1. Introduction
A cutaneous or skin cancerization field (SCF) refers to the medical condition where the chronic ultraviolet radiation (UVR)-damaged skin area around incident tumors or precancerous lesions harbors clonally expanding cell subpopulations with distinct pro-cancerous genetic alterations [
1]. As a result, this skin area becomes more susceptible to the development of new similar malignant lesions or the recurrence of existing ones [
2]. The identification of a SCF mainly relies on recognizing actinic keratosis (AK). AK, also known as keratosis solaris, is a common skin condition caused by long-term sun exposure. These pre-cancerous lesions not only serve as visible markers of chronic solar skin damage but, if left untreated, each of them can possibly progress into a potentially fatal cutaneous squamous cell carcinoma (CSCC) [
3,
4]. Since there are no established prognostic factors to predict which individual AK will progress into CSCC, early recognition, treatment, and follow-up are vital and endorsed by international guidelines to minimize the risk of invasive CSCC [
5,
6,
7]. On the other hand, AKs, more than being the hallmark lesions of UVR damaged skin at risk to develop skin cancers, because of their visual similarity to invasive neoplasia, also constitute a per se important macromorphological differential diagnostic challenge in a skin cancer screening setting.
Understanding the relationship between field cancerization and AK is crucial for identifying high-risk individuals, implementing early interventions, and preventing the progression to more aggressive forms of skin cancer [
8]. Currently, the clinical tools for grading SCF primarily involve assessing AK burden using systems like AKASI [
9] and AK-FAS [
10]. These systems depend on the subjective evaluation of preselected, target skin areas by trained physicians who count and evaluate AK lesions within them. In addition to their inherent subjectivity, these grading systems are limited by the high degree of spatial plasticity, including trend to recurrence of individual AK lesions. They are actually snapshots of the AK burden of a certain skin site, unable to track whether the recorded lesions are relapses of incident AK at the same site or are newly emerging lesions arising
de novo in nearby areas [
11].
Clinical photography is a low-cost, portable solution that efficiently stores 'scanned' visual information of an entire skin region. Considering the pathobiology of AK, Criscione et al. applied an image analysis-based methodology to analyze clinical images of chronically UV-damaged skin areas in a milestone study. [
12]. With this approach, they could estimate the risk of progression of AKs to keratinocyte skin carcinomas (KSCs) and assess the natural history of AK evolution dynamics for approximately six years. Since then, the evolving efficiency of deep learning algorithms in image recognition and the availability of extensive image archives have greatly accelerated the development of advanced computer-aided systems for skin disease diagnosis [
13,
14,
15,
16]. Particularly for the Aks, given the multifaced role of AK recognition in cutaneous oncology, the assignment of a distinct diagnostic label to this condition was also a core task in many studies that applied machine learning as a tool to differentiate skin diseases based on clinical images of isolated skin lesions [
17,
18,
19,
20,
21,
22,
23,
24,
25].
However, despite significant progress in image analysis of cropped skin lesions in recent years, the determination of AK burden in larger, sun-affected skin areas remains a still quite challenging diagnostic task. AKs present as pink, red, or brownish patches, papules, or flat plaques or may even have the same color as the surrounding skin. They vary in size from a few millimeters to 1-2 cm, and can be either isolated or more typically, numerous, sometimes even widely confluent in some patients. Moreover, the surrounding skin may show signs of chronic sun damage, including telangiectasias, dyschromia, and elastosis [
5] and also seborrheic keratoses and related lesions, notably lentigo solaris. These latter lesions are the most frequently encountered benign growths within a SCF and underlie most confusion related to the clinical differential diagnosis of AK lesions [
26]. All these visual features add substantial complexity to the task of automated AK burden determination.
Earlier approaches to automated AK recognition from clinical images were limited either by treating AK as a skin erythema detection problem [
27] or by restricting the detection to preselected smaller sub-regions of wider photographed skin areas, using a binary patch classifier for AK discrimination from healthy skin [
28]. Moreover, these approaches are prone to a large risk of false positive detections since erythema is present in various unrelated skin conditions, and the contamination of the images by diverse concurrent and confluent benign growths [
29,
30] seriously affects the accuracy of binary classification schemes to estimate AK burden in extended skin areas.
To tackle the demanding task of AK burden detection in UVR-chronically exposed skin areas, more recent work has proposed a superpixel-based, convolutional neural network (CNN) for AK detection (AKCNN) [
31]. The core engine of AKCNN is a patch classifier, a lightweight CNN, adequately trained to distinguish AK from healthy skin but also from seborrheic keratosis and solar lentigo. However, a main limitation of AKCNN is still the manual pre-selection of the area to scan, to exclude image parts with ‘disturbing’ visual features, like hairs, nose, lips, eyes and ears, that were found to result in false positive detections.
To improve the monitoring of AK burden in clinical settings with enhanced automation and precision, the present study evaluates the application of semantic segmentation using the U-Net architecture [
32], with transfer learning to compensate for the relatively small dataset of annotated images and a recurrent process to exploit efficiently contextual information and address the specific challenges related to low contrast and ambiguous boundaries of AK affected skin regions.
Although there has been considerable research on deep learning-based skin lesion segmentation, the primary focus has been delineating melanoma lesions in dermoscopic images [
33,
34]. The present study is a further development of our previous research on AK detection and, to the best of our knowledge, is the first study to employ semantic segmentation to clinical images for AK detection and SCF evaluation.
2. Materials and Methods
2.1. Methods
2.1.1. Overview of semantic segmentation and U-net architecture
Today, with the advent of deep learning and convolutional neural networks, semantic segmentation goes beyond traditional segmentation by associating a meaningful label to each pixel in an image [
35]. Semantic segmentation plays a crucial role in medical imaging, and it has numerous applications, including image analysis and computer-assisted diagnosis, monitoring the evolution of conditions over time, and assessing treatment responses.
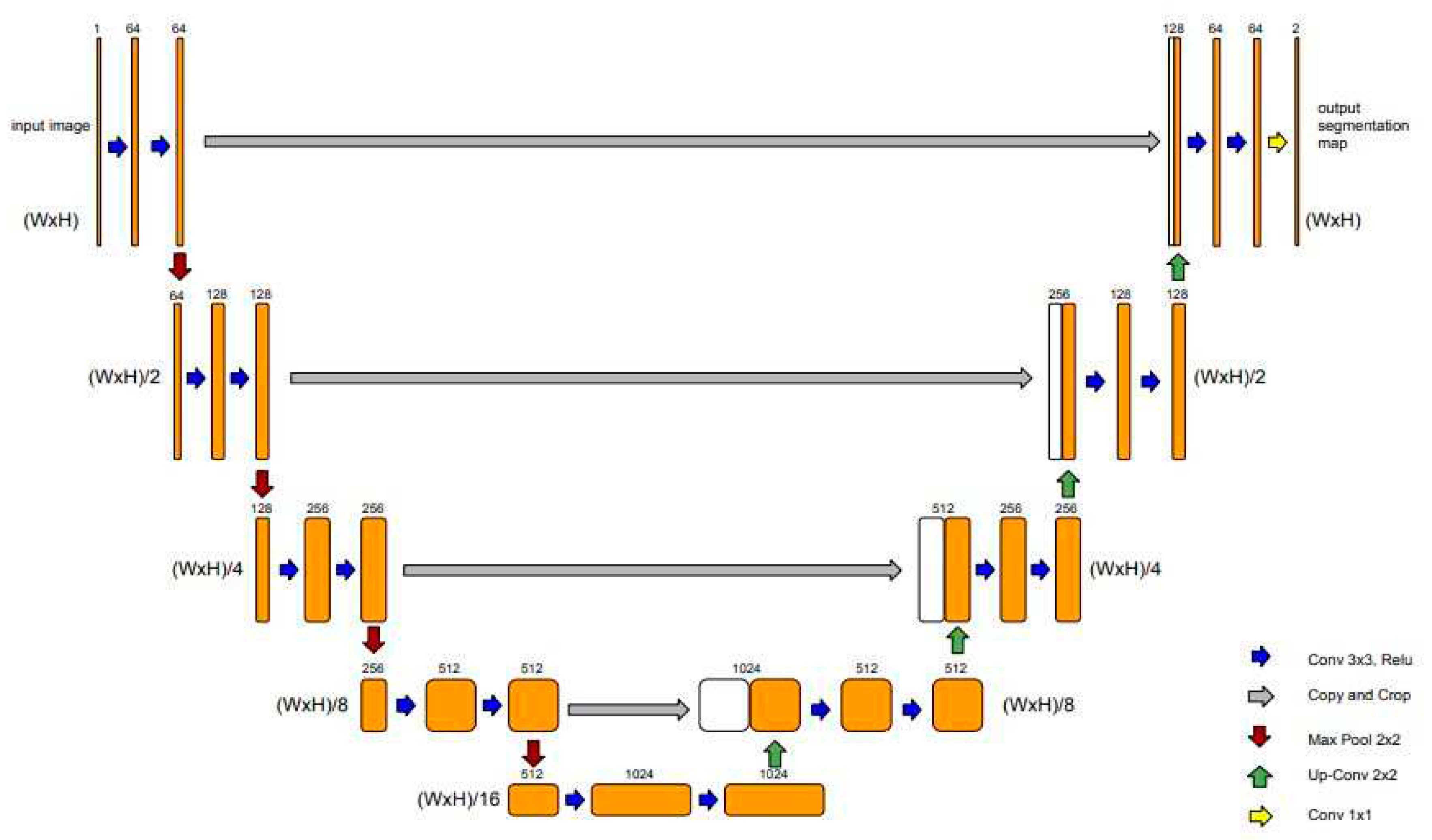
A groundbreaking application in the field of semantic segmentation is the U-net architecture, which emerged as a specialized variant of the fully convolutional network architecture. This network architecture excels in capturing fine-grained details from images and demonstrates effectiveness even with limited training data, making them well-suited for the challenges posed by the field of biomedical research [
32]. The U-Net architecture is characterized by its "U" shape, with symmetric encoder and decoder sections (
Figure 1).
The encoder section consists of a series of convolutional layers with 3x3 filters, followed by rectified linear unit (ReLU) activations. After each convolutional layer, a 2x2 max pooling operation is applied, reducing the spatial dimensions while increasing the number of feature channels. As the encoder progresses, the spatial resolution decreases while the number of feature channels increases. This allows the model to capture increasingly abstract and high-level features from the input image.
The decoder section of the U-Net starts with an up-sampling operation to increase the spatial dimensions of the feature maps. The up-sampling is typically performed using transposed convolutions. At each decoder step, skip connections are introduced. These connections directly connect the feature maps from the corresponding encoder step to the decoder, preserving fine-grained details and spatial information. The skip connections are achieved by concatenating the feature maps from the encoder with the up-sampled feature maps in the decoder.
In the following, the concatenation, the combined feature maps go through convolutional layers with 3x3 filters and ReLU activations to refine the segmentation predictions. Each decoder step typically reduces the number of feature channels to match the desired output shape.
Finally, a 1x1 convolutional layer is applied to produce the final segmentation map, with each pixel representing the predicted class label.
Since its introduction, U-Net has served as a foundation for numerous advancements in medical image analysis and has inspired the development of various modified architectures and variants [
36], [
37].
Transfer learning has significantly contributed to the success of U-Net in various medical image segmentation tasks [
38,
39,
40,
41]. Initializing the encoder part of U-Net with weights learned from a pre-trained model, enables U-Net to start with a strong foundation of learned representations, accelerating convergence, and improving segmentation performance, especially when the availability of annotated medical data is limited.
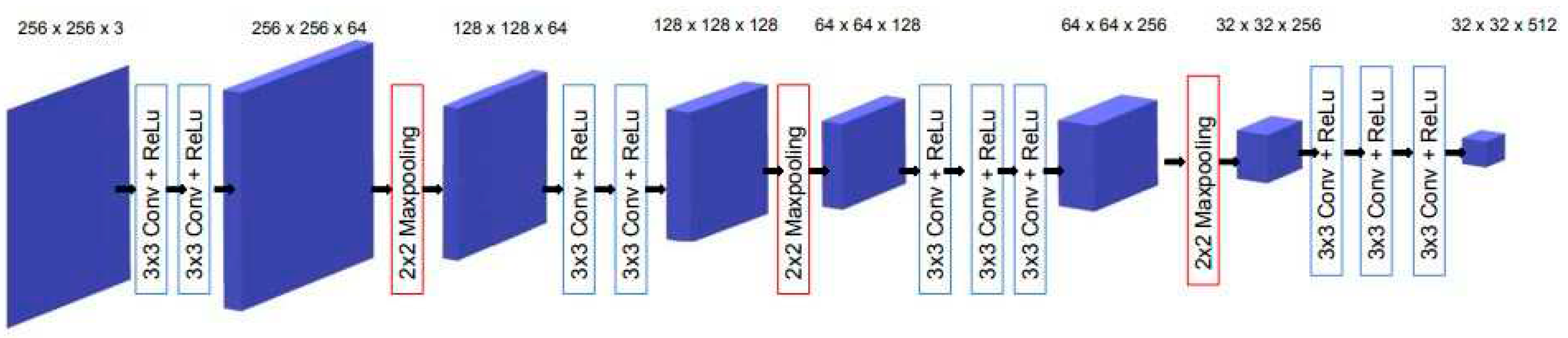
In this study, we used the VGG16 model [
42], pre-trained on ImageNet [
43], as the backbone for the encoder in the U-Net architecture. Both U-Net and VGG16 are based on the concepts of deep convolution neural networks, which are designed to encode hierarchical representations from images enabling them to capture increasingly complex features. Both architectures utilize convolutional layers with 3x3 filters followed by rectified linear unit activations and 2x2 max pooling operation for downsampling as their primary building blocks.
Figure 2 illustrates our transfer learning scheme, where four convolution blocks of pre-trained VGG16 were used for the U-Net encoder.
2.1.2. Batch Normalization
Batch normalization (BN), proposed by Sergey Ioffe and Christian Szegedy [
44], is a technique commonly used in neural networks to normalize the activations of a layer by adjusting and scaling them. It helps to stabilize and speed up the training process by reducing the internal covariate shift, which refers to the change in the distribution of the layer's inputs during training. The BN operation is typically performed by computing the mean and variance of the activations within a mini-batch during training. These statistics are then used to normalize the activations by subtracting the mean and dividing by the square root of the variance. Additionally, batch normalization introduces learnable parameters, known as scale and shift parameters, which allow the network to adaptively scale and shift the normalized activations [
44].
Since each mini-batch has a different mean and variance, this introduces some random variation or noise to the activations. As a result, the model becomes more robust to specific patterns or instances present in individual mini-batches and learns to generalize better to unseen examples.
In U-Net, the decoding layers are responsible for reconstructing the output image or segmentation map from the up-sampled feature maps. By applying BN to the decoding layers in our U-Net, the regularization effect is targeted to the reconstruction process, improving the model's generalization ability, and reducing the risk of overfitting in the output reconstruction.
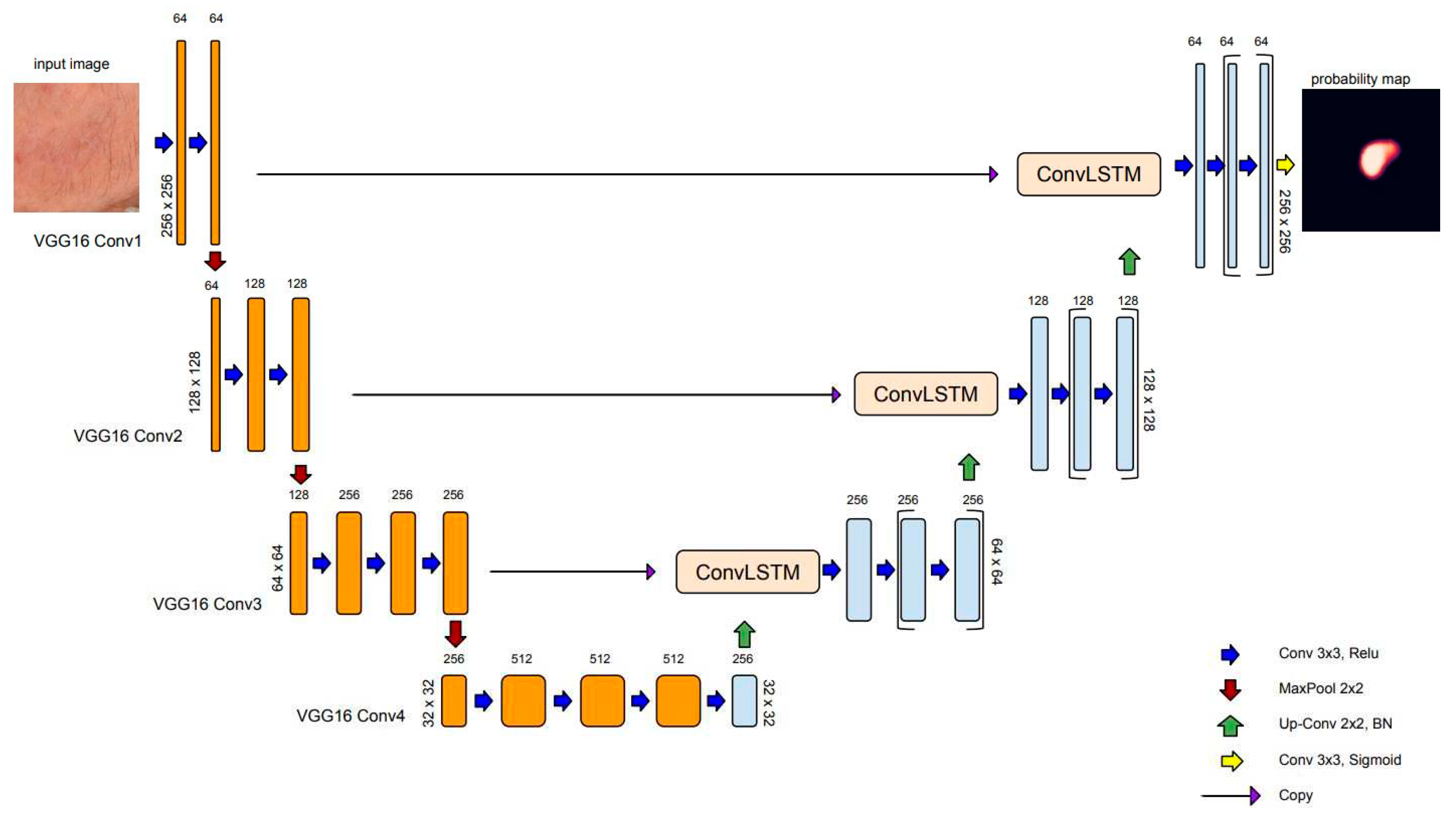
2.1.3. convLSTM: Spatial recurrent module in U-net architecture
The recurrent mechanisms in neural network architecture (RNN) have been adapted to work with sequential data, which decisions based on current and previous inputs. There are different kinds of recurrent units based on how the current and last inputs are combined, such as gated recurrent units and long short-term memory (LSTM)[
45]. LSTM module was designed by introducing three gating mechanisms that control the flow of information through the network: the input gate, the forget gate, and the output gate. These gates allow the LSTM network to selectively remember or forget information from the input sequence, which makes it more effective for long-term dependencies. For text, speech, and signal processing, plain RNNs are directly used, while for use in 2D or 3D data (e.g., image), RNNs have been extended correspondingly using convolutional structures. Convolutional LSTM (convLSTM) is the LSTM counterpart for long-term spatiotemporal predictions [
46].
In semantic segmentation, recurrent modules have been incorporated in various ways in U-net architecture. Alom et al. [
47] have introduced recurrent convolutional blocks at the backbone of the U-net architecture to enhance the ability of the model to integrate contextual information and improve feature representations for medical image segmentation tasks. In recent work, Arbelle et al. [
48] have proposed the integration of convLSTM blocks at every scale of the encoder section of the U-net architecture, enabling multi-scale spatiotemporal feature extraction and facilitating cell instants segmentation in time-lapse microscopy image sequences.
In the U-Net architecture, max pooling is commonly used in the encoding path to downsample the feature maps and capture high-level semantic information. However, max pooling can result in a loss of spatial information and details between neighboring pixels. Several researchers in the field of skin lesion segmentation using dermoscopic images have exploited recurrent layers as a mechanism to refine the skip connection process of U-Net [
49,
50,
51]. In the present study, to address the specific challenges related to low contrast and ambiguous boundaries of AK-affected skin regions, we employ convLSTM layers to bridge the semantic gap between the feature map extracted from the encoding path
and the output feature map after upsampling in the decoding path
. Assuming
is the concatenation of
and
, where
is the number of filters, and
are the height and width of the feature map.
is split into
patches
where:
and
Following, we will refer to
as
and replace subscripts i, j with the notion of processing step t. The input
is passed through convolution operation to compute three gates that regulate the spatial information flow: the input gate
, the forget gate
and the output gate
as follows:
ConvLSTM also maintains the cell state (
) and the hidden state (
):
and correspond to the 2D convolution kernel of the input and hidden states, respectively. * represents the convolution operation and bullet the Hadamard function (element-wise multiplication), respectively. are the bias terms and sigma is the sigmoid function.
Figure 3 depicts the model for AK detection based on U-net architecture (AKU-net) that incorporates transfer learning in the encoding path, the recurrent process in skip connections, and BN in the decoding path.
2.1.4. Loss function
In this study, we approached AK detection as a binary semantic segmentation task, where each pixel in the input image is classified as either foreground (belonging to the skin areas affected by AK) or background. For this purpose, we utilized the binary cross-entropy loss function, commonly used in U-Net, also known as the log loss or sigmoid loss.
Mathematically, the binary cross-entropy loss for a single pixel can be defined as:
Where is the predicted probability of the pixel belonging to the foreground class, obtained by applying a sigmoid activation function to the model's output, and is the ground truth label (0 for background, 1 for foreground) indicating the true class of the pixel.
The overall loss for the entire image is then computed as the average of the individual pixel losses. During training, the network aims to minimize this loss function by adjusting the model's parameters through backpropagation and gradient descent optimization algorithms.
2.1.5. Evaluation
To demonstrate the expected improvements in AK detection, AKU-net was compared with plain U-Net and U-Net
++ [
52]. U-Net
++ comprises an encoder and decoder connected through a series of nested dense convolutional blocks. The main concept behind U-Net
++ is to narrow the semantic gap between the encoder and decoder feature maps before fusion. The authors of U-Net
++ reported superior performance compared to the original U-Net in various medical image segmentation tasks, including electron microscopy cells, nuclei, brain tumors, liver, and lung nodules.
To ensure comparable results, the trained networks utilized the VGG16 as the backbone and BN in the decoding path. The segmentation models were assessed by means of the Dice coefficient and Intersection over Union (IoU).
The Dice coefficient measures the similarity or overlap between the predicted segmentation mask and the ground truth mask. The formula for the Dice coefficient is as follows:
The
measures the intersection between the predicted and ground truth masks relative to their union. The formula for
is as follows:
The notion represents the total number of pixels. are the true positive, false negative, and false positive prediction rates at the pixel level, respectively.
To provide comparison results with a recent work on AK detection, we also employed the adapted region-based
score
[
53]:
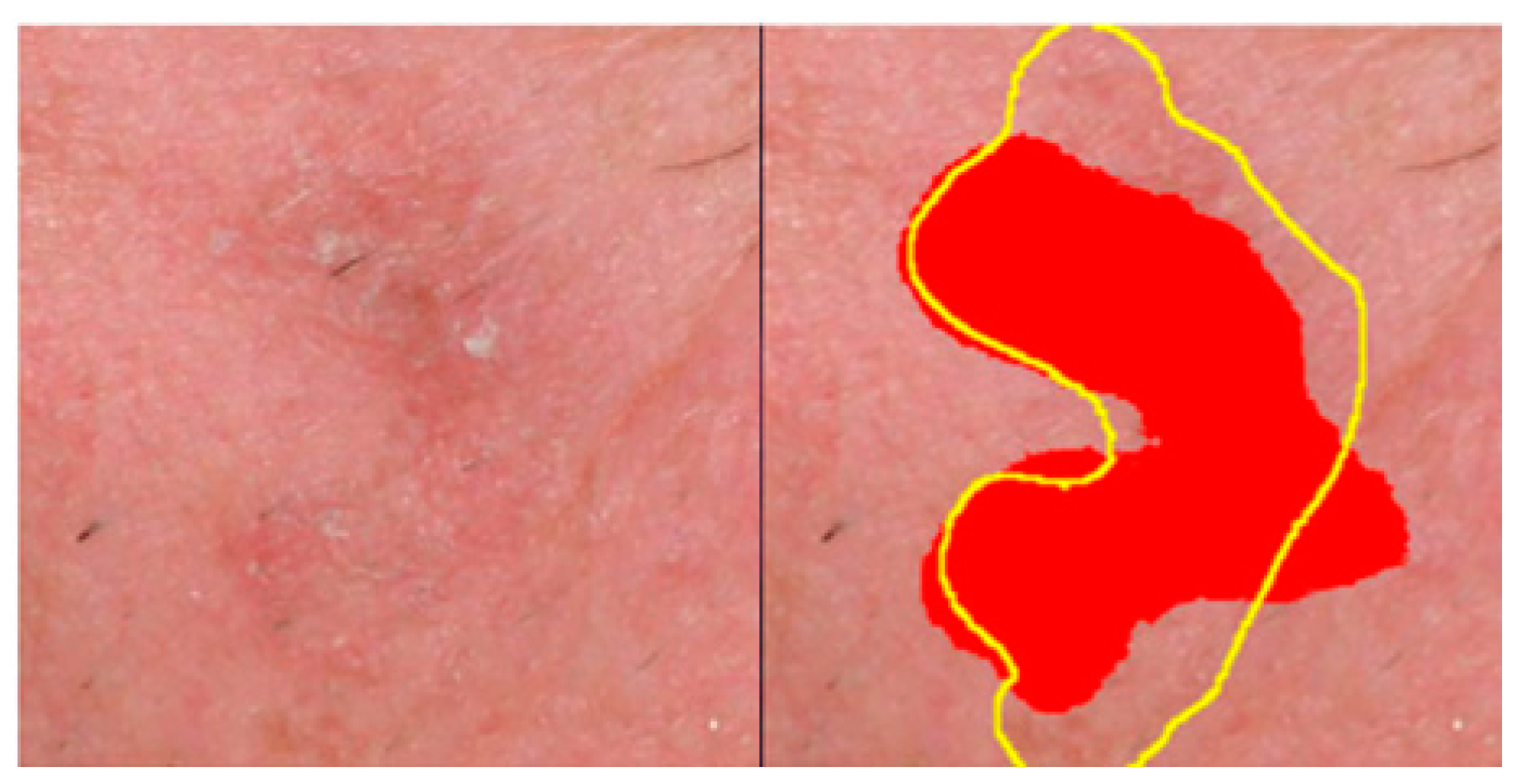
score was introduced by the authors to compensate for the fact that AK lesions often lack sharply demarcated borders, and experienced clinicians can provide only rough, approximate AK annotations in clinical imagesThe adapted estimators of Recall () and Precision () are estimated as follows:
Assuming a ground truth set of N annotated (labeled) areas,
, and the set of pixels predicted as AK by the system,
, we define:
and the
:
The adapted estimators
and
are given as follows:
All coefficients, the the and the range from 0 to 1. Higher values of these metrics indicate greater similarity between the predicted and ground truth masks and, consequently, better performance.
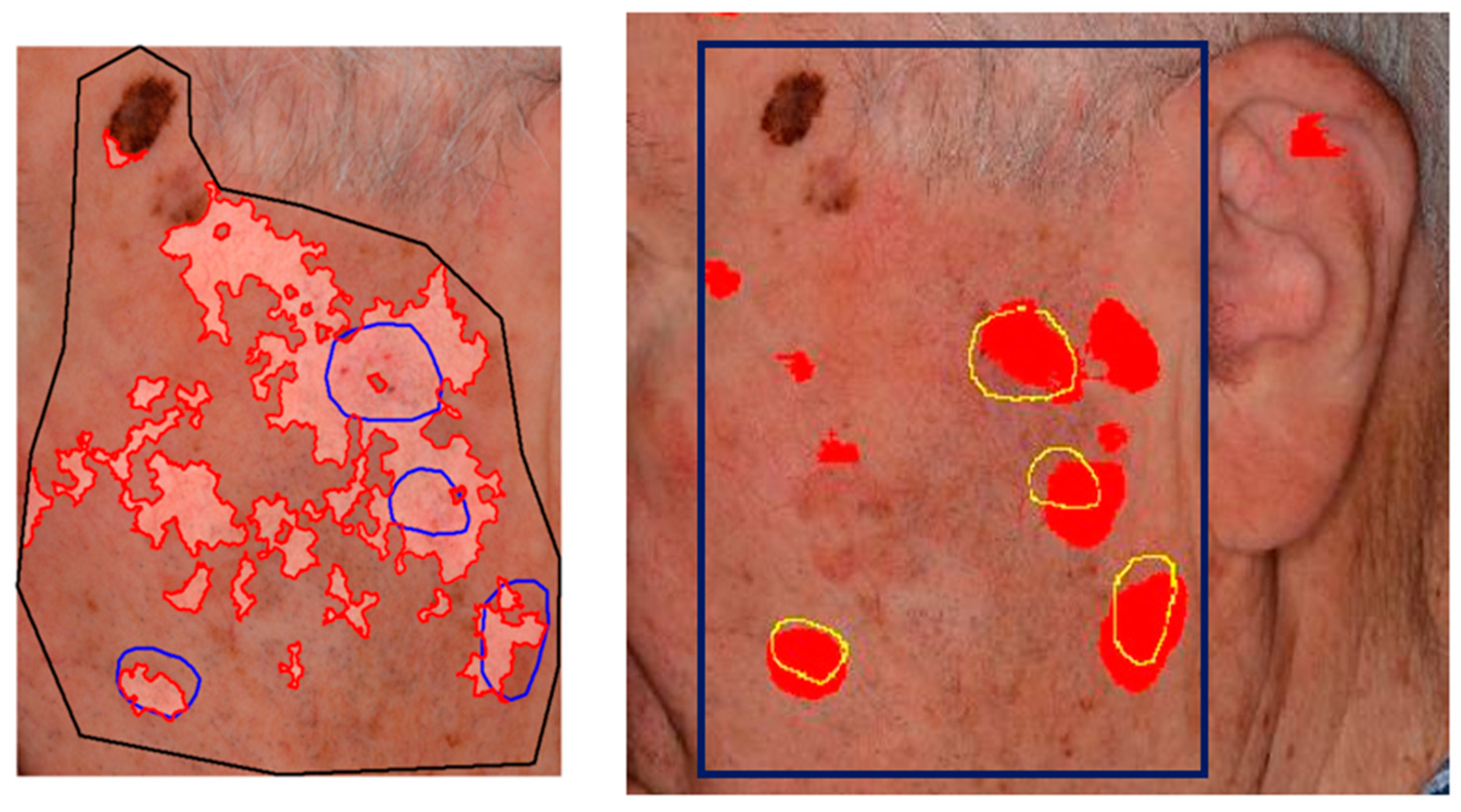
Figure 4 provides a qualitative example of the error tolerance in estimating
, favoring compensation for rough annotations
.
2.1.6. Implementation details
The segmentation models were implemented in Python 3, adopting an open-source framework Keras API with Tensorflow as the backend. The experimental environment is based on a Windows10 workstation configured with an AMD 9 series 3900X CPU@3.80 GHz processor, 64 GB of 3200MHz DDR4 ECC RDIMM, NVIDIA RTX 3070 GPU memory of 8 GB. The Adam optimizer was employed to train the networks. Considering the constraints of our computational environment, we set the batch size equal to 32, and the number of iterations to 100.
2.2. Material
The use of archival photographic material for this study was approved by the Human Investigation Committee (IRB) of the University Hospital of Ioannina (approval nr.: 3/17-2-2015[θ.17)]. The study included a total of 115 patients (60 males, 55 females; age range: 45-85 years) with facial AK who attended the specialized Dermato-oncology Clinic of the Dermatology Department.
Facial photographs were acquired with the camera axis perpendicular to the photographed region. The distance was adjusted to include the whole face from the chin to the top of the hair/scalp.
Digital photographs of 4016x6016 pixels spatial resolution were acquired according to a procedure adapted from Muccini et al. [
44], using a Nikon D610 (Nikon, Tokyo, Japan) camera with a Nikon NIKKOR
© 60mm 1:2.8G ED Microlens mounted on it. The camera controls were set at f18, shutter speed 1/80sec, ISO 400, autofocus, and white balance at auto-adjustment mode. A Sigma ring flash (Sigma, Fukushima, Japan) at TTL mode was mounted to the camera. In front of the lens and the flashlight, linear polarized filters were appropriately adapted to ensure a 90
o rotation of the ring flash polarization axis to the relevance of the lens-mounted polarizing filter. For the AK annotation, two physicians (GG and AZ) jointly discussed and reached an agreement on the affected skin regions.
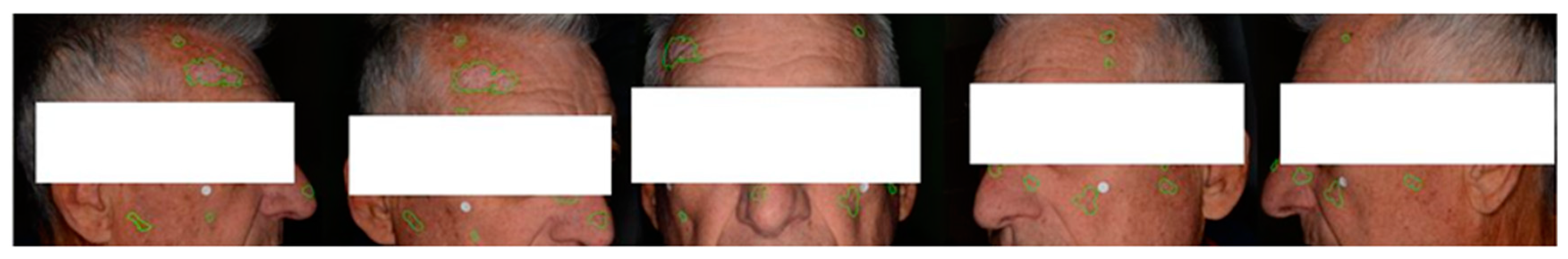
Multiple photographs were taken per patient to capture the presence of lesions across the entire face. Additionally, these photographs were intended to provide different views of the same lesions, resulting in 569 annotated clinical photographs (
Figure 5).
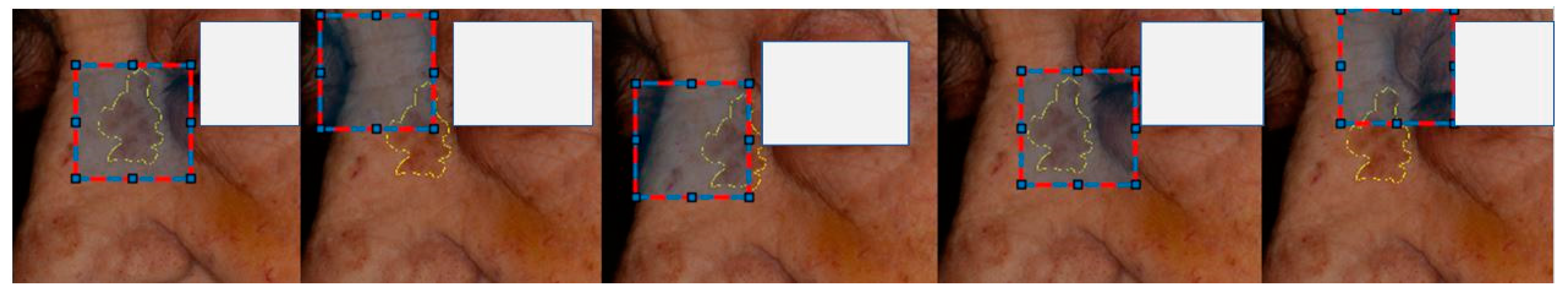
Patches of 512x512 pixels were extracted from each photograph using translation lesion boxes. These boxes encompassed lesions in various positions and captured different contexts of the perilesional skin. In total, 16891 lesion center and translation augmented patches were extracted. (
Figure 6).
2.2.1. Experimental settings
Since multiple samples (skin patches) from the same patient were used to prevent data leakage in train-validation-test data sets and ensure unbiased evaluations of the models' generalization, we performed data set splitting at the patient level. We used 510 photographs obtained from 98 patients for training, extracting 16,488 translation-augmented image patches. Among them, approximately 20% of the crops (3,298 patches) from 5 patients were reserved for validation. An independent set consisting of 17 patients (59 photographs) was used to assess the model's performance, yielding 403 central lesion patches (
Table 1).
It is important to note that since the available images had a high spatial resolution, we had to train the model using rectangular crops. The size of these crops was selected to include sufficient contextual information. To achieve this, the images were first rescaled by 0.5, and crops of size 512x512 were extracted. However, due to computational limitations, the patches were further rescaled by a factor of 0.5 (256x256 pixels). Using an internal fiducial marker [
14], we estimated that our model was ultimately trained with cropped images at a scale of approximately ~5 pixels/mm.
3. Results
and
coefficients were utilized to evaluate the segmentation accuracy of the model.
Table 2 summarizes the comparison results employing the standard U-net and U-net
++ architectures.
AKU-Net model demonstrated a statistically significant improvement in AK detection compared to U-Net
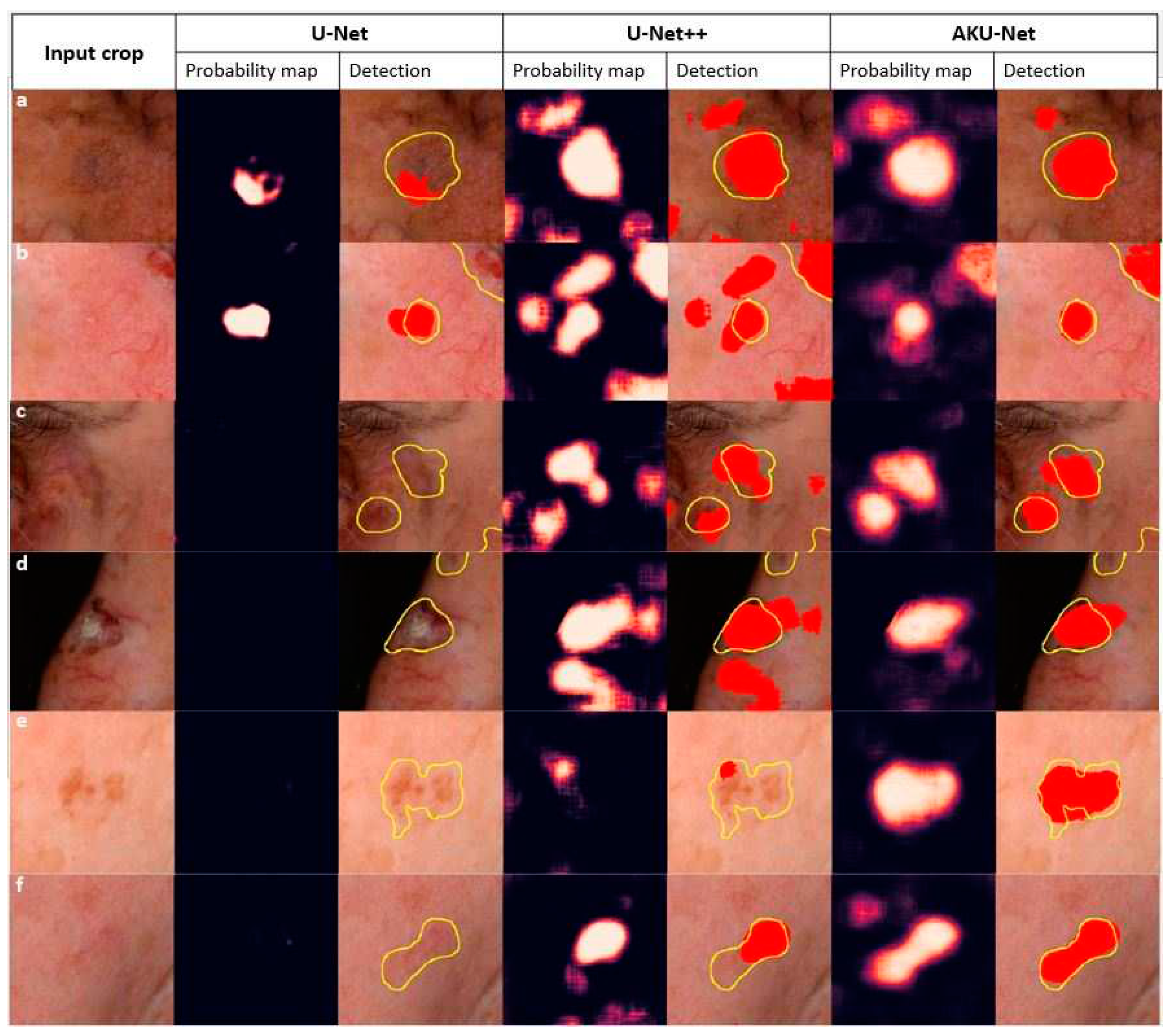
++ (p<.05; Wilcoxon signed-rank test). It is worth noting that the standard U-Net model had limitations in detecting AK areas, as it could not detect AK in 257 out of the total 403 testing crops, corresponding to approximately 63% of the testing cases. An exemplary qualitative comparison of the goodness of the segmentation of incident AK with the different model architectures is illustrated in
Figure 7.
In a recent study, we implemented the AKCNN model for efficient AK detection [
53] in manually preselected wide skin areas. To evaluate the performance of the AKU-Net model in broad skin areas and compare it with the AKCNN, we used the same evaluation set as the one employed in the previous study [
53].
The AKU-Net model was trained using patches of 256x256 pixels. However, to allow the present model to be evaluated and compared with the AKCNN, we performed the following steps:
Zero Padding: Appropriate zero-padding was applied to the input image to make it larger and suitable for subsequent cropping.
Image Cropping: The zero-padded image was divided into crops of 256x256 pixels. These crops were used as individual inputs to the AKU-Net model for AK detection.
Aggregating Results: The obtaining segmentation results for each 256x256 pixel crop were combined to obtain the overall AK detection for the entire broad skin area.
Table 3 compares the performances (accuracy measures) of the AKU-Net and AKCNN model architectures (for n=10 random frames). At a similar image scale of approximately ~7 pixels/mm, there were no significant differences in the level of model performance in terms of
(p=0.6),
and
Wilcoxon signed-rank test).
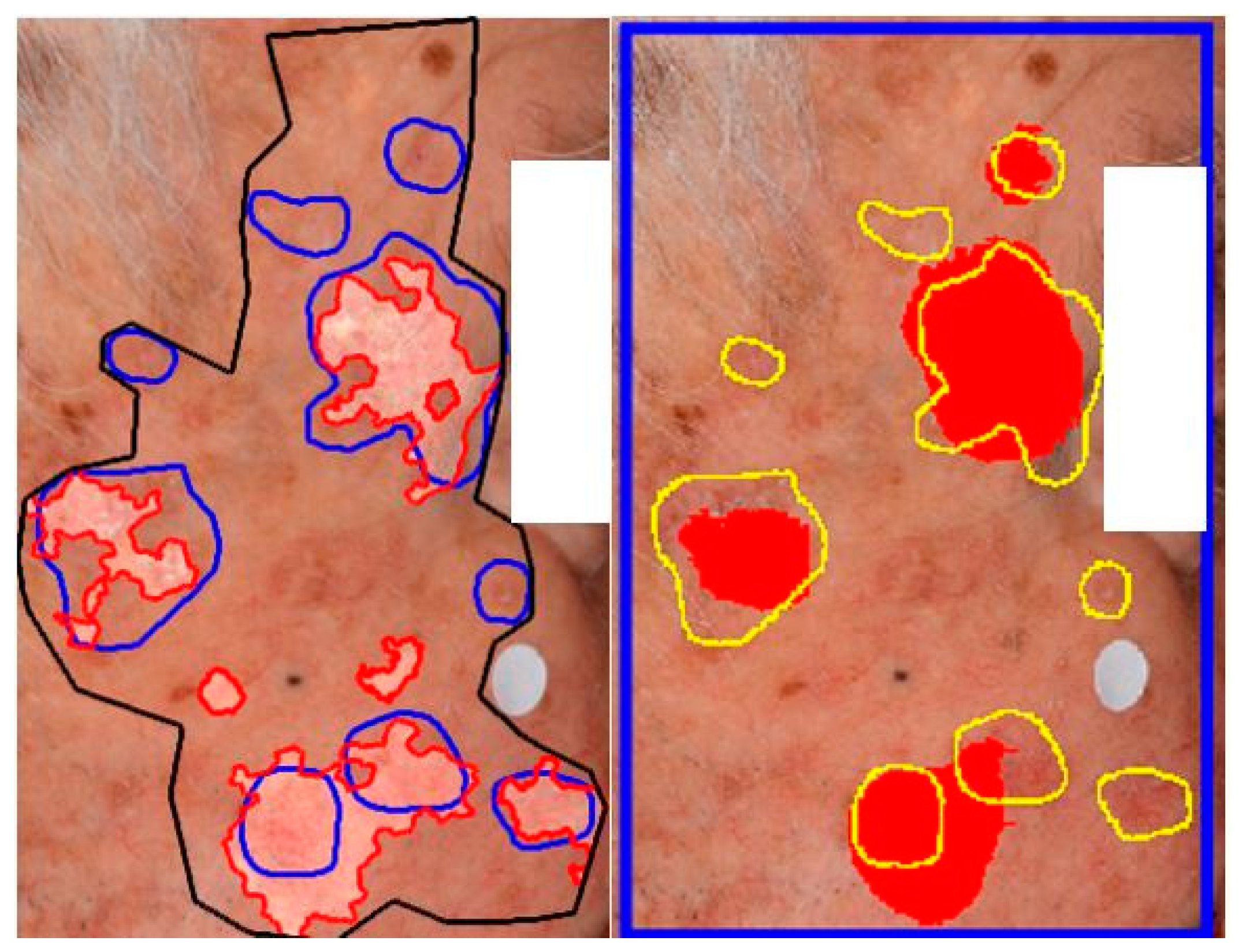
Although AKU-Net exhibited AK detection accuracy at the same level as that of AKCNN, the substantial advantage of AKU-Net is that the latter does not require manual preselection of scanning areas. Explanative examples are given in
Figure 8 and
Figure 9.
4. Discussion
In this study, we utilized deep learning, specifically in the domain of semantic segmentation, to enhance the detection of actinic keratosis (AK) on clinical photographs. In our previous approach we have introduced a CNN patch-classifier (AKCNN) to assess the burden of AK in large skin areas [
52]. Detecting AK in regions with field cancerization presents challenges, and a binary patch (regional) classifier, specifically "AK versus all," had serious limitations. AKCNN was implemented to effectively distinguish AK from healthy skin and differentiate it from seborrheic keratosis and solar lentigo, thereby reducing false positive detections. However, AKCNN is subject to manually predefined scanning areas which are necessary to exclude skin regions prone to falsely diagnosed as AK on clinical images (
Figure 8 and
Figure 9).
To enhance the monitoring of AK burden in real clinical settings with improved automation and precision, we utilized a semantic segmentation approach based on an adequately adapted U-Net architecture. Despite using a relatively small dataset of weakly annotated clinical images, AKU-Net exhibited a remarkably improved performance, particularly in challenging skin areas. The AKU-Net model outperformed the corresponding baseline models, the U-Net and U-Net++, highlighting the efficiency boost achieved through the spatial recurrent layers added in skip connections.
Considering the scanning performance, AKU-Net was evaluated by aggregating the scanning results from 256x256 pixel crops and compared with AKCNN (
Table 3). Both approaches exhibited a similar level of recall (true positive detection). However, AKU-Net is favourably tolerant on the selection of the scanning area, that can simply be a boxed area that includes the target region, providing false positive rates at least comparable to that obtained with the AKCNN on manually predefined scanning regions.
It is important to note that due to computational constraints, AKU-net was trained on image crops of 256x256 pixels, at a scale of about ~5 pixels/mm. For this, the original image was subsampled twice. This subsampling process resulted in a degradation of spatial resolution, which imposes limitations on the system's ability to detect AK lesions (recall level). This limitation is also supported by evidence from our previous studies: in a similar scale of ~7 pixels/mm, AKCNN experienced a drop in recall [
53]. We expect a significantly improved AK detection accuracy by subsampling the original image and training the network with crops of 512x512 pixels.
Future efforts will explore the optimal trade-off between image scale and the size of the input crop used to train the network, which could lead to superior detection of AK lesions. Moreover, implementing a system with known restrictions, that is, knowing the range of acceptable image scales, is essential for future studies to complement and validate the system’s generalization utilizing multicenter data sets from various cameras.
In future studies, it is also essential to evaluate the system's accuracy in relation to the variability among experts when recognizing AK using clinical photographs. This assessment is crucial as it will gauge the system's performance compared to the varying detection of human experts.
5. Conclusions
Understanding the relationship between the biology of a skin cancerization field and the burden of incident AK is crucial for identifying high-risk individuals, implementing early interventions, and preventing the development of more aggressive forms of skin cancer.
The present study evaluated the application of semantic segmentation based on the U-Net architecture to improve the monitoring of AK burden in clinical settings with enhanced automation and precision. Deep learning algorithms for semantic image segmentation are continuously evolving. However, the choice between different network architectures depends on the specific requirements of the segmentation task, the available computational resources, and the size and quality of the training dataset. Overall, the results from the present study indicated that the AKU-Net model is an efficient approach for AK detection, paving the way for more effective and reliable evaluation, continuous monitoring of condition progression, and assessment of treatment responses.
Author Contributions
Conceptualization, P.S., A.L. and I.B.; methodology, P.S.,P.D.; software, P.D, validation, P.S.,G.G.,and I.B. Formal analysis, P.S. and P.D; investigation, P.D. and P.S., data curation, G.G.,A.Z., and I.B.; writing—original draft preparation, P.S.; writing—review and editing, P.D.,G.G.,A.L., and I.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of University Hospital of Ioannina (approval nr.: 3/17-2-2015[θ.17)).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- T. J. Willenbrink, E. S. Ruiz, C. M. Cornejo, C. D. Schmults, S. T. Arron, and A. Jambusaria-Pahlajani, “Field cancerization: Definition, epidemiology, risk factors, and outcomes,” J. Am. Acad. Dermatol., vol. 83, no. 3, pp. 709–717, Sep. 2020.
- R. N. Werner, A. Sammain, R. Erdmann, V. Hartmann, E. Stockfleth, and A. Nast, “The natural history of actinic keratosis: a systematic review,” Br. J. Dermatol., vol. 169, no. 3, pp. 502–518, Sep. 2013.
- R. Gutzmer, S. Wiegand, O. Kölbl, K. Wermker, M. Heppt, and C. Berking, “Actinic Keratosis and Cutaneous Squamous Cell Carcinoma.,” Dtsch. Arztebl. Int., vol. 116, no. 37, pp. 616–626, Sep. 2019.
- D. de Berker, J. M. McGregor, M. F. Mohd Mustapa, L. S. Exton, and B. R. Hughes, “British Association of Dermatologists’ guidelines for the care of patients with actinic keratosis 2017,” Br. J. Dermatol., vol. 176, no. 1, pp. 20–43, Jan. 2017.
- R. N. Werner et al., “Evidence- and consensus-based (S3) Guidelines for the Treatment of Actinic Keratosis - International League of Dermatological Societies in cooperation with the European Dermatology Forum - Short version,” J. Eur. Acad. Dermatol. Venereol., vol. 29, no. 11, pp. 2069–2079, Nov. 2015.
- D. de Berker, J. M. D. de Berker, J. M. McGregor, M. F. Mohd Mustapa, L. S. Exton, and B. R. Hughes, “British Association of Dermatologists’ guidelines for the care of patients with actinic keratosis 2017,” Br. J. Dermatol., vol. 176, no. 1, pp. 20–43, Jan. 2017.
- D. B. Eisen et al., “Guidelines of care for the management of actinic keratosis,” J. Am. Acad. Dermatol., vol. 85, no. 4, pp. e209–e233, Oct. 2021.
- D. S. Rigel, L. F. D. S. Rigel, L. F. Stein Gold, and P. Zografos, “The importance of early diagnosis and treatment of actinic keratosis,” J. Am. Acad. Dermatol., vol. 68, no. 1 Suppl 1, Jan. 2013.
- T. Dirschka et al., “A proposed scoring system for assessing the severity of actinic keratosis on the head: actinic keratosis area and severity index,” J. Eur. Acad. Dermatology Venereol., vol. 31, no. 8, pp. 1295–1302, Aug. 2017.
- B. Dréno et al., “A novel actinic keratosis field assessment scale for grading actinic keratosis disease severity,” Acta Derm. Venereol., vol. 97, no. 9, pp. 1108–1113, Oct. 2017.
- T. Steeb et al., “How to Assess the Efficacy of Interventions for Actinic Keratosis? A Review with a Focus on Long-Term Results.,” J. Clin. Med., vol. 10, no. 20, Oct. 2021.
- V. D. Criscione, M. A. V. D. Criscione, M. A. Weinstock, M. F. Naylor, C. Luque, M. J. Eide, and S. F. Bingham, “Actinic keratoses: Natural history and risk of malignant transformation in the Veterans Affairs Topical Tretinoin Chemoprevention Trial,” Cancer, vol. 115, no. 11, pp. 2523–2530, Jun. 2009.
- A. Adegun and S. Viriri, “Deep learning techniques for skin lesion analysis and melanoma cancer detection: a survey of state-of-the-art,” Artif. Intell. Rev., vol. 54, no. 2, pp. 811–841, Jun. 2020.
- H. K. Jeong, C. H. K. Jeong, C. Park, R. Henao, and M. Kheterpal, “Deep Learning in Dermatology: A Systematic Review of Current Approaches, Outcomes, and Limitations,” JID Innov., vol. 3, no. 1, p. 100150, Jan. 2023.
- L.-F. Li, X. L.-F. Li, X. Wang, W.-J. Hu, N. N. Xiong, Y.-X. Du, and B.-S. Li, “Deep Learning in Skin Disease Image Recognition: A Review,” IEEE Access, vol. 8, pp. 208264–208280, 2020.
- M. A. Kassem, K. M. M. A. Kassem, K. M. Hosny, R. Damaševičius, and M. M. Eltoukhy, “Machine Learning and Deep Learning Methods for Skin Lesion Classification and Diagnosis: A Systematic Review,” Diagnostics, vol. 11, no. 8, p. 1390, Jul. 2021.
- L. Wang et al., “AK-DL: A shallow neural network model for diagnosing actinic keratosis with better performance than deep neural networks,” Diagnostics, vol. 10, no. 4, 2020.
- R. C. Maron et al., “Systematic outperformance of 112 dermatologists in multiclass skin cancer image classification by convolutional neural networks,” Eur. J. Cancer, vol. 119, pp. 57–65, 2019.
- P. Tschandl et al., “Expert-Level Diagnosis of Nonpigmented Skin Cancer by Combined Convolutional Neural Networks,” JAMA Dermatology, vol. 155, no. 1, pp. 58–65, Jan. 2019.
- A. G. C. Pacheco and R. A. Krohling, “The impact of patient clinical information on automated skin cancer detection,” Comput. Biol. Med., vol. 116, p. 103545, Jan. 2020.
- Y. Liu et al., “A deep learning system for differential diagnosis of skin diseases,” Nat. Med. 2020 266, vol. 26, no. 6, pp. 900–908, 20. 20 May.
- R. Karthik, T. S. R. Karthik, T. S. Vaichole, S. K. Kulkarni, O. Yadav, and F. Khan, “Eff2Net: An efficient channel attention-based convolutional neural network for skin disease classification,” Biomed. Signal Process. Control, vol. 73, no. 21, p. 103406, 2022. 20 October.
- S. S. Han, M. S. S. S. Han, M. S. Kim, W. Lim, G. H. Park, I. Park, and S. E. Chang, “Classification of the Clinical Images for Benign and Malignant Cutaneous Tumors Using a Deep Learning Algorithm,” J. Invest. Dermatol., vol. 138, no. 7, pp. 1529–1538, Jul. 2018.
- Y. Fujisawa et al., “Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis,” Br. J. Dermatol., vol. 180, no. 2, pp. 373–381, Feb. 2019.
- S. S. Han et al., “Keratinocytic Skin Cancer Detection on the Face Using Region-Based Convolutional Neural Network,” JAMA Dermatology, vol. 156, no. 1, pp. 29–37, Jan. 2020.
- S. Kato, S. M. S. Kato, S. M. Lippman, K. T. Flaherty, and R. Kurzrock, “The conundrum of genetic ‘Drivers’ in benign conditions,” J. Natl. Cancer Inst., vol. 108, no. 8, Aug. 2016.
- S. C. Hames et al., “Automated detection of actinic keratoses in clinical photographs,” PLoS One, vol. 10, no. 1, pp. 1–12, 2015.
- P. Spyridonos, G. P. Spyridonos, G. Gaitanis, A. Likas, and I. D. Bassukas, “Automatic discrimination of actinic keratoses from clinical photographs,” Comput. Biol. Med., vol. 88, pp. 50–59, Sep. 2017.
- A. P. South et al., “NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis,” J. Invest. Dermatol., vol. 134, no. 10, pp. 2630–2638, Jan. 2014.
- S. Durinck et al., “Temporal dissection of tumorigenesis in primary cancers,” Cancer Discov., vol. 1, no. 2, pp. 137–143, Jun. 2011.
- P. Spyridonos, G. P. Spyridonos, G. Gaitanis, A. Likas, and I. D. Bassukas, “A convolutional neural network based system for detection of actinic keratosis in clinical images of cutaneous field cancerization,” Biomed. Signal Process. Control, vol. 79, p. 104059, Jan. 2023.
- O. Ronneberger, P. O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 9351, pp. 234–241, 2015.
- Z. Mirikharaji et al., “A survey on deep learning for skin lesion segmentation,” Med. Image Anal., vol. 88, p. 102863, Aug. 2023.
- M. K. Hasan, M. A. M. K. Hasan, M. A. Ahamad, C. H. Yap, and G. Yang, “A survey, review, and future trends of skin lesion segmentation and classification,” Comput. Biol. Med., vol. 155, p. 106624, Mar. 2023.
- M. Aljabri and M. AlGhamdi, “A review on the use of deep learning for medical images segmentation,” Neurocomputing, vol. 506, pp. 311–335, Sep. 2022.
- N. Siddique, S. N. Siddique, S. Paheding, C. P. Elkin, and V. Devabhaktuni, “U-net and its variants for medical image segmentation: A review of theory and applications,” IEEE Access, 2021.
- R. Azad et al., “Medical Image Segmentation Review: The success of U-Net,” Nov. 2022.
- M. Ghafoorian et al., “Transfer Learning for Domain Adaptation in MRI: Application in Brain Lesion Segmentation BT - Medical Image Computing and Computer Assisted Intervention − MICCAI 2017,” 2017, pp. 516–524.
- R. Feng, X. R. Feng, X. Liu, J. Chen, D. Z. Chen, H. Gao, and J. Wu, “A Deep Learning Approach for Colonoscopy Pathology WSI Analysis: Accurate Segmentation and Classification,” IEEE J. Biomed. Heal. informatics, vol. 25, no. 10, pp. 3700–3708, Oct. 2021.
- A. Huang, L. A. Huang, L. Jiang, J. Zhang, and Q. Wang, “Attention-VGG16-UNet: a novel deep learning approach for automatic segmentation of the median nerve in ultrasound images,” Quant. Imaging Med. Surg., vol. 12, no. 6, pp. 3138–3150, Jun. 2022.
- N. Sharma et al., “U-Net Model with Transfer Learning Model as a Backbone for Segmentation of Gastrointestinal Tract,” Bioeng. 2023, Vol. 10, Page 119, vol. 10, no. 1, p. 119, Jan. 2023.
- K. Simonyan and A. Zisserman, “Very Deep Convolutional Networks for Large-Scale Image Recognition,” Int. Conf. Learn. Represent., pp. 1–14, Sep. 2015.
- J. Deng, W. J. Deng, W. Dong, R. Socher, L.-J. Li, K. Li, and L. Fei-Fei, “ImageNet: A large-scale hierarchical image database,” 2009 IEEE Conf. Comput. Vis. Pattern Recognit., pp. 248–255, Jun. 2009.
- S. Ioffe and C. Szegedy, “Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift,” 32nd Int. Conf. Mach. Learn. ICML 2015, vol. 1, pp. 448–456, Feb. 2015.
- Y. Yu, X. Y. Yu, X. Si, C. Hu, and J. Zhang, “A review of recurrent neural networks: Lstm cells and network architectures,” Neural Comput., vol. 31, no. 7, pp. 1235–1270, Jul. 2019.
- X. Shi, Z. X. Shi, Z. Chen, H. Wang, D. Y. Yeung, W. K. Wong, and W. C. Woo, “Convolutional LSTM Network: A Machine Learning Approach for Precipitation Nowcasting,” Adv. Neural Inf. Process. Syst., vol. 2015-Janua, pp. 802–810, Jun. 2015.
- M. Z. Alom, C. M. Z. Alom, C. Yakopcic, M. Hasan, T. M. Taha, and V. K. Asari, “Recurrent residual U-Net for medical image segmentation,” J. Med. imaging (Bellingham, Wash.), vol. 6, no. 1, p. 1, Mar. 2019.
- A. Arbelle, S. A. Arbelle, S. Cohen, and T. R. Raviv, “Dual-Task ConvLSTM-UNet for Instance Segmentation of Weakly Annotated Microscopy Videos,” IEEE Trans. Med. Imaging, vol. 41, no. 8, pp. 1948–1960, Aug. 2022.
- M. Attia, M. M. Attia, M. Hossny, S. Nahavandi, and A. Yazdabadi, “Skin melanoma segmentation using recurrent and convolutional neural networks,” in 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), 2017, pp. 292–296.
- R. Azad, M. R. Azad, M. Asadi-Aghbolaghi, M. Fathy, and S. Escalera, “Bi-Directional ConvLSTM U-Net with Densley Connected Convolutions,” in 2019 IEEE/CVF International Conference on Computer Vision Workshop (ICCVW), 2019, pp. 406–415.
- X. Jiang, J. X. Jiang, J. Jiang, B. Wang, J. Yu, and J. Wang, “SEACU-Net: Attentive ConvLSTM U-Net with squeeze-and-excitation layer for skin lesion segmentation,” Comput. Methods Programs Biomed., vol. 225, p. 107076, Oct. 2022.
- Z. Zhou, M. M. Z. Zhou, M. M. Rahman Siddiquee, N. Tajbakhsh, and J. Liang, “UNet++: A Nested U-Net Architecture for Medical Image Segmentation,” Deep Learn. Med. Image Anal. Multimodal Learn. Clin. Decis. Support 4th Int. Work. DLMIA 2018, 8th Int. Work. ML-CDS 2018, held conjunction with MICCAI 2018, Granada, Spain, S, vol. 11045, p. 3, 2018.
- P. Spyridonos, G. P. Spyridonos, G. Gaitanis, A. Likas, and I. D. Bassukas, A convolutional neural network based system for detection of actinic keratosis in clinical images of cutaneous field cancerization, vol. 79. Elsevier, p. 104059.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).