1. Introduction
A developmental disease, autism is characterized by communication and social interaction issues, and restricted or repetitive behaviors. Autism is a developmental disorder characterized by difficulties in social interaction, verbal and nonverbal communication, and repetitive behaviors. The problem of ASD is rising nowadays among many the human population. The World Health Organization (WHO) estimates that 1 in 68 children have ASD. This means there are more than 2 million people having ASD in the United States and more than 68 million people with ASD around the globe. There is no cure for autism, but early intervention can help kids with autism reach their full potential. For children with autism, the sooner treatment begins the better. Research in Machine Learning (ML) and Dl has been applied to the study of autism. Using the data points gathered from the parent surveys, the researchers classified the data into autistic and non-autistic groups using ML algorithms. Maintaining people’s physical and mental health is considerably aided by early diagnosis of this neurological condition. Predictions of many human diseases can benefit from the growing use of machine learning techniques. The preliminary results, based on a number of health and physiological variables, are currently thought to be feasible. This encouraged us to devote more resources to the diagnosis and investigation of ASD disorders, which in turn improves treatment strategies.
Autism is a brain-related disorder [
1]. A person who has autism spectrum disorder typically struggles with social interaction and communication. Consequences from this tend to last for the rest of a person’s life. The onset of symptoms could occur as early as the age of three and persist throughout an individual’s life. Although there is currently no cure for this condition, early discovery can help mitigate it. Although genetics may play a role in ASD, the underlying causes of the disorder are not yet understood by medical researchers. Low birth weight, having a sibling with autism spectrum disorder, having parents in their later years, etc., are all risk factors for the disorder? As an alternative, consider the following examples of difficulties with social contact and communication.
Sensory sensitivities: Individuals with ASD may experience hypersensitivity or hyposensitivity to sensory stimuli such as lights, sounds, textures, tastes, or smells.
Repetitive behaviors: This can come with repetitive movements (for example, hand flapping and rocking), or repetitive speech patterns.
Special interests: Many individuals with ASD develop intense interests in specific topics or objects, displaying extensive knowledge or fixation on particular subjects.
Difficulties in social communication: People with ASD may have trouble reading and responding to nonverbal signs like facial expressions and body language, which can make even everyday conversations difficult.
Restricted and repetitive interests: Alongside special interests, individuals with ASD may exhibit restricted interests or preoccupation with specific objects or topics.
Executive functioning difficulties: Difficulties in planning, organizing, and problem-solving can impact daily activities, academic performance, and independent living skills.
Sensitivity to change: Changes in routine, environment, or expectations can be particularly distressing for individuals with ASD, leading to anxiety or behavioral outbursts.
ASD is a tough neuro-developmental condition, which affects individuals in different ways. Understanding the causes of autism spectrum disorder can be aided by research into the brains of people with the illness. By gaining a better understanding of ASD, researchers aim to improve diagnosis, treatment, and support for individuals with the condition.
Based on our survey, it has been observed that a good number of methods have been introduced for accurate classification of ASD. Mostly, these methods have been developed using statistical or machine learning approaches. Among these deep learning methods forward more prominent and promising. However, with the evolving tool, library and technological support, there are ample scopes for further enhancements in terms of accuracy, which has motivated us to take up this challenge. Overall, the motivation for this research is likely driven by the goal of advancing the understanding and analysis of ASD using deep learning techniques. By exploring different models, datasets, and methods, researchers aim to contribute to the existing knowledge and potentially improve the diagnosis and treatment of individuals with ASD.
2. Materials and Methods
The main objective of this research is to evaluate and compare the performance of different deep learning models (VGG16, Inception v3, ResNet50,
DenseNet121, and MobileNet) in identifying and analyzing ASD problems in brain MRI data. The study aims to utilize a customized VGG16 architecture and compare its effectiveness with other pre-trained models in detecting ASD-related structural and functional distinctions in the brain. The research will focus on using the ABIDE dataset, which consists of unlabeled brain MRI data from individuals with ASD. The study will employ deep learning methods to cluster and analyze the unlabeled data, aiming to identify patterns or characteristics associated with ASD.
The results obtained from the proposed methods will be compared to the findings of previous studies conducted in the field of ASD research. By comparing the performance of various DL models, this research aims to contribute to the existing knowledge and potentially improve the accuracy and efficiency of ASD detection and analysis using brain MRI data. Overall, the objective of this research is to explore the application of DL techniques, specifically using customized various pre-trained models, in the analysis of brain MRI data for the identification and understanding of ASD, ultimately aiming to advance the understanding and diagnosis of this neuro developmental disorder.
The paper is based on a discussion of constructive method for classification of ASD. Within this frame the paper starts with a brief introduction of autism and its various symptoms followed by motivation of our work and the objective of the research. Further, it discusses the problem definition and overview of the ASD, continued by a comparison of the different DM/ML techniques for ASD classification. Further, it provides an elaborate discussion of proposed methodology module wise. Finally, the study shows the results and discussion and the future scope.
2.1. Problem Definition
For a given test f-MRI images Itest, the problem is to classify it with best possible accuracy with reference to a given set of training instances Itrains. We consider it two class (’Control’ and ’ASD’) problem and Itrains include instances from both the classes. Each f-MRI image is pre-processed and feature engineering is applied to extract ’n’ important features based on the contents of the MRI images and subsequently, out of n available features, ’k’ relevant and non-redundant features are selected to help achieve best possible classification accuracy.
2.2. Autism Spectrum Disorder: An Overview
Autism, a disorder of brain development, has negative effects on social interaction, communication, and behavior. Autistic people have trouble communicating verbally and nonverbally, and they also tend to engage in repeated patterns of behavior and interests. Functional magnetic resonance imaging (fMRI) has been essential in helping researchers understand the brain foundation of autism, even though its precise causes remain unknown. [
2]
fMRI is a powerful imaging technique that measures and maps brain activity by detecting changes in blood oxygenation levels. It has provided valuable insights into the neural functioning of individuals with autism. By comparing fMRI scans of individuals with autism to those without the disorder, researchers have identified several key findings:
2.2.1. Altered Brain Connectivity
fMRI studies have revealed differences in functional connectivity patterns in individuals with autism. Areas of the brain responsible for social interaction, language processing, and emotional regulation often show disrupted connectivity, leading to difficulties in these domains.
2.2.2. Reduced Mirror Neuron System Activation
Mirror neurons play a crucial role in empathy, imitation, and social cognition. Studies using fMRI have shown reduced activation of the mirror neuron system in individuals with autism, which may contribute to difficulties in understanding and imitating others’ actions and emotions.
2.2.3. Atypical Sensory Processing
Many individuals with autism experience sensory sensitivities or difficulties in processing sensory information. fMRI studies have revealed that these sensory processing differences are associated with altered activation patterns in sensory-related brain regions, providing a neurological basis for sensory symptoms in autism.
2.2.4. Enhanced Perceptual Processing
While individuals with autism often struggle with social communication, they may exhibit enhanced perceptual processing abilities. fMRI studies have shown increased activation in visual and auditory processing regions, suggesting that individuals with autism may rely more heavily on perceptual information in their interactions with the world.
2.2.5. Executive Functioning Deficits
Executive functions involve cognitive processes such as planning, decision-making, and impulse control. fMRI studies have demonstrated altered activation in brain regions associated with executive functioning in individuals with autism, providing insights into the difficulties they face in these areas.
It is important to note that fMRI findings in autism are diverse and complex, and individual differences among people with autism are significant. While fMRI research has contributed to our understanding of the neurologi cal underpinnings of autism, it is just one piece of the puzzle. Future studies combining fMRI with other techniques and methodologies hold promise for further unraveling the complexities of autism and developing targeted interventions to support individuals on the autism spectrum.
2.3. Corpus Callosum
The Corpus Callosum is a network of nerve fibres that provides a direct line of communication between the left and right sides of the brain. The corpus callosum is the connective tissue that links the two cerebral hemispheres. This connection permits interaction between the two components. The corpus callosum is a massive bundle of more than 200 million myelinated nerve fibres that allows the two hemispheres of the brain to communicate with one another. The corpus callosum, like most supratentorial structures, measures roughly 10 cm in length and has a C shape with a little upwardly convex architecture. The back is where it’s thickest. The corpus callosum’s primary function is to act as a connection between the two hemispheres of the brain, facilitating communication between, for example, the left and right frontal lobes. It is also speculated to have a pivotal role in vision, cognitive processes (including memory & learning), and motor control [
3].
The causes of ASD are not well understood, but likely involve Corpus Callosum, genetic, environmental, and neurological factors. Brain abnormalities, such as those affecting the Corpus Callosum, have been linked to autism; however, these changes alone do not account for the spectrum condition. Some research suggests that the Corpus Callosum in people with autism may be smaller, have a different structure, or have less connectivity than in neuro typical people. While some people with autism do show these distinctions, the majority does not, and there is wide diversity in the brain findings of those with autism.
2.4. Brain MRI Slices of Corpus Callosum
Brain MRI slices of the Corpus Callosum has revealed significant differences in individuals with ASD. Studies using MRI have shown that individuals with ASD often exhibit abnormalities in size, shape, & connectivity of Corpus Callosum. These alterations are believed to contribute to the atypical brain connectivity patterns observed in individuals with autism. The Corpus Callosum’s involvement suggests that disruptions in inter hemispheric communication may play a role in the social and cognitive challenges experienced by individuals with ASD. Further research into these findings may provide valuable insights into the neural mechanisms underlying autism spectrum disorders.
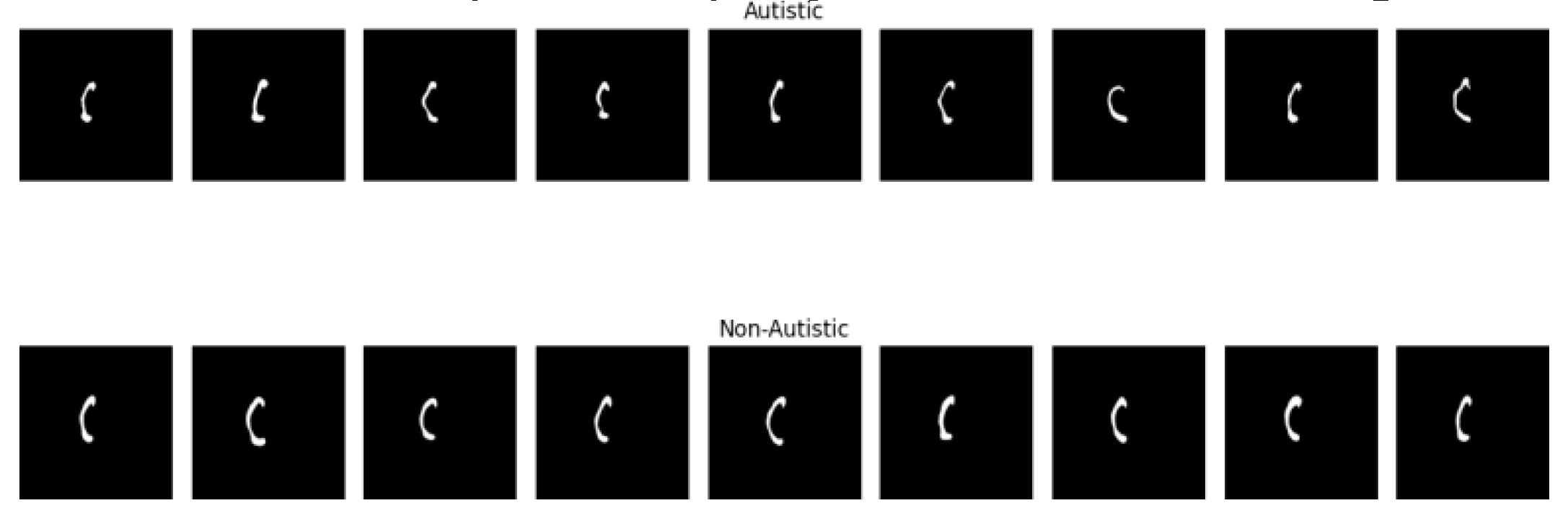
Figure 1 shows f-MRI slices of autistic and non-autistic brains.
2.5. Related Works
This section presents a comprehensive literature survey on the applications of various DL techniques for the classification of ASD. The use of DL algorithms in the field of ASD research has gained significant attention due to their capability to automatically extract relevant features from complex image data and provide accurate classification results. In the field of ASD analysis and classification, several studies have been conducted using DL and ML techniques.
The paper Vandewouw, Marlee et al. (2023) [
4] aimed to identify data dependent transdiagnostic subgroups among children & adolescents with and without neurodevelopmental problems. They used SNF technique and clustering algorithms, applying the Calinsk Harabasz index to determine the optimal cluster size. The study analyzed measures obtained from the brains functional networks and used the POND and HBN datasets.
Deba Kanta, Jyotismita et al. (2023) [
5], which I have worked on, provides a comparative assessment of ML classifiers used for analyzing & classify ing ASD in toddlers & adolescents. It reviews the existing literature on this topic and evaluates the strengths and limitations of various classifiers, including SVM, RF, NB, LR. The study also discusses the performance metrics used to assess the classifiers efficiency and highlights the importance of addressing challenges such as limited sample sizes and imbalanced datasets. Additionally, future directions, including the exploration of hybrid models and improving interpretability, are suggested.
(2022) [
6] focused on the classification of neuro developmental disorders using structural MRI data. They utilized CNN and Siamese networks, achieving an AUC of 91%. The study employed the NDAR dataset.
Bayram, Muhammed Ali, et al. [
7], 2021, uses RNN and ACC of 74.74% are achieved and SENS is 72.95%. This study presents a promising approach for the early detection of ASD using rs-fMRI data, employing a combination of long short-term memory network (LSTM), CNN, and hybrid models on the dataset.
Subah, Faria Zarin, et al. [
8] in 2021 uses Deep neural network (DNN) models and Acc of 88% is archived. This paper proposes a deep neural network model utilizing functional connectivity features from resting-state f-MRI data and multiple brain atlases to detect ASD, achieving higher accuracy than state-of-the-art methods and demonstrating the superiority of the Bootstrap Analysis of Stable Clusters (BASC) atlas for classification.
Devika, K et al. (2021) [
9] developed a machine learning framework using a Support Vector Machine (SVM) classifier to categorize neurological disorders. Long term rs-fMRI data was utilized, resulting in an accuracy of 80.76%. The research used the ABIDE 2 dataset.
Zhan, Yafeng et al. (2021) [
10] aimed to find brain markers linked to DSM diagnoses and symptom importance using noninvasive neuroimaging. They employed a Sparse Logistic Regression (LR) classifier, reporting an accuracy of 82.14%, sensitivity of 79.70%, and specificity of 83.74%. The study utilized the ABIDE 1 dataset.
Husna, R. Nur Syahindah, et al. (2021) [
11] uses CNN models, VGG-16 and ResNet-50 and Acc 87.0%. This research investigates the use of deep learning models, specifically VGG-16 and ResNet-50. ABIDE dataset is used.
Yin, Wutao, Sakib Mostafa, et al. (2021) [
12] uses DNN and Acc 79.2% accuracy is achieved and AUC of 82.4%. This study presents deep learning methods for diagnosing ASD using functional brain networks constructed from fMRI data. They uses ABIDE 1 dataset.
Leming, Matthew et. al. (2020) [
13] uses CNN and Acc = 57.1150%. This paper presents a deep learning approach using a CNN trained on a large fMRI dataset to classify ASD versus TD controls, achieving high accuracy and providing insights into the brain connections involved in the classification. Where largest multi-source fMRI conectomic dataset is used.
Huang, Zhi-An, et al. (2020) [
14] uses Deep Belief Networks (DBN) model and Acc of 76.4% is archived. In this work, a novel graph-based classification model using DBN and the ABIDE database is proposed, achieving superior performance compared to existing models, with the ability to identify subtypes within ASD and provide interpretability of the neural correlation patterns. Where ABIDE dataset is used.
Sherkatghanad, Zeinab et al. (2020) [
15] proposed the use of CNN for automated ASD finding. They achieved an acc of 70.22% using the ABIDE dataset.
Ali, Nur et al. (2020) [
16] proposed a DL model based on a 6-layer CNN architecture for ASD detection. The model achieved a max accuracy of 80%, although the specific dataset used was not mentioned.
Li, Jing et al. (2020) [
17] utilized machine learning models, specifically SVM and LSTM networks, for ASD detection, achieving an accuracy of 92.6%. The dataset used in their study was not specified.
Tao, Yudong et al. (2019) [
18] employed SP-ASDNet, a deep learning model, to identify ASD in patients. They reported an accuracy of 74.22% using the Saliency 4 ASD data.
Akhavan Aghdam, Maryam et (2018) al. [
19] uses DBN and Acc of 65.56% is achieved. This paper explores the application of a DBN utilizing data from the datasets to classify ASDs from typical controls (TCs) by combining resting-state fMRI, gray matter, and white matter data, achieving improved accuracy compared to previous methods. Where ABIDE I and ABIDE II is used.
Heinsfeld, Anibal S’olon, et al. (2018) [
20] uses DNN, RF, SVM models and acc of 70% is achieved. This paper applies deep learning algorithms to identify ASD patients based on brain activation patterns, and differentiating ASD from typically developing controls and revealing disrupted anterior-posterior brain connectivity in ASD. Where ABIDE dataset is used.
Thomas, Monisha et al. (2018) [
21] presented a novel method for detecting ASD using ANN, without specifying performance metrics or datasets.
Heinsfeld, Anibal S´olon et al. (2018) [
20] applied deep learning algorithms including a two-layered denoising auto-encoder, for ASD identification using the ABIDE dataset. The deep neural network achieved 70% accuracy.
These studies highlight the advancements in utilizing deep learning techniques for ASD classification using MRI images. The application of various deep learning architectures, including 3D CNNs, auto encoders, attention-guided models, and multi-view frameworks, showcases the diversity of approaches in this field. The findings emphasize the potential of deep learning in providing accurate and automated diagnosis of ASD, assisting clinicians and researchers in early detection and intervention strategies.
Overall, the literature survey demonstrates the growing interest in utilizing deep learning methods for ASD classification using MRI images.
Table 1: The Results are presented here.
3. Discussion
Based on our limited survey, it has been observed that
A good number of methods have been developed using DL approaches for ASD classification.
Other classifiers most commonly used are SVM, NB, etc.
ABIDE and its versions 1 & 2 are commonly used for validation.
Highest accuracy achieved in ABIDE repository is 93.69%.
3.1. Proposed Methodology
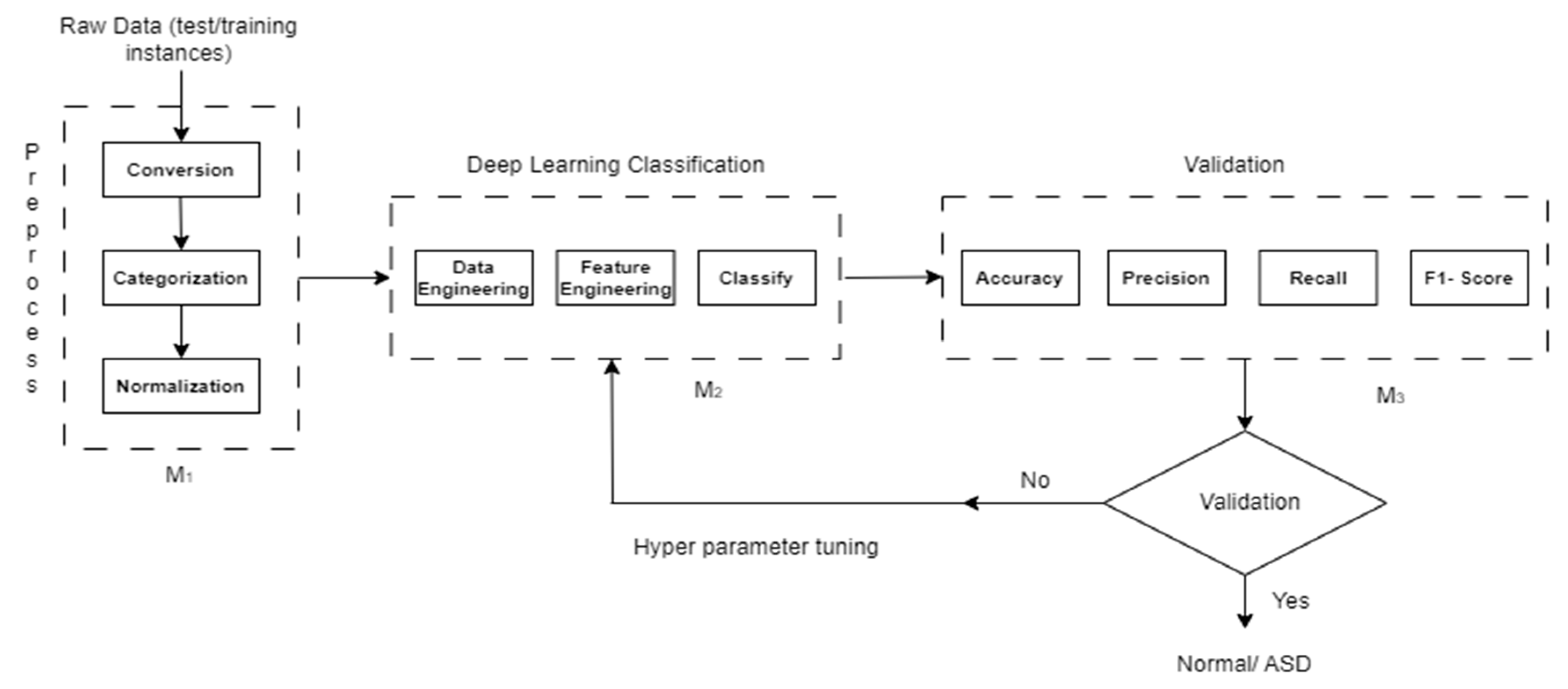
Figure 2.
Block diagram of the proposed methodology.
Figure 2.
Block diagram of the proposed methodology.
We have considered three modules as Module 1 (M1), Module 2 (M2), and Module 3 (M1) in our proposed methodology.
3.2. Module 1
In Module 1, the raw data is typically processed through three steps: conversion, categorization, and normalization.
Conversion: The conversion step involves transforming the raw data into a suitable format for further analysis. This could include converting data from one data type to another, such as converting categorical data into numerical representations.
Categorization: Categorization is the process of grouping data into meaningful categories or classes based on certain criteria or attributes. Categorization can be done manually by assigning labels or automatically using techniques such as clustering or classification algorithms.
Normalization: Normalization aims to standardize the data by scaling it to a specific range or distribution. Common normalization techniques include min-max scaling, where the data is transformed to a predefined range (e.g., between 0 and 1), and z-score normalization (standardization), where the data is transformed to have zero mean and unit variance.
3.3. Module 2
Data Engineering: This step focuses on preparing and organizing the data. It includes tasks such as data cleaning, preprocessing, and augmentation to ensure data quality and compatibility with deep learning models.
Feature Engineering: Feature engineering aims to extract meaningful representations from the data. Techniques like dimensionality reduction, feature selection, and creation of new features are used to provide relevant and informative inputs to the deep learning model.
Classification: In this step, a deep learning model is trained and used for prediction. Models such as CNNs, or transformers are commonly employed. The trained model can then classify new data instances into different classes or provide probabilities for each class.
3.4. Module 3
In Module 3, classification includes train and testing instances typically includes three evaluation steps: Accuracy, Precision, Recall, and F1-Score.
3.5. Dataset
The
Table 2 shows the dataset description.
We use a benchmark dataset from preprocessed neuro imaging ABIDE 1 to validate our method.
3.6. Training, Validation and Testing
When working with datasets, it is crucial to split the data into three distinct parts: training, validation, & testing. The dataset is divided into training, validation, & testing sets with predetermined ratios. In this case, the dataset has been split into three parts with a ratio of 70:20:10, respectively.
4. Proposed Model Architecture
4.1. VGG-16
The specified neural network layer is a convolutional layer. It is denoted by “Conv2D” as a data type. This layer’s output has the shape (None, 224, 224, 64), which indicates a batch size of “None” (which will be chosen at runtime), a height and width of 224 pixels, and 64 channels or filters. There are 1,792 adjustable parameters in the layer. Later layers, including convolutional and max pooling ones, are used to glean more complex features from the input data. At its conclusion, the network consists of completely connected layers (Dense layers) that are used for classification. There are 435 million total trainable parameters over the entire network.
Figure 3
VGG16 is a CNN architecture that is widely considered to be one of the best deep learning models available today. The 16 in VGG16 refers to that it has 16 layers that have weight.
Architecture of VGG-16: The input to VGG-16 is a fixed size 244X244 RGB image. Each pixel in a picture is deducted from its mean RGB value in a pre-processing phase.
After the initial processing is complete, the images are sent through a series of convolutional layers, each with 33-sized small receptive-field filters. When the filter size is (11) it means that the input channels have been linearly converted (then non linearity has been applied).
The convolution operation’s stride is set at 1 by default. Max-pooling layers is 5, which come after countless convolutional layers, are used to do spatial pooling.
All the layers of convolution and the non-linear activation function, a rectified linear unit (ReLU), are depicted in blue boxes up top. A total of 13 blue rectangles reflects the number of convolution layers, while the 5 red rectangles show the number of max-pooling layers. In addition, three green rectangles represent three nested layers. Because of its 16 layers, of which 13 are convolution layers and 3 are fully connected layers with tweakable parameters VGG-16 was given this moniker. Each image category in the imagenet dataset generates 1000 outputs from a softmax layer used as the final output layer.
In this design, we begin with a small channel size of 64 and raise it by a factor of 2 after each max-pooling layer, all the way up to a maximum of 512.
4.2. Inception v3
Given an input image of H x W x C (224, 224, 3) size, here H shows height, W shows the width, and C represents the number of channels. The Inception v3 architecture consists of multiple Inception modules stacked together. Each Inception module is composed of multiple parallel branches. Within each branch, there are different operations applied to the input feature maps. These operations include 1x1 convolutions, 3x3 convolutions, and 5x5 convolutions, with the aim of capturing features at different scales.
Specifically, let’s denote the input feature maps to the Inception module as X. Within each branch, the feature maps are processed differently. For example, a branch might apply a 1x1 convolution to X, followed by a 3x3 convolution, while another branch might apply a 1x1 convolution followed by a 5x5 convolution. The outputs from all branches are then concatenated along the channel dimension, resulting in a fused feature map. This fused feature map is passed to the next Inception module or to other layers in the network.
4.3. ResNet50
The ResNet50 design begins with a convolutional layer followed by a max pooling layer to extract low-level features from an input image with dimensions H x W x C (224, 224, 3), where H represents the height, W represents the width, and C denotes the number of channels. Residual blocks are the framework upon which ResNet50 is built. Separate convolutional layers, batch normalization, and ReLU activations make up each residual block. Additionally, the residual block has a connection that skips over the convolutional layers and simply adds the input to the final output. This helps to prevent the identification information deterioration that can occur in deep networks and preserves privacy.
In order to limit the number of parameters and computational cost, ResNet50 uses a variety of residual block configurations, including bottle-neck blocks that use 1x1, 3x3, and 1x1 convolutions. The spatial dimensions of the feature maps are shrunk by using global average pooling after many stacked residual blocks have been generated. The next step is a fully linked layer, followed by a softmax activation layer, which is used for classification.
4.4. DenseNet121
DenseNet121′s design starts with a convolutional layer followed by a max pooling layer to extract initial features from an input image of size H x W x C (224, 224, 3), where H denotes the height, W represents the width, and C represents the number of channels. An essential part of DenseNet121 is the dense block. Layers within the same thick block are all connected to one another through feed-forward connections. Thus, the results of one layer are used as input for the next levels in the block. This high degree of interconnection facilitates communication and encourages the sharing of features.
Each dense block is built from several layers of convolutional processing, batch normalization, and ReLU activations. Each thick block can have any number of layers within it, depending on the design. Convolution and pooling techniques are used to minimize the spatial dimensions of the feature maps, and transition layers are introduced between dense blocks to further this effect. After multiple dense blocks, global average pooling is used to shrink the feature maps in terms of space. The next step is a fully linked layer, followed by a softmax activation for classification.
4.5. MobileNet
MobileNet’s architecture begins with a depth wise separable convolutional layer given an input image of size H x W x C (224, 224, 3), where H indicates the height, W represents the width, and C denotes the number of channels. There are two basic parts to the depth wise separable convolution, and they are the depth convolution and the point convolution. The computational cost of depth wise convolutions is lower than that of regular convolutions since they convolve each channel of the input with its own set of filters. In this process, spatial details are recorded.
Next, the point wise convolution is applied to perform a 1x1 convolution across all channels, which helps to create new features by linearly combining the information obtained from the depth-wise convolution. This step helps to capture inter-channel relationships. Multiple depth-wise separable convolution layers are stacked together to form the backbone of the MobileNet architecture. The depth wise separable convolutions allow for parameter reduction while preserving the representational capacity of the network. To further reduce the spatial dimensions of the feature maps, the MobileNet architecture includes stride convolutions or pooling layers. These layers down sample the feature maps, reducing their spatial dimensions and capturing more global information. The final part of the MobileNet architecture typically includes fully connected layers followed by a softmax activation for classification.
4.6. Performance Assessment Matrix
Classification model evaluation uses measures like Accuracy, Precision, Recall, and F1-score in machine learning and deep learning tasks. Precision assesses how well the model avoids producing false positives, while Accuracy counts how often it gets predictions right. Measures of precision & recall are combined into a score called F1-score, which evaluates a model’s overall performance. Using these measures to evaluate models is essential for making comparisons and well-informed decisions about which categorization algorithms or architectures to use. Here, T denotes True, F denoted False. For performance evaluation above performance measures are used:
5. Results
In this study, we employed autism brain MRI datasets consisting of both autistic and non-autistic brain images for classification purposes. As a first stage in our study, we used the DBSCAN method to thoroughly label the dataset, which is crucial for achieving meaningful classifications later on. With this carefully annotated dataset in hand, we dove into our main objective, which was to evaluate several DL models’ capabilities in terms of classification, paying particular attention to how well-known architectures like VGG16, ResNet50, and DenseNet121 fared. We evaluated the performance of several deep learning models, including VGG16, ResNet50, and DenseNet121, using performance metrics such as Accuracy, Precision, Recall, and F1-score.
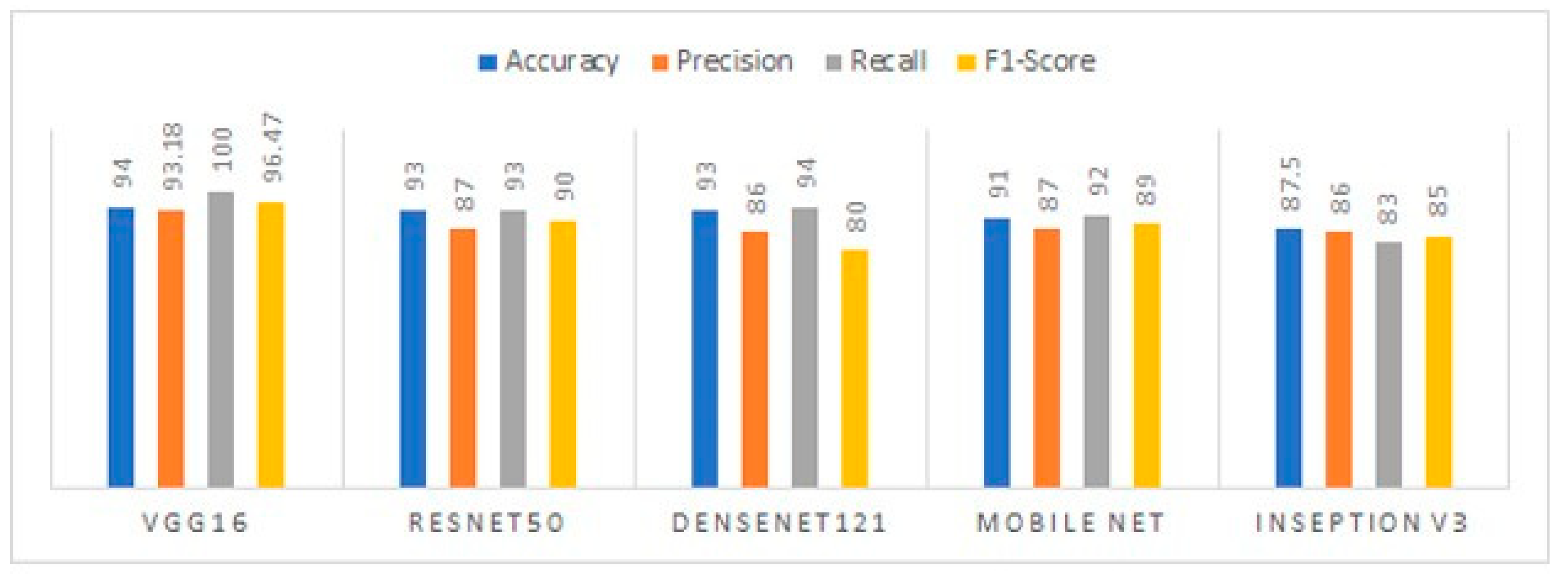
We have tested with autism brain MRI datasets which presents autistic and non-autistic brain images. For classifying datasets VGG16 with custom layered DL classifier is performing relatively higher results, shown in
Table 3.
VGG16 achieved the highest accuracy of 94 %. We have considered the performances parameters such as Accuracy, Precision, Recall, F1-score. ResNet50 and DenseNet121 model is performing second highest accuracy of 93% as shown in
Figure 4.
When compared to other models for the classification of autism brain MRI datasets, VGG16′s 94% accuracy using a unique deep learning classifier was far and away the best. This shows how crucial architectural decisions are for completing such projects successfully. VGG16′s competitive performance was further validated by study of performance parameters like Precision, Recall, and F1-score, which demonstrated the model’s capacity to properly recognize positive samples while reducing the number of false positives. ResNet50 and DenseNet121 came in second place, with a total performance of 93%. Implications for better understanding, diagnosis, and treatment of Autism Spectrum Disorder are highlighted by these findings, which highlight the potential of deep learning models, in particular VGG16, ResNet50, and DenseNet121, in the accurate classification of autism brain MRI datasets. Deeper exploration of the models’ learnt properties and their implications for autism classification is necessary.
Figure 4, shows the model testing accuracy, precision, recall, F1-score metrics for all five deep learning classification models.
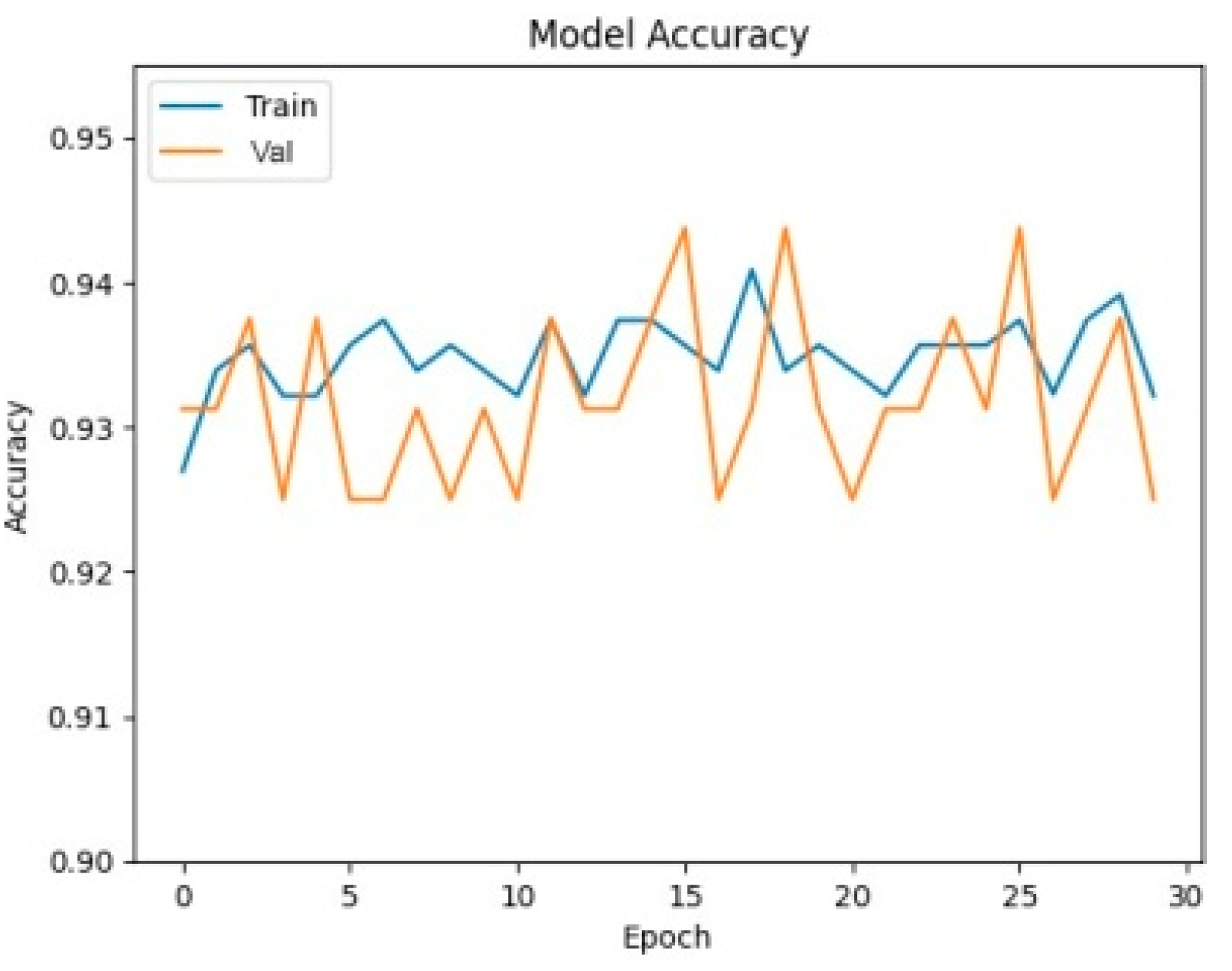
In order to get a visual understanding of how the deep learning models were doing during training, accuracy curves were created, shown in
Figure 5 and
Figure 6.
The curves illuminate the models’ convergence and generalization abilities, enabling a comparison. VGG16′s rapid convergence and consistent performance are highlighted by its accuracy curve, which reveals the extraordinary efficiency with which it reaches high accuracy levels early in training. The accuracy curves of ResNet50 and DenseNet121 are also encouraging, indicating that these networks are able to learn and generalize within the autistic brain MRI dataset. These curves provide visible evidence that the models are effective and provide important feedback on how well they are being trained and whether or not they are stable.
Figure 7 present a visual depiction of the model output, showcasing f-MRI brain images classified as either autistic or non-autistic. These images provide a visual representation of the model’s ability to distinguish between the two classes relies on distinctive features extracted from the brain f-MRI data. Images in the first row, the autistic brain images are displayed, show casing the regions and patterns that the model identifies as indicative of autism. These images may exhibit specific structural or functional characteristics that are associated with Autism Spectrum Disorder. The model’s accurate classification of these images signifies its capability to recognize and discern the unique features present in autistic brains. On the other hand, second row illustrates the non-autistic brain images, representing the normative brain structures or functional patterns typically observed in individuals without Autism Spectrum Disorder. The model’s ability to correctly classify these images reaffirms its proficiency in discerning the absence of autistic characteristics and accurately identifying non-autistic brain patterns.
The visual representation provided by these figures offers valuable insights into the model’s performance and its capacity to differentiate between autistic and non-autistic brain images. It aids in understanding the model’s decision-making process by showcasing the regions of interest or distinctive features that contribute to the classification results. Overall, these figures serve as valuable tools for visualizing and interpreting the model’s output in the context of autism classification using brain f-MRI data.
6. Discussion
In
Table 4, we show the best results achieved by previous studies and our own model for fMRI-based ASD detection utilizing brain atlases. When compared to prior research, the results of the current study are striking. However, there are constraints that must be overcome. As a result, future research may benefit from including data from other imaging modalities, such as structural MRI, in addition to functional MRI data, when examining ASD.
7. Conclusion
This paper has reported an effected deep learning-based method for detection of ASD with best possible accuracy. It establishes the potential of DL models, especially a customization of VGG16 architecture in accurate classification of ASD using f-MRI images. The method has been validated using benchmark dataset shared through ABIDE 1 and using several validity measures. The experimental results reveal that DL based methods can address the issue of accurate classification of ASD after necessary customization. The experimental comparisons with other competing methods also establish the superiority of our method.
As a future work, this evolving complex disease can be more successfully handled using an integrative approach. The approach should consider not only medical images but also omics scale and clinical data while deciding the class label of test case after applying an appropriate consensus function. Further, distributed computational approach could be explored to yield real-time responses without compromising accuracy.
Author Contributions
Conceptualization, Jyotismita Talukdar and Deba Gogoi; Data curation, Jyotismita Talukdar and Deba Gogoi; Methodology, Jyotismita Talukdar and Deba Gogoi; Resources, Deba Gogoi; Supervision, Jyotismita Talukdar, Dhruba Bhattacharyya and Thipendra Singh; Validation, Dhruba Bhattacharyya and Thipendra Singh; Visualization, Deba Gogoi; Writing – original draft, Deba Gogoi; Writing – review & editing, Jyotismita Talukdar, Deba Gogoi and Dhruba Bhattacharyya.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The research data used is ABIDE I from the neuroimaging website.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alarifi, H.S.; Young, G. Using multiple machine learning algorithms to predict autism in children, in: Proceedings on the International Conference on Artificial Intelligence (ICAI), The Steering Committee of The World Congress in Computer Science, Computer 2018, pp. 464–467.
- Lord, C.; Elsabbagh, M.; Baird, G.; Veenstra-Vanderweele, J. Autism spectrum disorder. The lancet 2018, 392, 10146. [Google Scholar] [CrossRef] [PubMed]
- Unterberger, I.; Bauer, R.; Walser, G.; Bauer, G. Corpus callosum and epilepsies. Seizure 2016, 37, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Vandewouw, M.M.; Brian, J.; Crosbie, J.; Schachar, R.J.; Iaboni, A.; Georgiades, S.; Nicolson, R.; Kelley, E.; Ayub, M.; Jones, J.; et al. Identifying replicable subgroups in neurodevelopmental conditions using resting-state functional magnetic resonance imaging data. JAMA Network Open 2023, 6, e232066. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, J.; Gogoi, D.K.; Singh, T.P. A comparative assessment of most widely used machine learning classifiers for analysing and classifying autism spectrum disorder in toddlers and adolescents. Healthcare Analytics 2023, 3, 100178. [Google Scholar] [CrossRef]
- Gao, K.; Sun, Y.; Niu, S.; Wang, L. Unified framework for early stage status prediction of autism based on infant structural magnetic resonance imaging. Autism Research 2021, 14, 2512–2523. [Google Scholar] [CrossRef] [PubMed]
- Bayram, M.A.; İlyas, Ö.Z.E.R.; Temurtaş, F. Deep learning methods for autism spectrum disorder diagnosis based on fmri images. Sakarya University Journal of Computer and Information Sciences 2021, 4, 142–155. [Google Scholar] [CrossRef]
- Subah, F.Z.; Deb, K.; Dhar, P.K.; Koshiba, T. A Deep Learning Approach to Predict Autism Spectrum Disorder Using Multisite Resting-State fMRI. Appl. Sci. 2021, 11, 3636. [Google Scholar] [CrossRef]
- Devika, K.; Oruganti, V.R.M. A machine learning approach for diagnosing neurological disorders using longitudinal resting-state fmri, in: 2021 11th International Conference on Cloud Computing, Data Science & Engineering (Confluence), IEEE, 2021, pp. 494–499.
- Zhan, Y.; Wei, J.; Liang, J.; Xu, X.; He, R.; Robbins, T.W.; Wang, Z. Diagnostic classification for human autism and obsessive-compulsive disorder based on machine learning from a primate genetic model. American Journal of Psychiatry 2021, 178, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Husna, R.N.S.; Syafeeza, A.; Hamid, N.A.; Wong, Y.; Raihan, R.A. Functional magnetic resonance imaging for autism spectrum disorder detection using deep learning. Jurnal Teknologi 2021, 83, 45–52. [Google Scholar] [CrossRef]
- Yin, W.; Mostafa, S.; Wu, F.-X. Diagnosis of autism spectrum disorder based on functional brain networks with deep learning. Journal of Computational Biology 2021, 28, 146–165. [Google Scholar] [CrossRef]
- Leming, M.; G´orriz, J.M.; Suckling, J. Ensemble deep learning on large, mixed-site fmri datasets in autism and other tasks. International journal of neural systems 2020, 30, 2050012. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-A.; Zhu, Z.; Yau, C.H.; Tan, K.C. Identifying autism spectrum disorder from resting-state fmri using deep belief network. IEEE Transactions on neural networks and learning systems 2020, 32, 2847–2861. [Google Scholar] [CrossRef]
- Sherkatghanad, Z.; Akhondzadeh, M.; Salari, S.; Zomorodi-Moghadam, M.; Abdar, M.; Acharya, U.R.; Khosrowabadi, R.; Salari, V. Automated detection of autism spectrum disorder using a convolutional neural network. Frontiers in neuroscience 2020, 13, 1325. [Google Scholar] [CrossRef]
- Ali, N.A.; Syafeeza, A.; Jaafar, A.; Alif, M.; Ali, N. Autism spectrum disorder classification on electroencephalogram signal using deep learning algorithm. IAES International Journal of Artificial Intelligence 2020, 9, 91–99. [Google Scholar] [CrossRef]
- Li, J.; Zhong, Y.; Han, J.; Ouyang, G.; Li, X.; Liu, H. Classifying asd children with lstm based on raw videos. Neurocomputing 2020, 390, 226–238. [Google Scholar] [CrossRef]
- Tao, Y.; Shyu, M.-L. Spasdnet: Cnnlstm based asd classification model using observer scanpaths, in: 2019 IEEE International conference on multimedia & expo workshops (ICMEW), IEEE, 2019, pp. 641–646.
- Aghdam, M.A.; Sharifi, A.; Pedram, M.M. Combination of rs-fmri and smri data to discriminate autism spectrum disorders in young children using deep belief network. Journal of digital imaging 2018, 31, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Heinsfeld, A.S.; Franco, A.R.; Craddock, R.C.; Buchweitz, A.; Meneguzzi, F. Identification of autism spectrum disorder using deep learning and the abide dataset. NeuroImage: Clinical 2018, 17, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Chandran, A. Artificial neural network for diagnosing autism spectrum disorder, in: 2018 2nd International Conference on Trends in Electronics and Informatics (ICOEI), IEEE, 2018, pp. 930–933.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).