Submitted:
02 September 2023
Posted:
05 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Gene expression and Kaplan-Meier analyses
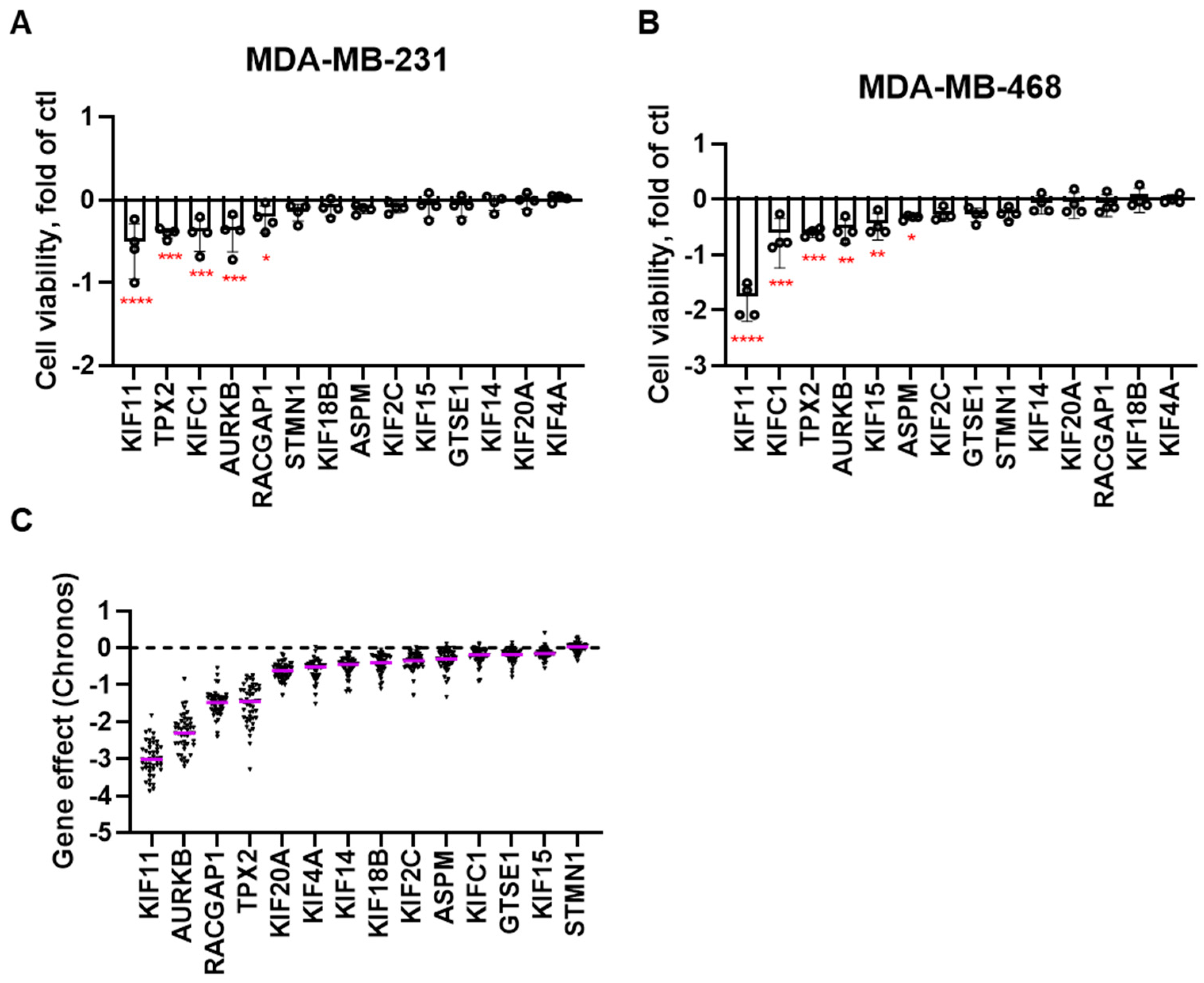
RNAi screen and Cancer dependency map
Systems biology
3. Results
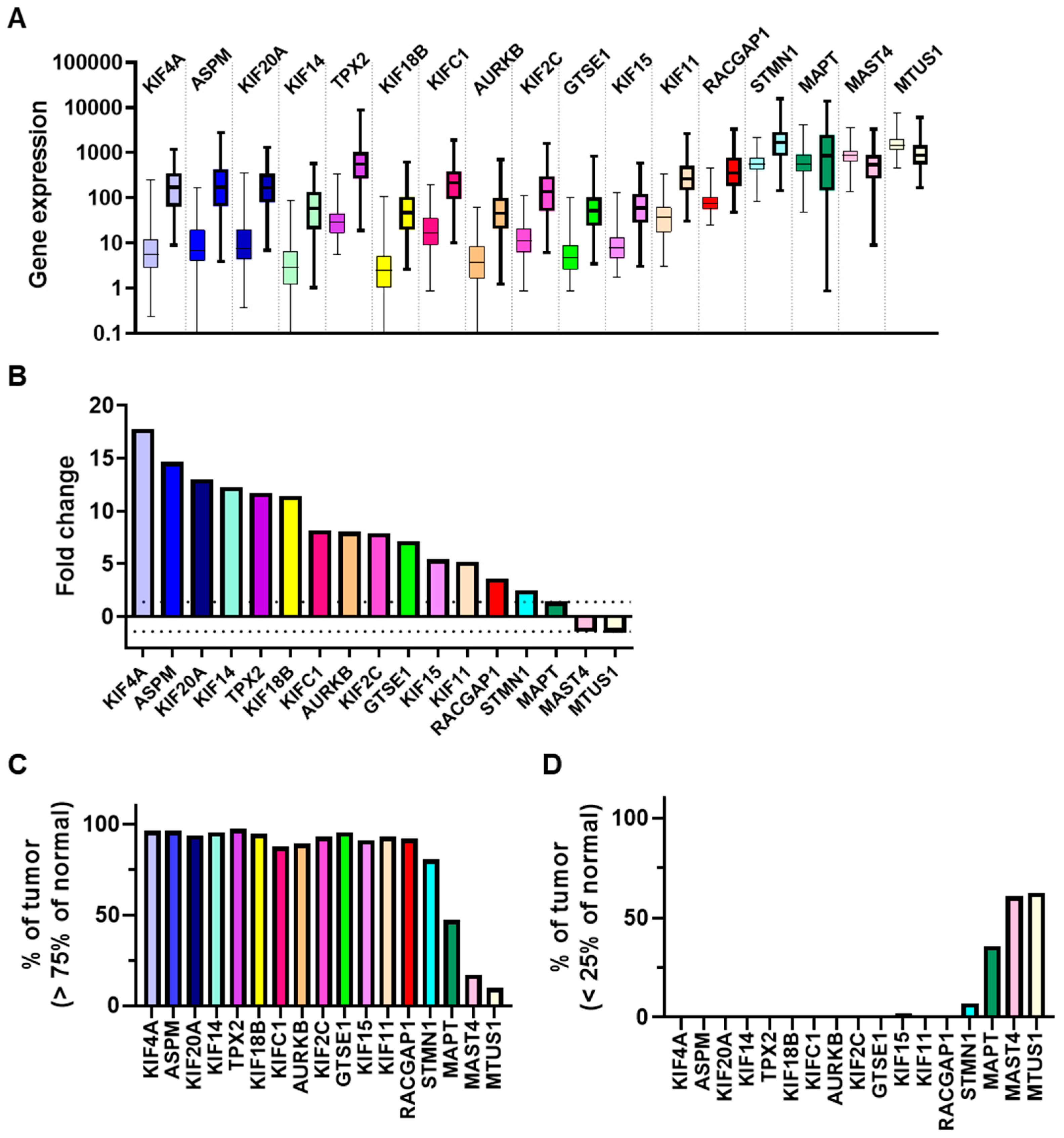
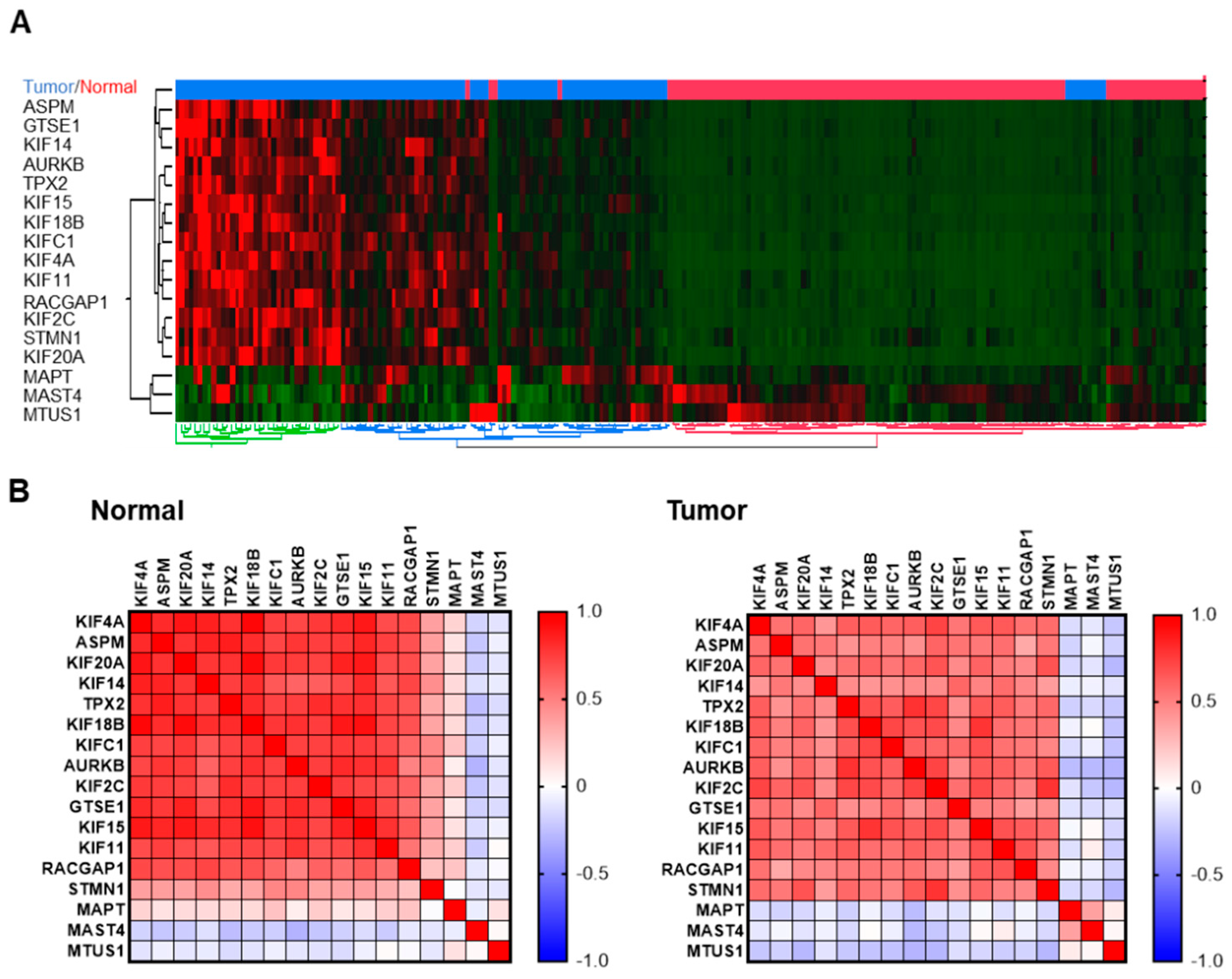
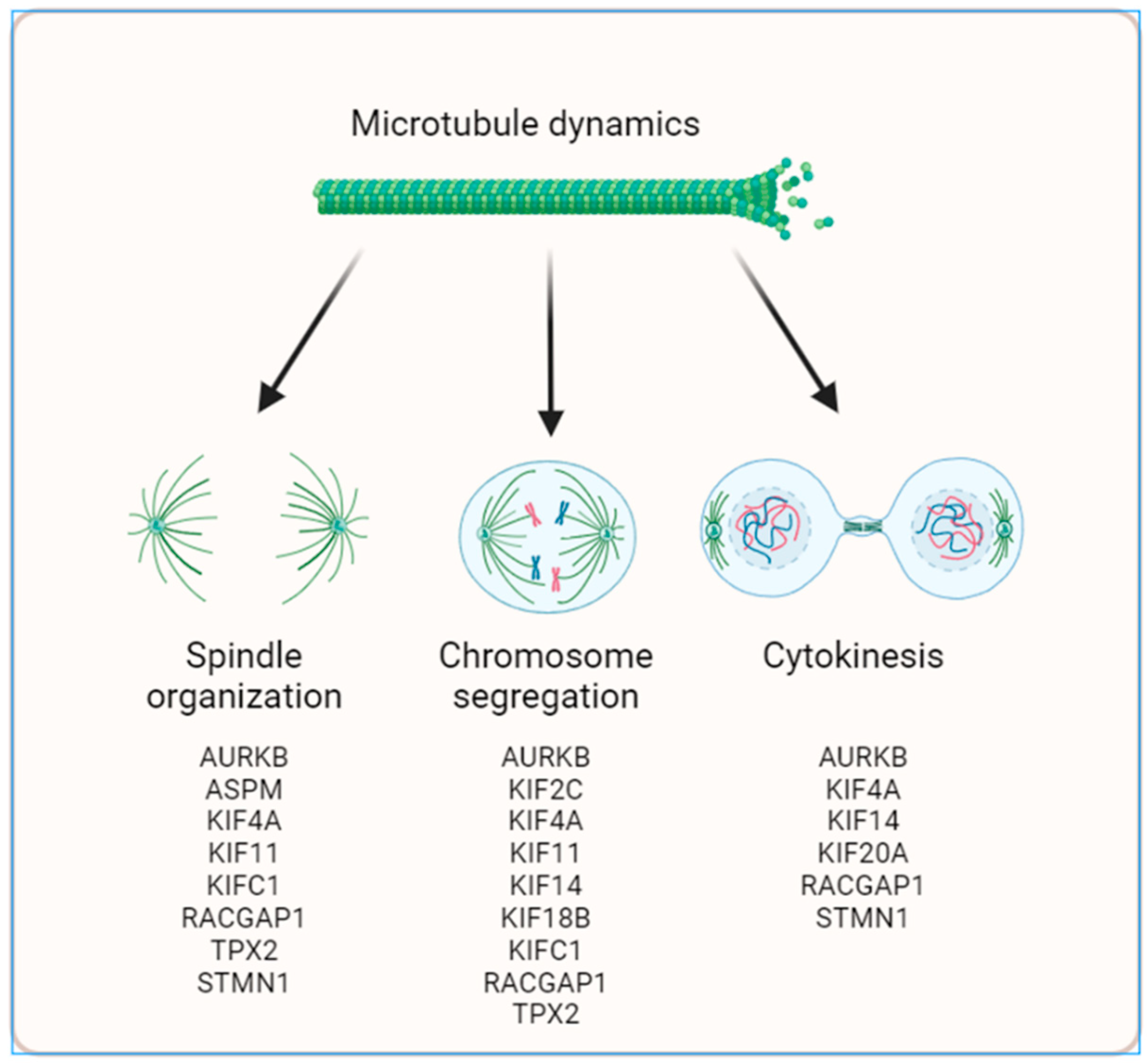
3.1. Regulation of expression of 17 MT-Rel genes in breast tumors
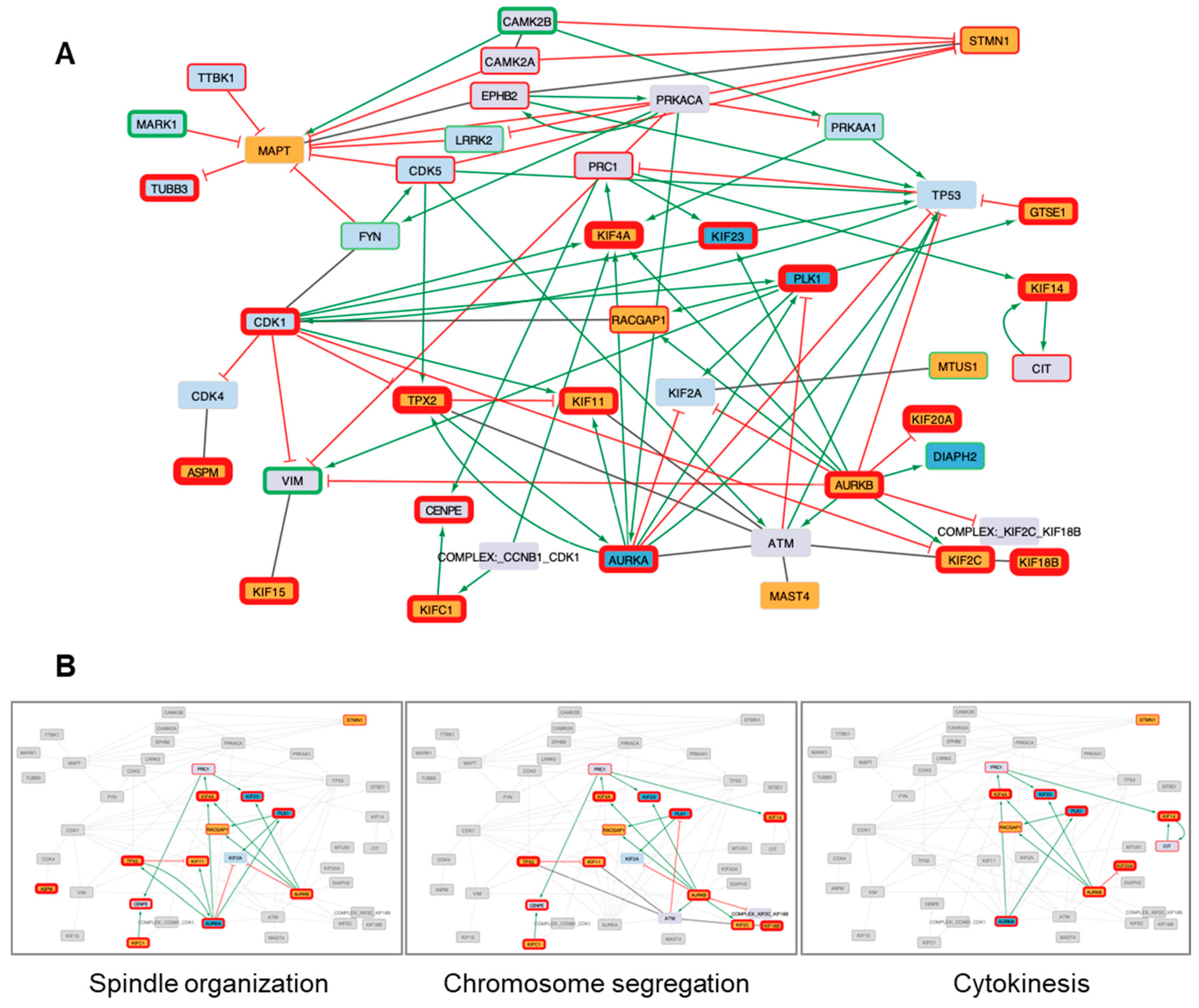
3.2. Systems biology analysis of functional interplays and gene networks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brenton, J.D.; Carey, L.A.; Ahmed, A.A.; Caldas, C. Molecular Classification and Molecular Forecasting of Breast Cancer: Ready for Clinical Application? J. Clin. Oncol. 2005, 23, 7350–7360. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Natarajan, E.; Balaji Raghavendran, H.R.; Markandan, U.D. Molecular Classification, Treatment, and Genetic Biomarkers in Triple-Negative Breast Cancer: A Review. Technol. Cancer Res. Treat. 2023, 22, 153303382211452. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Ferreira, S.; Nahmias, C. Predictive Biomarkers for Personalized Medicine in Breast Cancer. Cancer Lett. 2022, 545, 215828. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.; Kirschner, M. Dynamic Instability of Microtubule Growth. Nature 1984, 312, 237–242. [Google Scholar] [CrossRef]
- Brouhard, G.J. Dynamic Instability 30 Years Later: Complexities in Microtubule Growth and Catastrophe. Mol. Biol. Cell 2015, 26, 1207–1210. [Google Scholar] [CrossRef]
- Hirokawa, N. Microtubule Organization and Dynamics Dependent on Microtubule-Associated Proteins. Curr. Opin. Cell Biol. 1994, 6, 74–81. [Google Scholar] [CrossRef]
- Akhmanova, A.; Steinmetz, M.O. Control of Microtubule Organization and Dynamics: Two Ends in the Limelight. Nat. Rev. Mol. Cell Biol. 2015, 16, 711–726. [Google Scholar] [CrossRef]
- Shigematsu, H.; Imasaki, T.; Doki, C.; Sumi, T.; Aoki, M.; Uchikubo-Kamo, T.; Sakamoto, A.; Tokuraku, K.; Shirouzu, M.; Nitta, R. Structural Insight into Microtubule Stabilization and Kinesin Inhibition by Tau Family MAPs. J. Cell Biol. 2018, 217, 4155–4163. [Google Scholar] [CrossRef]
- Bhat, K.M.R.; Setaluri, V. Microtubule-Associated Proteins as Targets in Cancer Chemotherapy. Clin. Cancer Res. 2007, 13, 2849–2854. [Google Scholar] [CrossRef]
- Khwaja, S.; Kumar, K.; Das, R.; Negi, A.S. Microtubule Associated Proteins as Targets for Anticancer Drug Development. Bioorganic Chem. 2021, 116, 105320. [Google Scholar] [CrossRef]
- Wattanathamsan, O.; Pongrakhananon, V. Emerging Role of Microtubule-Associated Proteins on Cancer Metastasis. Front. Pharmacol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Ferreira, S.; Nehlig, A.; Moindjie, H.; Monchecourt, C.; Seiler, C.; Marangoni, E.; Chateau-Joubert, S.; Dujaric, M.-E.; Servant, N.; Asselain, B.; et al. Improving Breast Cancer Sensitivity to Paclitaxel by Increasing Aneuploidy. Proc. Natl. Acad. Sci. 2019, 116, 23691–23697. [Google Scholar] [CrossRef] [PubMed]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Survival Analysis across the Entire Transcriptome Identifies Biomarkers with the Highest Prognostic Power in Breast Cancer. Comput. Struct. Biotechnol. J. 2021, 19, 4101–4109. [Google Scholar] [CrossRef]
- Rodrigues-Ferreira, S.; Di Tommaso, A.; Dimitrov, A.; Cazaubon, S.; Gruel, N.; Colasson, H.; Nicolas, A.; Chaverot, N.; Molinié, V.; Reyal, F.; et al. 8p22 MTUS1 Gene Product ATIP3 Is a Novel Anti-Mitotic Protein Underexpressed in Invasive Breast Carcinoma of Poor Prognosis. PloS One 2009, 4, e7239. [Google Scholar] [CrossRef]
- Molina, A.; Velot, L.; Ghouinem, L.; Abdelkarim, M.; Bouchet, B.P.; Luissint, A.-C.; Bouhlel, I.; Morel, M.; Sapharikas, E.; Di Tommaso, A.; et al. ATIP3, a Novel Prognostic Marker of Breast Cancer Patient Survival, Limits Cancer Cell Migration and Slows Metastatic Progression by Regulating Microtubule Dynamics. Cancer Res. 2013, 73, 2905–2915. [Google Scholar] [CrossRef]
- Nehlig, A.; Molina, A.; Rodrigues-Ferreira, S.; Honoré, S.; Nahmias, C. Regulation of End-Binding Protein EB1 in the Control of Microtubule Dynamics. Cell. Mol. Life Sci. 2017, 74, 2381–2393. [Google Scholar] [CrossRef]
- Lo Surdo, P.; Iannuccelli, M.; Contino, S.; Castagnoli, L.; Licata, L.; Cesareni, G.; Perfetto, L. SIGNOR 3.0, the SIGnaling Network Open Resource 3.0: 2022 Update. Nucleic Acids Res. 2023, 51, D631–D637. [Google Scholar] [CrossRef]
- Chen, G.; Yu, M.; Cao, J.; Zhao, H.; Dai, Y.; Cong, Y.; Qiao, G. Identification of Candidate Biomarkers Correlated with Poor Prognosis of Breast Cancer Based on Bioinformatics Analysis. Bioengineered 2021, 12, 5149–5161. [Google Scholar] [CrossRef]
- Alam, M.S.; Sultana, A.; Wang, G.; Haque Mollah, M.N. Gene Expression Profile Analysis to Discover Molecular Signatures for Early Diagnosis and Therapies of Triple-Negative Breast Cancer. Front. Mol. Biosci. 2022, 9, 1049741. [Google Scholar] [CrossRef]
- Lucanus, A.J.; Yip, G.W. Kinesin Superfamily: Roles in Breast Cancer, Patient Prognosis and Therapeutics. Oncogene 2018, 37, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-F.; Zeng, H.-J.; Shan, Z.; Ye, R.-Y.; Cheang, T.-Y.; Zhang, Y.-J.; Lu, S.-H.; Zhang, Q.; Shao, N.; Lin, Y. Overexpression of Kinesin Superfamily Members as Prognostic Biomarkers of Breast Cancer. Cancer Cell Int. 2020, 20, 123. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, C.; Gurard-Levin, Z.A.; Andre, F.; Pusztai, L.; Rouzier, R. Predictive and Prognostic Value of the TauProtein in Breast Cancer. Anticancer Res. 2015, 35, 5179–5184. [Google Scholar] [PubMed]
- Darlix, A.; Hirtz, C.; Thezenas, S.; Maceski, A.; Gabelle, A.; Lopez-Crapez, E.; De Forges, H.; Firmin, N.; Guiu, S.; Jacot, W.; et al. The Prognostic Value of the Tau Protein Serum Level in Metastatic Breast Cancer Patients and Its Correlation with Brain Metastases. BMC Cancer 2019, 19. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Ferreira, S.; Nahmias, C. From Tumorigenesis to Cell Death: The Aneuploidy Paradox. Mol. Cell. Oncol. 2020, 7, 1709390. [Google Scholar] [CrossRef]
- Prezel, E.; Elie, A.; Delaroche, J.; Stoppin-Mellet, V.; Bosc, C.; Serre, L.; Fourest-Lieuvin, A.; Andrieux, A.; Vantard, M.; Arnal, I. Tau Can Switch Microtubule Network Organizations: From Random Networks to Dynamic and Stable Bundles. Mol. Biol. Cell 2018, 29, 154–165. [Google Scholar] [CrossRef]
- Holmfeldt, P.; Sellin, M.E.; Gullberg, M. Upregulated Op18/stathmin Activity Causes Chromosomal Instability through a Mechanism That Evades the Spindle Assembly Checkpoint. Exp. Cell Res. 2010, 316, 2017–2026. [Google Scholar] [CrossRef]
- Hunter, A.W.; Caplow, M.; Coy, D.L.; Hancock, W.O.; Diez, S.; Wordeman, L.; Howard, J. The Kinesin-Related Protein MCAK Is a Microtubule Depolymerase That Forms an ATP-Hydrolyzing Complex at Microtubule Ends. Mol. Cell 2003, 11, 445–457. [Google Scholar] [CrossRef]
- Stout, J.R.; Yount, A.L.; Powers, J.A.; LeBlanc, C.; Ems-McClung, S.C.; Walczak, C.E. Kif18B Interacts with EB1 and Controls Astral Microtubule Length during Mitosis. Mol. Biol. Cell 2011, 22, 3070–3080. [Google Scholar] [CrossRef]
- Tanenbaum, M.E.; Macurek, L.; van der Vaart, B.; Galli, M.; Akhmanova, A.; Medema, R.H. A Complex of Kif18b and MCAK Promotes Microtubule Depolymerization and Is Negatively Regulated by Aurora Kinases. Curr. Biol. 2011, 21, 1356–1365. [Google Scholar] [CrossRef]
- McHugh, T.; Welburn, J.P.I. Potent Microtubule-Depolymerizing Activity of a Mitotic Kif18b–MCAK–EB Network. J. Cell Sci. 2023, 136. [Google Scholar] [CrossRef] [PubMed]
- Tipton, A.R.; Wren, J.D.; Daum, J.R.; Siefert, J.C.; Gorbsky, G.J. GTSE1 Regulates Spindle Microtubule Dynamics to Control Aurora B Kinase and Kif4A Chromokinesin on Chromosome Arms. J. Cell Biol. 2017, 216, 3117–3132. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Schmidt, N.; Müller, F.; Bange, T.; Bird, A.W. Destabilization of Long Astral Microtubules via Cdk1-Dependent Removal of GTSE1 from Their Plus Ends Facilitates Prometaphase Spindle Orientation. Curr. Biol. 2021, 31, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Gai, M.; Bianchi, F.T.; Vagnoni, C.; Vernì, F.; Bonaccorsi, S.; Pasquero, S.; Berto, G.E.; Sgrò, F.; Chiotto, A.M.; Annaratone, L.; et al. <span Style=“font-Variant:small-caps;”>ASPM</span> and <span Style=“font-Variant:small-caps;”>CITK</span> Regulate Spindle Orientation by Affecting the Dynamics of Astral Microtubules. EMBO Rep. 2016, 17, 1396–1409. [Google Scholar] [CrossRef]
- Jiang, K.; Rezabkova, L.; Hua, S.; Liu, Q.; Capitani, G.; Altelaar, A.F.M.; Heck, A.J.R.; Kammerer, R.A.; Steinmetz, M.O.; Akhmanova, A. Microtubule Minus-End Regulation at Spindle Poles by an ASPM–katanin Complex. Nat. Cell Biol. 2017, 19, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Ogren, A.; Parmar, S.; Mukherjee, S.; Gonzalez, S.J.; Plooster, M.; McClellan, M.; Mannava, A.G.; Davidson, E.; Davis, T.N.; Gardner, M.K. Kinesin-14 Motors Participate in a Force Balance at Microtubule plus-Ends to Regulate Dynamic Instability. Proc. Natl. Acad. Sci. 2022, 119. [Google Scholar] [CrossRef]
- Andrews, P.D.; Ovechkina, Y.; Morrice, N.; Wagenbach, M.; Duncan, K.; Wordeman, L.; Swedlow, J.R. Aurora B Regulates MCAK at the Mitotic Centromere. Dev. Cell 2004, 6, 253–268. [Google Scholar] [CrossRef]
- Lan, W.; Zhang, X.; Kline-Smith, S.L.; Rosasco, S.E.; Barrett-Wilt, G.A.; Shabanowitz, J.; Hunt, D.F.; Walczak, C.E.; Stukenberg, P.T. Aurora B Phosphorylates Centromeric MCAK and Regulates Its Localization and Microtubule Depolymerization Activity. Curr. Biol. 2004, 14, 273–286. [Google Scholar] [CrossRef]
- Jang, C.-Y.; Coppinger, J.A.; Seki, A.; Yates, J.R.; Fang, G. Plk1 and Aurora A Regulate the Depolymerase Activity and the Cellular Localization of Kif2a. J. Cell Sci. 2009, 122, 1334–1341. [Google Scholar] [CrossRef]
- Damodaran, A.P.; Vaufrey, L.; Gavard, O.; Prigent, C. Aurora A Kinase Is a Priority Pharmaceutical Target for the Treatment of Cancers. Trends Pharmacol. Sci. 2017, 38, 687–700. [Google Scholar] [CrossRef]
- Kovacs, A.H.; Zhao, D.; Hou, J. Aurora B Inhibitors as Cancer Therapeutics. Molecules 2023, 28, 3385. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.C.; Husted, S.; Pilrose, J.; Ems-McClung, S.C.; Stout, J.R.; Carpenter, R.L.; Walczak, C.E. MCAK Inhibitors Induce Aneuploidy in Triple-Negative Breast Cancer Models. Cancers 2023, 15, 3309. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




