Submitted:
14 September 2023
Posted:
15 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Cell Culture and Media
2.3. Bacterial Infection and Adherence
2.4. Isolation of Bacterial RNA from Infected Cells and mRNA Enrichment
2.5. Illumina Library Preparation and RNA Sequencing
2.6. Sequence Analysis and Detection of Differential Gene Expression
2.7. Statistical Analysis, Software, and Data Preparation
2.8. Data Availability
3. Results
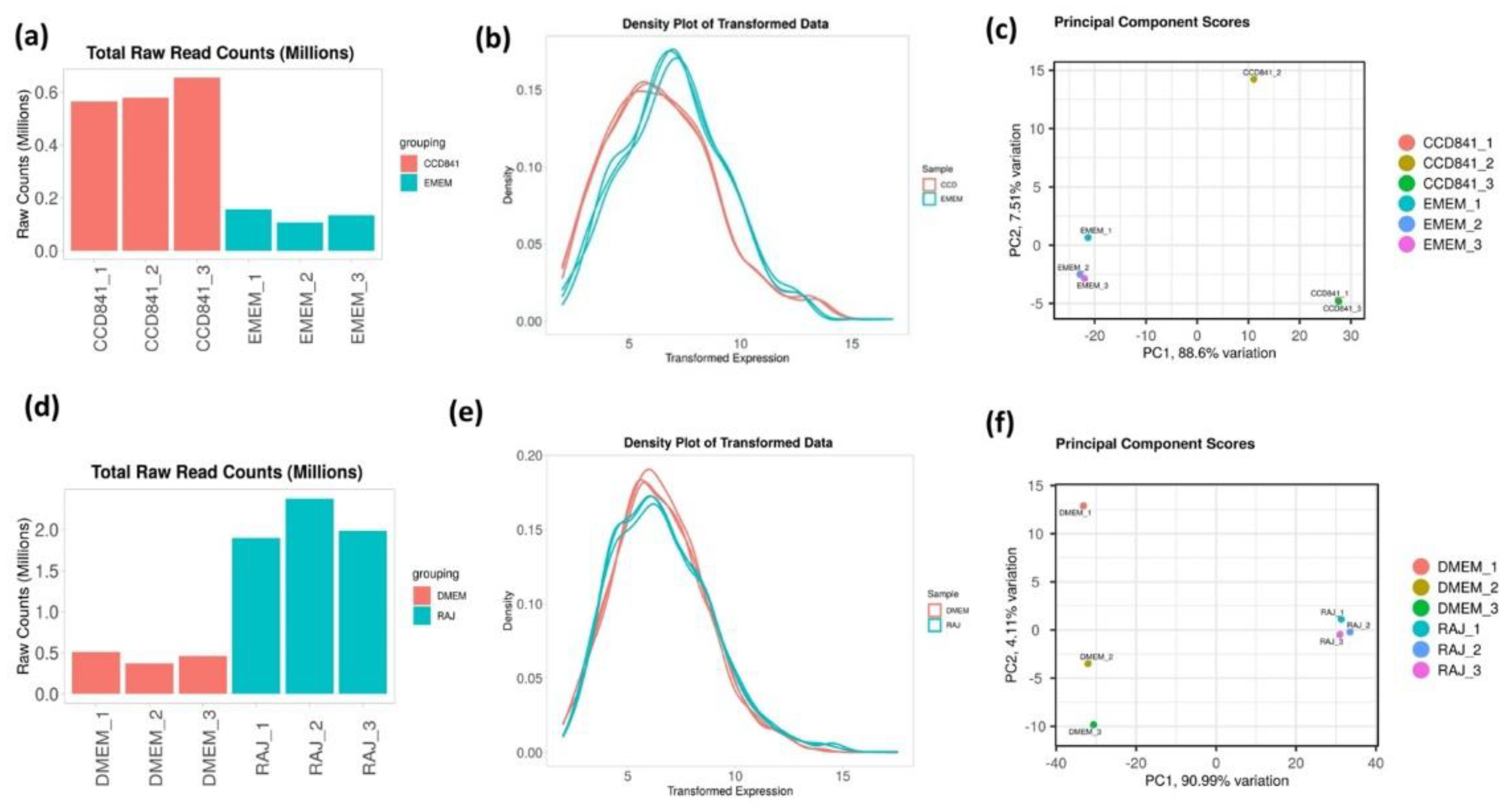
3.1. Overall transcriptomics profiles of E. coli O157:H7 cultured with CCD CoN 841 and RAJ cells during initial adherence
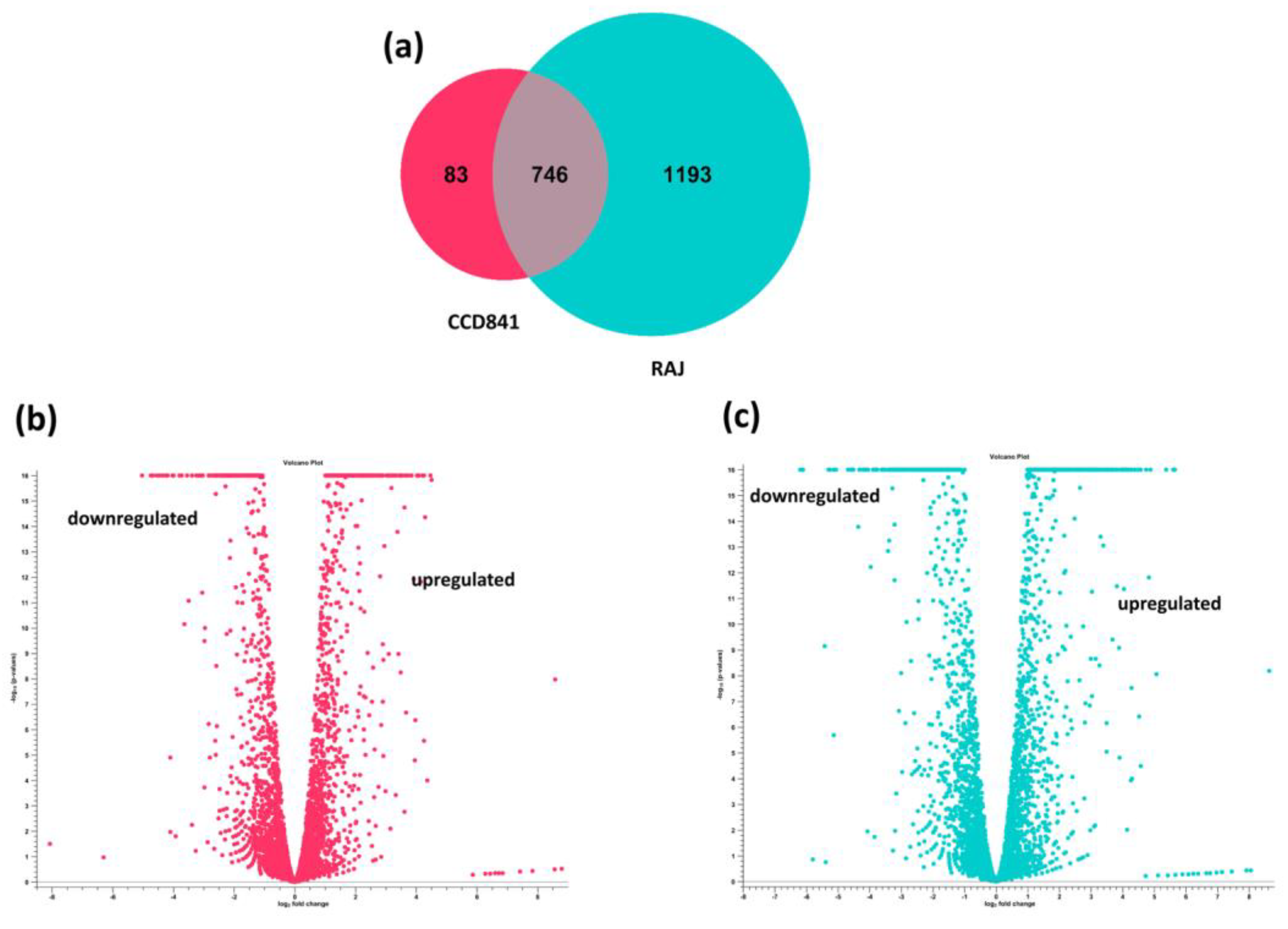
3.2. Overall Transcriptomics Differences of E. coli O157:H7 cultured with CCD CoN 841 and RAJ Cells during Initial Adherence
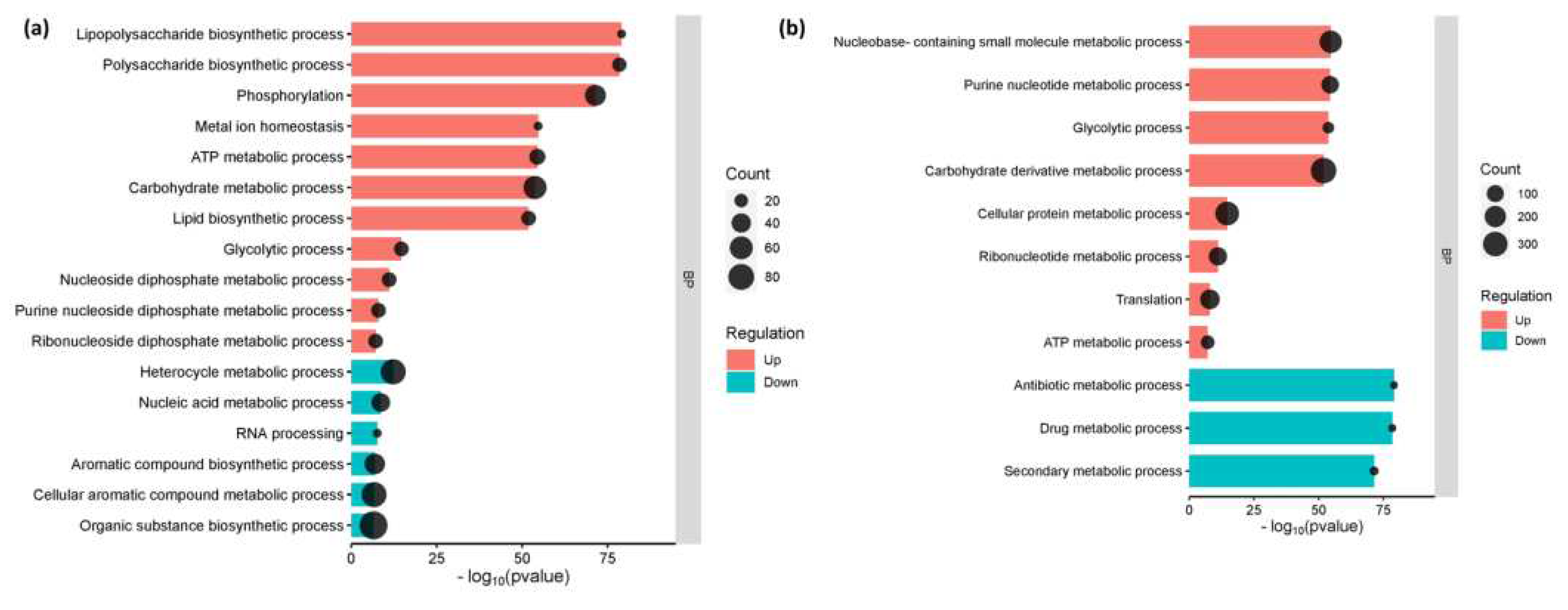
3.3. The Enriched Biological Pathways During the Initial Adherence of E. coli O157:H7 in CCD CoN 841 and RAJ cells
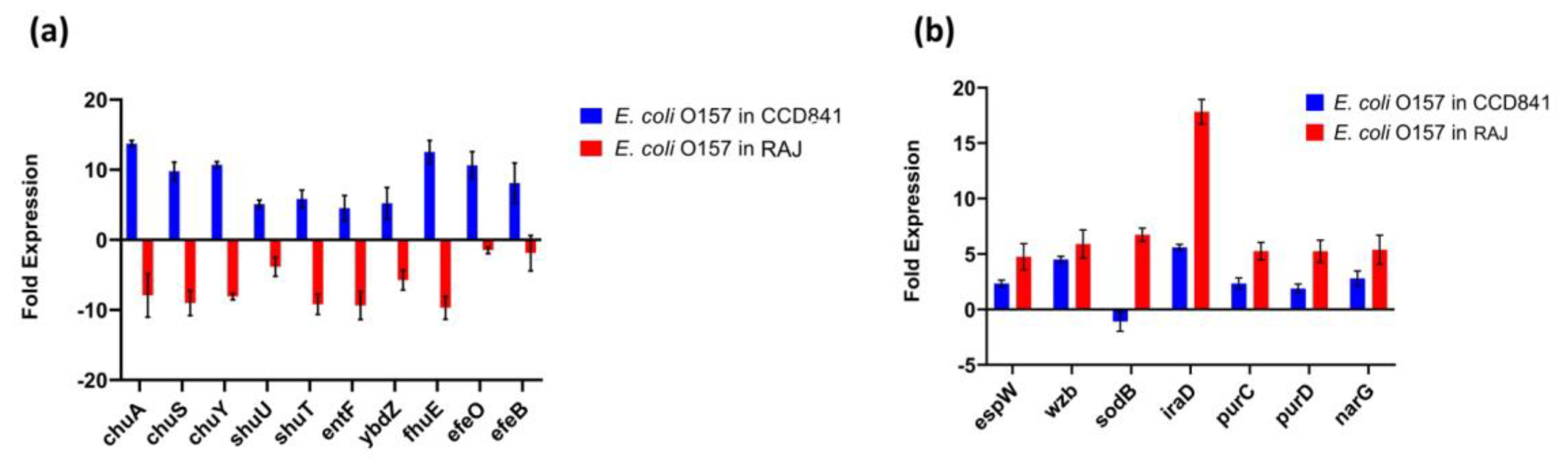
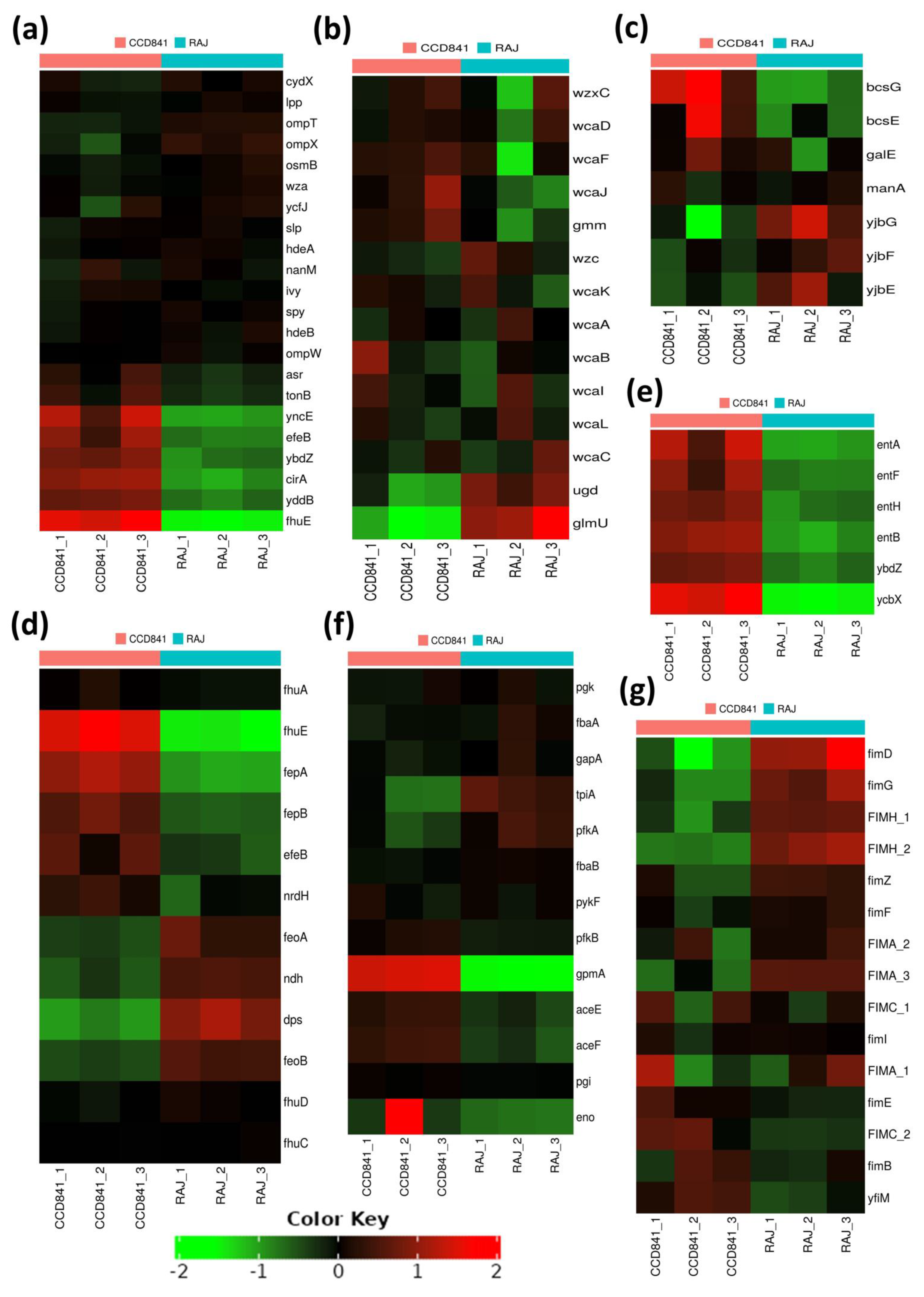
3.4. Functional Annotation of E. coli O157:H7 Genes Differentially Expressed in Two Different Host Cell Types
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
References
- Rangel, J. M.; Sparling, P. H.; Crowe, C.; Griffin, P. M.; Swerdlow, D. L. Epidemiology of Escherichia coli O157:H7 Outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 2005, 11(4), 603–609. [Google Scholar] [CrossRef]
- Tarr, P. I.; Gordon, C. A.; Chandler, W. L. Shiga-Toxin-Producing Escherichia coli and Haemolytic Uraemic Syndrome. Lancet 2005, 365(9464), 1073–1086. [Google Scholar] [CrossRef]
- Karmali, M. A.; Steele, B. T.; Petric, M.; Lim, C. Sporadic Cases of Haemolytic-Uraemic Syndrome Associated with Faecal Cytotoxin and Cytotoxin-Producing Escherichia coli in Stools. Lancet 1983, 1(8325), 619–620. [Google Scholar] [CrossRef]
- Matthews, L.; Low, J. C.; Gally, D. L.; Pearce, M. C.; Mellor, D. J.; Heesterbeek, J. A.; Chase-Topping, M.; Naylor, S. W.; Shaw, D. J.; Reid, S. W.; Gunn, G. J.; Woolhouse, M. E. Heterogeneous Shedding of Escherichia coli O157 in Cattle and Its Implications for Control. Proc. Natl. Acad. Sci. U. S. A. 2006, 103(3), 547–552. [Google Scholar] [CrossRef]
- Friedrich, A. W.; Bielaszewska, M.; Zhang, W. L.; Pulz, M.; Kuczius, T.; Ammon, A.; Karch, H. Escherichia coli Harboring Shiga Toxin 2 Gene Variants: Frequency and Association with Clinical Symptoms. J. Infect. Dis. 2002, 185(1), 74–84. [Google Scholar] [CrossRef]
- Lim, J. Y.; Yoon, J.; Hovde, C. J. A Brief Overview of Escherichia coli O157:H7 and Its Plasmid O157. J. Microbiol. Biotechnol. 20(1), 5–14. [CrossRef]
- Bosilevac, J. M.; Guerini, M. N.; Kalchayanand, N. , et al. Prevalence and Characterization of Non-O157 Shiga Toxin-Producing Escherichia coli Isolates from Commercial Ground Beef in the United States. Appl. Environ. Microbiol. 2007, 73 (18), 6365–6371.
- Mir, R. A.; Schaut, R. G.; Looft, T.; Allen, H. K.; Sharma, V. K.; Kudva, I. T. Recto-Anal Junction (RAJ) and Fecal Microbiomes of Cattle Experimentally Challenged with Escherichia coli O157:H7. Front. Microbiol. 11, 693. [CrossRef]
- Naylor, S. W.; Low, J. C.; Besser, T. E.; Mahajan, A.; Gunn, G. J.; Pearce, M. C.; McKendrick, I. J.; Smith, D. G.; Gally, D. L. Lymphoid Follicle-Dense Mucosa at the Terminal Rectum Is the Principal Site of Colonization of Enterohemorrhagic Escherichia Coli O157:H7 in the Bovine Host. Infect. Immun. 2003, 71(3), 1505–1512. [Google Scholar] [CrossRef]
- Hancock, D. D.; Besser, T. E.; Rice, D. H.; Ebel, E. D.; Herriott, D. E.; Carpenter, L. V. Multiple Sources of Escherichia coli O157 in Feedlots and Dairy Farms in the Northwestern USA. Prev. Vet. Med. 1998, 35(1), 11–19. [Google Scholar] [CrossRef]
- Hancock, D. D.; Besser, T. E.; Kinsel, M. L. , et al. The Prevalence of Escherichia coli O157. H7 in Dairy and Beef Cattle in Washington State. Epidemiol. Infect. 2001, 127(2), 293–302. [Google Scholar]
- Garmendia, J.; Frankel, G.; Crepin, V. F. Enteropathogenic and Enterohemorrhagic Escherichia Coli Infections: Translocation, Translocation, Translocation. Infect. Immun. 2005, 73(5), 2573–2585. [Google Scholar] [CrossRef]
- Lawley, T. D.; Clare, S.; Walker, A. W. , et al. Targeted Restoration of the Intestinal Microbiota with a Simple, Defined Bacteriotherapy Resolves Relapsing Clostridium difficile Disease in Mice. PLOS Pathog. 2013, 9 (10), e1002995.
- Govindarajan, D. K.; Viswalingam, N.; Meganathan, Y.; Kandaswamy, K. Adherence Patterns of Escherichia coli in the Intestine and Its Role in Pathogenesis. Medicine in Microecology 2020, 5. [Google Scholar] [CrossRef]
- Pakbin, B.; Brück, W. M.; Rossen, J. W. A. Virulence Factors of Enteric Pathogenic Escherichia coli: A Review. Int. J. Mol. Sci. 2021 , 22 (18), 9922. 14 September. [CrossRef]
- Nguyen, Y.; Sperandio, V. Enterohemorrhagic E. coli (EHEC) Pathogenesis. Front. Cell. Infect. Microbiol. 2012 , 2, 90. 12 July. [CrossRef]
- Lai, Y.; Rosenshine, I.; Leong, J. M.; Frankel, G. Intimate Host Attachment: Enteropathogenic and Enterohaemorrhagic Escherichia Coli. Cell. Microbiol. 15(11), 1796–1808. [CrossRef]
- Galán, J. E.; Wolf-Watz, H. Protein Delivery into Eukaryotic Cells by Type III Secretion Machines. Nature 2006, 444(7119), 567–573. [Google Scholar] [CrossRef]
- Fedorchuk, C.; Kudva, I. T.; Kariyawasam, S. The Escherichia coli O157:H7 Carbon Starvation-Inducible Lipoprotein Slp Contributes to Initial Adherence In Vitro via the Human Polymeric Immunoglobulin Receptor. PLOS ONE 14(6), e0216791. [CrossRef]
- Barh, D.; Kumar, A.; Tiwari, S. , et al. A Novel Comparative Genomics Analysis for Common Drug and Vaccine Targets in Corynebacterium pseudotuberculosis and Other CMN Group of Human Pathogens. Chem. Biol. Drug Des. 2013, 81(4), 60–69. [Google Scholar]
- Grin, I.; Linke, D.; Schütz, M. S. Controlling the Message: A Structural Analysis of Protein Sorting to the TpsB/Omp85 Superfamily. Curr. Opin. Struct. Biol. 2014, 29, 1–8. [Google Scholar]
- Kudva, I. T.; Dean-Nystrom, E. A. Bovine Recto-Anal Junction Squamous Epithelial (RSE) Cell Adhesion Assay for Studying Escherichia coli O157 Adherence. J. Appl. Microbiol. 2011, 111(5), 1283–1294. [Google Scholar] [CrossRef]
- Kudva, IT. In vitro adherence patterns of Shigella serogroups to bovine recto-anal junction squamous epithelial (RSE) cells are similar to those of Escherichia coli O157. Foodborne Pathog Dis. 2012;9(4):346-351. [CrossRef]
- Pawitan, Y.; Michiels, S.; Koscielny, S.; Gusnanto, A.; Ploner, A. False Discovery Rate, Sensitivity and Sample Size for Microarray Studies. Bioinformatics 21(13), 3017–3024. [CrossRef]
- Ge, S. X.; Son, E. W.; Yao, R. iDEP: An Integrated Web Application for Differential Expression and Pathway Analysis of RNA-Seq Data. B.M.C. Bioinformatics 2018, 19(1), 534. [Google Scholar] [CrossRef]
- Blankenship, H. M.; Mosci, R. E.; Dietrich, S.; Burgess, E.; Wholehan, J.; McWilliams, K.; Pietrzen, K.; Benko, S.; Gatesy, T.; Rudrik, J. T.; Soehnlen, M.; Manning, S. D. Population Structure and Genetic Diversity of Non-O157 Shiga Toxin-Producing Escherichia coli (STEC) Clinical Isolates from Michigan. Sci. Rep. 2021, 11(1), 4461. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S. E.; Scallan, E.; Jones-Bitton, A.; Sargeant, J. M.; Stapleton, J.; Angulo, F. J.; Yeung, D. H.; Kirk, M. D. Global Incidence of Human Shiga Toxin-Producing Escherichia coli Infections and Deaths: A Systematic Review and Knowledge Synthesis. Foodborne Pathog. Dis. 11(6), 447–455. [CrossRef] [PubMed]
- Hedican, E. B.; Medus, C.; Besser, J. M.; Juni, B. A.; Koziol, B.; Taylor, C.; Smith, K. E. Characteristics of O157 Versus Non-O157 Shiga Toxin-Producing Escherichia coli Infections in Minnesota, 2000–2006. Clin. Infect. Dis. 2009, 49(3), 358–364. [Google Scholar] [CrossRef]
- Adegbola, R. A. Bacterial Adhesion and Pathogenicity. Afr. J. Med. Med. Sci. 1988, 17(2), 63–69. [Google Scholar] [PubMed]
- Rahal, E. A.; Kazzi, N.; Nassar, F. J.; Matar, G. M. Escherichia coli O157:H7—Clinical Aspects and Novel Treatment Approaches. Front. Cell. Infect. Microbiol. 2012, 2, 138. [Google Scholar] [CrossRef]
- Paquin-Veillette, J.; Dozois, C. M.; Harel, J. The ecological habitat and transmission of Escherichia coli O157:H7. Hekabab SM. F.E.M.S. Microbiol. Lett. 2013, 341(1), 1–12. [Google Scholar] [CrossRef]
- Segura, A.; Bertin, Y.; Durand, A.; Benbakkar, M.; Forano, E. Transcriptional Analysis Reveals Specific Niche Factors and Response to Environmental Stresses of Enterohemorrhagic Escherichia Coli O157:H7 in Bovine Digestive Contents. B.M.C. Microbiol. 2021, 21(1), 284. [Google Scholar] [CrossRef]
- Dresen, M.; Valentin-Weigand, P.; Berhanu Weldearegay, Y. Role of Metabolic Adaptation of Streptococcus suis to Host Niches in Bacterial Fitness and Virulence. Pathogens 2023, 12, 541. [Google Scholar] [CrossRef]
- Torres, A. G.; Payne, S. M. Haem Iron-Transport System in Enterohaemorrhagic Escherichia Coli O157:H7. Mol. Microbiol. 1997, 23(4), 825–833. [Google Scholar] [CrossRef]
- Sheldon, J. R.; Laakso, H. A.; Heinrichs, D. E. Iron Acquisition Strategies of Bacterial Pathogens. Microbiol. Spectr. 2016 April, 4 (2). [CrossRef]
- Seyedsayamdost, M. R. Toward a Global Picture of Bacterial Secondary Metabolism. J. Ind. Microbiol. Biotechnol. 2019 March, 46 (3–4), 301–311. [CrossRef]
- Baral, B.; Akhgari, A.; Metsä-Ketelä, M. Activation of Microbial Secondary Metabolic Pathways: Avenues and Challenges. Synth. Syst. Biotechnol. 2018 , 3 (3), 163–178. 12 September. [CrossRef]
- Hammer, N. D.; Skaar, E. P. Molecular Mechanisms of Staphylococcus aureus Iron Acquisition. Annu. Rev. Microbiol. 2011, 65, 129–147. [Google Scholar] [CrossRef]
- Nagy, G.; Dobrindt, U.; Kupfer, M.; Emödy, L.; Karch, H.; Hacker, J. Expression of Hemin Receptor Molecule ChuA Is Influenced by RfaH in Uropathogenic Escherichia coli strain 536. Infect. Immun. 69(3), 1924–1928. [CrossRef] [PubMed]
- Gao, Q.; Wang, X.; Xu, H.; Xu, Y.; Ling, J.; Zhang, D.; Gao, S.; Liu, X. Roles of Iron Acquisition Systems in Virulence of Extraintestinal Pathogenic Escherichia coli: Salmochelin and Aerobactin Contribute More to Virulence than Heme in a Chicken Infection Model. B.M.C. Microbiol. 2012, 12, 143. [Google Scholar] [CrossRef]
- Chandrangsu, P.; Rensing, C.; Helmann, J. D. Metal Homeostasis and Resistance in Bacteria. Nat. Rev. Microbiol. 15(6), 338–350. [CrossRef] [PubMed]
- Burcham, L. R.; Hill, R. A.; Caulkins, R. C.; Emerson, J. P.; Nanduri, B.; Rosch, J. W.; Fitzkee, N. C.; Thornton, J. A. Streptococcus pneumoniae Metal Homeostasis Alters Cellular Metabolism. Metallomics 2020 , 12 (9), 1416–1427. 23 September. [CrossRef]
- Sandu, P.; Crepin, V. F.; Drechsler, H.; McAinsh, A. D.; Frankel, G.; Berger, C. N. The Enterohemorrhagic Escherichia Coli Effector EspW Triggers Actin Remodeling in a Rac1-Dependent Manner. Infect. Immun. 85(9), e00244–17. [CrossRef] [PubMed]
- Kudva, I. T.; Hovde, C. J.; John, M. Adherence of Non-O157 Shiga Toxin-Producing Escherichia coli to Bovine Recto-Anal Junction Squamous Epithelial Cells Appears to Be Mediated by Mechanisms Distinct from Those Used by O157. Foodborne Pathog. Dis. 10(4), 375–381. [CrossRef]
- Hagelueken, G.; Huang, H.; Mainprize, I. L.; Whitfield, C.; Naismith, J. H. Crystal Structures of Wzb of Escherichia coli and CpsB of Streptococcus pneumoniae, Representatives of Two Families of Tyrosine Phosphatases That Regulate Capsule Assembly. J. Mol. Biol. 2009 , 392 (3), 678–688. 25 September. [CrossRef]
- Nie, D.; Hu, Y.; Chen, Z.; Li, M.; Hou, Z.; Luo, X.; Mao, X.; Xue, X. Outer Membrane Protein A (OmpA) as a Potential Therapeutic Target for Acinetobacter baumannii Infection. J. Biomed. Sci. 2020, 27(1), 26. [Google Scholar] [CrossRef]
- Lin, J.; Huang, S.; Zhang, Q. Outer Membrane Proteins: Key Players for Bacterial Adaptation in Host Niches. Microbes Infect. 4(3), 325–331. [CrossRef]
- Balducci, E.; Papi, F.; Capialbi, D. E.; Del Bino, L. Polysaccharides’ Structures and Functions in Biofilm Architecture of Antimicrobial-Resistant (AMR) Pathogens. Int. J. Mol. Sci. 2023, 24(4), 4030. [Google Scholar] [CrossRef]
- Limoli, D. H.; Jones, C. J.; Wozniak, D. J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015 June, 3 (3). [CrossRef]
- Davey, M. E.; O’Toole, G. A. Microbial Biofilms: From Ecology to Molecular Genetics. Microbiol. Mol. Biol. Rev. 64(4), 847–867. [CrossRef]
- Wang, R.; Bono, J. L.; Kalchayanand, N.; Shackelford, S.; Harhay, D. M. Biofilm Formation by Shiga Toxin-Producing Escherichia coli O157:H7 and Non-O157 Strains and Their Tolerance to Sanitizers Commonly Used in the Food Processing Environment. J. Food Prot. 2012, 75(8), 1418–1428. [Google Scholar] [CrossRef]
- Raymond, K. N.; Dertz, E. A.; Kim, S. S. Enterobactin: An Archetype for Microbial Iron Transport. Proc. Natl. Acad. Sci. U. S. A. 100(7), 3584–3588. [CrossRef] [PubMed]
- Saha, P.; Xiao, X.; Yeoh, B. S.; Chen, Q.; Katkere, B.; Kirimanjeswara, G. S.; Vijay-Kumar, M. The Bacterial Siderophore Enterobactin Confers Survival Advantage to Salmonella in Macrophages. Gut Microbes 2019, 10(3), 412–423. [Google Scholar] [CrossRef] [PubMed]
- Cassat, J. E.; Skaar, E. P. Iron in Infection and Immunity. Cell Host Microbe 13(5), 509–519. [CrossRef] [PubMed]
- Mandlik, A.; Swierczynski, A.; Das, A.; Ton-That, H. Pili in Gram-Positive Bacteria: Assembly, Involvement in Colonization and Biofilm Development. Trends Microbiol. 16(1), 33–40. [CrossRef]
- Zeiner, S. A.; Dwyer, B. E.; Clegg, S. FimA, FimF, and FimH Are Necessary for Assembly of Type 1 Fimbriae on Salmonella enterica serovar Typhimurium. Infect. Immun. 80(9), 3289–3296. [CrossRef]
- Gahlot, D. K.; Taheri, N.; MacIntyre, S. Diversity in Genetic Regulation of Bacterial Fimbriae Assembled by the Chaperone Usher Pathway. Int. J. Mol. Sci. 24(1), 161. [CrossRef]
- Schwan, W. R. Regulation of fim Genes in Uropathogenic Escherichia coli. World J. Clin. Infect. Dis. 1(1), 17–25. [CrossRef]
- Katani R, Kudva, IT, Srinivasan S, Stasko JB, Schilling M, Li L, Cote R, DebRoy C, Arthur TM, Sokurenko EV, and Kapur V. Strain and host-cell type dependent role of type 1 fimbriae genes in the adherence phenotype of super-shedder strains of Escherichia coli O157:H7. Int. J. Med. Micro. 2021. [CrossRef]
- Kendall, M.M. , Gruber, C., Parker, C., & Sperandio, V. (2012). Ethanolamine Controls Expression of Genes Encoding Components Involved in Interkingdom Signaling and Virulence in Enterohemorrhagic Escherichia coli O157:H7. mBio, 3.
- Rowley CA, Anderson CJ, Kendall MM. Ethanolamine Influences Human Commensal Escherichia coli Growth, Gene Expression, and Competition with Enterohemorrhagic E. coli O157:H7. mBio. 2018 Oct 2;9(5):e01429-18. [CrossRef]
| E. coli O157:H7 adhering to CCD CoN 841 cells | ||||
|---|---|---|---|---|
| Symbol | Log2 FC | Location | Type | Gene Name |
| ndhF | 4.224748 | membrane | dehydrogenase | Type II NADH:quinone oxidoreductase |
| ynfM | 3.996075 | membrane | transporter | Inner membrane transport protein YnfM |
| yagU | 3.99386 | membrane | unknown | Inner membrane protein YagU |
| wcaJ | 3.745511 | membrane | transferase | UDP-glucose: undecaprenyl-phosphate glucose-1-phosphate transferase |
| fhuE | 3.649405 | cell outer membrane | Iron transporter | Hydroxamate siderophore receptor FhuE |
| yjbF | 3.632319 | membrane | Unknown | Uncharacterized lipoprotein YjbF |
| wza | 3.417282 | cell outer membrane | transporter | Putative polysaccharide export protein Wza |
| efeO | 3.411274 | periplasmic space | transporter | Iron uptake system component EfeO |
| fdhF | 3.283415 | cytosol | dehydrogenase | Formate dehydrogenase H |
| feoB | 3.260966 | membrane | transporter | Fe(2+) transporter FeoB |
| wcaD | 3.235622 | membrane | Unknown | Putative colanic acid polymerase |
| slp | 3.206504 | cell outer membrane | Unknown | Outer membrane protein Slp |
| wzc | 3.199261 | membrane | Kinase | Tyrosine-protein kinase wzc |
| adhE | 3.186165 | cytosol | acetaldehyde dehydrogenase | Bifunctional aldehyde-alcohol dehydrogenase AdhE |
| yohK | 3.087307 | membrane | unknown | Inner membrane protein YohK |
| pflB | 3.080942 | cytoplasm | transferase | Formate acetyltransferase 1 |
| hycC | 3.043606 | membrane | dehydrogenase | Formate hydrogenlyase subunit 3 |
| efeB | 3.017162 | periplasmic space | ferrochelatase | Deferrochelatase |
| glnK | 2.907097 | membrane | enzyme regulator | Nitrogen regulatory protein GlnK |
| amtB | 2.852313 | membrane | transporter | Ammonium transporter AmtB |
| E. coli O157:H7 adhering to RAJ cells | ||||
| yohJ | 4.832419 | membrane | unknown | UPF0299 membrane protein YohJ |
| fdhF | 4.422977 | cytosol | dehydrogenase | Formate dehydrogenase H |
| yagU | 4.28355 | membrane | Unknown | Inner membrane protein YagU |
| hycC | 4.249511 | membrane | dehydrogenase | Formate hydrogenlyase subunit 3 |
| yohK | 4.207073 | membrane | unknown | Inner membrane protein YohK |
| ynfM | 4.193856 | membrane | transporter | Inner membrane transport protein YnfM |
| pflB | 4.165908 | cytoplasm | transferase activity | Formate acetyltransferase 1 |
| yjbF | 4.156806 | membrane | unknown | Uncharacterized lipoprotein YjbF |
| hycD | 3.936897 | membrane | unknown | Formate hydrogenlyase subunit 4 |
| wza | 3.806629 | cell outer membrane | transporter | Putative polysaccharide export protein Wza |
| ndh | 3.8042 | membrane | dehydrogenase | Type II NADH:quinone oxidoreductase |
| btsT | 3.674308 | membrane | transporter | Pyruvate/proton symporter BtsT |
| adiC | 3.625606 | membrane | transporter | Arginine/agmatine antiporter |
| wcaJ | 3.532739 | membrane | transferase | UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferase |
| hycF | 3.352964 | membrane | dehydrogenase | Formate hydrogenlyase subunit 6 |
| tpiA | 3.309538 | cytoplasm | isomerase | Triosephosphate isomerase |
| gapA | 3.309281 | cytoplasm | dehydrogenase | Glyceraldehyde-3-phosphate dehydrogenase A |
| wzc | 3.280554 | membrane | kinase | Tyrosine-protein kinase wzc |
| ynfF | 3.279045 | periplasmic space | reductase | Probable dimethyl sulfoxide reductase chain YnfF |
| ptsG | 3.210761 | membrane | transporter | PTS system glucose-specific EIICB component |
| slp | 2.011981 | cell outer membrane | unknown | Outer membrane protein Slp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





