Submitted:
31 August 2023
Posted:
04 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
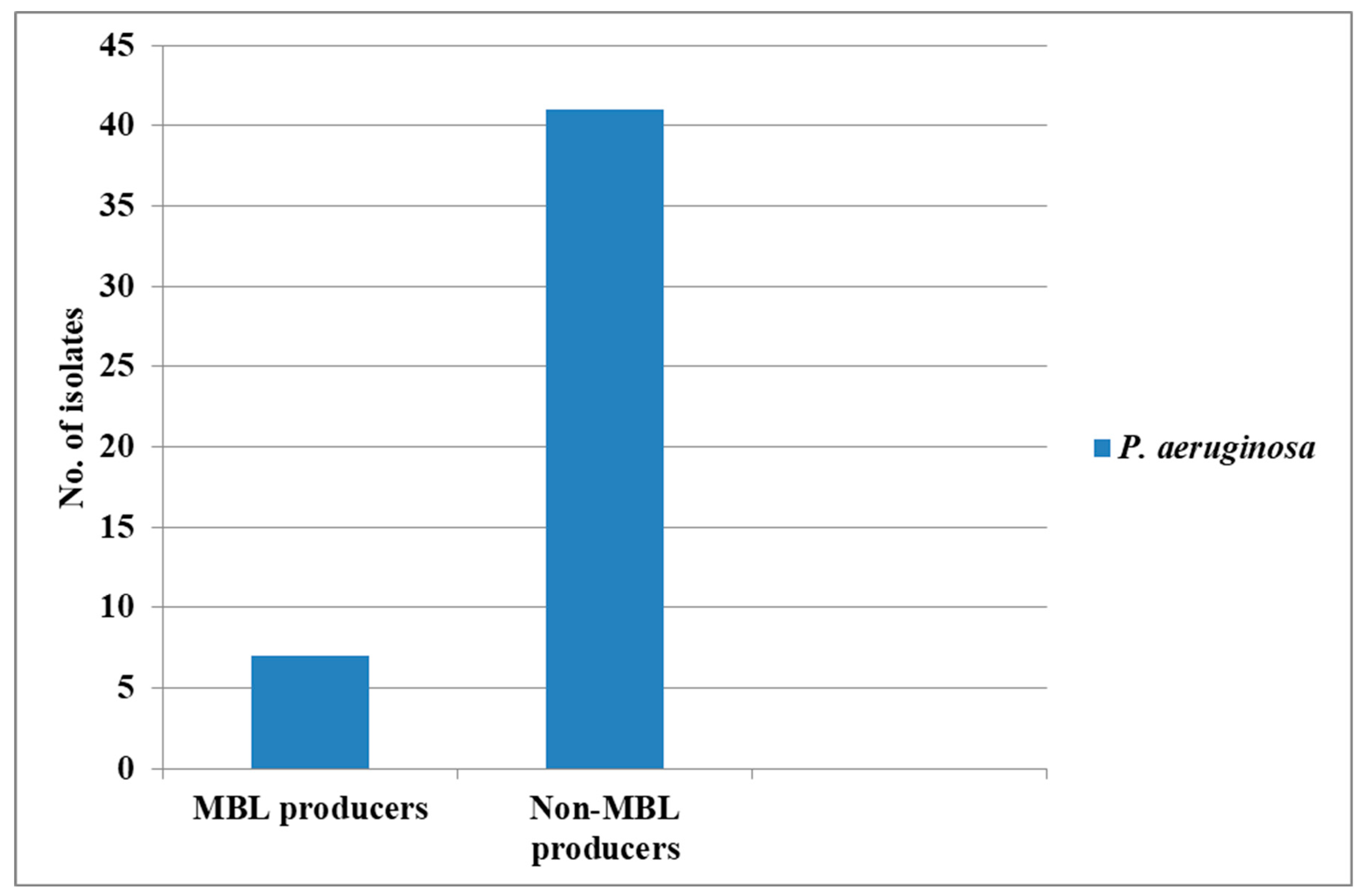
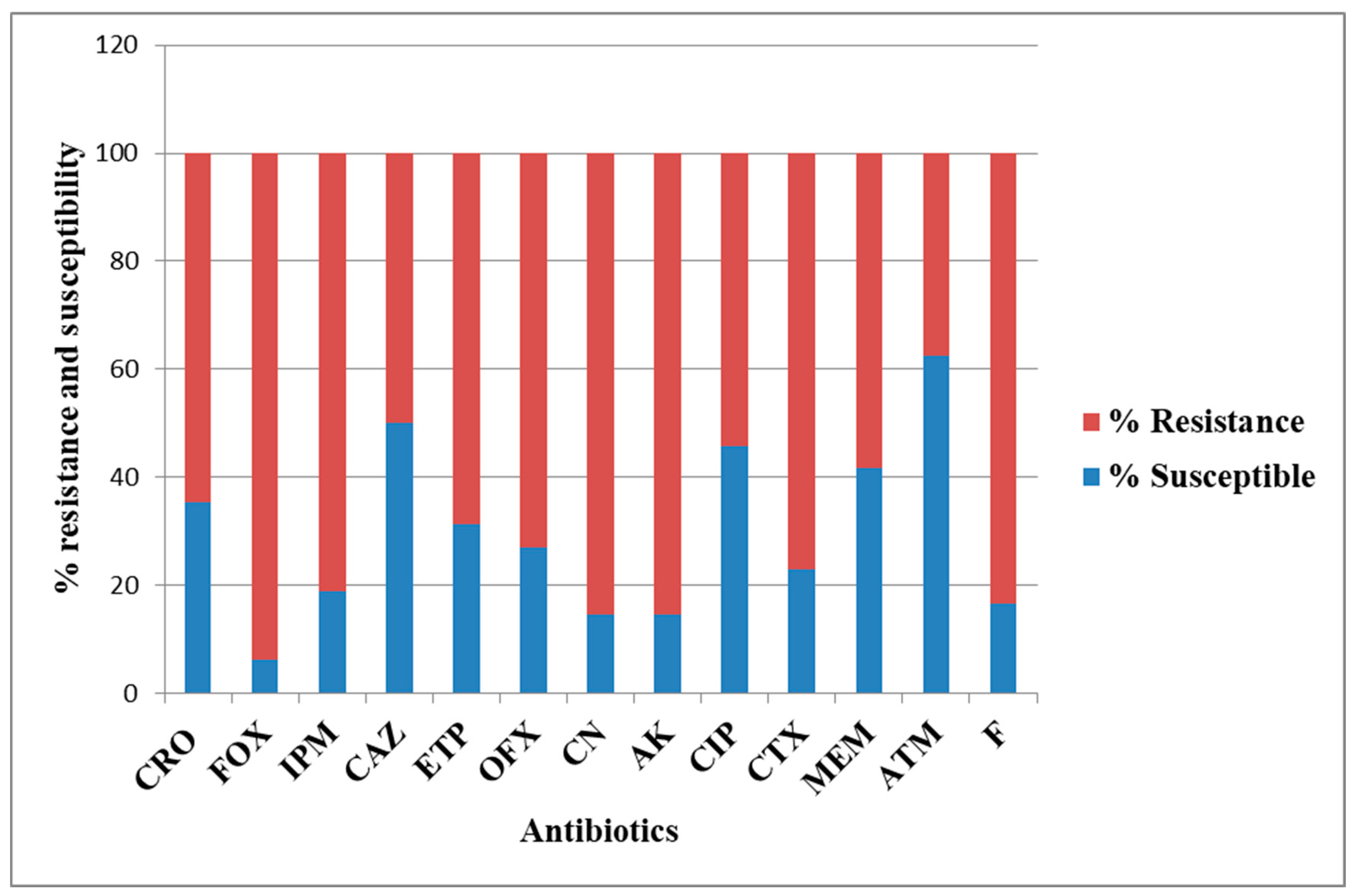
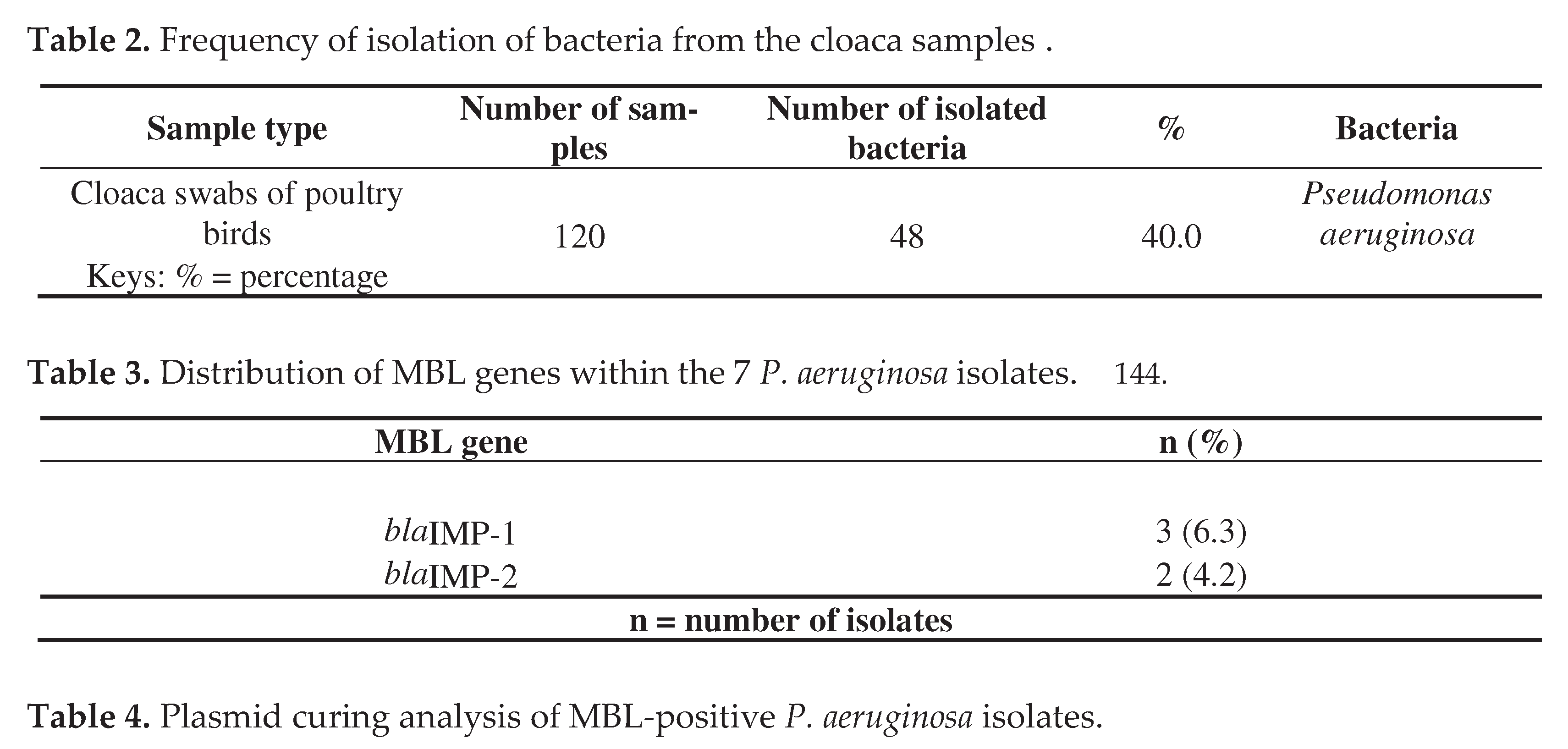
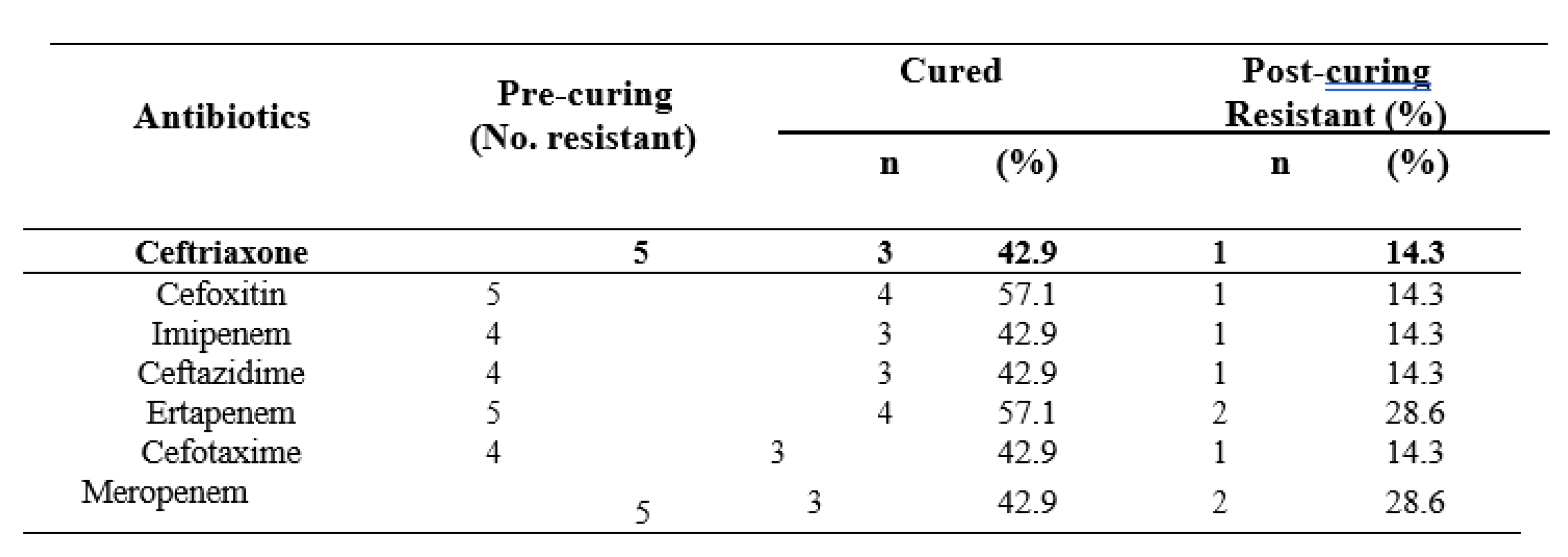
3. Discussion
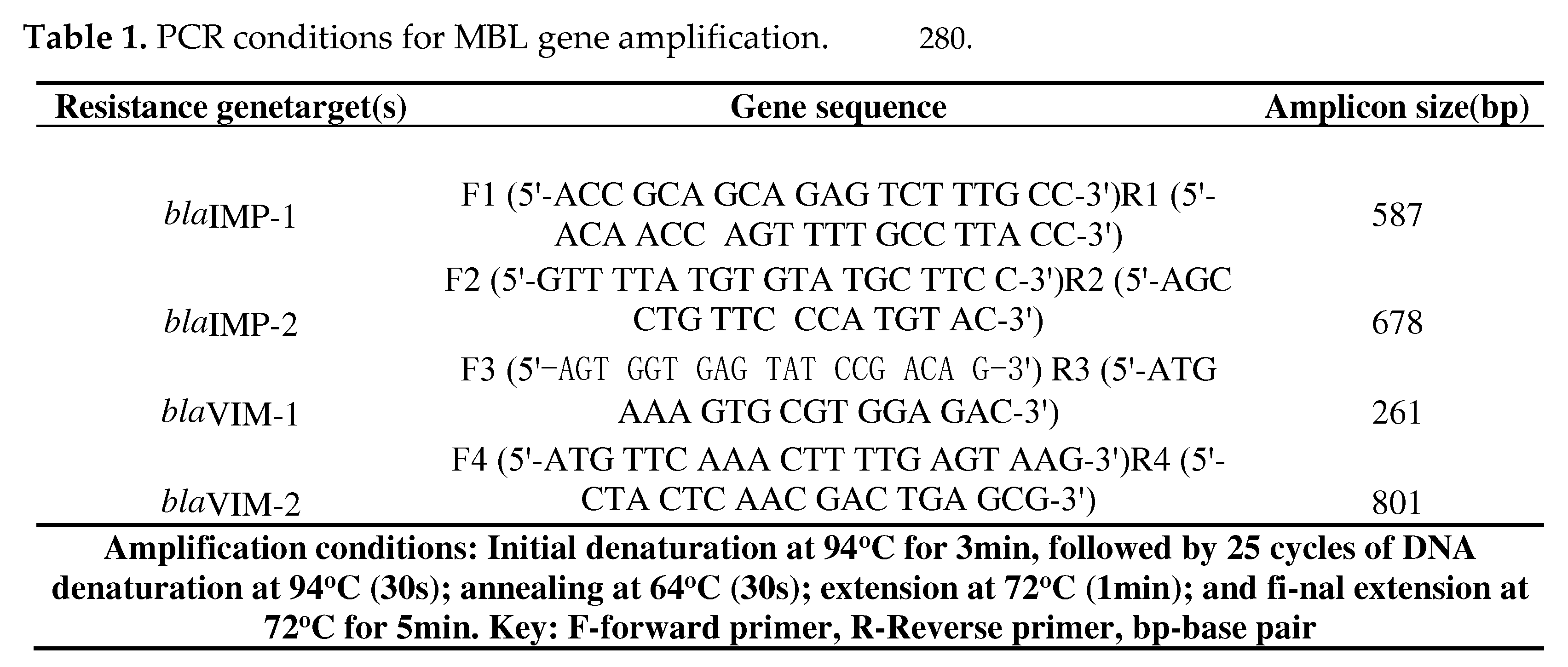
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abbas FM (2021). Metallo-b-lactamases: A review. Ann. Rom. Soc. Cell Biol. 25, 1308–1316.
- Hernando-Amado S, Coque TM, Baquero F, Martinez JL (2019). Defining and combating antibiotic resistance from One Health and global health perspectives. Nat. Microbiol. 4, 1432–1442. [CrossRef]
- Agarwal S, Durlabhji P, Gupta S, Mittal J, Dalela G (2017). Incidence of metallo-β-lactamase producing Pseudomonas aeruginosa isolates and their antimicrobial susceptibility pattern in clinical samples from a tertiary care hospital. International Journal of Research and Review, 4(1):92-98.
- Ejikeugwu C, Ugwu M, Iroha I, Gugu T, Duru C, Eze P, Esimone C (2013). Detection and antimicrobial susceptibility of some Gram negative bacteria producing carbapenemases and extended spectrum beta lactamases. International Journal of Microbiology and Immunology Research, 2(6):064-069.
- Zhang R, Liu Z, Li J, Lei L, Yin W, Li M, Wu C, Walsh TR, Wang Y, Wang S, Wu Y (2017). Presence of VIM-positive Pseudomonas species in chickens and their surrounding environment. Antimicrob Agents Chemother, 61:e00167-17. [CrossRef]
- Falodun OS and Musa IB (2020). Pseudomonas species from cattle dung producing extended spectrum and metallo beta-lactamases. European Journal of Biological Research, 10(1):1-10. [CrossRef]
- Woolhouse M, Ward M, Van Bunnik B, Farrar J (2015). Antimicrobial resistance in humans, livestock and the wider environment. Phil Trans Royal Soc B Lond Ser B, Biol Sci. 370: 1-7. [CrossRef]
- Ejikeugwu C, Okoro Nworie, Morteza Saki, Hussein O.M. Al-Dahmoshi, Noor S.K. Al-Khafaji, Chika Ezeador, Emmanuel Nwakaeze, Peter Eze, Eniola Oni, Chidiebere Obi, Ifeanyichukwu Iroha, Charles Esimone, Michael U. Adikwu (2021). Detection of metallo-β-lactamase and AmpC genes in Escherichia coli, Klebsiella species, and Pseudomonas aeruginosa isolates from abattoir and poultry origin in Nigeria. BMC Microbiology, 21(124):1-9. [CrossRef]
- Walsh T.R., Toleman M.A., Poirel L and Nordmann P (2005). Metallo β – Lactamases: the Quiet before the Storm? Clinical Microbiology Review, 18(2):306-325. [CrossRef]
- Saderi H., Karimi Z., Owlia P., Bahar M.A., Rad S.M (2008). Phenotypic Detection of Metallo – beta – lactamase producing Pseudomonas aeruginosa strains isolated from Burned Patients. Iranian Journal of Pathology, 3(1):20-24.
- Fang L, Liu Z, Lu Z, Huang R, Xiang R (2022). Identification and characterization of a novel metallo β-lactamase, SZM-1, in Shenzhen Bay, South China. Front. Microbiol. 13:996834. [CrossRef]
- Cheesbrough M (2006). District Laboratory Practice in Tropical Countries. Part 2, 2nd Ed. Cambridge University Press, 2006.
- Wayne PA (2006). Performance Standards for Antimicrobial Susceptibility Testing. In CLSI Approved Standard M100. (Clinical and Laboratory Standards Institute CLSI, 2019).
- Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, Kato H, Kai K and Arakawa Y (2003). PCR typing of genetic determinants for metallo-β-lactamases and integrases carried by Gram-negative bacteria isolated in Japan, with focus on the class 3 integron. Journal of Clinical Microbiology, 41(12):5407-5413. [CrossRef]
- Ejikeugwu C, Morteza S, Nwakaeze E, Eze P, Orinya C, Carissa D, Edeh C, Esimone C, Iroha I, Adikwu M (2019). Characterization of blaIMP-1 MBL genes among Klebsiella species from abattoir samples by multiplex PCR. Gene Reports, 16:100428.
- Onyeadi DJ and Agbagwa OE (2019). Plasmid curing in multidrug resistant hospital and community uropathogenic Escherichia coli. J. Appl. Sci. Environ. Manage. 23(1):29-34. [CrossRef]
- Ejikeugwu C, Onele S, Okonkwo E, Onu E, Afiukwa N, Nwakaeze E, Udu-Ibiam O, Iroha C, Edeh C, Iroha I (2022). Antimicrobial drug resistance in strains of Salmonella isolated from pig effluents in Abakaliki, Nigeria. Nigerian Journal of Microbiology, 36(2):6229-6235.
- Elmonir W, Abd El-Aziz NK, Tartor YH, Moustafa SM, Abo Remela EM, Eissa R, Saad HA, and Tawab AA (2021). Emergence of colistin and carbapenem resistance in extended-spectrum beta-lactamase producing Klebsiella pneumoniae isolated from chickens and humans in Egypt. Biology (Basel). 10:373. [CrossRef]
- Zhai R, Fu B, Shi X, Sun C, Liu Z, Wang S, Shen Z, Walsh TR, Cai C, Wang Y, and Wu C (2020). Contaminated in-house environment contributes to the persistence and transmission of NDM-producing bacteria in a Chinese poultry farm. Environ. Int.139:105715. [CrossRef]
- Kelly AM, Mathema B and Larson EL (2017). Carbapenem-resistant Enterobacteriaceae in the community: A scoping review. Int. J. Antimicrob. Agents, 50, 127–134. [CrossRef]
- Chaalal N, Touati A, Bakour S, Aissa MA, Sotto A, Lavigne JP, and Pantel A (2021). Spread of OXA-48 and NDM-1-Producing Klebsiella pneumoniae ST48 and ST101 in Chicken Meat in Western Algeria. Microb. Drug Resist, 27:492–500. [CrossRef]
- Sarhangi M, Motamedifar M, Sarvari J. Dissemination of Pseudomonas aeruginosa producing blaIMP1, blaVIM2, blaSIM1, blaSPM1 in Shiraz, Iran. Jundishapur J Microbiol, 2013; 6:e6920–5. [CrossRef]
- Toraman Z, Yakupogullari Yand Kizirgil A (2004). Detection of metallo β-lactamase production and antibiotic resistance with E-test method in Pseudomonas, Acinetobacter and Klebsiella strains, in Turkey. J Infect hemother, 10:257-261. [CrossRef]
- Mojica MF, Rossi MA, Vila AJ and Bonomo RA. (2022). The urgent need for metallo-beta-lactamase inhibitors: An unattended global threat. Lancet Infect. Dis, 22, e28–e34. [CrossRef]
- Pongchaikul P and Mongkolsuk P (2022). Comprehensive analysis of imipenemase (IMP)-type metallo-beta-lactamase: A global distribution threatening asia. Antibiotics, 11:236. [CrossRef]
- Zwanzig M (2021). The ecology of plasmid-coded antibiotic resistance: a basic framework for experimental research and modeling. Computational and Structural biotechnology Journal, 19:586-599. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




