posed a major threat to treatment of bacterial infections [
12].
In CC10 isolates, the incidence of CTX-M was highest, at 98.4% of the total and most dominant ESBL gene was blaCTX-M-15 (56%, 32/57). Majority of CTX-M-15 producing isolates have common features in that they belonging to serotype O25:H16 (53%, 17/32). Our finding indicated the common features that the CTX-M-15 producing E. coli O25:H16 in CC10 isolates presented to have emerged and expanded in the distribution during the last decade. These strains were sporadically isolated from 10 regions but clonally related with less than 70 SNPs separating them.
Also, our study contributes to the highlighting that plasmid acquisition is probably an important mechanism for the dissemination of CTX-M-producing pathogenic
E. coli. Resistance to third generation cephalosporins is caused by the acquisition of ESBL genes, primarily
blaCTX-M gene [
13]. Conjugation experiments were performed to confirm the horizontal transmission of plasmid-borne
blaCTX-M genes and identified such transfer in 78.9 % (45/57) CC10 isolates. This suggested that the high incidence of CC10 isolates is caused by horizontal transfer of ESBL genes between bacteria seems as the best way of transmission. The predominant genotype of plasmid-mediated
blaCTX-M gene were CTX-M-15 (64.4%, 29/45) and CTX-M-14 (22.2%, 10/45). The mobile elements located upstream of
blaCTX-M-14 and
-15 gene mainly included
ISEcp1 (complete or incomplete). Downstream of
blaCTX-M-14 and
-15 genes
IS903 and
orf477 was found, respectively.
Here, we found the persistent occurrence of
blaCTX-M-15 gene and observed the highest proportion in 2018. The
blaCTX-M-15 gene was known as the most widely distributed
blaCTX-M gene in the world [
14]. Previous studies in the Republic of Korea have indicated that
blaCTX-M-15-harboring
E. coli have isolated from raw vegetables and food animals in 2018, respectively [
15,
16]. It suggested
blaCTX-M-
15-producing
E. coli may circulate among food, food animals, and humans that might contribute to the acquisition of resistance.
To compare with the WGS-based population structure of our isolates in the context of the international CC10 lineages, five isolates is highly similar that of ETEC isolates from diarrhoea patients in China collected from 2017 to 2019 [
10]. Among five isolates, one isolate was collected in 2012, other four isolates were isolated in 2018. SNP analysis of those isolates found 6 to 91 SNPs, thus indicating a close relationship among the isolates, even though they were identified from distinct countries. These isolates were possessed
blaCTX-M-15 or
blaCTX-M-14 genes, genetic determinants, plasmids, and serotype. This study from China described
E. coli isolated from patients, suggesting that spread from unknown source could possibly disseminate the presence of the clone. These results revealed that the circulating CC10 from Republic of Korea, as well as in other countries were genetically close related, which are suggesting expansion of global or endemic population.
Based on these observations, CC10 may become the most important strain in the Republic of Korea. During last decade, CC10 of ESBL-producing pathogenic
E. coli have steadily isolated, especially, it has almost doubled recently. This result suggested that CC10 clone is emerging as the one of the important clones in human clinical cases for its association with third cephalosporins resistance. Several previous studies reported that
E. coli CC10 have a predominant clonal group associated with extraintestinal disease in both animals and humans [
17,
18] Some regional monitoring investigations from Italy, Spain, and Portugal showed that CC10 strains from humans, bird, and swine were associated with multiple CTX-M-type genes [
19,
20,
21].
Pathogenic E. coli has emerged as a major cause of food- and water-borne diseases in Korea. Currently, most of the data on ESBL-producing pathogenic E. coli CC10 are from studies conducted in the Republic of Korea and may reflect a local situation. Therefore, the necessary data for the national management of pathogenic E. coli infections and the management of related fields were secured. Future studies surveying for the presence of CC10 clone will provide information as to whether this is the case in other countries worldwide.
4. Materials and methods
4.1. Background
A total of 187 third-generation cephalosporin-resistant
E. coli isolates were collected by the national surveillance system (Enter-Net and Pulse-Net Korea) between 2009 and 2018. These isolates were collected from stool or rectal swabs from patients with gastrointestinal symptoms, including diarrhea, abdominal pain, vomiting, nausea, and fever. Initial investigation was performed to determine sequence types (STs) and clonal complexes (CCs) based on seven housekeeping genes (
https://enterobase.warwick.ac.uk/species/ecoli/allele_st_search) and the 187 resistance isolates were grouped into 77 STs. CC10 was the most prevalent clonal complex, comprising 57 (31.4%, 57/187) isolates with 9 different STs (4, 10, 34, 218, 752, 1201, 1312, 1491, 6955). 57 CC10 strains were selected for analysis of molecular characterization such as serotype, other antibiotic resistance genes, genetic environments, plasmid profiles and genetic correlation between the CC10 strains isolated in the Republic of Korea.
4.2. Whole genome sequencing (WGS)
Genomic DNA was isolated using a Blood and Tissue kit (Qiagen, Stockach, Germany) according to the manufacturer’s protocol. The purified total DNA quality was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher, DE, USA). The concentration was determined with a Qubit 4 fluorometer using a high-sensitivity kit (Invitrogen, CA, USA). Library fragment lengths were assessed through the use of a Bioanalyzer TapeStation with DNA 1000 kit (Agilent Technologies, Inc., CA, USA). A paired-end sequencing library was constructed with an Illumina DNA prep kit (Illumina, San Diego, CA, USA) following the manufacturer’s protocol. Sequencing was performed using a 500-cycle (2×250-bp paired-end) MiSeq reagent kit version 2 with an MiSeq sequencer.
4.3. Data analysis and molecular characterization
Raw sequences generated by Illumina MiSeq were quality filtered using FastQC, with average quality set at Q30. Raw reads from the Illumina sequencing were quality trimmed using CLC genomic workbench 22 (Qiagen, Hilden, Germany). Contigs of genomic sequences were assembled with a minimum size threshold of 200 bp using the de novo assembler in CLC genomic workbench 22 (Qiagen, Hilden, Germany). Assembled sequences were analyzed for antimicrobial resistance genes (ResFinder 4.1), plasmid replicon types (PlasmidFinder 2.1), serotypes (SerotypeFinder 2.0), and
fimH and
fumC (CHTyper 1.0) using web tools available from the Center for Genomic Epidemiology (CGE) (
http://www.genomicepidemiology.org/) [
22]. Single-nucleotide polymorphisms (SNPs) were identified using CSI phylogeny 1.4 (
https://cge.cbs.dtu.dk/services/CSIPhylogeny/) by comparison with
E. coli K-12 MG1655 (GenBank accession no. U00096) as a reference strain with 265
E. coli genomes with the same sequence type (CC10), retrieved from EnteroBase (listed in Supplementary Table S1). Selection of SNPs used default parameters in CSI Phylogeny, which included a minimum distance of 10 bp between SNPs, a minimum of 10% of the average depth, mapping quality above 25, and SNP quality above 30. All insertions and deletions (INDELs) were excluded.
4.4. Plasmid transfer by bacterial conjugation
For strains with results for cefotaxime resistance was examined by conjugation experiments using azide-resistant
E. coli J53 as recipient strain to confirm the transmission capacity of the
bla genes [
23]. Transconjugants were selected on MacConkey agar plates (Difco, USA) supplemented with cefotaxime (1mg/L) and sodium azide (200mg/L). The acquisition of the
bla gene was confirmed by PCR and sequencing analysis.
4.5. Nucleotide sequence accession numbers
The whole-genome sequences of these strains were deposited with the NCBI Sequence Read Archive (SRA) under the Bio-Project PRJNA628558.
5. Conclusions
CC10 of ESBL-producing pathogenic E. coli isolates have been a steady increase for the past decade. Presently, ESBL-producing pathogenic E. coli CC10 may become the most important strain in the Republic of Korea. Among these isolates, CTX-M-15-producing pathogenic E. coli O25:H16 isolates were the major type in CC10 clones. There have been a few studies that address this issue in CC10 isolates both in the Republic of Korea and in other countries. Identification of CC10 isolates highlights the possibility of the emergence of resistant isolates with epidemic potential within this CC. Therefore, continuous monitoring will be required to prevent further spread of resistant ESBL-producing E. coli CC10 strains.
Author Contributions
JP and JuK conceived of the study and participated in its design and draft the manuscript. ES and JH collected samples and identified isolates. JP carried out the experiments and analyzed the data. JY, JsY and DHR contributed to experiment conception. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the Korea Disease Control and Prevention Agency (grant number 4847-311-210).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated for this study are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- CDC ANTIBIOTIC RESISTANCE THREATS IN THE UNITED STATES, 2019. 2019, 10. [CrossRef]
- Watkins, R.R.; Bonomo, R.A. Overview: Global and Local Impact of Antibiotic Resistance. Infectious disease clinics of North America 2016, 30, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, S.; Fatima, J.; Shakil, S.; Rizvi, S.M.D.; Kamal, M.A. Antibiotic Resistance and Extended Spectrum Beta-Lactamases: Types, Epidemiology and Treatment. Saudi journal of biological sciences 2015, 22, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Malik, F.; Figueras, A. Continuous Rise in Cephalosporin and Fluoroquinolone Consumption in Pakistan: A 5 Year Analysis (2014–18). JAC-Antimicrobial Resistance 2019, 1, 1–4. [Google Scholar] [CrossRef]
- Yu, F.; Chen, X.; Zheng, S.; Han, D.; Wang, Y.; Wang, R.; Wang, B.; Chen, Y. Prevalence and Genetic Diversity of Human Diarrheagenic Escherichia Coli Isolates by Multilocus Sequence Typing. International Journal of Infectious Diseases 2018, 67, 7–13. [Google Scholar] [CrossRef]
- Zhong, Y.; Guo, S.; Seow, K.L.G.; Ming, G.O.H.; Schlundt, J. Characterization of Extended-Spectrum Beta-Lactamase-Producing Escherichia Coli Isolates from Jurong Lake, Singapore with Whole-Genome-Sequencing. International journal of environmental research and public health 2021, 18. [Google Scholar] [CrossRef]
- Merino, I.; Hernández-García, M.; Turrientes, M.-C.; Pérez-Viso, B.; López-Fresneña, N.; Diaz-Agero, C.; Maechler, F.; Fankhauser-Rodriguez, C.; Kola, A.; Schrenzel, J.; et al. Emergence of ESBL-Producing Escherichia Coli ST131-C1-M27 Clade Colonizing Patients in Europe. The Journal of antimicrobial chemotherapy 2018, 73, 2973–2980. [Google Scholar] [CrossRef]
- Park, S.H.; Byun, J.-H.; Choi, S.-M.; Lee, D.-G.; Kim, S.-H.; Kwon, J.-C.; Park, C.; Choi, J.-H.; Yoo, J.-H. Molecular Epidemiology of Extended-Spectrum β-Lactamase-Producing Escherichia Coli in the Community and Hospital in Korea: Emergence of ST131 Producing CTX-M-15. BMC Infectious Diseases 2012, 12, 149. [Google Scholar] [CrossRef]
- Fagerström, A.; Mölling, P.; Khan, F.A.; Sundqvist, M.; Jass, J.; Söderquist, B. Comparative Distribution of Extended-Spectrum Beta-Lactamase-Producing Escherichia Coli from Urine Infections and Environmental Waters. PloS one 2019, 14, e0224861. [Google Scholar] [CrossRef]
- Yang, C.; Li, Y.; Zuo, L.; Jiang, M.; Zhang, X.; Xie, L.; Luo, M.; She, Y.; Wang, L.; Jiang, Y.; et al. Genomic Epidemiology and Antimicrobial Susceptibility Profile of Enterotoxigenic Escherichia Coli From Outpatients With Diarrhea in Shenzhen, China, 2015-2020. Frontiers in microbiology 2021, 12, 732068. [Google Scholar] [CrossRef]
- Greig, D.R.; Jenkins, C.; Gharbia, S.; Dallman, T.J. Comparison of Single-Nucleotide Variants Identified by Illumina and Oxford Nanopore Technologies in the Context of a Potential Outbreak of Shiga Toxin-Producing Escherichia Coli. GigaScience 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sun, H.; Bai, X.; Fu, S.; Fan, R.; Xiong, Y. Occurrence of Multidrug-Resistant and ESBL-Producing Atypical Enteropathogenic Escherichia Coli in China. Gut Pathogens 2018, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Bartoloni, A.; Pallecchi, L.; Riccobono, E.; Mantella, A.; Magnelli, D.; Di Maggio, T.; Villagran, A.L.; Lara, Y.; Saavedra, C.; Strohmeyer, M.; et al. Relentless Increase of Resistance to Fluoroquinolones and Expanded-Spectrum Cephalosporins in Escherichia Coli: 20 Years of Surveillance in Resource-Limited Settings from Latin America. Clinical Microbiology and Infection 2013, 19, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global Epidemiology of CTX-M β-Lactamases: Temporal and Geographical Shifts in Genotype. The Journal of antimicrobial chemotherapy 2017, 72, 2145–2155. [Google Scholar] [CrossRef]
- Song, J.; Oh, S.-S.; Kim, J.; Shin, J. Extended-Spectrum β-Lactamase-Producing Escherichia Coli Isolated from Raw Vegetables in South Korea. Scientific Reports 2020, 10, 19721. [Google Scholar] [CrossRef]
- Song, J.; Oh, S.-S.; Kim, J.; Park, S.; Shin, J. Clinically Relevant Extended-Spectrum β-Lactamase–Producing Escherichia Coli Isolates From Food Animals in South Korea. Frontiers in Microbiology 2020, 11. [Google Scholar] [CrossRef]
- Usein, C.-R.; Papagheorghe, R.; Oprea, M.; Condei, M.; Strãuţ, M. Molecular Characterization of Bacteremic Escherichia Coli Isolates in Romania. Folia microbiologica 2016, 61, 221–226. [Google Scholar] [CrossRef]
- Giufrè, M.; Graziani, C.; Accogli, M.; Luzzi, I.; Busani, L.; Cerquetti, M. Escherichia Coli of Human and Avian Origin: Detection of Clonal Groups Associated with Fluoroquinolone and Multidrug Resistance in Italy. The Journal of antimicrobial chemotherapy 2012, 67, 860–867. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, N.; Dias, D.; Sousa, M.; Capelo-Martinez, J.L.; Brito, F.; Caniça, M.; Igrejas, G.; Poeta, P. Clonal Diversity of ESBL-Producing Escherichia Coli in Pigs at Slaughter Level in Portugal. Foodborne pathogens and disease 2013, 10, 74–79. [Google Scholar] [CrossRef]
- Okeke, I.N.; Wallace-Gadsden, F.; Simons, H.R.; Matthews, N.; Labar, A.S.; Hwang, J.; Wain, J. Multi-Locus Sequence Typing of Enteroaggregative Escherichia Coli Isolates from Nigerian Children Uncovers Multiple Lineages. PLOS ONE 2010, 5, e14093. [Google Scholar] [CrossRef]
- Oteo, J.; Pérez-Vázquez, M.; Campos, J. Extended-Spectrum Beta-Lactamase Producing Escherichia Coli: Changing Epidemiology and Clinical Impact. Current Opinion in Infectious Diseases 2010, 23, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinformatics 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kim, J.; Jeon, S.E.; Kim, S.J.; Kim, N.O.; Hong, S.; Kang, Y.H.; Han, S.; Chung, G.T. Complete Nucleotide Sequence of the IncI1 Plasmid PSH4469 Encoding CTX-M-15 Extended-Spectrum β-Lactamase in a Clinical Isolate of Shigella Sonnei from an Outbreak in the Republic of Korea. International Journal of Antimicrobial Agents 2014, 44, 533–537. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).