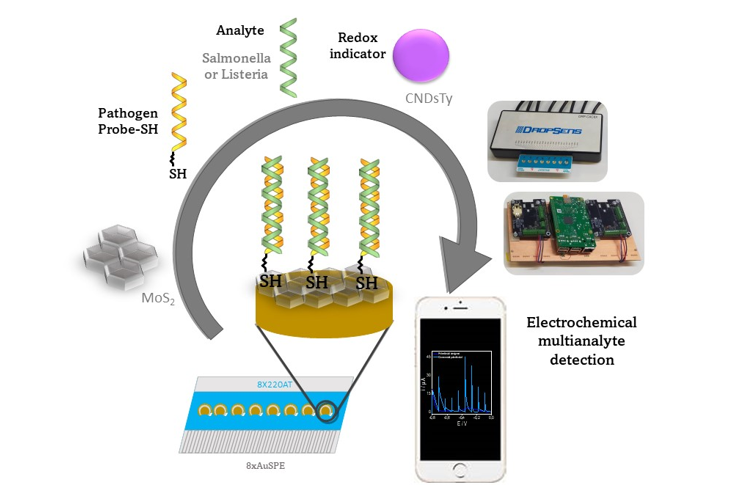
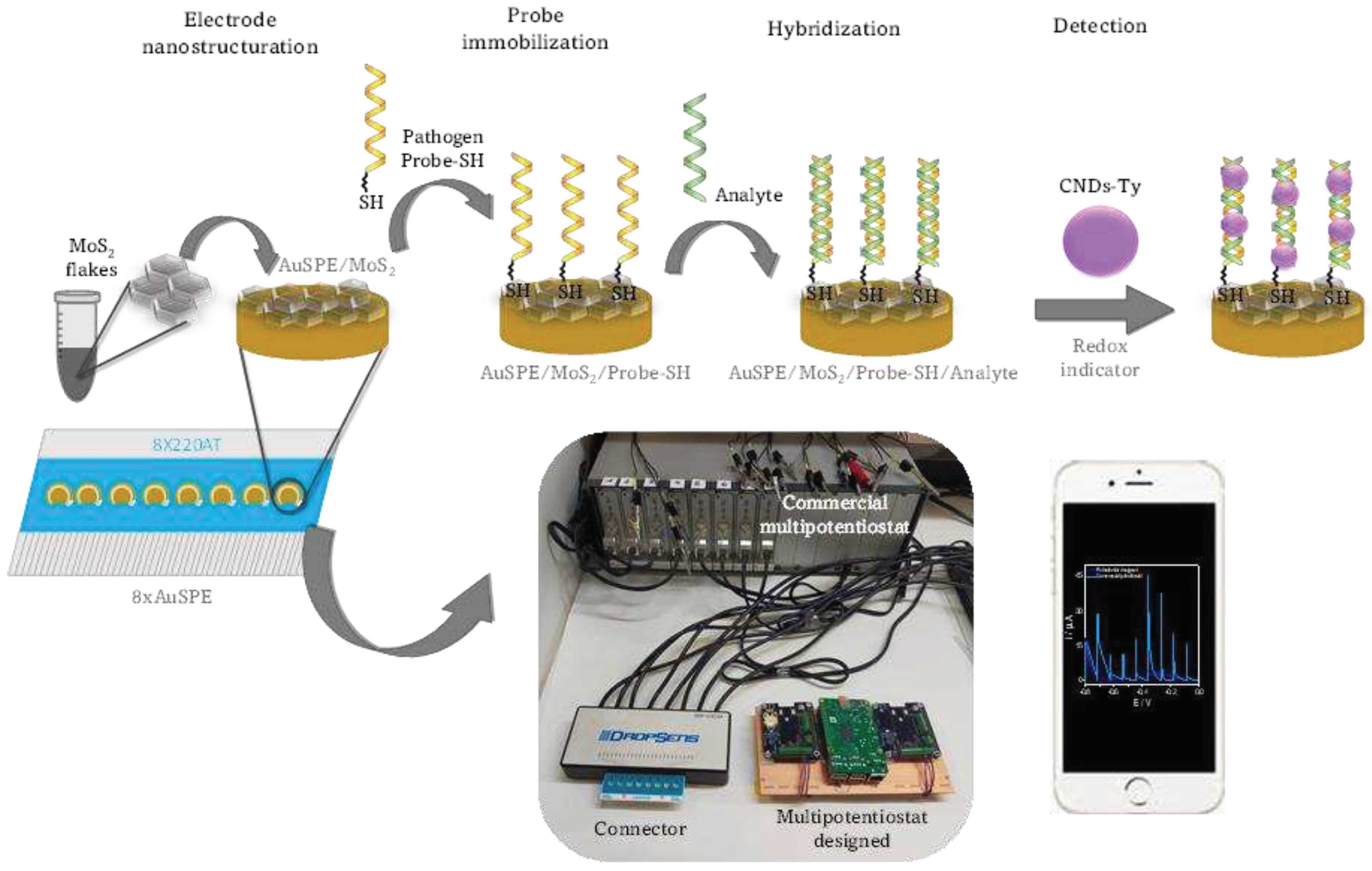
2.3.4. Biosensing platform development
Nanostructuration of the array of screen-printed electrodes (SPE)
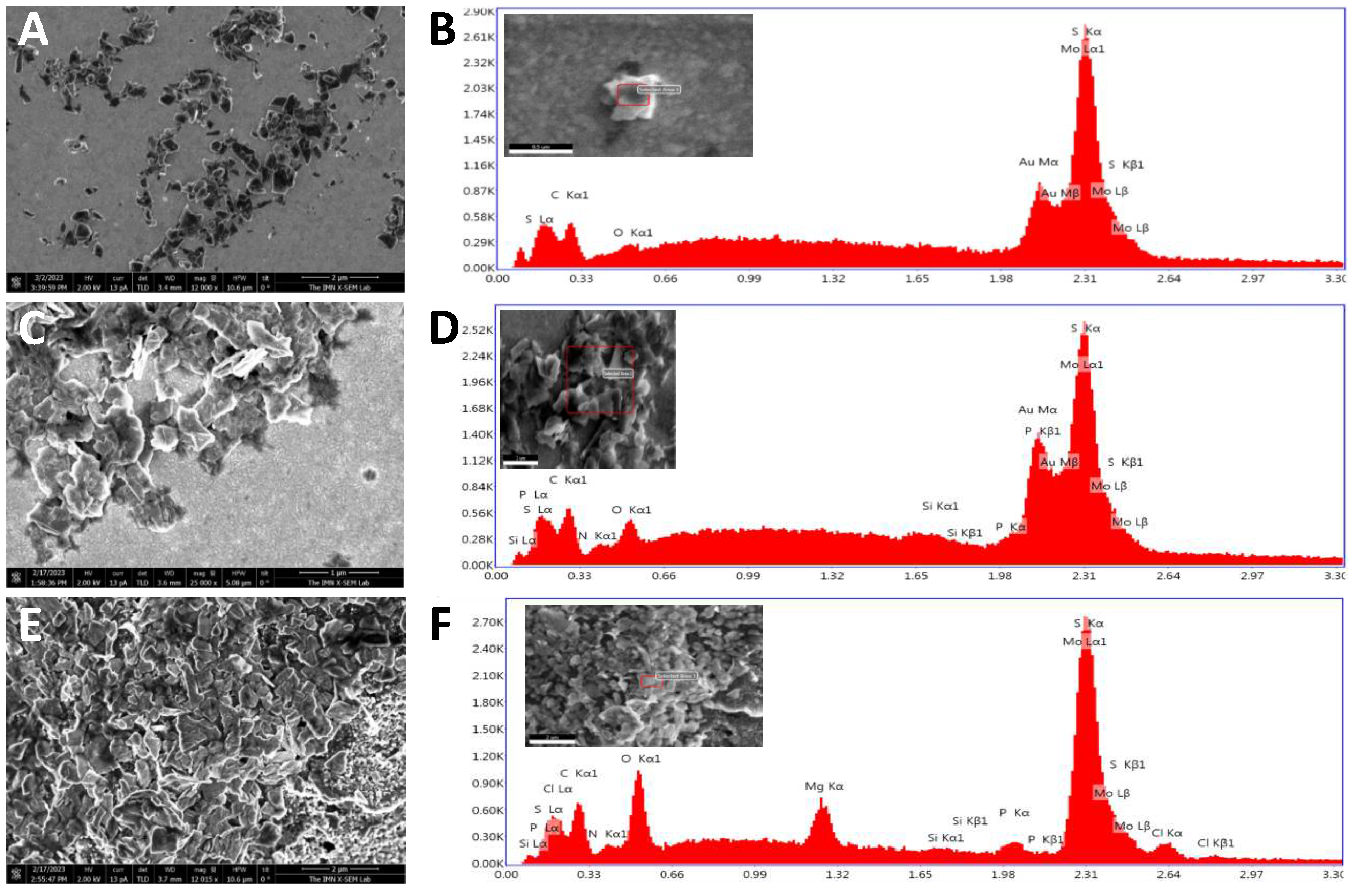
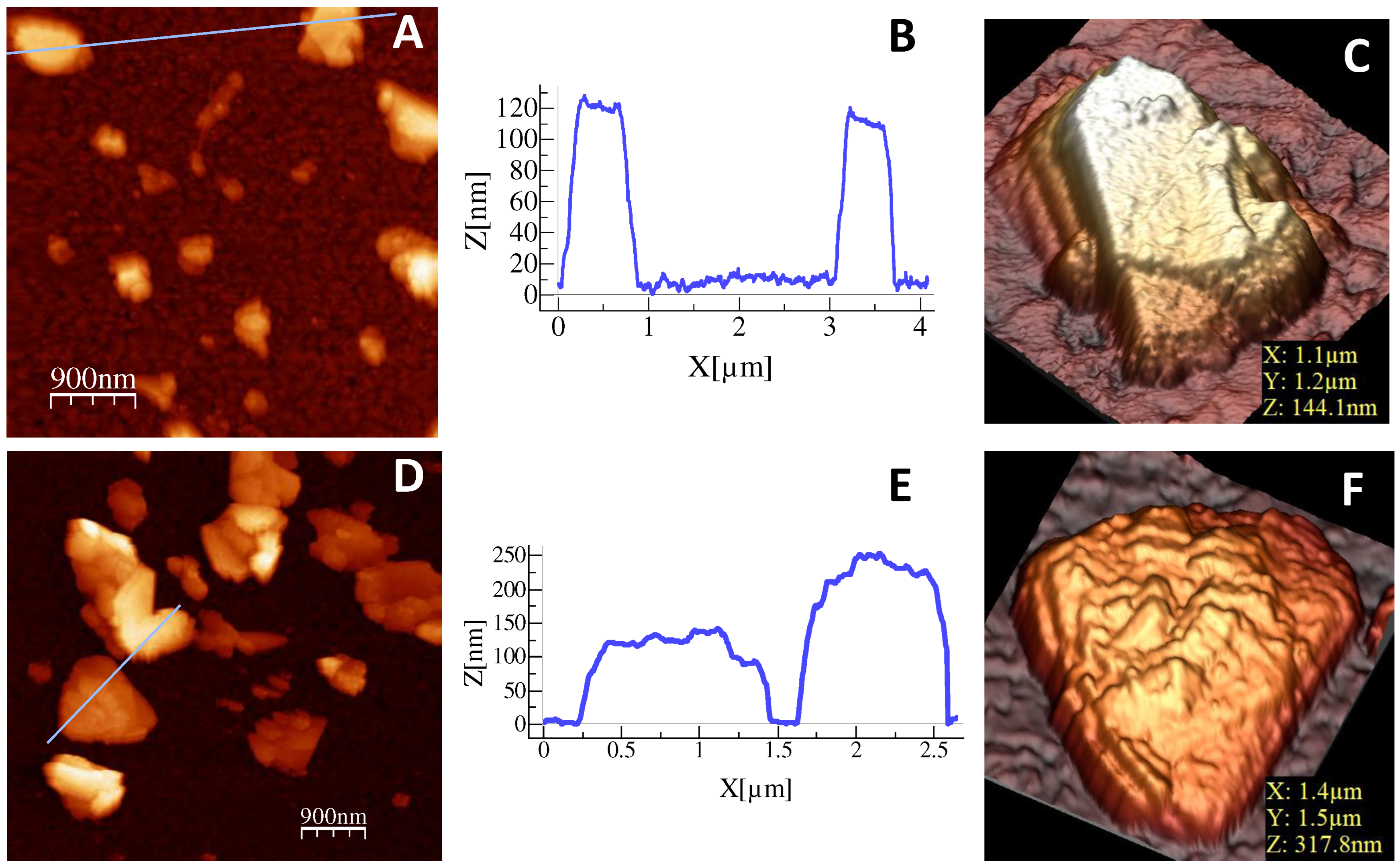
Screen-printed electrochemical array formed by eight electrodes (8xSPE) was nanostructured with MoS2 flakes by drop-casting to improve the analytical properties of the biosensor. For this purpose, 5 µL of a colloidal dispersion of MoS2 in water was deposited on the surface of the gold working electrode by drop-casting over a hot plate to evaporate the organic solvent. The resulting platform is denoted as 8xSPE/MoS2.
DNA capture probe immobilization
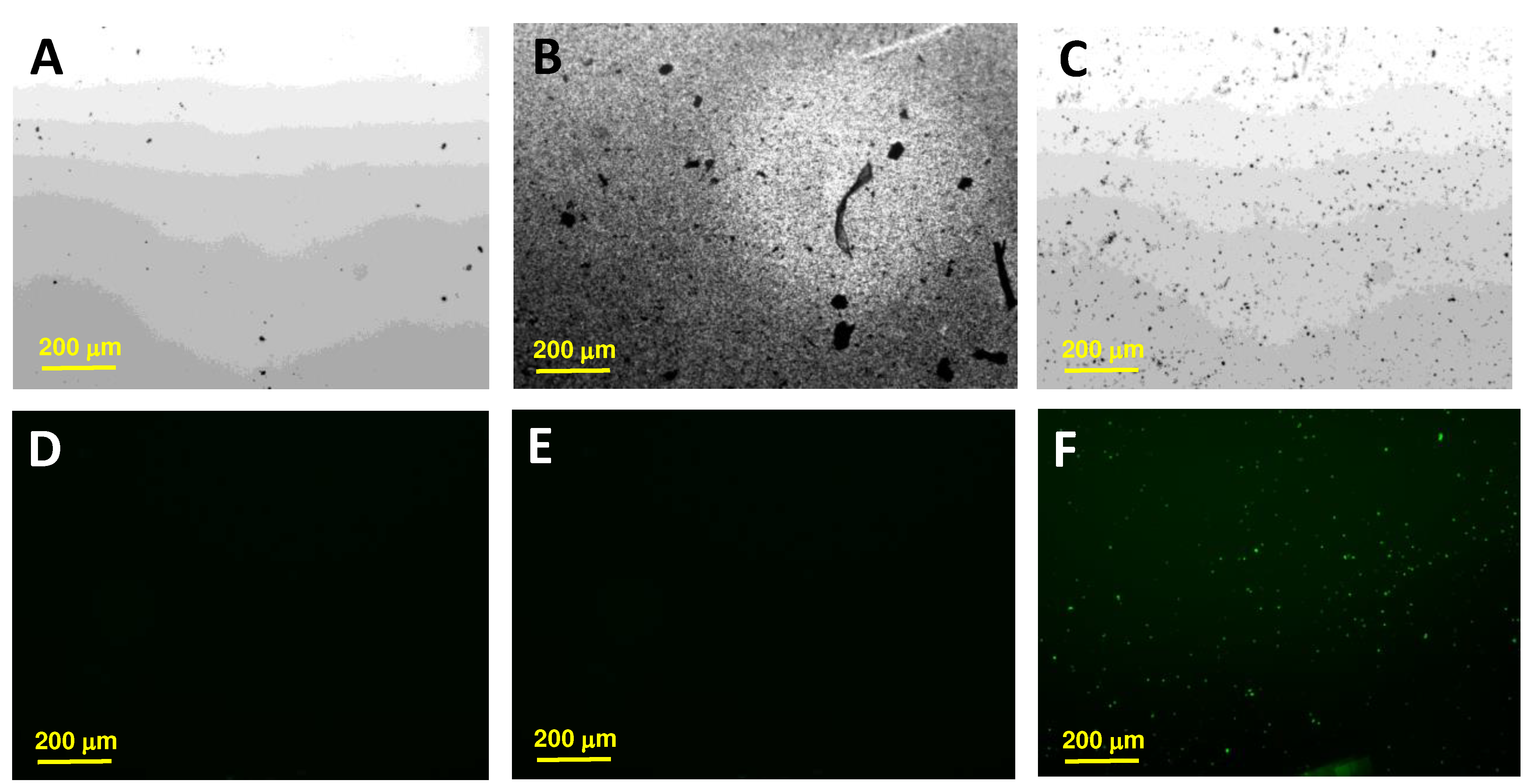
8xSPE/MoS2 platform was then modified with 5 µL of 10 µM thiolated DNA Listeria and Salmonella capture probe by drop-casting and was kept for 24 hours at room temperature (SPE/MoS2/List-SH or SPE/MoS2/Salm-SH). The platform was then washed with Milli-Q water.
Hybridization reaction and electrochemical detection
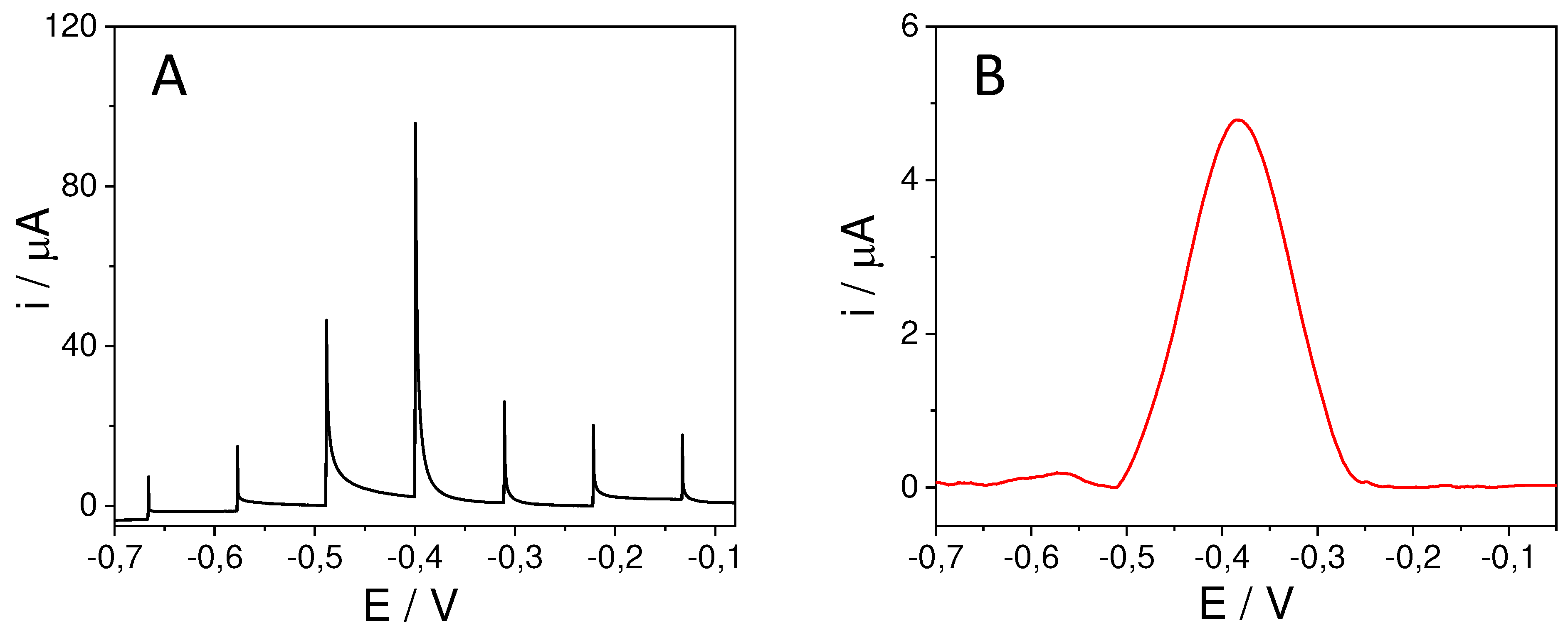
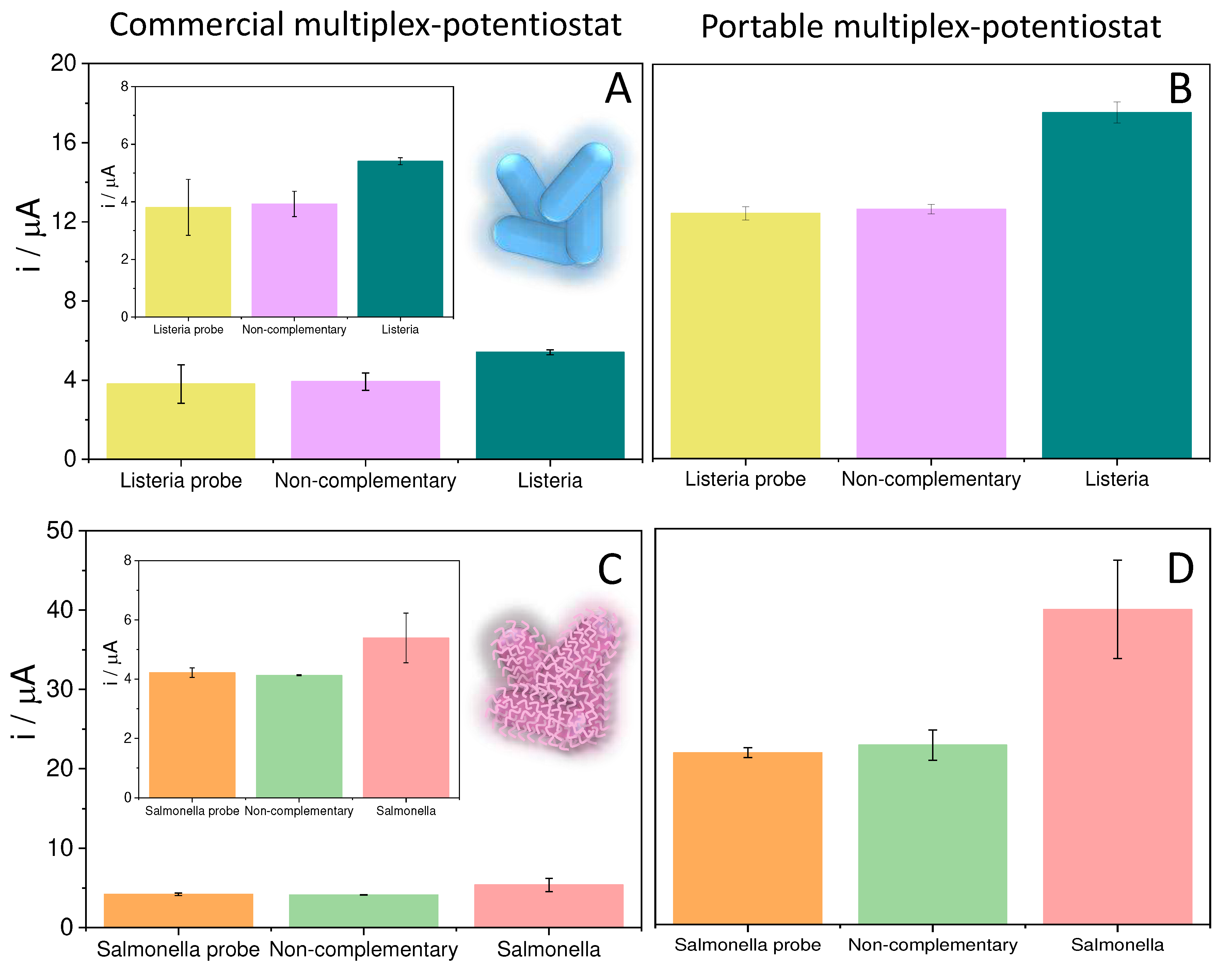
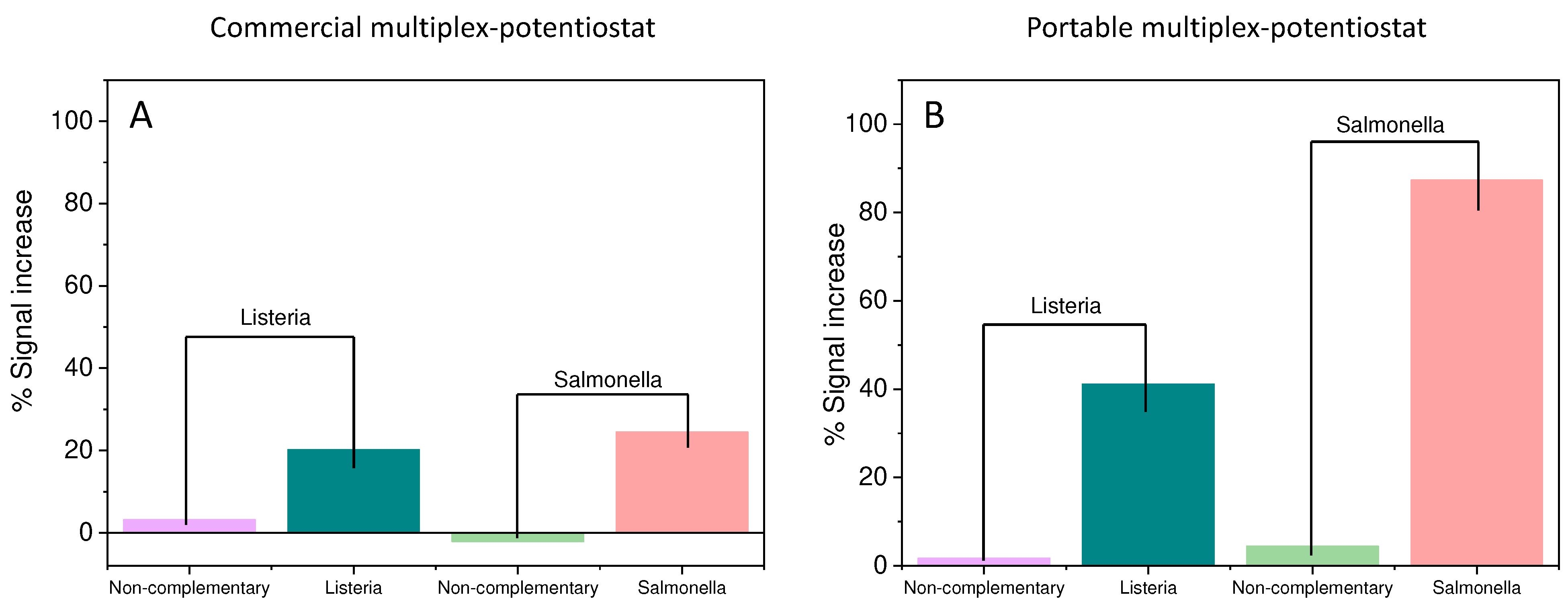
The platform with the immobilized probe was then hybridized with 5 µL of 100 pM of the target sequences of Listeria or Salmonella, for 1 hour at 40°C. To detect the hybridization evet carbon nanodots modified with thionine (Ty-CDs) were used as redox indicator. For this purpose, 5 µL of 2.83 mg/mL Ty-CDs solution were incubated for 1 hour at room temperature over the working electrode. Finally, the electrodes were rinsed with Milli-Q water and the differential pulse voltammograms (DPV) were recorded in each case using PB 0.1M pH 7.0 as the supporting electrolyte. Measurements were carried out on the designed portable multiplex-potentiostat (Single-pulse potential measurement) and on the commercial Autolab multiplex-potentiostat to compare the results.
Study of the selectivity of the biosensor
To study the selectivity of the biosensor, 8xSPE/MoS2/List-SH and 8xSPE/MoS2/Salm-SH platforms were incubated in the same conditions of time and temperature (1hour, 40°C) with Salmonella and Listeria target sequences respectively, acting then as non-complementary sequences. Then, in the same way as in the previous step, they were incubated for 1 hour with Ty-CDs, the electrodes were washed with sterile water and the DPVs were recorded in 0.1M PB pH 7.0.
2.3.5. Multiplex-potentiostat design and development
The developed multiplex-potentiostat presents a comprehensive device for conducting end-to-end measurements. The design has been meticulously executed at both the hardware and software levels.
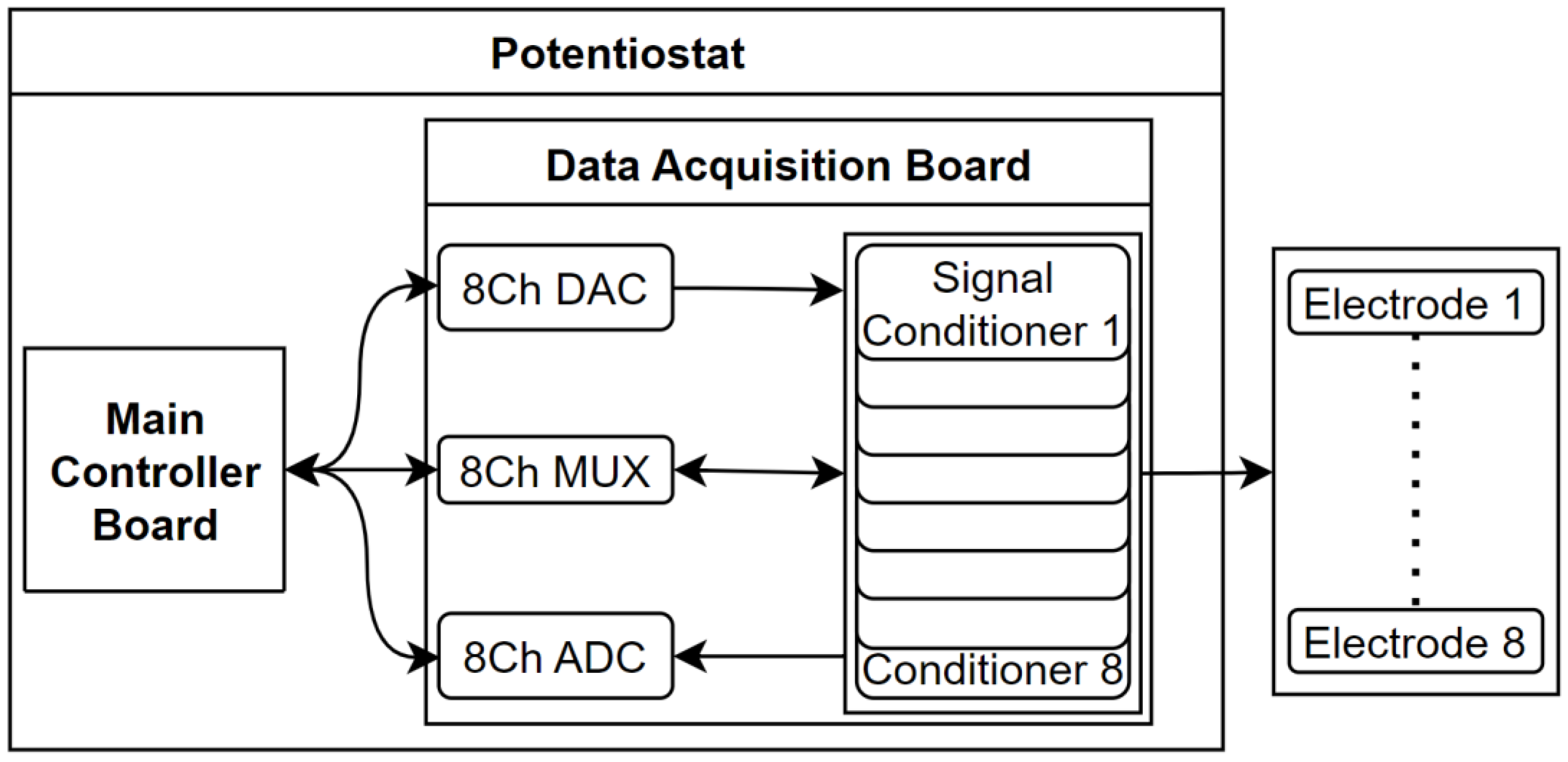
At the hardware level, the design led to the creation of two distinct modules: the Main Controller Board and the Data Acquisition Board. The first one acts as a repository for all necessary software and takes charge of overseeing the entire system's functioning, running all the necessary algorithms and commanding the second module, which, in turn, can be seen as a group of other three modules, this is, the Data Multiplexing Module, the Signal Conversion Module and the Signal Acquisition Module. The first one is the core part that allows to perform multiple measures, by routing properly the signals between all chemical cells and the Main Controller Module. The second one incorporates all the essential electronic components required to process the signal, including analog-to-digital and digital-to-analog converters and a set of filters that allows to achieve a low electrical noise level in the system. At last, the third one is composed of all the essential electronic components and signal conditioners required to acquire the raw signal from the electrochemical cell. A scheme of the hardware design can be seen in the
Scheme 1.
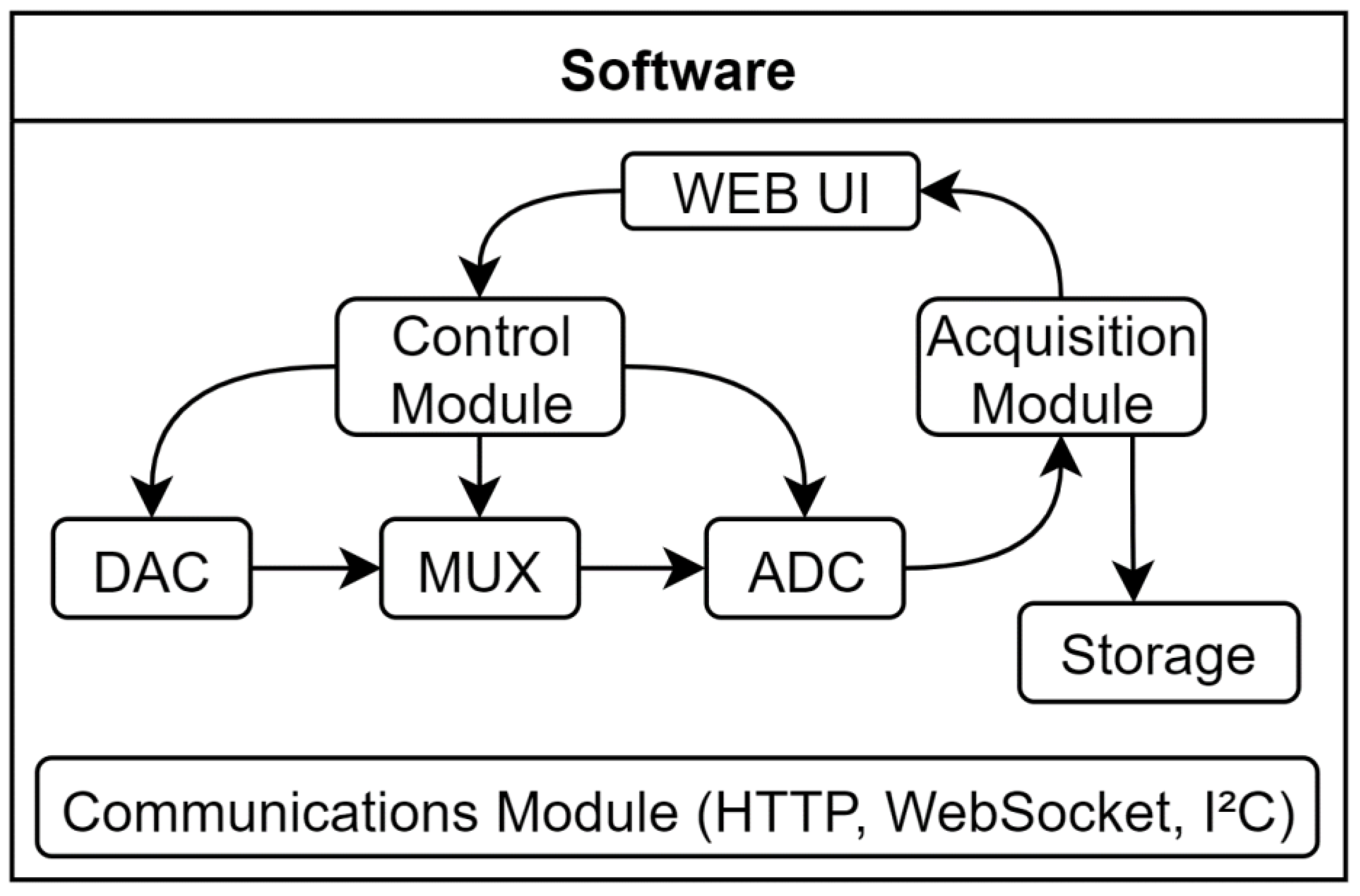
Conversely, the software-level design yielded five modules: the Communications Module, which is responsible for managing the data transfer among all system components; the Control Module, that governs the behaviour of each individual component within the system; the Digital Filter Module is dedicated to digitally processing the acquired signal using the Savitzky-Golay algorithm; the Data Acquisition Module obtains and processes the signal from the biosensor; and, lastly, the User Web Interface Module, which purpose is serving as the user interface for the operator, allowing for seamless interaction with the multiplex-potentiostat system. A scheme of the software design can be seen in the
Scheme 2.
2.3.5.1. Main Controller Board
The primary objective of the Main Controller Board is to offer an affordable and portable device. To achieve this goal, the design emphasizes modularity, aiming to utilize commercially available modules as much as possible. This approach reduces the need for custom electronic boards, thus lowering manufacturing costs. Among the various modules within the system, the controller module is particularly complex and expensive. In the prototyping phase, a complete computer was initially used. However, to reduce costs, a commercial development board was chosen as the controller.
Several development boards, including
Arduino YUN,
BeagleBone Black, and
Raspberry Pi model 3B+ [
17,
18,
19], were evaluated to determine the most suitable option for the project. Considerations such as communication and processing capabilities, as well as flexibility in software development and maintenance, were considered during the selection process. Based on the analysis, the
Raspberry Pi 3B+ development board emerged as the most suitable choice due to its high core speed, communication capabilities, and cost-effectiveness. Although it lacks analog ports, this issue can be easily resolved by incorporating an Analog-to-Digital converter, which will be discussed later.
An additional advantage of the selected development board is that it features HDMI and USB ports, enabling standalone usage by simply connecting a screen, mouse, and keyboard to it. To utilize the Raspberry Pi 3B+ board, a few additional elements are required, including a μSD card with a minimum capacity of 4 GB and a 5V μUSB power adapter with a current output of 2.5A is also necessary. Additionally, an Ethernet cable can be used to connect to the board through SSH protocol, or alternatively, an HDMI screen with a USB mouse and keyboard can be employed.
2.3.5.2. Data Acquisition Board
The Data Acquisition Board serves as an integration point for essential electronic components involved in the sensing and physical treatment of the signal generated during the chemical reaction. These components are configured and controlled by the Controller Board, and the captured signal is subsequently returned to it for further processing. The primary modules housed on the Data Acquisition Board includes the Data Multiplexing Module, which enables the routing of the signal from the electrochemical cell to the main controller board using the TDM technique; the Signal Acquisition module, that consists on several signal conditioners; The Signal Conversion module, which includes analog-to-digital converters (ADC) and digital-to-analog converters (DAC); and a set of non-configurable components that contributes to maintaining a low noise level within the system.
To facilitate the required functionalities and interactions with the Controller Board, the Data Acquisition Board incorporates I²C communication bus [
20] through the SCL (Serial Clock) and SDA (Serial Data) ports. This communication enables the programming of the components and the retrieval of measurement data from the sensor.
The multiplexing module is the core circuit that allows performing multiple measures using the same device. It is based on a key component that is the Texas Instruments PCA9548 8-channel I²C-bus multiplexer. This chip plays a crucial role as it provides up to eight channel communications, which enables having up to eight electrochemical cells and, thus, perform up to eight measures on distinct analytes at the same execution. This enables the system to analyze multiple samples simultaneously, understanding the simultaneity not as parallel execution but as obtaining all results at the end of an operational window, with a maximum capacity of 8 samples. The multiplexer works using a TDM technique, by selectively exposing one of the connected signal conditioners, connected to a chemical cell associated with it, routing the I²C bus signals of the desired channel to the Controller Board. This functionality allows for the parallel processing and analysis of multiple samples, providing greater efficiency and throughput in the system. By leveraging the capabilities of the PCA9548 multiplexer, the system gains the flexibility to handle a higher number of samples and perform several measurements on them, enhancing the overall performance and productivity of the electrochemical sensing setup. One additional reason to use the multiplexing technique is that the signal acquisition module, LMP91000, has a non-configurable I²C address, which means that without a multiplexing technique, the Controller Board would be able to work only with one electrochemical cell.
The signal acquisition module is based on a Texas Instruments LMP91000 configurable AFE Potentiostat, specially designed for low-power chemical sensing applications, was chosen as the signal conditioner. This component plays a critical role in conditioning and preparing the raw signal for further processing. It may involve amplification, filtering and any necessary adjustments to ensure the signal is suitable for subsequent conversion and analysis. Furthermore, the signal conditioner requires a reference voltage, which is supplied through the Vref port. This reference voltage is essential for performing a potential sweep on the chemical sample, enabling precise control and measurement of the electrochemical reaction. To ensure the integrity and quality of the acquired signal, the signal conditioner incorporates a capacitor connected to ports C1 and C2. This capacitor plays a vital role in analog filtering, effectively attenuating electrical noise and disturbances from the signal. The result is an output signal, available at Vout port, that is as clean and noise-free as possible, enhancing the accuracy and reliability of the measurements.
The Raspberry Pi 3B+ development board chosen for the system lacks analog GPIOs, which means it can only handle digital signals. To overcome this limitation, the signal conversion module is designed using analog-to-digital converters (ADC) and digital-to-analog converters (DAC). The first one, Texas Instruments ADS1115, is responsible for converting the analog signal received from the biosensor, Vout port of the signal conditioner, into a digital format that can be processed by the Controller Board. This component stands out for its 16-bit resolution, which translates to greater precision in working with the signals, and sigma-delta conversion procedure, which offers several advantages, including inherent linearity, monotonicity, and minimal requirements for external anti-aliasing filters. The digital nature of this conversion method ensures that the performance remains stable over time and temperature. However, it requires complex digital circuitry and operates at an oversampled rate significantly higher than the maximum signal bandwidth. Conversely, the second one, Microchip MCP4728, is utilized to convert digital signals from the Controller Board into analog signals, Vref signal, used by the signal conditioners. These integrated circuits are specifically designed to operate with high precision and low noise, which are crucial factors for handling weak signals and minimizing electrical noise susceptibility. This component incorporates a high-precision output amplifier, enabling precise and stable output voltage generation.
Additionally, the Data Acquisition Board incorporates various non-configurable components that contribute to maintaining a low noise level in the overall system. These components work in tandem to reduce or eliminate unwanted electrical noise, interference, or disturbances that could adversely affect the quality and accuracy of the acquired signal. Capacitors are involved in the power line to ensure a smooth and clean power signal throughout the circuit, also, decoupling capacitors are placed at the power entry of each integrated circuit on the board to clean the power signal for each chip. Moreover, Texas Instruments OPA388 Precision Operational Amplifiers (OPAs), are placed in buffer configuration. These OPAs are strategically positioned between the signal conditioners and the ADC/DAC integrated circuits. This component is designed to meet high precision and ultra-low noise requirements, and it integrates filters to mitigate electromagnetic and radiofrequency interference.
Together, these components as a whole serve as a vital intermediary between the physical sensing of the chemical reaction and the digital processing performed by the Controller Board. They enable efficient signal capture, conditioning and conversion, ensuring that the acquired signal is of sufficient quality and fidelity for subsequent analysis and interpretation by the system.
2.3.5.4. Software design
The software implementation of the device can be divided into two main blocks: the backend and the frontend. The backend comprises all the software necessary for managing the system but is not directly accessed by the operator. This includes the communication protocols between the various components of the device, internal controls of each chip, digital filtering algorithms, and data acquisition algorithms. Thus, it ensures that the commands are correctly interpreted and executed. On the other hand, the frontend encompasses the software that allows the operator to interact with and manage the system. This includes a user web-based interface where the operator can visualize the results and control various aspects of the POC device. The frontend module is designed to be user-friendly and accessible, enabling operators to work from any device, such as computer, tablet, or mobile phone.
Together, these software modules collectively contribute to the efficient functioning and usability of the system, enabling seamless communication, precise signal treatment, accurate data acquisition, and an intuitive user experience.
2.3.5.4.1. Communications Module
The Communications Module plays a crucial role in facilitating communication between different components of the POC device and enabling interaction with the operator through the user interface. This module utilizes various protocols to ensure effective and reliable data transfer.
The I²C protocol is utilized for communication between the Controller Board and the Data Acquisition Board. It enables the Controller Board to configure and control the multiplexer, signal conditioner, ADC, and DAC on the Data Acquisition Board. Through I²C, the Controller Board can send commands and receive data from these components, facilitating the overall operation of the POC device.
TCP (Transmission Control Protocol) is employed for data transfer between the operator's user interface (UI) and the device. HTTP (Hypertext Transfer Protocol) is specifically used within TCP to enable the operator to interact with the POC device through a web-based UI. The HTTP protocol allows the operator to send commands, configure device settings, and retrieve information from the device in a user-friendly manner.
In addition to HTTP, the Communications Module utilizes WebSocket, which is built on top of TCP, to achieve real-time data display on the UI plot part. WebSocket enables a persistent, full-duplex communication channel between the UI and the device. This allows the acquired data to be streamed and displayed on the UI plot in real time, providing the operator with immediate feedback and visualization of the measurements.
By leveraging these protocols, the Communications Module ensures seamless communication between the different components of the POC device and facilitates user interaction through a web-based UI. This enhances the usability and functionality of the device, enabling efficient configuration, control, and real-time data visualization for the operator.
2.3.5.4.2. Control Module
The Control Module of the software is responsible for handling the commands received from the operator through the web-based UI and configuring the internal registers of the components on the Data Acquisition Board accordingly. Here's an overview of the configuration requirements:
Multiplexer (PCA9548): The Control Module configures the multiplexer to select the desired channel corresponding to a specific signal conditioner on the Data Acquisition Board. This allows the system to choose which signal conditioner to expose for measurement, allowing analyzing up to 8 samples simultaneously, remember that simultaneously means obtaining all results at the end of the same operational window.
-
Signal Conditioner (LMP91000): The Control Module configures internal registers of the signal conditioner based on the operator's commands. This configuration includes:
- -
Transimpedance Control Register (TIACN): This register sets the amplifier gain (RTIA) and the load resistance (RLOAD).
- -
Reference Control Register (REFCN): This register determines the voltage reference (Vref) source (internal or external), sets the internal zero value as a percentage of Vref, and configures the bias sign and value as a percentage of Vref.
- -
Mode Control Register (MODECN): This register enables or disables the FET feature and selects the operation mode of the sensor.
The Control Module ensures that the signal conditioner is properly configured to provide the desired amplification, reference voltage, biasing, and operating mode for the electrochemical cell.
DAC (Digital-to-Analog Converter): The DAC integrated circuit on the Data Acquisition Board has a single register that needs to be configured by the Control Module. This register sets the desired output voltage, which will be used as the Vref (reference voltage) for the signal conditioner. Having a variable voltage reference makes the device very flexible as it can be configured to analyze a large diversity of samples.
ADC (Analog-to-Digital Converter): The ADC integrated circuit on the Data Acquisition Board also requires configuration, but these settings are handled automatically and are not accessible for operator modification. The Control Module ensures that the ADC is properly calibrated and set up during system startup, allowing it to accurately convert the analog signal obtained from the signal conditioner.
By properly configuring the registers of each component, the Control Module ensures that the Data Acquisition Board is correctly set up for signal conditioning, conversion and measurement of the electrochemical cell's output.
2.3.5.4.3. Digital Filter Module
The Digital Filter Module in the software implementation is embedded in the Data Acquisition Module, but it is relevant enough to dedicate this paragraph to it. This module utilizes a
Savitzky-Golay [
21,
22,
23,
24,
25] filter to enhance the precision of the data obtained from the ADC. The
Savitzky-Golay filter is a type of digital
Finite Impulse Response (FIR) filter that aims to improve the accuracy of the data without significantly distorting the signal trend.
The filter operates through a process known as convolution, where successive subsets of adjacent data points are fitted with a low-degree polynomial using the method of linear least squares. By finding a set of "convolution coefficients," which can be determined analytically when the data points are equally spaced, the filter estimates the smoothed signal at the central point of each subset.
The original work by Savitzky and Golay demonstrated that the smoothed output value obtained by sampling the fitted polynomial is equivalent to a fixed linear combination of the local set of input samples. This means that the output samples can be computed through discrete convolution.
The Savitzky-Golay filter has two key parameters: the window length, which determines the number of samples used for the filtering process, and the polynomial order, which specifies the degree of the fitting polynomial. These parameters are calibrated and set up in the Control Module of the software, ensuring optimal filtering performance. However, they are not accessible for modification by the operator.
By applying the Savitzky-Golay filter, the Digital Filter Module enhances the representation of the signal, providing a smoother and more precise visualization of the data for the operator.
2.3.5.4.4. Data Acquisition Module
The Data Acquisition Module is responsible for sampling the Vout output signal from the signal conditioner and processing it for various purposes.
Firstly, the module is responsible for presenting the acquired data on the user interface (UI), allowing the operator to monitor the measurements and observe any changes or trends in the data.
Additionally, the Data Acquisition Module facilitates data storage for future reference and analysis. The acquired data is saved in a CSV (Comma-Separated Values) file format, which is a common format that can be easily accessed and processed by various third-party applications such as spreadsheet software or data analysis tools. Storing the data in a CSV file enables the operator to retrieve and utilize the data outside of the POC device, enhancing its usability and compatibility with other applications or analysis workflows.
2.3.5.4.5. User Web Interface Module
The User Web Interface Module provides a user-friendly and intuitive control panel to the operator, enabling them to configure various parameters of the system with ease. The interface offers a simple and interactive interface where the operator can make selections or click buttons to adjust settings or choose from pre-set configurations.
The User Web Interface Module also facilitates the visualization of the analysis results on a graph, allowing the operator to easily interpret and analyze the data obtained from the biosensor. The graph provides a visual representation of the measurements, trends, and changes over time, aiding in the understanding of the analyzed samples.
One of the key advantages of the User Web Interface Module is its accessibility. Being a web-based interface, it can be accessed from any device that is connected to the same local network as the POC device. This eliminates the dependency on a specific computer or the need for installing third-party drivers, making it highly convenient to use. The operator can access and control the system from their computer, laptop, tablet, or even a mobile phone, providing flexibility and ease of use.
The User Web Interface Module enhances the overall user experience by offering a user-friendly interface, real-time visualization of results, and accessibility from various devices within the local network. It simplifies the control and monitoring of the system, making it more convenient and efficient for the operator.