1. Introduction
Damage to the optic nerve can lead to axonal degeneration, which in turn results in a gradual decline in retinal ganglion cells (RGCs) and ultimately irreversible vision loss [
1]. Such optic neuropathy can stem from various factors, including blunt head trauma, ischemia, metabolic disorders, genetic mitochondrial diseases, autoimmune inflammation, and infiltrative conditions [
2]. The optic nerve crush (ONCr) model, involving a deliberate crushing of the optic nerve to induce RGC apoptosis, has been broadly utilized to study neuronal death and survival mechanisms, as well as to evaluate potential therapeutic strategies for various types of optic neuropathy [3, 4]. This model has been used in conjunction with pharmacological and molecular approaches to test and identify therapeutic agents. Previous studies have proven the efficacy of cellular treatments in the ONCr model, created using a self-clamping protocol with forceps [5, 6]. Notably, this model has facilitated research into RGC regeneration [
7]. Thus, previous our studies also demonstrated the effectiveness of cellular treatment using the ONCr model [8, 9].
In recent studies, the optic nerve compression (ONCo) injury model has emerged as a less hazardous alternative to the ONCr model, due to its minimal disruption to the ophthalmic artery and its blood flow. This method involves the use of Carlson DSEK graft forceps as opposed to self-clamping to temper the severity of the crush [10, 11]. Investigations into optic nerve transection (ONT) and ONCo have shed light on the origin and migration of myeloid and microglial cells in the optic nerve and optic nerve head (ONH). ONT, which uses scissors to sever the nerve while maintaining blood flow to the retina, is more effective than ONCo for inducing RGC axon loss and degeneration in the retina. The latter leaves approximately 50% of the axons intact, allowing the corresponding RGCs to survive [
11]. Moreover, the higher concentration of microglia and myeloid cell proliferation in the optic nerve compared to ONT suggests that complete transection of the optic nerve may impede the migration of reactive myeloid cells to the retina [
11].
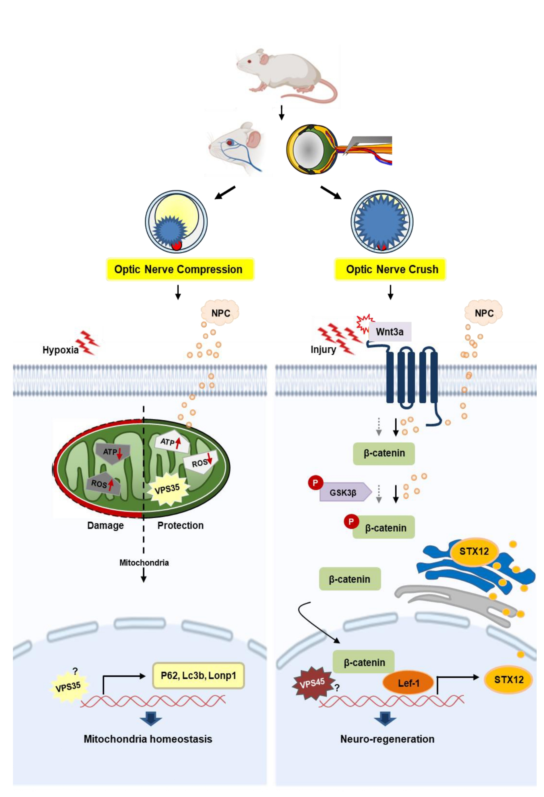
To the best of our knowledge, no study to date has performed a comparative analysis of different types of optic nerve injuries. Therefore, we examined the patterns of damage and recovery using two distinct models of optic nerve injury, both of which avoid full transection of the optic nerve. We compared the ONCo and ONCr models to evaluate changes in essential mitochondrial function, RGC regeneration, and axoplasmic flow in the optic nerve before and after injury, considering the injury pressure severity. In addition, we investigated the therapeutic potential of neural progenitor cells (NPCs) in both types of optic nerve injury.
2. Materials and methods
2.1. Cell culture and hypoxic damage conditioning
Immortalized R28 retinal precursor cells were cultured as previously described [
9]. H9 human ESCs (WiCell Research Institute, Madison, WI, USA) were routinely maintained on Matrigel-coated culture dishes (BD Biosciences, San Jose, CA, USA) in TeSR™-E8™ medium (STEMCELL Technologies, Vancouver, BC, Canada). The culture process for NPCs is detailed in a recent publication [
8]. A hypoxic environment was created in R28 cells using cobalt chloride (CoCl
2). R28 cells were seeded at a density of 2 × 10
5 and then treated with 300 µM CoCl
2 for 3 h. Following this, NPCs were added to the damaged R28 cells. After 24 h, the cells were harvested and prepared for analysis.
2.2. Construction of the optic nerve compression (ONCo) and crush (ONCr) models
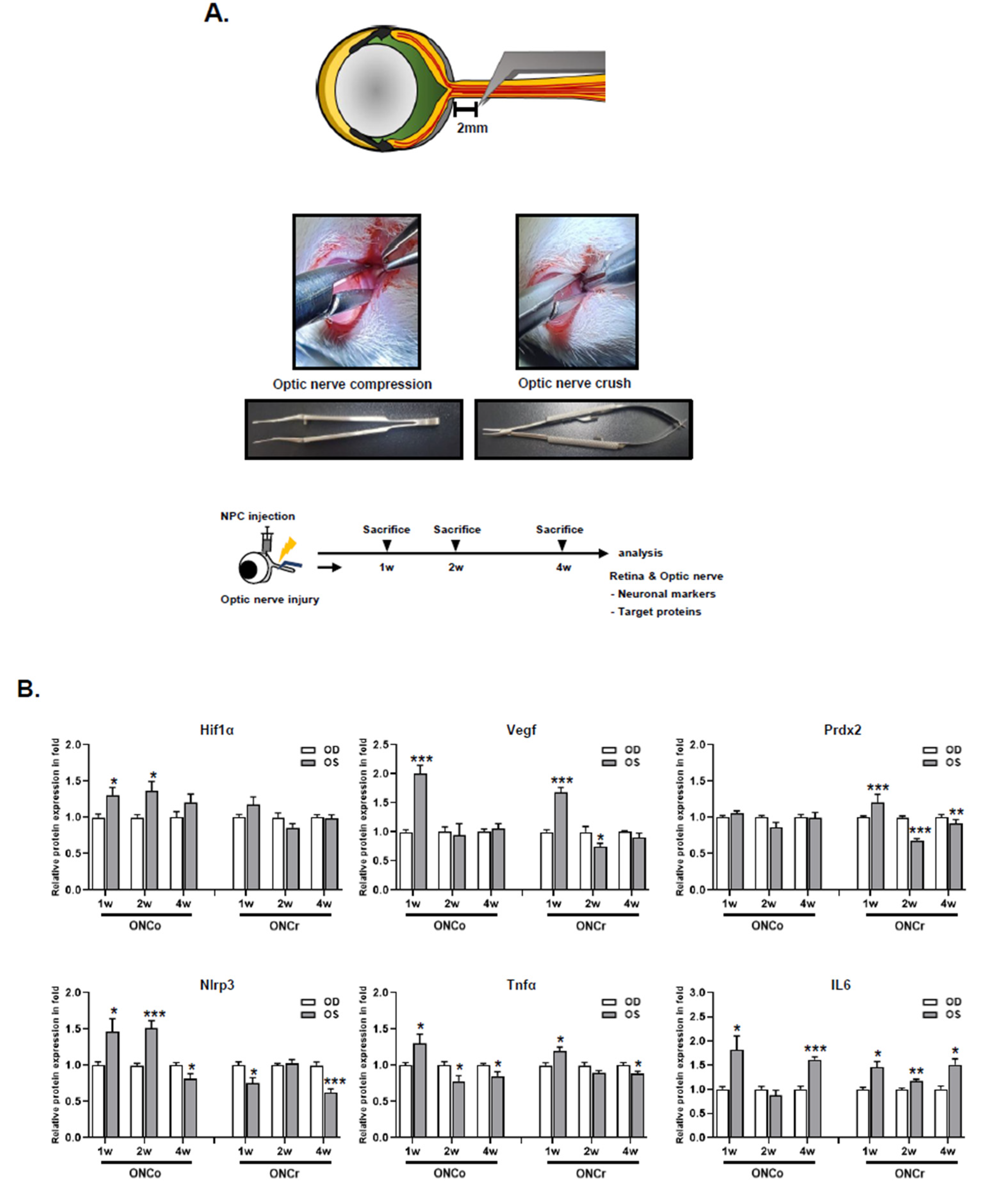
A 6-week-old female Sprague-Dawley (SD) rats (Orient Bio, Gyeonggido, Republic of Korea) were housed in standard animal facilities where food and water were provided at constant temperatures of 21°C. All animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of Bundang CHA Medical Center (IACUC No. 220101). ONCo and ONCr were performed in the left eye (oculus sinister; OS) and the right eye (oculus dexter; OD) was used as control. The rats were anesthetized via an intraperitoneal injection of Zoletil and Rompun. A lateral canthotomy and conjunctival incision were made after the topical application of 0.5% proparacaine hydrochloride. Subsequently, the tissues surrounding the optic nerve were carefully dissected to expose the optic nerve without damaging the adjacent blood supply. The ONCo model was constructed using Calison DSEK graft forceps, 3 1/4”, non-self-closing (Ambler Surgical, Exton, PA, USA) to apply mild compression to the nerve 2 mm behind the globe for 5 s [10, 11]. Conversely, the ONCr model was created by applying strong crush pressure using extra-fine self-closing forceps. After thoroughly suturing the canthal site, a subtenon injection of NPCs was performed on the nasal side of the eyeballs. The animals were euthanized after 1, 2, and 4 weeks, and the tissue was collected for analysis. The rats were grouped as follows: the Sham group (received a balanced salt solution [BSS] injection after optic nerve compression) and the NPC group (received a 2 × 106/0.06 mL injection after optic nerve compression).
2.3. Immunoblot analysis
Total proteins from cells or at least three of tissues were extracted using either RIPA or PRO-PREP buffer (iNtRON Biotechnology, Gyeonggi-do, Republic of Korea). Protein concentration was determined using the BCA method (Thermo Fisher Scientific, Waltham, MA, USA). Target proteins were separated via SDS-PAGE and then transferred to PVDF membranes (GE Healthcare, Chicago, Il, USA). The membranes were incubated overnight at 4°C with primary antibodies (Table S1). After a series of washing steps, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit or mouse secondary antibodies at 1:10,000 dilution (GeneTex), again overnight at 4°C. Target bands were visualized using enhanced chemiluminescence solutions (Bio-Rad Laboratories, Hercules, CA, USA) and were detected on an ImageQuant™ LAS 4000 (GE Healthcare).
2.4. Proteomics
The comprehensive process for proteomics analysis is described in a previous paper [
9]. Briefly, proteomic analyses were conducted on R28 cells treated with PBS (control), with CoCl
2, and with CoCl2 and NPCs to elucidate the extensive effects of NPCs on undamaged cells. Protein digestion was performed using the filter-aided sample preparation (FASP) protocol [
12] with Ultracel® YM-30 centrifugal filters (Merck Millipore, Darmstadt, Germany).
Sample analysis was performed on an LC-MS/MS system, specifically a Dionex Ultimate 3000 HPLC coupled with a Q Exactive™ Hybrid Quadrupole-Orbitrap MS (Thermo Fisher Scientific). This was equipped with an Acclaim™ PepMap™ 100 C18 nano-trap column (75 μm × 2 cm, 3 μm particles, 100 Å pores, Thermo Fisher Scientific) and an Acclaim™ PepMap™ C18 100A RSLC nano-column (75 μm × 50 cm, 2 μm particles, 100 Å pores, Thermo Fisher Scientific). The sample loading flow rate was 2.5 μL/min. Peptide mixtures were separated with solvent A and solvent B (0.1% FA/80% acetonitrile) at a flow rate of 300 nL/min. The gradient setup for solvent B was as follows: 4% (14 min), 4–20% (61 min), 20–50% (81 min), 50–96% (1 min), 96% (10 min), 96–4% B (1 min), and 4% (17 min). The nano-electrospray ionization source was operated in positive mode with a spray voltage of 2.0 kV. Additional parameters included a capillary temperature of 320°C, an isolation width of ± 2 m/z, a scan range of 400–2000 m/z, and resolutions in full-MS scans and MS/MS scans (at 200 m/z) of 70,000 and 17,500, respectively. MS was performed using a data-dependent acquisition method. The top ten precursor ions with the highest intensity were isolated in the quadrupole and fragmented by higher-energy collisional dissociation with 27% normalized collisional energy. Dynamic exclusion was set at 20 s to minimize repeated analyses of the same abundant precursor ions.
2.5. Data processing and bioinformatics
Raw MS/MS data files were analyzed against a SwissProt human protein database using Proteome Discoverer (version 2.4) equipped with Sequest HT. The search parameters were set as follows: 10 ppm for precursor ion mass tolerances, 0.02 Da for fragment ion mass, and a maximum of 2 missed cleavages with the trypsin enzyme. Peptide sequence modifications included static carbamidomethylation of cysteine (+57.012 Da), dynamic modifications of methionine oxidation (+15.995 Da), carbamylation of protein at the N-terminal (+43.006 Da), and acetylation of protein at the N-terminal (+42.011 Da). A false discovery rate (FDR) cutoff of 1% was applied. Statistical comparisons of protein abundances across groups were made using the Student’s T-test. Differentially expressed proteins (DEPs) were filtered with a cutoff p-value ≤ 0.05 and log2FC ≥ 1 (fold-change). A heatmap was generated using Proteome Discoverer. Protein–protein interactions were analyzed using the STRING database (
https://string-db.org/). Volcano plots were prepared using R software (version 3.6.1). Gene ontology analysis was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) (doi:10.1093/nar/gkac194).
2.6. Immunofluorescence of retinal whole mounts and RGC survival analysis
To prepare retinal whole mounts, both normal and injured eyes were immersion-fixed overnight in 4% paraformaldehyde (PFA), after which the retina was isolated. The retina was sectioned into four parts to allow it to lay flat on the slide. Then the whole mounts were rinsed three times for 15 min each in phosphate-buffered saline (PBS; pH 7.4). For enhanced permeability, the retinas were frozen for 15 min at -80 ºC in PBS/0.5% Triton® X-100. The retinas were incubated with primary antibodies overnight at 4°C (Table S1), and after rinsing, they were incubated with Alexa–Fluor conjugated secondary antibodies (Rabbit: Alexa 555, Invitrogen) for 2 h at room temperature. DAPI was used as a counterstain.
Then the retinas were transferred onto a slide, with the inner retina facing upwards, using a paintbrush. PBS was removed and a mounting medium (Dako, Glostrup, Denmark) was added. A small piece of broken coverslip glass was placed between the slide and coverslip glass to prevent crushing and damaging the retina. After the coverslip was positioned, images were captured using confocal microscopy (LSM 880; Carl Zeiss, Jena, Germany) for fluorescence quantification.
2.7. Optic nerve histological analysis
Optic nerves were fixed with 4% PFA, dehydrated with ethanol, and embedded in paraffin. For histological analysis, paraffin blocks were sectioned into 5 μm slices and Hematoxylin & Eosin (H&E) staining was performed using standard methods. To conduct immunofluorescence staining, sections were deparaffinized in xylene and rehydrated in ethanol. For antigen retrieval, slides were heated in a microwave using 0.01 M citric acid buffer (pH6.0) (Biosesang, Gyeonggi-do, Republic of Korea) for 10 min, and subsequently rinsed with PBS. Then the slides were incubated with a blocking solution (Protein Block Serum-Free, Dako) for 1 h. Subsequently, slides were incubated with primary antibodies overnight at 4℃. The following day, the samples were incubated with secondary antibodies conjugated with Alexa Fluor 488 and 555 (Invitrogen) for 1 h at room temperature. DAPI was used as a counterstain, followed by mounting with a fluorescence mounting medium (Dako). Images were captured using an Axio slide scanner and processed using ZEN 3.1 software.
2.8. Small-interfering RNA for Vps35 protein
For the knockdown of the VPS35 protein, we used a target sequence of siRNA supplied by Precaregene (Seoul, Republic of Korea). The sequence used for siRNA rat VPS35 was 5′- ACA GUG GAG AUA UUC AAU AAA CUT A. As a negative control, we used siRNA Negative Control (scramble) provided by Precaregene. R28 cells were transfected using Lipofectamine 3000 (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions.
2.9. Statistical analyses
All results are presented as the mean ± standard error of the mean (SEM). Data analyses were performed using GraphPad Prism 9 software (GraphPad Software, Inc, La Jolla, CA, USA). The statistical significance criteria used for data analysis are detailed in the figure legends.
4. Discussion
The optic nerve injury model necessitates meticulous construction to prevent excessive force or extended crushing, as these can damage the ophthalmic artery and induce retinal ischemia. Furthermore, extreme care is needed to avoid harming other blood vessels surrounding the animal’s eye. The level of precision required renders this model challenging to implement in research.
The ONCo model, which entails a compression injury, can progressively reduce blood flow to the optic nerve, leading to ischemia and nerve tissue damage. This form of injury can trigger axonal degeneration, myelin loss, and eventually the death of RGCs. Although a compression injury may still permit some nutrients and organelles to traverse the compressed region, it can disrupt axonal transport due to nerve deformation [
8]. In comparison, a crush injury completely severs the axons, causing a total cessation of axonal transport and subsequent degeneration of the axons and neighboring cells. Therefore, it is anticipated that axoplasmic flow in the optic nerve would be more severely disrupted in crush injuries than in compression injuries.
The ONCr model, conversely, involves physical harm to the optic nerve due to excessive pressure or force. This type of injury can cause immediate axonal damage and swift loss of RGCs [
13]. Furthermore, the mechanical force of the crush injury can lead to tissue distortion and swelling, resulting in additional inflammation and fibrosis [
14].
VPS35 is a component of the retromer complex, which plays an instrumental role in the endosomal sorting and trafficking of cargo proteins [
15]. While its precise role in hypoxic injury to neuronal tissue remains to be fully elucidated, certain evidence points towards a possible role in mediating cellular responses to hypoxia. For instance, studies have shown that reducing VPS35 expression in neuronal cells can impede autophagic flux and increase vulnerability to hypoxic injury [
16]. In addition, VPS35 deficiency can intensify hypoxic-ischemic brain injury in animal models [
17].
VPS35 also plays a role in the transport and preservation of mitochondria in neurons [
18]. Specifically, it is associated with the retrograde transport of damaged mitochondria from axons to the cell body for degradation and recycling. This process is vital for maintaining mitochondrial health and avoiding the accumulation of damaged mitochondria [
19]. Mutations or dysfunction in VPS35 have been connected to impaired mitochondrial transport and clearance, leading to mitochondrial dysfunction and neuronal degeneration [
20]. However, further research is necessary to thoroughly understand the function of VPS35 in the context of hypoxic injury to neuronal tissue.
In this study, we also identified a novel target, Syntaxin12, which is regulated by NPCs during the RGCs recovery process. Syntaxin12 belongs to the SNARE (soluble NSF attachment protein receptor) protein family, which plays a crucial role in vesicle fusion events during intracellular trafficking [
21]. This protein localizes to endosomal compartments in various cell types and is involved in the process of vesicle fusion between synaptic vesicles and the presynaptic membrane during neurotransmitter release [
22].
Dysfunction of Syntaxin12 has been linked with neurodegenerative diseases, including Alzheimer’s and Parkinson’s disease [
23]. Syntaxin16, another member of the same family, has also been implicated in the regulation of autophagy, a process by which damaged cellular components are degraded and recycled [
24]. Therefore, understanding the role of Syntaxin12 in response to NPCs during the RGC recovery process could provide valuable insights for potential therapeutic strategies.
Evidence suggests that Syntaxin12 may play a role in the Wnt signaling pathway. One study demonstrated that Syntaxin12 interacts with Dishevelled-3 (Dvl3), a pivotal component of the Wnt signaling pathway [
25]. This interaction is critical for the proper localization of Dvl3 to the plasma membrane and for the activation of downstream Wnt signaling events, which may have implications for the regulation of Wnt signaling in neuronal tissue. However, further research is required to fully comprehend the relationship between Syntaxin12 and the Wnt signaling pathway in neuronal tissue.
Concerning the regulation of mitochondrial dynamics in neurons, some evidence suggests that Syntaxin4 interacts with proteins implicated in mitochondrial fission and fusion, such as dynamin-related protein 1 (Drp1) and Syntaxin17 with Mfn1, respectively [26, 27]. In addition, the knockdown of Syntaxin1A impairs mitochondrial function and leads to increased oxidative stress in neurons [
28]. Recently, it was found that Syntaxin12 and the COMMD3/CCC complex interact physically and functionally with disease-associated proteins VPS33B and VPS16B in the α-granule biogenesis of platelets [
21]. These findings suggest that Syntaxin12 may play a role in maintaining the health and proper functioning of mitochondria in neurons.
Various studies have indicated a relationship between VPS35 and the Wnt pathway. The Wnt pathway plays a crucial role in several biological processes, including neuronal development and synaptic plasticity. VPS35 participates in the Wnt signaling pathway by regulating the degradation of Wnt receptors and modulating the activity of β-catenin, a key downstream effector of the Wnt pathway [29, 30]. In particular, it is involved in endocytosis and recycling of Wnt receptors, including Frizzled and LRP6, which are necessary for Wnt signaling [
31]. These studies suggest that VPS35 plays a role in regulating the Wnt pathway in neuronal tissue. However, further research is needed to fully understand the relationship between hypoxia and VPS35 in nerve tissues.
There is limited research on the specific impact of inflammation on VPS35 in nerves but inflammation could potentially influence VPS35 function in other cell types. Moreover, studies have demonstrated that inflammation-induced oxidative stress can lead to VPS35 dysfunction, contributing to the pathogenesis of neurodegenerative diseases such as Parkinson’s disease [
32]. Thus, it is plausible that inflammation might also affect the function of VPS35 in nerves. However, more research is required to fully elucidate this relationship.
Inflammation can be triggered by various stimuli, which in turn activate intracellular signaling pathways such as NF-κB and MAPK. Other pathways and mechanisms might also be involved under certain conditions, such as autoimmune diseases, viral infections, and cancer. For instance, the release of TNFα might be triggered by toll-like receptors (TLRs) or other pattern recognition receptors (PRRs) on immune cells, as well as cytokine signaling and other pathways [
33]. In addition, the activation of intracellular protein complexes known as inflammasomes can also instigate inflammation. Inflammasomes are typically activated by pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), which stimulate cytosolic pattern recognition receptors (PRRs), such as NOD-like receptors (NLRs) [
34]. This can result in the activation of caspase-1, leading to a robust inflammatory response by cleaving and activating pro-inflammatory cytokines such as IL-1β and IL-18 [
35]. However, hypoxia counteracts inflammation through the downregulation of the binding of mTOR and NLRP3 and the activation of autophagy, which is protective in mouse models of colitis [
36]. This study significantly contributes to the field of molecular neurodegeneration research, offering novel insights and potential therapeutic strategies for neural injuries and related degenerative conditions. And it paves the way for future investigations and translational applications, providing a foundation for developing targeted therapies that harness the regenerative power of neural progenitor cells and the identified proteins.
Figure 1.
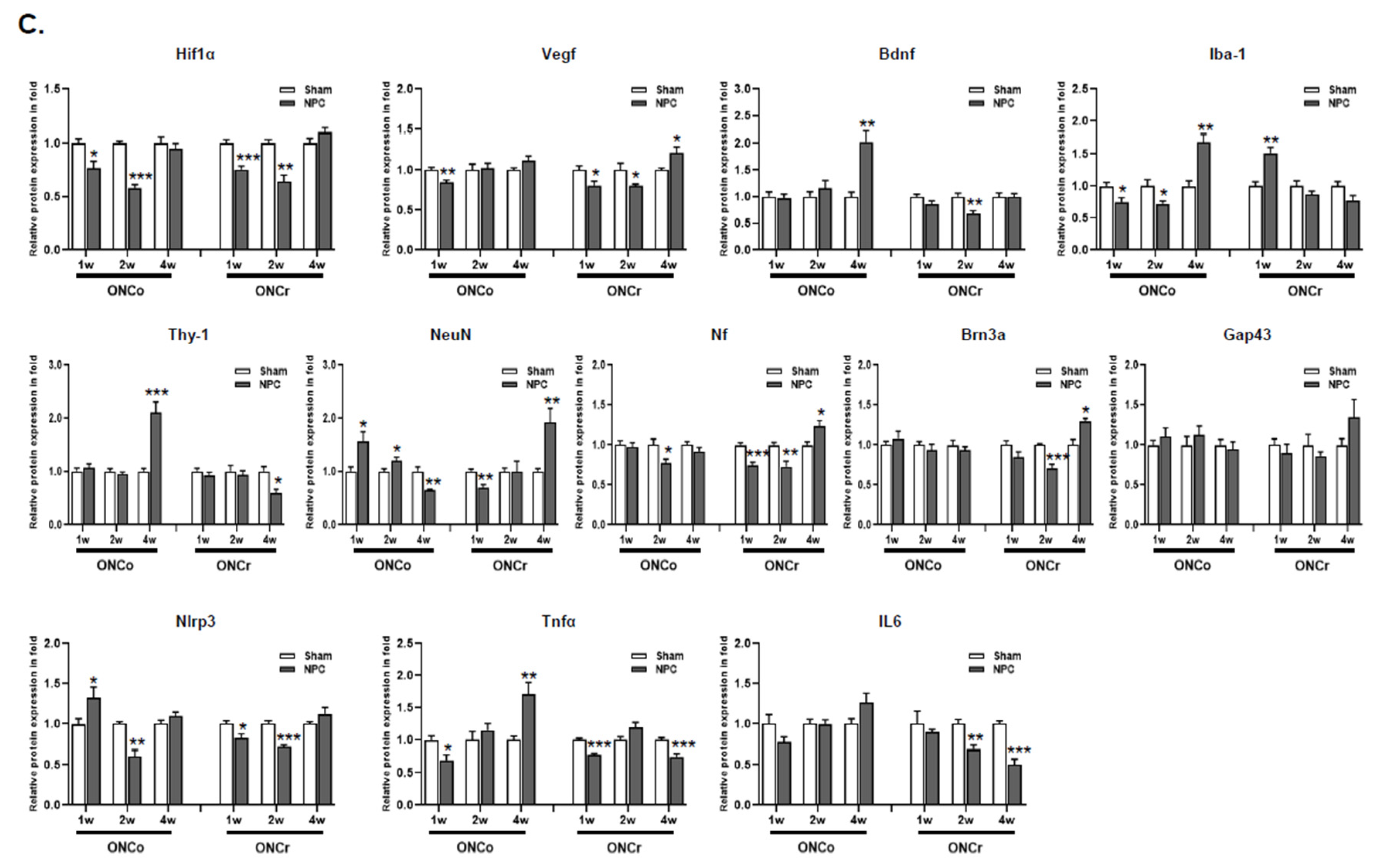
Analysis of ONCo and ONCr in vivo models. (A) The scheme of in vivo optic nerve injury model construction. Changes in target proteins were assessed by immunoblot analyses of rat retina extracts. The samples were analyzed 1, 2, and 4 weeks after injection with optic nerve injury. (B) Comparison with two types of optic nerve disease models. Quantified values of Hif-1α, Vegf, Prdx2, Nlrp3, Tnfα, and IL6 expression in OS compared with OD are presented. (C) After neural progenitor cells (NPCs) injection, the retinas were also analyzed. Expression levels were normalized to β-actin and the values of OS were divided OD. The results are expressed as the mean ± standard error of the mean (SEM) of the independent retina and optic nerve analyses and are expressed as fold changes compared to the control (*p<0.05, **p<0.01, ***p<0.001 vs. the age-matched sham [BSS]) OD, oculus dexter; OS, oculus sinister.
Figure 1.
Analysis of ONCo and ONCr in vivo models. (A) The scheme of in vivo optic nerve injury model construction. Changes in target proteins were assessed by immunoblot analyses of rat retina extracts. The samples were analyzed 1, 2, and 4 weeks after injection with optic nerve injury. (B) Comparison with two types of optic nerve disease models. Quantified values of Hif-1α, Vegf, Prdx2, Nlrp3, Tnfα, and IL6 expression in OS compared with OD are presented. (C) After neural progenitor cells (NPCs) injection, the retinas were also analyzed. Expression levels were normalized to β-actin and the values of OS were divided OD. The results are expressed as the mean ± standard error of the mean (SEM) of the independent retina and optic nerve analyses and are expressed as fold changes compared to the control (*p<0.05, **p<0.01, ***p<0.001 vs. the age-matched sham [BSS]) OD, oculus dexter; OS, oculus sinister.
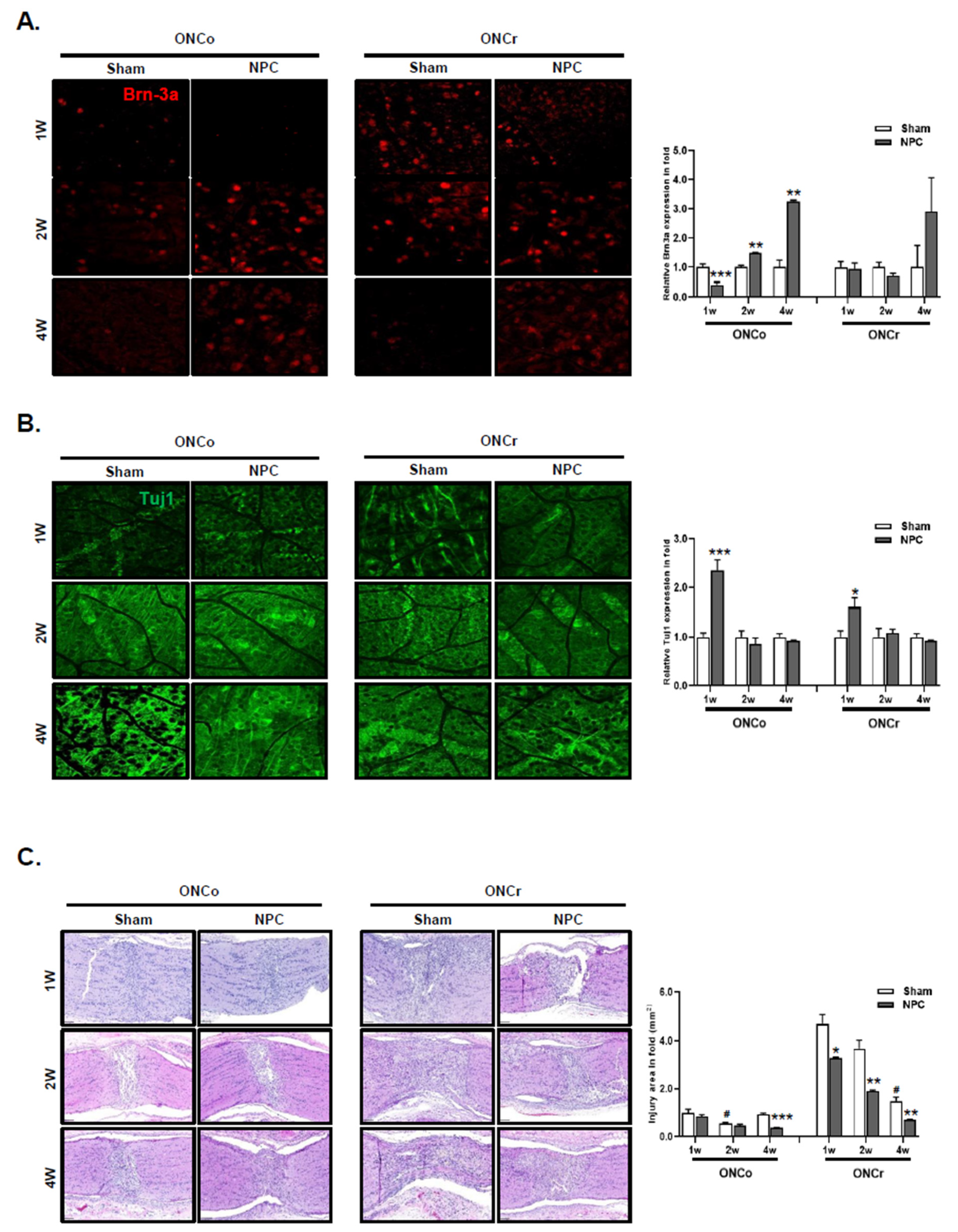
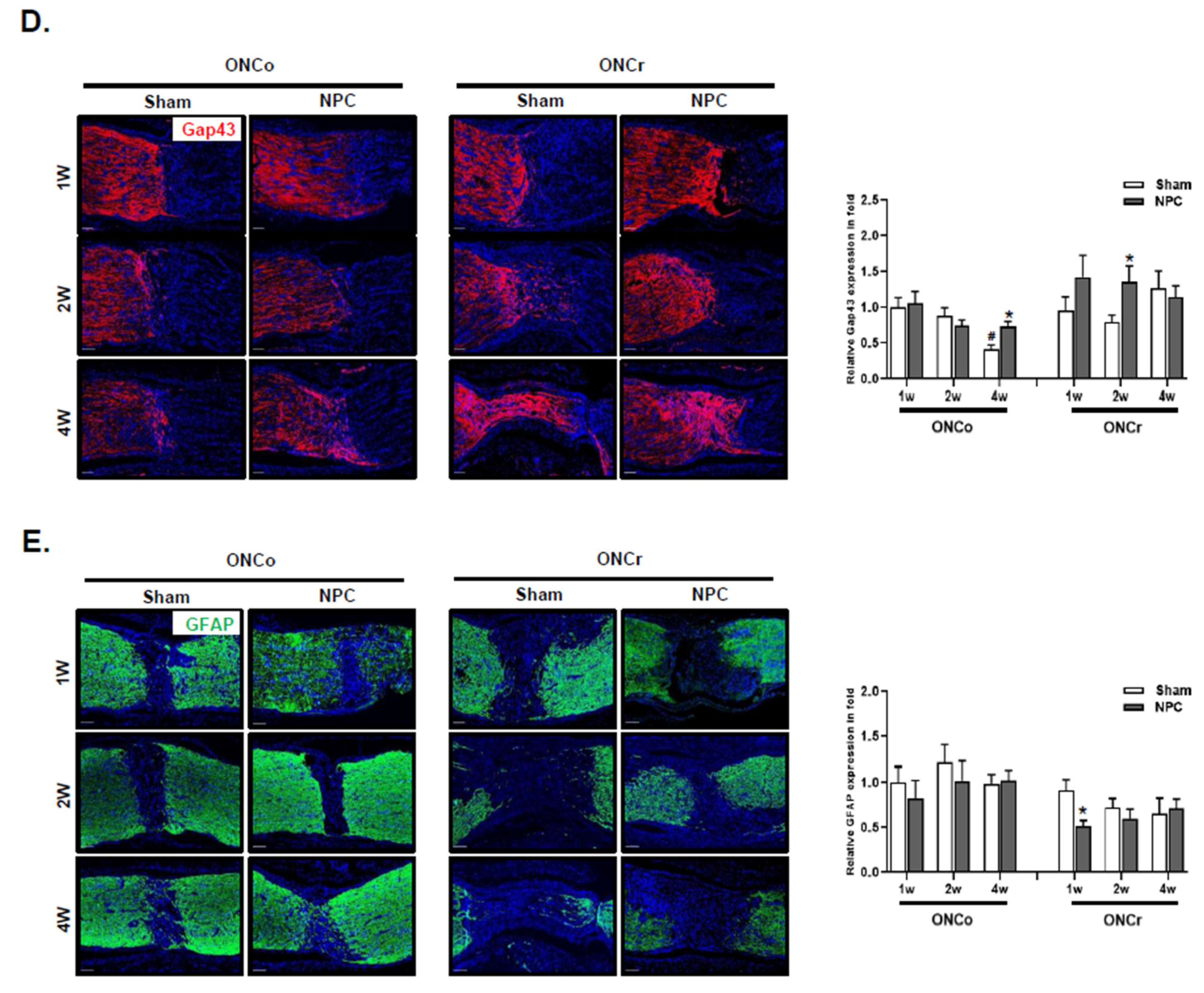
Figure 2.
NPCs promote retinal ganglion cell and axon regeneration in the optic nerve injury animal model. Representative confocal microscopy-based fluorescence images following (A) Brn-3a and (B) Tuj1 staining (original magnification: x 400) of NPCs injection in the optic nerve-injured animal model. (C) H&E-stain of an injured area on the optic nerve (scale bar: 100 μm). (D) Gap43 and (E) Gfap fluorescence quantification were measured in a box with a 7,690 mm2 including the injured area (scale bar: 100 μm). Total Gap43 and Gfap positive cells were measured using ZEN software. Total of two retinas and optic nerves from each group were used. The results are presented as the mean ± SEM (*p<0.05, **p<0.01, ***p<0.001 vs. the age-matched sham; #p<0.05 vs. 1 w sham of each group).
Figure 2.
NPCs promote retinal ganglion cell and axon regeneration in the optic nerve injury animal model. Representative confocal microscopy-based fluorescence images following (A) Brn-3a and (B) Tuj1 staining (original magnification: x 400) of NPCs injection in the optic nerve-injured animal model. (C) H&E-stain of an injured area on the optic nerve (scale bar: 100 μm). (D) Gap43 and (E) Gfap fluorescence quantification were measured in a box with a 7,690 mm2 including the injured area (scale bar: 100 μm). Total Gap43 and Gfap positive cells were measured using ZEN software. Total of two retinas and optic nerves from each group were used. The results are presented as the mean ± SEM (*p<0.05, **p<0.01, ***p<0.001 vs. the age-matched sham; #p<0.05 vs. 1 w sham of each group).
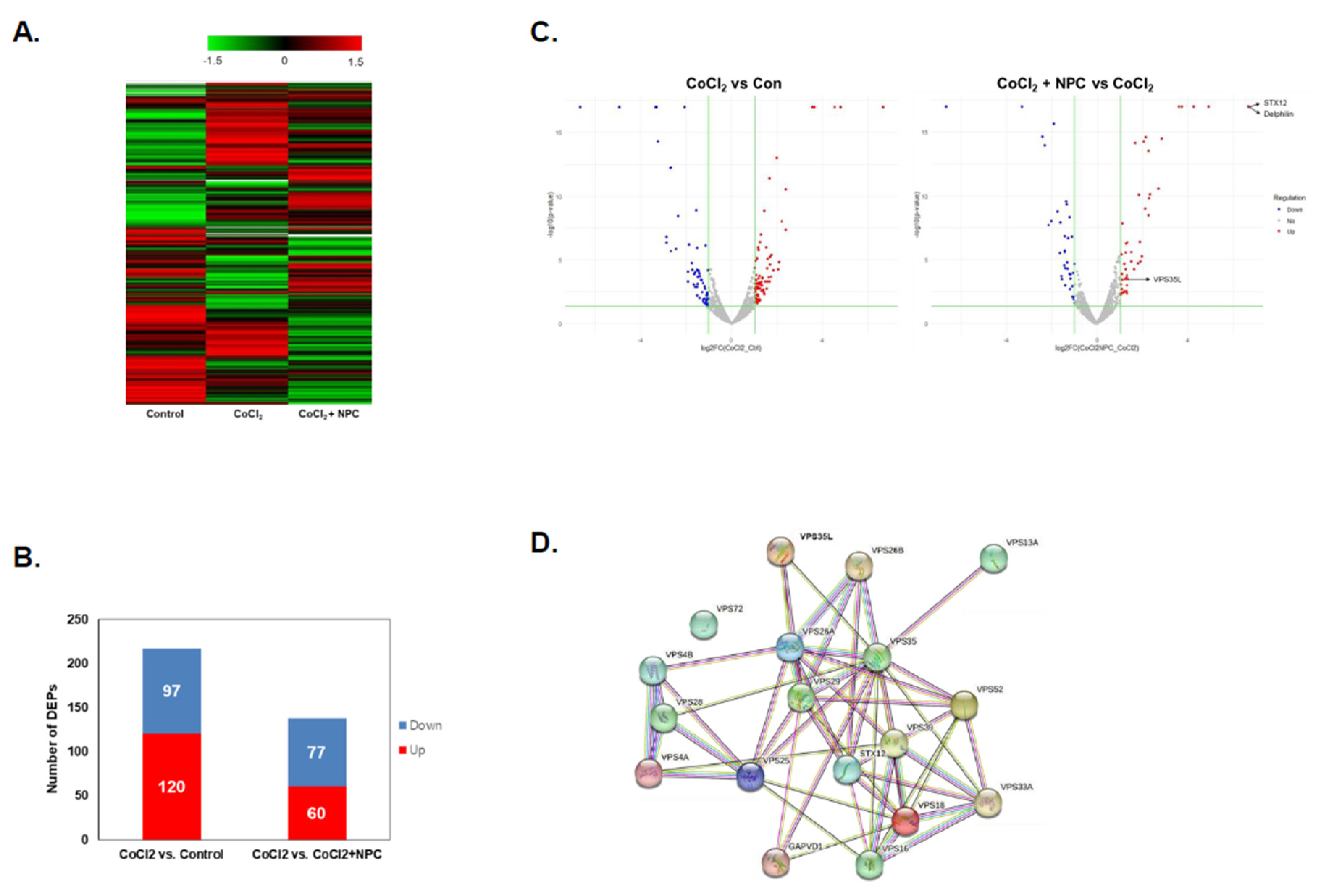
Figure 3.
Cluster analysis of DFEs by NPCs in hypoxia-damaged R28 cells. Identification of differentially expressed proteins in treated groups (CoCl2, and CoCl2 + NPCs) and PBS-treated control cells. (A) Heatmap of differentially expressed proteins (p ≤ 0.05 and log2FC ≥ 1) in lysates of three groups (Control, CoCl2, and CoCl2 + NPCs) analyzed by hierarchical clustering. High expression is shown in red; low expression is shown in green. (B) The graph showing the number of up and down-expressed proteins identified in proteomic analysis of each group in R28 cells. (C) Volcano plot of DEPs of each group. Red dots described up-regulated proteins and blue dots present down-regulated proteins. Proteins with significantly different expressions were presented. (D) Identification of protein–protein interaction of DEF in experimental groups of R28 cells.
Figure 3.
Cluster analysis of DFEs by NPCs in hypoxia-damaged R28 cells. Identification of differentially expressed proteins in treated groups (CoCl2, and CoCl2 + NPCs) and PBS-treated control cells. (A) Heatmap of differentially expressed proteins (p ≤ 0.05 and log2FC ≥ 1) in lysates of three groups (Control, CoCl2, and CoCl2 + NPCs) analyzed by hierarchical clustering. High expression is shown in red; low expression is shown in green. (B) The graph showing the number of up and down-expressed proteins identified in proteomic analysis of each group in R28 cells. (C) Volcano plot of DEPs of each group. Red dots described up-regulated proteins and blue dots present down-regulated proteins. Proteins with significantly different expressions were presented. (D) Identification of protein–protein interaction of DEF in experimental groups of R28 cells.
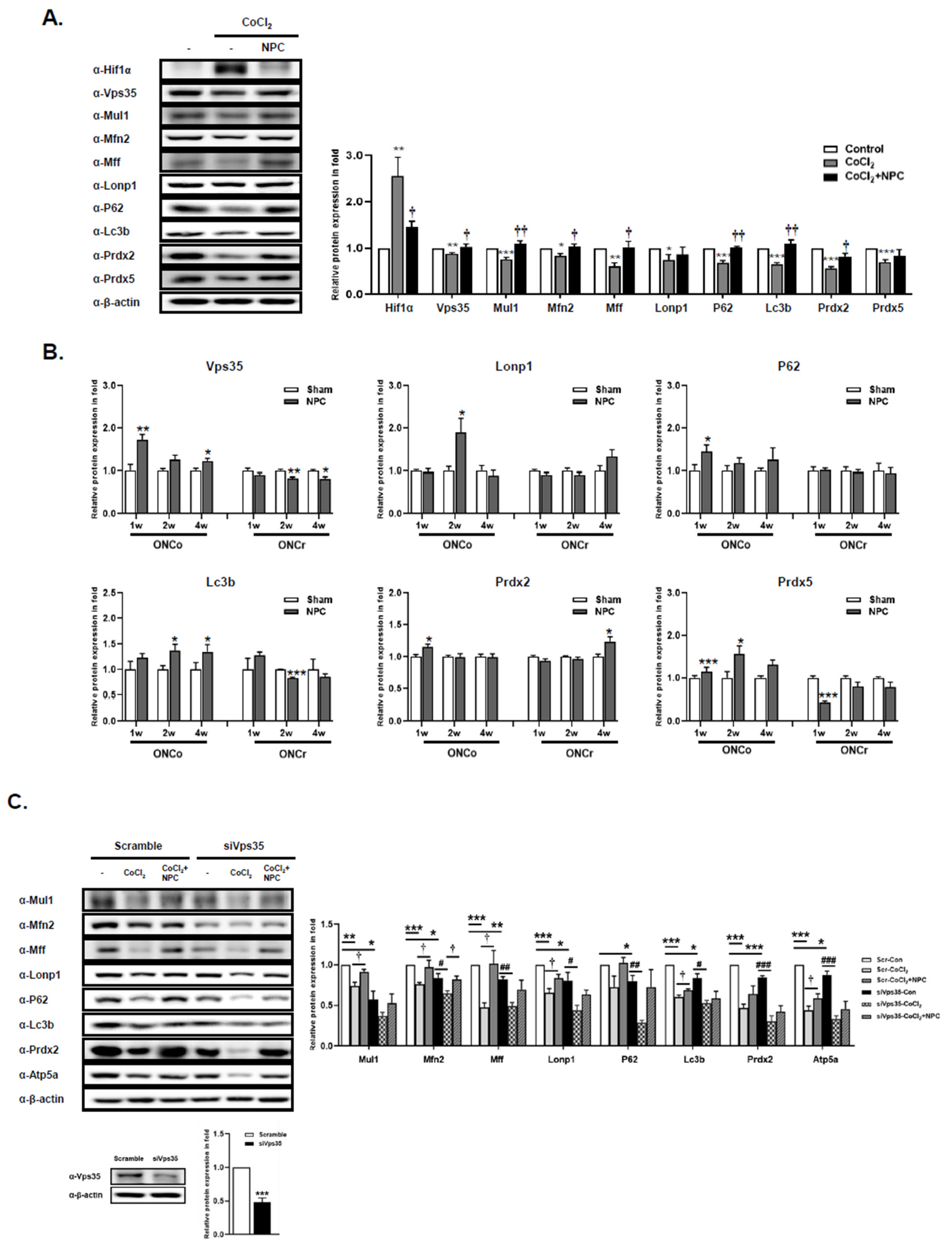
Figure 4.
NPCs have a function to rescue mitochondria quality control of hypoxia damaged in vitro/in vivo models. The R28 cells were treated with CoCl2 (300 μM). Then, R28 cells were cultured with NPCs after 3 h of CoCl2 treatment. (A) Mitochondrial homeostasis-related protein expressions were also determined after 24 h. (B) Using in vivo retina tissues, immunoblot analyses of mitochondrial-regulated protein expression levels were also evaluated. (C) Scramble and siRNA targeting Vps35 were transfected into R28 cells. After incubation, Vps35 deficient cells were exposed to CoCl2 (300 μM). After incubation for 3 h, the hypoxia-induced cells were co-cultured with NPCs. Then, Western blot analyses were performed. (*p<0.05, **p<0.01, ***p<0.001 vs. the control or the age-matched sham or scramble_control; #p<0.05, ##p<0.01, ###p<0.001 vs. siVps35_control; †p<0.05; ††p<0.01vs. the CoCl2 of each group).
Figure 4.
NPCs have a function to rescue mitochondria quality control of hypoxia damaged in vitro/in vivo models. The R28 cells were treated with CoCl2 (300 μM). Then, R28 cells were cultured with NPCs after 3 h of CoCl2 treatment. (A) Mitochondrial homeostasis-related protein expressions were also determined after 24 h. (B) Using in vivo retina tissues, immunoblot analyses of mitochondrial-regulated protein expression levels were also evaluated. (C) Scramble and siRNA targeting Vps35 were transfected into R28 cells. After incubation, Vps35 deficient cells were exposed to CoCl2 (300 μM). After incubation for 3 h, the hypoxia-induced cells were co-cultured with NPCs. Then, Western blot analyses were performed. (*p<0.05, **p<0.01, ***p<0.001 vs. the control or the age-matched sham or scramble_control; #p<0.05, ##p<0.01, ###p<0.001 vs. siVps35_control; †p<0.05; ††p<0.01vs. the CoCl2 of each group).
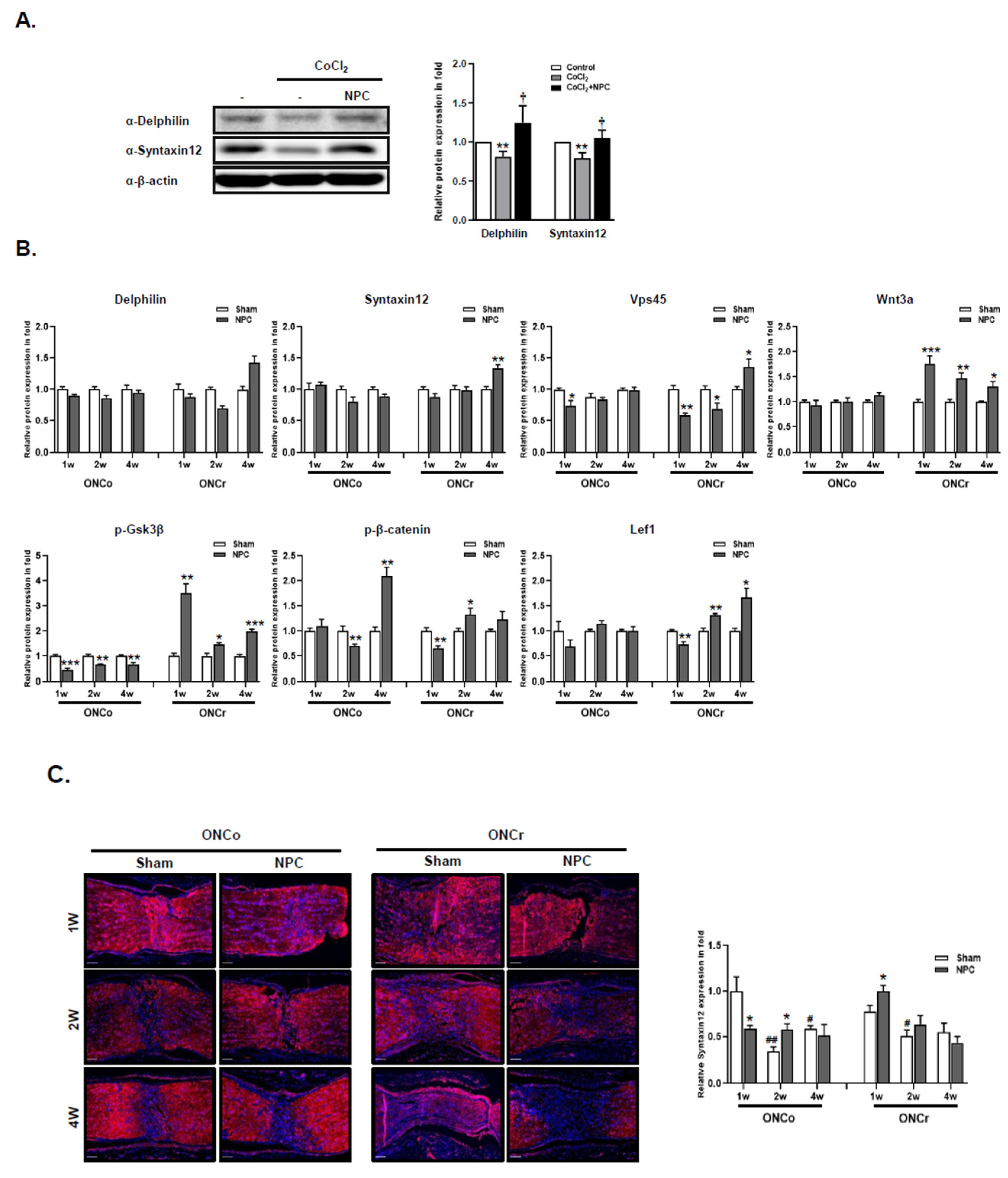
Figure 5.
NPCs recover neuro-regeneration of hypoxia damaged in vitro/in vivo models. The R28 cells were treated with CoCl2 (300 μM). Then, R28 cells were cultured with NPCs after 3 h of CoCl2 treatment. (A) Delphilin and Syntaxin12 protein expressions were also determined after 24 h. (**p<0.01 vs. control; †p<0.05 vs. CoCl2) (B) Using in vivo retina tissues, immunoblot analyses of target protein expression levels were also evaluated. (C) Syntaxin12 fluorescence quantification was measured in the injured area (scale bar: 100 μm). (*p<0.05,**p<0.01,***p<0.001 vs. the control or the age-matched sham; #p<0.05, ##p<0.01 vs. 1w sham of each group).
Figure 5.
NPCs recover neuro-regeneration of hypoxia damaged in vitro/in vivo models. The R28 cells were treated with CoCl2 (300 μM). Then, R28 cells were cultured with NPCs after 3 h of CoCl2 treatment. (A) Delphilin and Syntaxin12 protein expressions were also determined after 24 h. (**p<0.01 vs. control; †p<0.05 vs. CoCl2) (B) Using in vivo retina tissues, immunoblot analyses of target protein expression levels were also evaluated. (C) Syntaxin12 fluorescence quantification was measured in the injured area (scale bar: 100 μm). (*p<0.05,**p<0.01,***p<0.001 vs. the control or the age-matched sham; #p<0.05, ##p<0.01 vs. 1w sham of each group).
Table 1.
GO enrichment analysis of genes up-regulated or down-regulated in R28.
Table 1.
GO enrichment analysis of genes up-regulated or down-regulated in R28.
| GO term |
Definition |
P-value |
No. up- or down regulated |
Total in category |
| CoCl2 vs. Control |
|
|
|
|
| Up |
|
|
|
|
| GO:0007080 |
mitotic metaphase plate congression |
9.36E-05 |
5 |
111 |
| GO:0006915 |
apoptotic process |
1.68E-04 |
13 |
111 |
| GO:0005829 |
cytosol |
9.57E-12 |
66 |
115 |
| GO:0005654 |
nucleoplasm |
1.64E-08 |
49 |
115 |
| GO:0005634 |
nucleus |
3.94E-08 |
62 |
115 |
| GO:0005874 |
microtubule |
2.48E-07 |
13 |
115 |
| GO:0005737 |
cytoplasm |
9.53E-06 |
54 |
115 |
| GO:0072562 |
blood microparticle |
1.57E-04 |
7 |
115 |
| GO:0070062 |
extracellular exosome |
4.06E-04 |
26 |
115 |
| GO:0072686 |
mitotic spindle |
0.00109782 |
6 |
115 |
| GO:0000776 |
kinetochore |
0.00172263 |
6 |
115 |
| GO:0005871 |
kinesin complex |
0.00283831 |
4 |
115 |
| GO:0005515 |
protein binding |
6.20E-09 |
102 |
113 |
| GO:0008017 |
microtubule binding |
2.60E-05 |
10 |
113 |
| GO:0005524 |
ATP binding |
2.47E-04 |
22 |
113 |
| GO:0042802 |
identical protein binding |
9.06E-04 |
22 |
113 |
| GO:0005198 |
structural molecule activity |
0.00104832 |
7 |
113 |
| GO:0031267 |
small GTPase binding |
0.00151412 |
8 |
113 |
| GO:0031625 |
ubiquitin protein ligase binding |
0.00230867 |
8 |
113 |
| Down |
|
|
|
|
| GO:0005654 |
nucleoplasm |
1.34E-04 |
34 |
93 |
| GO:0005829 |
cytosol |
1.82E-04 |
42 |
93 |
| GO:0005515 |
protein binding |
3.32E-05 |
79 |
92 |
| |
|
|
|
|
| CoCl2 vs. CoCl2+NPC |
|
|
|
|
| up |
|
|
|
|
| GO:0031508 |
pericentric heterochromatin assembly |
1.43E-04 |
3 |
75 |
| GO:0005829 |
cytosol |
4.03E-04 |
34 |
73 |
| GO:0005515 |
protein binding |
1.12E-04 |
61 |
70 |
| GO:0031492 |
nucleosomal DNA binding |
4.49E-04 |
4 |
70 |
| Down |
|
|
|
|
| GO:0072562 |
blood microparticle |
5.96E-05 |
6 |
60 |
| GO:0070062 |
extracellular exosome |
1.11E-04 |
18 |
60 |
| GO:0005737 |
cytoplasm |
2.97E-04 |
30 |
60 |
| GO:0005829 |
cytosol |
5.41E-04 |
29 |
60 |
| GO:0005634 |
nucleus |
9.02E-04 |
30 |
60 |
| GO:0071682 |
endocytic vesicle lumen |
0.00119946 |
3 |
60 |
| GO:0005615 |
extracellular space |
0.0029718 |
14 |
60 |
| GO:0045095 |
keratin filament |
0.0038826 |
4 |
60 |