1. Introduction
Proso millet (
Panicum miliaceum L.) also known as common millet (USA) or broomcorn millet (China) belongs to the genus
Panicum, tribe
Paniceae, family
Poaceae and order
Poales [
1]. Millet is a small-grained self-pollinated allotetraploid cereal crop and has short growing season period. The plant was domesticated about 10,000 years ago in Central and Eastern Asia [
2,
3]. Proso millet is a diploid (2n = 36) annual herbaceous plant and currently cultivated in Asia, Australia, North America, Europe, and more rarely Africa [
4,
5,
6]. Asian countries use millet as a food crop, and the United States actively grows the plant to produce valuable fodder for birds and livestock [
7].
Although the grain yield of proso millet is lower than the major crops, it is highly adapted to a semi-arid climate [
8]. Today, the world production of millet is at a fairly high level and occupies a significant area. According to the FAO millet ranks 6th among cereals worldwide in terms of cultivated area (34.7 million hectares) and gross grain harvest (31.6 million tons), following wheat, rice, barley, corn and sorghum. Proso millet is cultivated in 30 countries [
9,
10], on about 0.82 million ha in Russia, 0.32 million ha in China [
11], 0.20 million ha in USA [
12], 0.03 million ha in India [
13] and 0.002 million ha in Korea [
14].
Since ancient times millet was worshipped by the people of Central Asia. So, in the diet of the Kazakhs there have always been such foods as tary and talcane, suitable for long-term storage and providing valuable nutritional elements in a nomadic way of life. Now in the Republic of Kazakhstan proso millet is one of the main cereal crops owing to its high drought resistance, salt resistance and less dependence on sowing dates of the dry-steppe zone. Due to the development of virgin lands of the country millet sown area reaches 1.7 million hectares. The largest areas designated for the cultivation of this crop arein the agricultural regions: Pavlodar, Akmola, Aktobe, West Kazakhstan (Ural), Kostanay. In Western Kazakhstan millet is the main cereal crop [
15]. Millet grain contains all essential amino acids, 10 to 15% protein, 55-65% starch, more than 5% fat, 0.3-0.9 mg-% carotenoids and a relatively small amount of fiber. Millet contains various vitamins such as PP, B
1 and B
2, large amounts of potassium, magnesium, phosphorus, molybdenum, magnesium, iodine, zinc, sodium and bromine [
16,
17,
18].
Assessment of genetic diversity in genbank collections has significance for plant breeding programs and genetic resources conservation [
19,
20]. The proso millet genome is relatively small and sized at 923 megabases [
21,
22]. Population structure studies of agricultural crop gene pools based on genotyping is important for the genetic improvement of economically valuable traits [
23].
Currently, the range of markers used for molecular genetic analysis is very extensive, but studies on genetic diversity based on molecular markers of proso millet worldwide collections are limited [
24]. Molecular studies of the DNA polymorphism of the proso millet germplasm are mainly based on the following markers: RAPD [
25,
26], ISSR [
27,
28], AFLP [
29], and SSR [
30,
31]. Among all PCR-based molecular markers, simple sequence repeat (SSR) markers have been proven to be the most widely used in germplasm collection characterization. They are highly reproducible, co-dominant, multi-allelic, numerous, relatively low cost, evenly distributed in the plant genome and present high degree of polymorphism and information value [
32,
33]. Most of the SSR markers in proso millet were developed using the genomic resources of related grass species. Hu et al. identified 46 SSR markers from rice, wheat, oats, and barley to analyze the genetic diversity of millet [
34]. Cho et al. (2010) developed 25 SSR markers from proso millet BAC library [
35], but they have not yet been widely utilized with
P. miliaceum. [
36]. Currently, about 100 SSR markers are available for genetic and breeding studies of millet [
37]. A population-level analysis was conducted by Rajput and Santra, 100 SSR markers were used on 90 samples of millet, and the results discovered a relationship between genetic clustering and geographic origin [
38]. The first case of breeding of waxy proso millet in Kazakhstan was presented by Zhirnova et. al. [
39], the dCAPS (9bF/15delRB) molecular marker was applied. There is limited information on SSR marker-based genetic diversity analysis of the Kazakhstan genbank, which is important for its genetic improvement [
40]. The objectives of this study were to evaluate the genetic diversity in proso millet collection consisting of 100 accessions from different ecological and geographical zones using SSR markers; and identify SSR markers associated with main agronomic traits to improve millet breeding in the future.
2. Materials and Methods
2.1. Plant Material
A total of 100 proso millet (
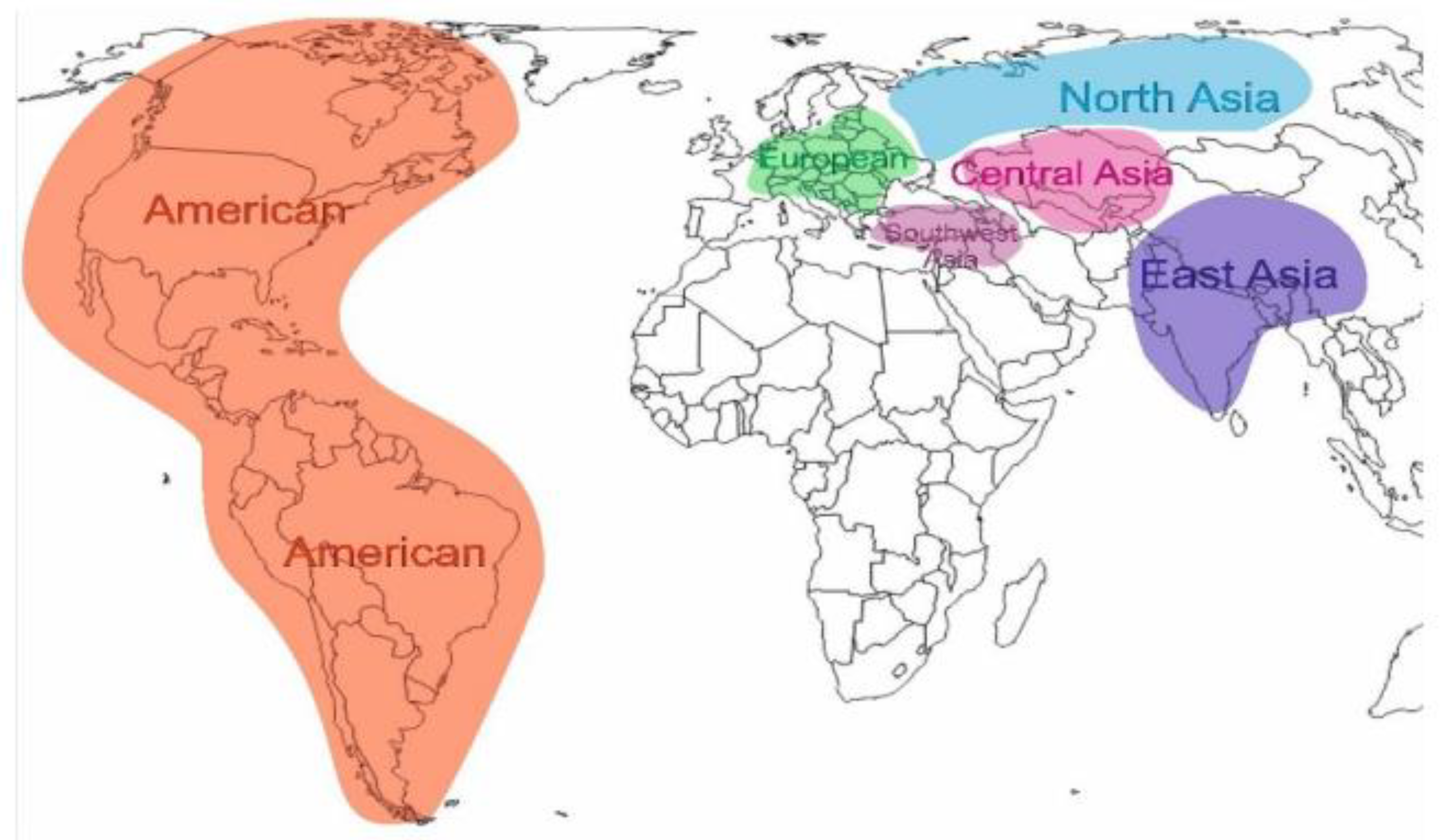
Panicum miliaceum L.) varieties and accessions of various ecological and geographical origins served as experimental material. Approximately 50 accessions were sourced from the germplasm collection of the Peginal Plant Introduction Station (the world collection of USDA), Iowa State University (USA). The germplasm collection was divided into 6 groups based on origin: American, European, East Asian, Southwest Asian, Central Asian and North Asian (
Figure 1).
American - 4; European - 8; East Asian - 10; Southwest Asian - 11; Central Asian – 26; North Asian – 41.
The experiments were carried out in the laboratory as well as in the field. The laboratory studies were performed at the Scientific Research Platform of Agricultural Biotechnology (RPAB) at the Saken Seifullin Kazakh Agrotechnical Research University, Astana, Republic of Kazakhstan.
For the identification of molecular markers that are associated with main agronomic traits field experiments were conducted in the plant nursery of the A.I. Baraev Scientific Production Center of Grain Farming (Shortandy village-1, Shortandy district, Akmola region, Republic of Kazakhstan) in the dry steppe zone of the Akmola region (51°41'58" N; 70°59'41" E; DD: 51.6995, 70.9946; height above mean sea level, in meters: 375). The field observations and the collection characterization were conducted from May to September for three consecutive years i.e. 2020, 2021 and 2022 growing seasons (
Table 1).
The experiment was performed according to the All-Russian Institute of Plant Growing guidelines and the Field Experiment Methodology [
41]. The observations of yield and yield contributing traits were recorded for each accession and for every replication: seed weight per panicle (g), thousand seed weight (g), productive tillering (pcs), grain yield (g/m
2).
2.2. DNA Extraction
The leaves of proso millet germplasm seedling were collected and used for the isolation of DNA for SSR marker genotyping. The genomic DNA from each genotype was isolated from 7 days old chlorophyll-free seedlings following modified cetyl trimethylammonium bromide (CTAB) method [
42]. The concentration of the genomic DNA samples was determined with a UV-Vis spectrophotometer (NanoDrop 2000, Thermo Fisher Scientific, Waltham, Massachusetts, USA). The integrity of the DNA was evaluated by electrophoresis on 1% agarose gel.
2.3. SSR Marker Analysis
Genotyping proso millet germplasm was performed using a set of twenty (20) microsatellite or SSR markers (
Table 2).
The PCR amplifications were performed in a total volume of 15 μL containing 8 μL 2×Master Mix for PCR (BioRad, USA), 5.2 μL ddH2O, 10 μM 1 μL each primer (F, R) (Lumiprobe Corporation (Americas)) and 100-150 ng DNA template. The PCR amplification was run using a VeritiPro™ Thermal Cycler (Applied Biosystems, Singapore) with the following program: denaturation at 95°C for 5 min, followed by 35 cycles of 40 sec at 95°C, 40 sec at the annealing temperature, 35 sec at 72°C, and then finally by extension at 72°C for 10 min. The PCR products were separated on a 6% polyacrylamide gel followed by ethidium bromide and visualized using a gel documenting system (Doc-Print CX3, Vilber, France, 2020). The marker alleles for all SSRs were recorded manually using the PAAG gels. SSR fragments were assessed for the presence of bands in the gel profile. When two DNA bands of different sizes were observed at least in two genotypes the marker was considered polymorphic. The DNA fragments of different sizes were considered different alleles, while DNA fragments of same size were considered as the same allele.
2.4. Data Analysis
The obtained field data was analyzed with Microsoft Excel software. Based on SSR scoring data, calculations for the number of different alleles (Na), number of effective alleles (Ne), Shannon's information index (I), expected heterozygosity (He), unbiased expected heterozygosity (uHe), polymorphic information content (PIC value) were performed using GenAlEx 6.5 application for MS-Excel [
44,
45]. The population structure analysis was carried out using STRUCTURE 2.2 software [
46]. The number of subgroups (K) in the population was determined by running the software with 10 independent replicate runs per K value (number of clusters) ranging from 1 to 10. Each run involved a burning period of 100,000 iterations, and a post burning simulation length of 1,000,000. The principal coordinate analysis (PCoA) was performed using GenAlEx 6.5. Neighbor-joining phylogenetic tree was constructed by the unweighted pair group method with arithmetic average (UPGMA) using PAST v.3.25 software [
47].
3. Results
3.1. Agronomic Traits in the Studied Groups of Proso Millet by Origin
Under climate conditions of North Kazakhstan, proso millet typically planted in late May or early June and harvested in early September. In general, four agronomic traits, seed weight per panicle (SWP, g), thousand seed weight (TSW, g), productive tillering (PT, pcs), grain yield (GY, g/m
2) were studied. Values of minimum/maximum (max/min), range, mean, coefficient of variation (CoV) and standard deviation (SD) for the four traits under study for each origin group were presented in
Table 3.
High yield and related components are important targeted traits in proso millet. Field trials for three years revealed a marked difference in the GY between the studied groups by origin. The highest average GY values were revealed for North Asian and American groups, average yield accounted for 535.2 and 474.2 g/m2 respectively. SWP and PT traits did not show significant results by group, SWP ranged from 2.6-2.8 g, PT from 1.1 to 1.3. As for TSW, the highest average values were for American and Central Asia groups (6.6 and 6.5 g respectively), the lowest ones were for European and East Asian groups (5.4 and 5.3 g respectively). Coefficient of variation was the highest for SWP and GY in the Central Asian group, 246.1 and 242.5 respectively.
3.2. Genotyping by SSR Markers of Proso Millet Collection
A set of 20 SSR markers was used to analyze the genetic diversity of 100 millet accessions collected from 16 various ecological and geographical areas. 9 SSR markers out of 20 SSR markers were polymorphic: SSR 67, SSR 82, SSR 85, SSR 86, SSR 92, SSR 100, SSR 109, SSR 142 and SSR 146, other 11 were monomorphic. These 20 primers amplified a total of 47 alleles, among them, 31 were polymorphic. The sizes of observed fragments ranged from 132 to 580 bp (
Table 4).
A maximum of 8 alleles was observed at the SSR 82, 6 alleles at the SSR 85 and SSR 109, 4 alleles at the SSR 67, 3 alleles at the SSR 86 and SSR 142, 2 alleles at the SSR 92, SSR 100 and SSR 146. the other 11 SSRs had one allele (SSR 70, SSR 71, SSR 120, SSR 121, SSR 127, SSR 128, SSR 129, SSR 131, SSR 143, SSR 144 and SSR 182). The highest percentage of polymorphism (100%) was observed by SSR 67, SSR 82, SSR 85 and SSR 109, while SSR 86, SSR 92, SSR 100, SSR 142 and SSR 146 showed medium level (50-67%) of polymorphism.
Among the 100 proso millet accessions, a total of 47 alleles was identified, with average of 2.35 alleles per marker. Genetic diversity analysis showed that the mean number of different alleles (Na) per SSR locus ranged from 1.77 to 3.66, for which no unique alleles were obtained (
Table 5).
The mean number of effective alleles was higher in European, Central Asian and North Asian groups, (2.209; 2.153 and 2.093 respectively), while it was lower in American, Southwest Asian and East Asian groups (1.904; 1.636 and 1.584 respectively). The number of effective alleles (Ne) ranged from 1.584 to 2.209, with an average of 1.929. Shannon’s information index (I) ranged from 0.370 to 0.827, with an average of 0.628. Expected heterozygosity (He) ranged from 0.222 to 0.441, with mean value of 0.360. The average value of unbiased expected heterozygosity (uHe) was 0.380, ranging from 0.234 for East Asian group, 0.296 for Southwest Asian, 0.397 for American, 0.430 for North Asian, 0.456 for Central Asia to 0.471 for European group, respectively.
The average of polymorphic information content (PIC value) generated by SSR primers was moderate at 0.424, ranging from 0.125 for SSR-67 to 0.795 for SSR-86 (
Table 6).
Out of 9 SSR markers four showed high PIC values: SSR 67 (0.536), SSR-82 (0.756), SSR-85 (0.795) and SSR-109 (0.758), that indicates exceeding the critical value of 0.5.
Evaluation of the amount of genetic variation in proso millet germplasm is one of the important tasks for breeding. T-test analysis was used to detect the correlations between SSR markers and the studied agronomic traits (
Table 7).
Markers SSR 85 and SSR 86 showed a significant relationship to variance in PT trait, p-value was 0.013 and 0.008 respectively. Marker SSR-85 also was associated with the mean grain yield, at p < 0.05.
3.3. Population Structure, UPGMA Cluster and PCoA Analysis
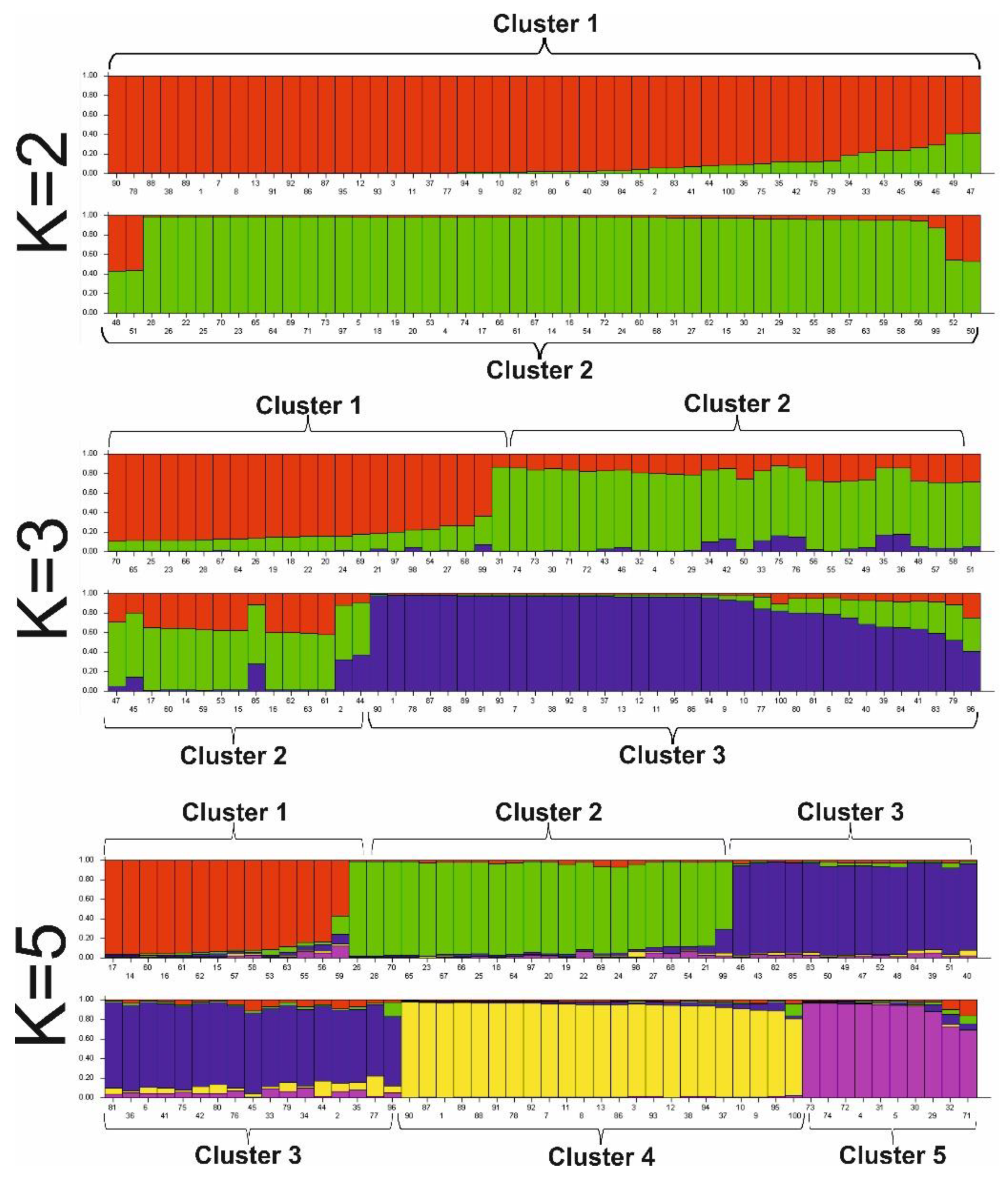
The relationship among the genotypes from different origin groups based on SSR genotyping of 100 accessions was determined by genetic structure analysis, principal coordinates analysis (PCoA) analysis, and UPGMA clustering analysis. Structure analysis of proso millet germplasm was performed using the model-based software program STRUCTURE 2.3.1 and was evaluated using the results from K=2 to K=10. Each accession in the core collection was grouped to a specific cluster by its K value resulted from population structure analysis (
Figure 2).
According to the K=2 model, the proso millet germplasm were differentiated into two clusters, the first one consisted mostly of groups originated from Central Asia, North Asia and Europe. The second cluster, in general, included the East Asian, Southwest Asian and American groups. Some of the accessions of different origins were assigned to both the first and second clusters with different degrees of probability, which indicates their intermediate position. The overall assignments of the genotypes for clusters 1 and 2 were 52% and 48%, respectively. At K=3, we observed that cluster 1 (red color), cluster 2 (green color) and cluster 3 (blue color) contained 22, 43, and 35 accessions respectively. The cultivars from the American, European and Southwest Asian groups were mostly assigned to the clusters 1 and 3, while the East Asia group accessions were mostly included in clusters 2 and 3. The accessions from Central Asian and North Asian groups were detected in all the three clusters. Similarly, at K=4, the same phenomenon occurred as at K=3. At K=5, most accessions of the East Asian group were detected in cluster 1, the gene pool of the Southwest Asian group in cluster 2 and genotypes of the European group mainly were included in cluster 3. Genotypes from the Central Asian and North Asian groups dominated cluster 4, these groups were also distributed all over the cluster. Cluster 1 had 14 genotypes, cluster 2 – 22 genotypes, cluster 3 – 31 genotypes and cluster 4 – 23 genotypes. Only 10 genotypes from European, Southwest Asian, Central Asian and North Asian groups were included in cluster 5. The genotypes of American origin were included in groups 2 and 4.
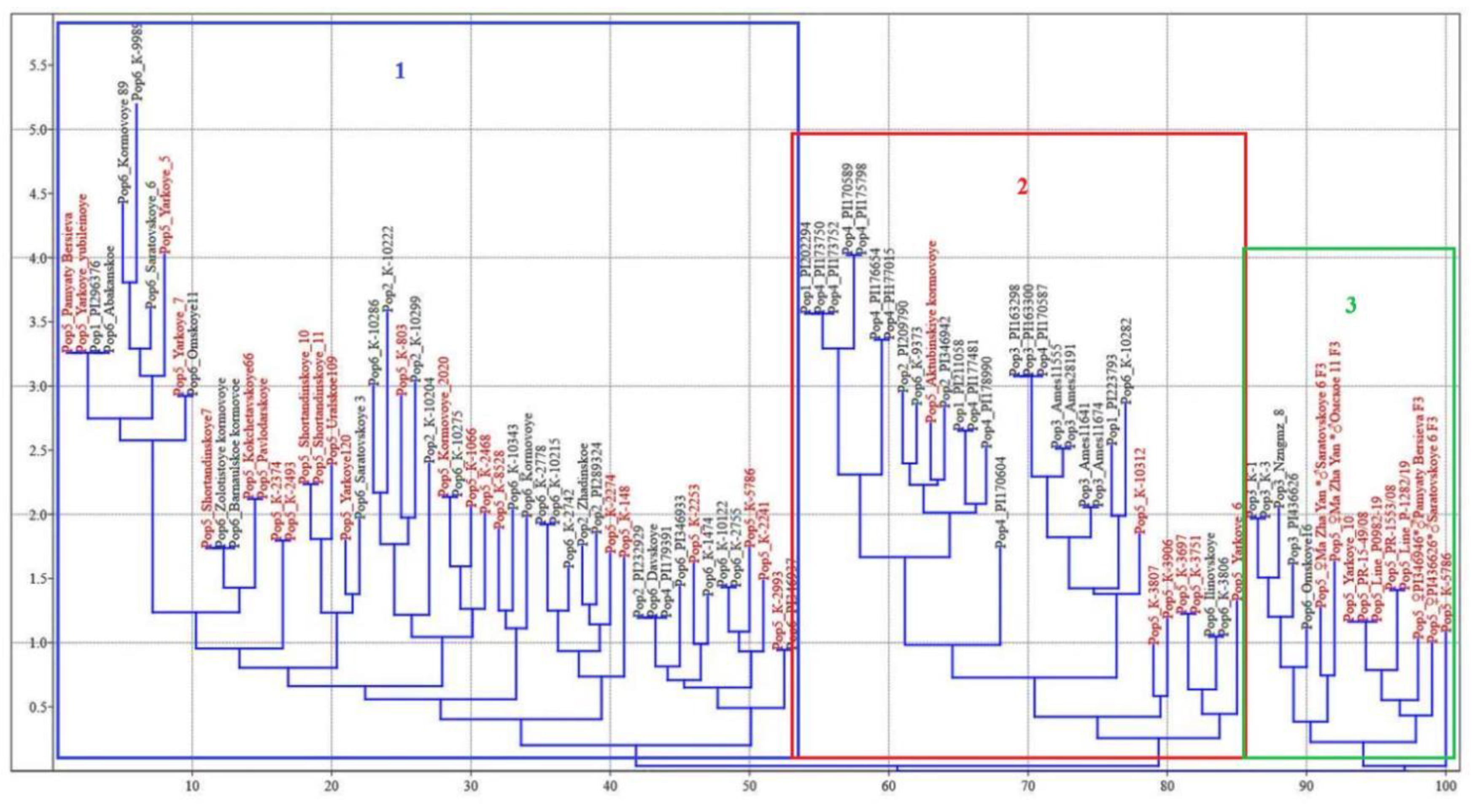
All 100 proso millet accessions were discriminated successfully by SSR markers based on the UPGMA (
Figure 3).
The results of UPGMA clustering based on similarity index found that 100 proso millet accessions were divided into 3 groups (Groups 1, 2 and 3). Group 1 (blue colour) included 53 accessions, while Group 2 (red colour) consisted of 32 accessions and Group 3 (green colour) of 15 accessions. The first group mostly included the genotypes from Central Asia (24), North Asia (21), Europe (6), Southwest Asia (1) and America (1). The second cluster contained accessions from Southwest Asia (10), Central Asia (7), East Asia (6), North Asia (4), America (3) and Europe (2), while the third group mainly included germplasm from Central Asia (10), East Asia (4) and North Asia (1). Group 1 was further subdivided into three subgroups. Subgroup I consisted of 22 genotypes, which belong to Central Asia, North Asia and America. Subgroup II comprised of 19 genotypes, most of them belong to Central Asia, North Asia and Europe. 12 cultivars were categorized into subgroup III, which constituted Central Asia, North Asia, European and Southwest Asia. Although the accessions from Central and North Asian groups dominated in the first cluster, some varieties of these origins were distributed to all three clusters.
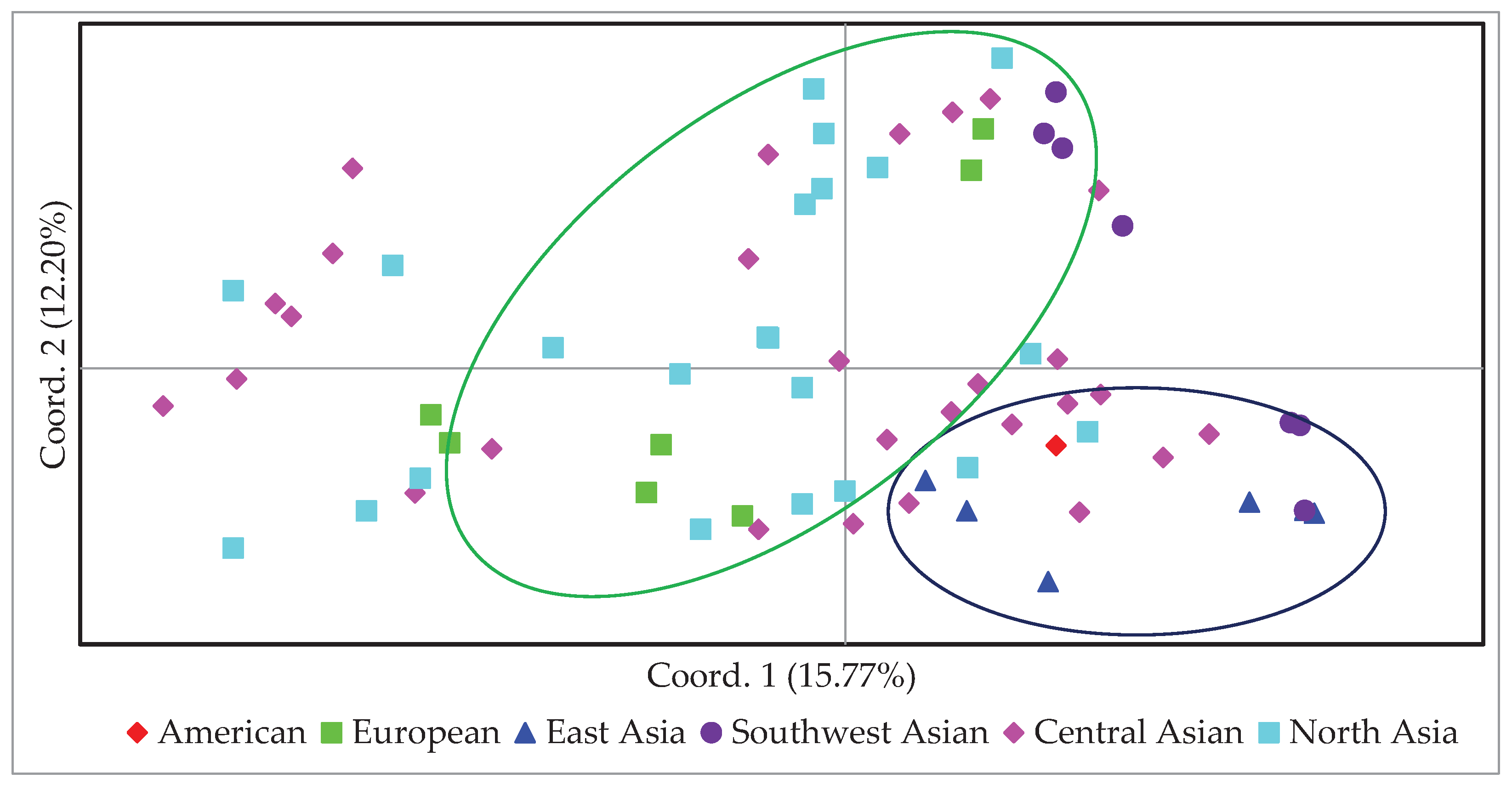
In order to obtain an alternative view of the phylogenetic relationships among 100 proso millet accessions PCoA was performed. The principal coordinates accounted for 15.77 and 12.20 of variation, respectively (
Figure 4).
A score plot of Coord.1 and Coord.2 showed the relationships among the 100 accessions. The PCoA clearly separated the East Asian group from others. The majority of the East Asia accessions clustered together on the bottom right of the plot. Southwest Asian genotypes are located along with the accessions from Central and Eastern Asia (top and bottom right). European and American groups presented an intermediate position between the Central and North Asian groups. The accessions from Central Asia and Northern Asia were distributed across the whole spectrum which was similar to the pattern shown by the population structure and UPGMA dendrogram analysis. PCoA using origin data revealed that most of genotypes of Central Asian origin were genetically closer to the North Asian group.
4. Discussion
Despite the importance of proso millet, the available information of its genetic diversity, genetic relationships, phylogenetic relationships, population structure and core collection are still limited. There are many conclusions regarding the genetic diversity of proso millet based on RAPDs [
48], AFLPs [
49], intersimple sequence repeats, single nucleotide polymorphisms [
50], and SSRs [
51] molecular markers analyses. It was discovered that proso millet accessions demonstrated high polymorphism levels and grouped together according to their geographical origins based on RAPDs markers [
52]. Genetic diversity analysis was performed by Santosh G. et. al. using 100 simple-sequence repeat (SSR) markers in the United States proso millet genotypes (landraces and cultivars). Highly genetically diverse proso millet have been found in the US germplasm [
53]. We analyzed the genetic diversity between different origins of proso millet using SSR markers. 20 pairs of SSR primers were used for initial analysis of proso millet germplasm and 9 of them demonstrated polymorphism. The percentage of polymorphic bands across the SSR markers varied from 50 to 100%, with an average of 76%. For further investigation, we selected only those markers which showed polymorphism. The 20 SSR markers in our study possessed 47 alleles with an average of 2.35 alleles per locus. The PIC value ranged from 0.125 to 0.795 with an average of 0.424, which indicates that the SSR markers used in this study could be useful for genetic diversity studies of the proso millet gene pool. Similar results were obtained in proso millet germplasm by using 25 polymorphic microsatellite markers [
54]. Also, these results are in accordance with Minxuan Liu et al., according to their research a total of 179 alleles were detected, with an average of 2.7 alleles per locus, the mean PIC and He were 0.376 and 0.445, respectively [
55].
Genetic diversity analysis based on SSR markers can provide insights into the origin and evolution of proso millet. We evaluated population structure and differentiation of the 100 accessions of different origins using STRUCTURE 2.2 software. The delta K analysis including 100 accessions from 6 origin groups suggested that the most likely number of clusters was at K=5. Hierarchical levels of population structure could hardly be recognized at K=2 and K=3. Both clusters have mixed group and no well-assigned population designations could therefore be recognized. In our study, population structure analysis showed that accessions from European, Central Asian and North Asian groups were mainly classified as mixed populations. A dendrogram based on Neighbor-joining phylogenetic tree of the SSR data was constructed among the 100 proso millet accessions belonging to six origin groups. According to the UPGMA cluster analysis, all accessions were separated into three main clusters, but the genotypes from one origin did not form separate clusters. Genotypes of Central and North Asian origin investigated in this study showed broader genetic diversity. The PCoA based on SSR genotyping of the 100 proso millet accessions was conducted using 9 SSR markers. The accessions of the studied collection were divided into groups depending on their attribution to species and place of origin, respectively.
5. Conclusions
SSR marker-based evaluation is a very effective tool for assessing genetic diversity in proso millet genotype population of different origins. In this research, 20 SSR markers were used to genotype 100 accessions of proso millet from different origins. The Neighbor joining phylogenetic tree divided the 100 proso millet accessions into three main clusters, where clusters 1 and 3 were mainly represented by Central Asian genotypes. The application of SSR markers suggested that Central and North Asian accessions have wide genetic differences from other groups. Particularly, the Principal Coordinate plot showed that accessions from Central and North Asian groups were distributed across the whole plot spectrum. The application of the t-test indicated that SSR 85 and SSR 86 were associated with some agronomic traits, such as productive tillering (PT, pcs) and grain yield (GY, g/m2). Obtained results can be useful in the future breeding processes for the increase of yield productivity in proso millet.
Author Contributions
Conceptualization, A.O. and A.Z.; methodology, E.D., I.Z. and G.Y.; investigation, G.Y. and A.R.; formal analysis, A.Z., I.Z. and A.R.; resources, E.D., A.R.; writing-original draft preparation, E.D., A.R. and A.O.; writing-review and editing, A.R. and E.D.; supervision, A.R. and E.D.; funding acquisition, A.R. All authors have read and agreed to the published version of the manuscript.
Funding
The work was carried out within the framework of the scientific project AP14870014 "Application of DNA technologies in breeding and genetic studies of millet culture when creating new domestic drought-resistant varieties" (2022-2024), grant funding for research work was provided by the "Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan" State Institution.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This work was supported by RUDN University Strategic Academic Leadership Program.
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- Gomeshe S. Proso millet, Panicum miliaceum (L.): Genetic improvement and research needs. In: Patil JV, editor. Millets and sorghum biology and genetic improvement. Hoboken: Wiley; 2016. p. 150-79. [CrossRef]
- Bandyopadhyay, T., Muthamilarasan, M. & Prasad, M. Millets for next generation climate-smart agriculture. Front. Plant Sci., 2017, 8, 1266. [CrossRef]
- Baltensperger D.D. Foxtail and proso millet/In: JANICK, J. (Ed.) Progress in new crops. Alexandria, VA: ASHS Press, 1996. P. 182-190.
- D. D. Baltensperger, “Progress with Proso, Pearl and Other Millets,” In: J. Janick and A. Whipkey, Eds., Trends in New Crops and New Uses, ASHS Press, Alexandria, 2002, pp. 100-103.
- E. A. Oelke, E. S. Oplinger, D. H. Putnam, B. R. Durgan, J. D. Doll and D. J. Undersander Millets, “Alternative Field Crops Manual,” University of Wisconsin, Cooperative Extension and University of Minnesota, Center for Alternative Plants & Animal Products and Minnesota Extension Service, 1990.
- Santosh G. Rajput, Tammy Plyler-Harveson, Dipak K. Santra Development and Characterization of SSR Markers in Proso Millet Based on Switchgrass Genomics. American Journal of Plant Sciences, 2014, 5, 175-186 Published Online January 2014 (http://www.scirp.org/journal/ajps). [CrossRef]
- Santra D.K., Rose D. “Alternative Uses of Proso Millet”, University of Nebraska-Lincoln Neb Gude G2218, 2013.
- Saseendran, S. A., D. C. Nielsen, D. J. Lyon, L. Ma, D. G. Felter, D. D. Baltensperger, G. Hoogenboom, and L. R. Ahuja. Modeling responses of dryland spring triticale, proso millet and foxtail millet to initial soil water in the High Plains. Field Crops Research, 2009, 113: 48–63. [CrossRef]
- 9. M. Vetriventhan, Vania C.R. Azevedo, H.D. Upadhyaya, A.Nirmalakumari, Joanna Kane-Potaka, S.Anitha, S. Antony Ceasar, M. Muthamilarasan, B. Venkatesh Bhat, K. Hariprasanna, Amasiddha Bellundagi, Deepika Cheruku, C. Backiyalakshmi, Dipak Santra, C. Vanniarajan,·Vilas A.Tonapi Genetic and genomic resources, and breeding for accelerating improvement of small millets: Current status and future interventions. Nucleus, 2020, 63:217–239. [CrossRef]
- ZotikovV.I., SidorenkoV.S., Bobkov S.V. Area and Production of Proso Millet (Panicum miliaceum L.) in Russia. Advances in Broomcorn Millet Research. Proceedings of the 1st International Symposium on Broomcorn Millet. Northwest A&F University (NWSUAF), 25–31 August. – Yangling, Shaanxi, People‟s Republic of China, 2012, P. 3-9.
- Diao X. Production and genetic improvement of minor cereals in China. Crop J. 2017;5:103–14. [CrossRef]
- Habiyaremye C., Matanguihan J.B., Guedes J.D., Ganjyal G.M., Whiteman M.R., Kidwell K.K.; et al. Proso millet (Panicum miliaceum L.) and its potential for cultivation in the Pacifc Northwest, U.S.: A Review. Front Plant Sci, 2017;8:1961. [CrossRef]
- Bhat B.V., Tonapi V.A., Rao B.D., Singode A., Santra D. Production and utilization of millets in India. In: Santra D.K., Johnson J.J., editors. International millet symposium on 3rd international symposium on broomcorn millet (3rd ISBM). 2018, p. 24–6.
- Park C.H. Production and utilization of broomcorn millet in korea. In: Santra D.K, Johnson J.J., editors. International millet symposium 3rd international symposium on broomcorn millet (3rd ISBM) Progr Abstr. 2018, p. 27.
- Tsygankov, I.G., Tsygankov, V.I., Tsygankov, and M.Yu. Millet in the dry steppe zone of Western Kazakhstan. Agricultural Sciences Volume, 2004, 2: 91-95 (in Russian).
- Hegde P.S., Rajasekaran N.S., Chandra T.S. Effects of the antioxidant properties of millet species on oxidative stress and glycemic status in alloxan induced rats. Nutr. Res.,2005, V.25. P. 1109-1120. [CrossRef]
- Saleh A.S.M., Zhang Q., Chen J., Shen Q. Millet grains :nutritional quality, processing, and potential health benefits. Compr. Rev.Food Sci.Food Saf., 2013, V.12. P. 281-295. [CrossRef]
- Devi P.B., Vijayabharathi R., Sathyabama S., Malleshi N.G., Priyadarisini V. B. Health benefits of finger millet (Eleusine coracana L.) polyphenols and dietary fiber: A Review. J. Food Sci.Technol., 2014, V. 51, P. 1021-1040. [CrossRef]
- Upadhyaya, H. D., Dwivedi, S. L., Singh, S., Vetriventhan, M., & Sharma, S. Forming core collections in barnyard, kodo, and little millets using morphoagronomic descriptors. Crop Science, 2014, 54(6), 2673–2682. [CrossRef]
- Jiang, Y., H. Li, J. Zhang, J. Xiang, R. Cheng and G. Liu. Whole genomic EST-SSR development based on high-throughput transcript sequencing in proso millet (Panicum miliaceum). Int. J. Agric. Biol., 2018, 20: 617-620. [CrossRef]
- Zou, C., Li, L., Miki, D., Li, D., Tang, Q., Xiao, L., Rajput, S., Deng, P.,Peng, L., Jia, W., Huang, R., Zhang, M., Sun, Y., Hu, J., Fu, X.,Schnable, P. S., Chang, Y., Li, F., & Zhang, H. The genome of broomcorn millet. Nature Communications, 2019, 10(1). [CrossRef]
- Rituraj Khound, Guangchao Sun, Ravi V. Mural, James C. Schnable, Dipak K. Santra SNP discovery in proso millet (Panicum miliaceum L.) using lowpass genome sequencing Plant Direct., 2022 Sep 13;6(9):e447. doi: 10.1002/pld3.447. eCollection 2022 Sep. PMID: 36176305 PMCID: PMC9470529. [CrossRef]
- Johnson, M., Deshpande, S., Vetriventhan, M., Upadhyaya, H. D., & Wallace, J. G. Genome-wide population structure analyses of three minor millets: Kodo millet, little millet, and proso millet. The Plant Genome, 2019, 12(3), 190021. [CrossRef]
- Sameh Boukail , Mercy Macharia, Mara Miculan, Alberto Masoni, Alessandro Calamai, Enrico Palchetti and Matteo Dell’Acqua Genome wide association study of agronomic and seed traits in a world collection of proso millet (Panicum miliaceum L.). BMC Plant Biol, 2021, 21:330. [CrossRef]
- Khound, R., & Santra, D. K. Omics for proso millet genetic improvement. Nucleus, 2020, 63(3), 241–247. Springer. [CrossRef]
- Santra, D. K., Khound, R., & Das, S. Proso millet (Panicum miliaceum L.) breeding: Progress, challenges and opportunities. In Advances in plant breeding strategies: Cereals, 2019, Vol. 5, pp. 223–257. Springer International Publishing. [CrossRef]
- Zdislav aDvořákováa, Petra Hlásná Čepkováa, Dagmar Janovskáb, Iva Viehmannováa, Eva Svobodováa, Eloy Fernández Cusimamania, Luigi MilellacComparative analysis of genetic diversity of 19 millet genera revealed by ISSR markers. Emirates Journal of Food and Agriculture, 2015, 27(8): 617-628 http://www.ejfa.me/. [CrossRef]
- SmitaShingane, J. V. Patil, Sunil Gomasheand Dinesh Chand Assessing Genetic Diversity among Foxtail millet (Setariaitalica (L.) P. Beauv.) Accessions Using RAPD and ISSR Markers. International Journal of Bio-resource and Stress Management, 2018, 9(1):001-006.
- Karam D., Westra P., Niessen S.J., Ward S.M., Figueiredo J.E.F. Assessment of silver-stained AFLP markers for studying DNA polymorphism in proso millet (Panicum miliaceum L.). Rev Bras Bot, 2006;29:609-15. [CrossRef]
- Rajput S, Plyler-Harveson T, Santra D. Development and characterization of SSR markers in proso millet based on switchgrass genomics. Am J. Plant Sci., 2014;05:175–86. [CrossRef]
- K. Trivedi,L. Arya M. Verma, S. K. Verma, R. K. Tyagi,A. Hemantaranjan Genetic variability in proso millet [Panicum miliaceum] germplasm of Central Himalayan Region based on morpho-physiological traits and molecular markers. Acta Physiol Plant, 2015, 37:23. [CrossRef]
- W.Powell, M. Morgante and C. Andre The Comparison of RFLP, RAPD, AFLP and SSR (Microsatellite) Markersfor Germplasm analysis. Molecular Breeding, 1996, Vol. 2, No. 3, pp. 225-238. [CrossRef]
- Gupta and R. K. Varshney, The Development and Use of Microsatellite Markers Fore Genetic Analysis and Plant Breeding with Emphasis on Bread Wheat,” Euphytica, 2000, Vol. 113, No. 3, pp. 163-185. [CrossRef]
- X. Hu, J. Wang, P. Lu and H. Zhang Assessment of Genetic Diversity in Broomcorn Millet (Panicum miliaceumL.) Using SSR Markers, Journal of Genetics and Genomics, 2009, Vol. 36, No. 8, pp. 491-500. [CrossRef]
- C. Young-II, J.-W. Chung, G.-A. Lee, K.-H. Ma, A. Dixit, J.-G. Gwag and Y.-J. Park, “Development and Characterization of Twenty-Five New Polymorphic Microsatellite Markers in Proso Millet (Panicum miliaceumL.). Genes & Genomics, 2010, Vol. 32, No. 3, pp. 267-273. [CrossRef]
- Harriet V. Hunt, Michael G. Campana, Matthew C. Lawes, Yong-Jin Park, Mim A. Bower, Christopher J. Howe and Martin K. Jones Genetic diversity and phylogeography of broomcorn millet (Panicum miliaceum L.) across Eurasia. Molecular Ecology, 2011, 20, 4756-4771. [CrossRef]
- Santosh G. Rajput, Tammy Plyler-Harveson, Dipak K. Santra Development and Characterization of SSR Markers in Proso Millet Based on Switchgrass Genomics. American Journal of Plant Sciences, 2014, 5, 175-186 Published Online January 2014 (http://www.scirp.org/journal/ajps). [CrossRef]
- Santosh G. Rajput, Tammy Plyler-Harveson, Dipak K. Santra Development and Characterization of SSR Markers in Proso Millet Based on Switch grass Genomics. American Journal of Plant Sciences, 2014, 5, 175-186 Published Online January 2014 (http://www.scirp.org/journal/ajps). [CrossRef]
- Irina Zhirnova, Aiman Rysbekova, Elmira Dyussibayeva, Aiym Zhakenova, Bekzak Amantaev, Yin-Gang Hu, Bai-Li Feng, and Zhazira ZhunusbayevaPre-breeding for waxy proso millet by phenotyping and marker-assisted selection. Chilean Journal Of Agricultural Research, 2021, 81(4). P.518-526. [CrossRef]
- Dyusibaeva E.N., Esenbekova G.T., Zhirnova I.A., Rysbekova A.B., Makhmudova C.K., Seitkhozhaev A.I. and Zhakenova A.E. Assessment of millet genetic variability using molecular-genetic approach for increasing the efficiency of breeding. Eco. Env. & Cons. 2019, 25 (1): pp. (410-415).
- Dospekhov B.A. Methods of field experience (with the basics of statistical processing of research results). 2011, M.: Alliance (http://www.vir.nw.ru/).
- Murray, M.G., and Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8:4321-4325.
- Xiao-Han Wang, Myung-Chul Lee, Yu-Mi Choi, Seong-Hoon Kim, Seahee Han, Kebede Taye Desta Hye-Myeong Yoon, Yoon-Jung Lee , Mi-Ae Oh, Jung-Yoon Yi and Myoung-Jae Shin Phylogeography and Antioxidant Activity of Proso Millet (Panicum miliaceum L.). Plants, 2021, 10, 2112. [CrossRef]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press, 1987, 512.
- Peakall, R. and Smouse P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics, 2012, 28, 2537-2539. [CrossRef]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics, 2000, 155:945–959. [CrossRef]
- Hammer Ø., Harper D.A.T., Ryan P.D. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron., 2001;4(1):9.
- M'Ribu, H.K.; Hilu, K.W. Detection of interspecific and intraspecific variation in Panicum millets through random amplified polymorphic DNA. Theor. Appl. Genet., 1994, 88, 412-416. [CrossRef]
- Karam, D.; Westra, P.; Nissen, S.J.; Ward, S.M.; Figueiredo, J.E.F. Genetic diversity among proso millet (Panicum miliaceum) biotypes assessed by AFLP techniques. PlantaDaninha, 2004, 22, 167-174. [CrossRef]
- Lágler, R.; Gyulai, G.; Humphreys, M.; Szabó, Z.; Horváth, L.; Bittsánszky, A.; Kiss, J.; Holly, L.; Heszky, L. Morphological and molecular analysis of common millet (P. miliaceum) cultivars compared to a DNA sample from the 15th century (Hungary). Euphytica, 2005, 146, 77–85.6. [CrossRef]
- Hunt, H.V.; Campana, M.G.; Lawes, M.C.; Park, Y.J.; Bower, M.A.; Howe, C.J.; Jones, M.K. Genetic diversity and phylogeography of broomcorn millet (Panicum miliaceum L.) across Eurasia. Mol. Ecol, 2011, 20, 4756–4771. [CrossRef]
- M'Ribu, H.K.; Hilu, K.W. Detection of interspecific and intraspecific variation in Panicum millets through random amplified polymorphic DNA. Theor. Appl. Genet. 1994, 88, 412–416. [CrossRef]
- Santosh G. Rajput, Dipak K. Santra Evaluation of Genetic Diversity of Proso Millet Germplasm Available in the United States using Simple-Sequence Repeat Markers. Crop Science, 2016, V. 56, Issue 5, P.2401-2409. [CrossRef]
- Cho, Y.I., Chung, J.W., Lee, G.A. et al. Development and characterization of twenty-five new polymorphic microsatellite markers in proso millet (Panicum miliaceum L.). Genes Genom, 2010, 32, 267–273. [CrossRef]
- Minxuan Liu , Yue Xu, Jihong He, Shuang Zhang, Yinyue Wang and Ping Lu Genetic Diversity and Population Structure of Broomcorn Millet (Panicum miliaceum L.) Cultivars and Landraces in China Based on Microsatellite Markers. Int. J. Mol. Sci., 2016, 17, 370. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).