Submitted:
18 September 2023
Posted:
19 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
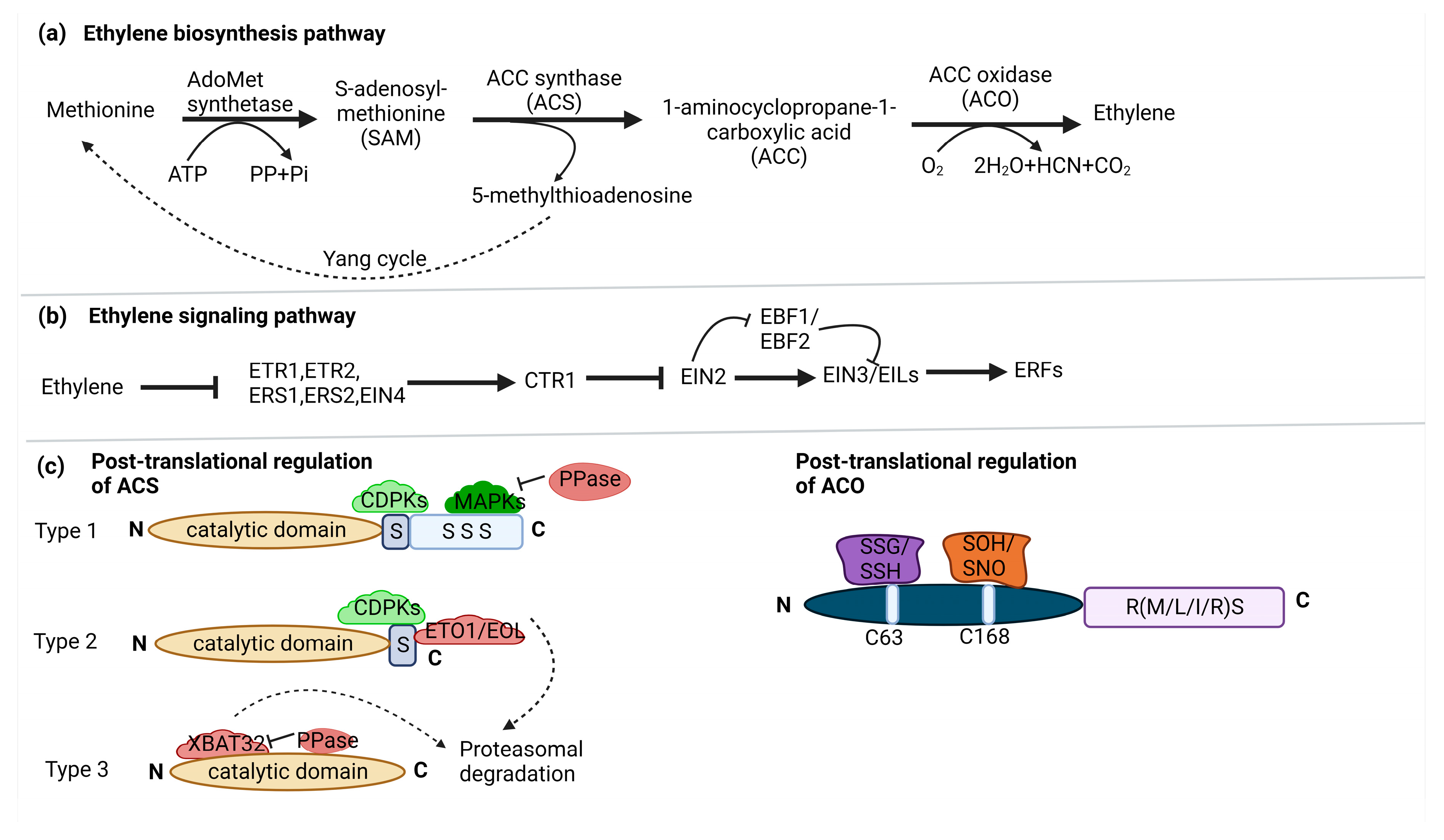
2. Ethylene biosynthesis and signaling
3. Snapshot of regulation of ethylene biosynthesis
4. Involvement of ethylene in plant developmental processes
4.1. Cell division and elongation
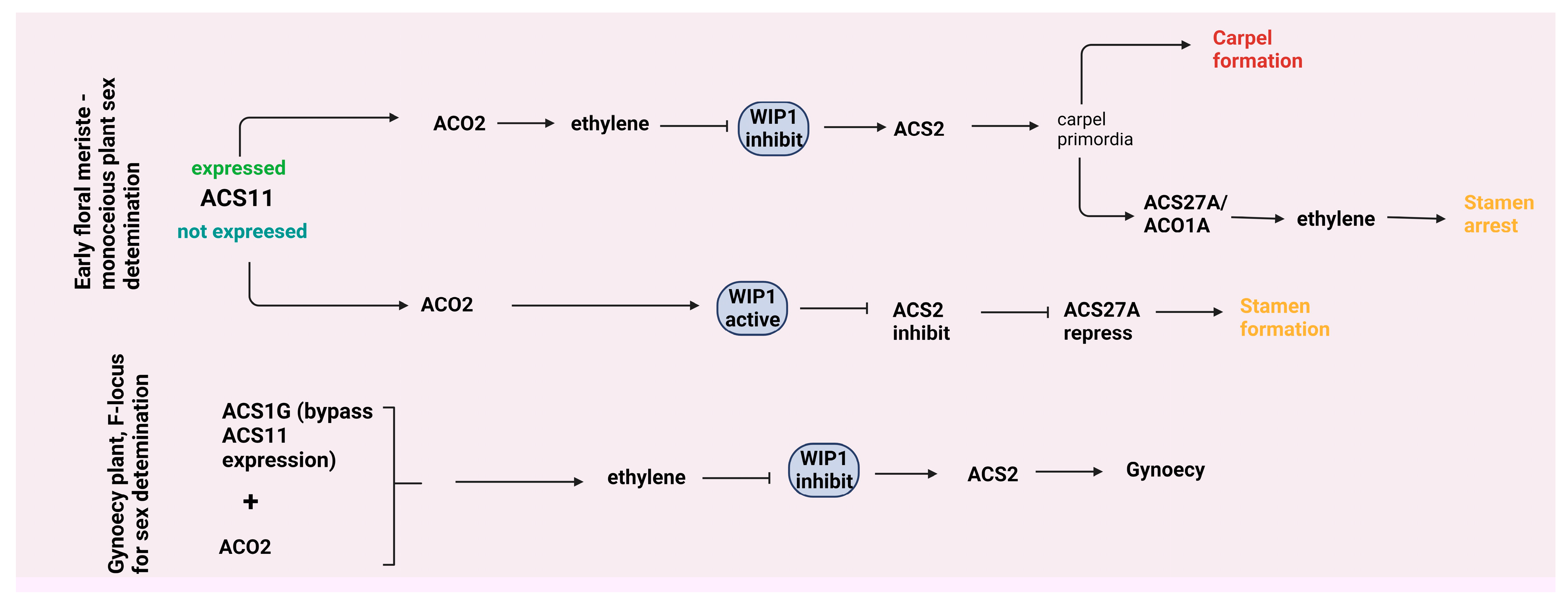
4.2. Leaf growth and flower development
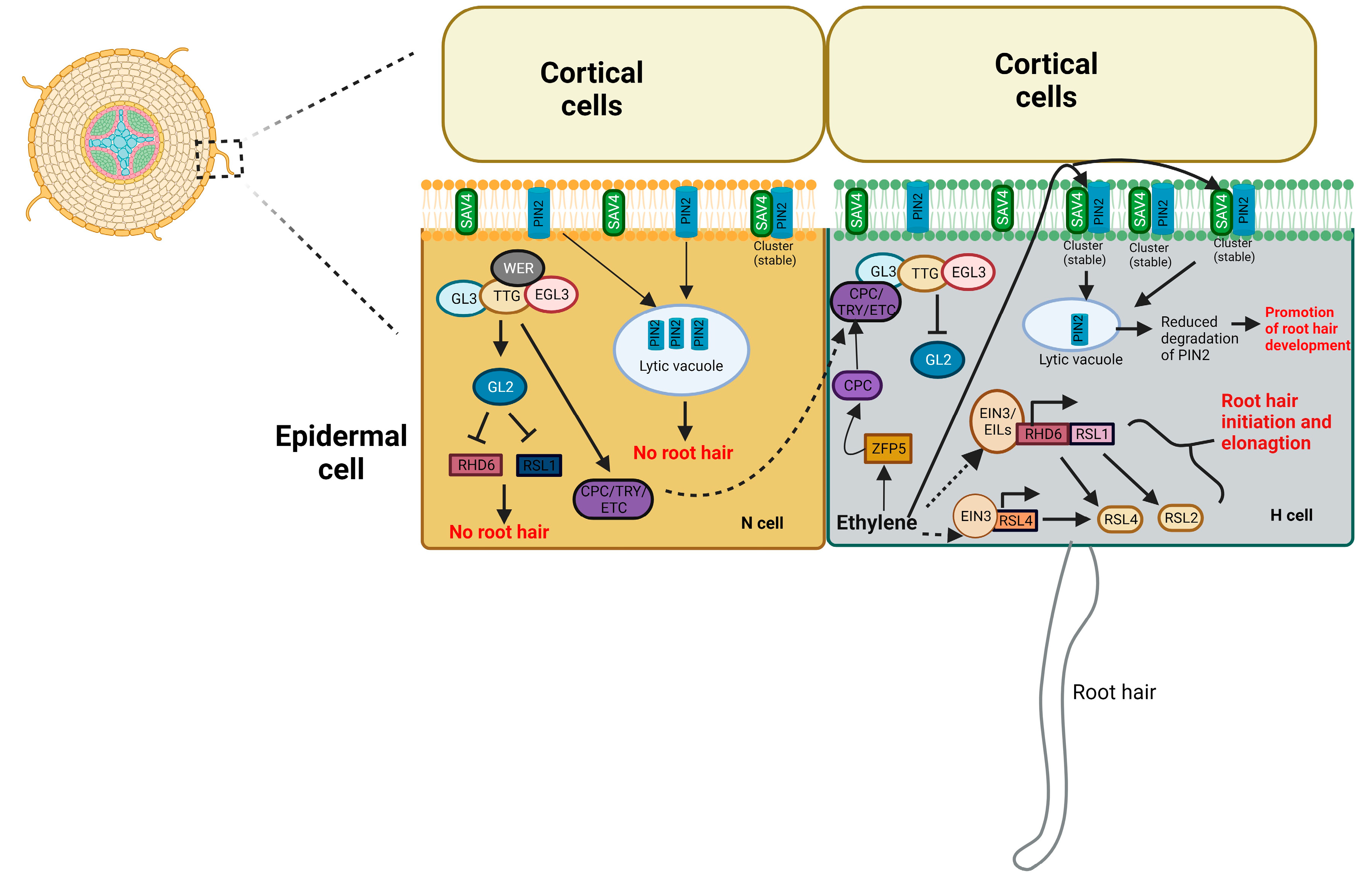
4.3. Root hair development
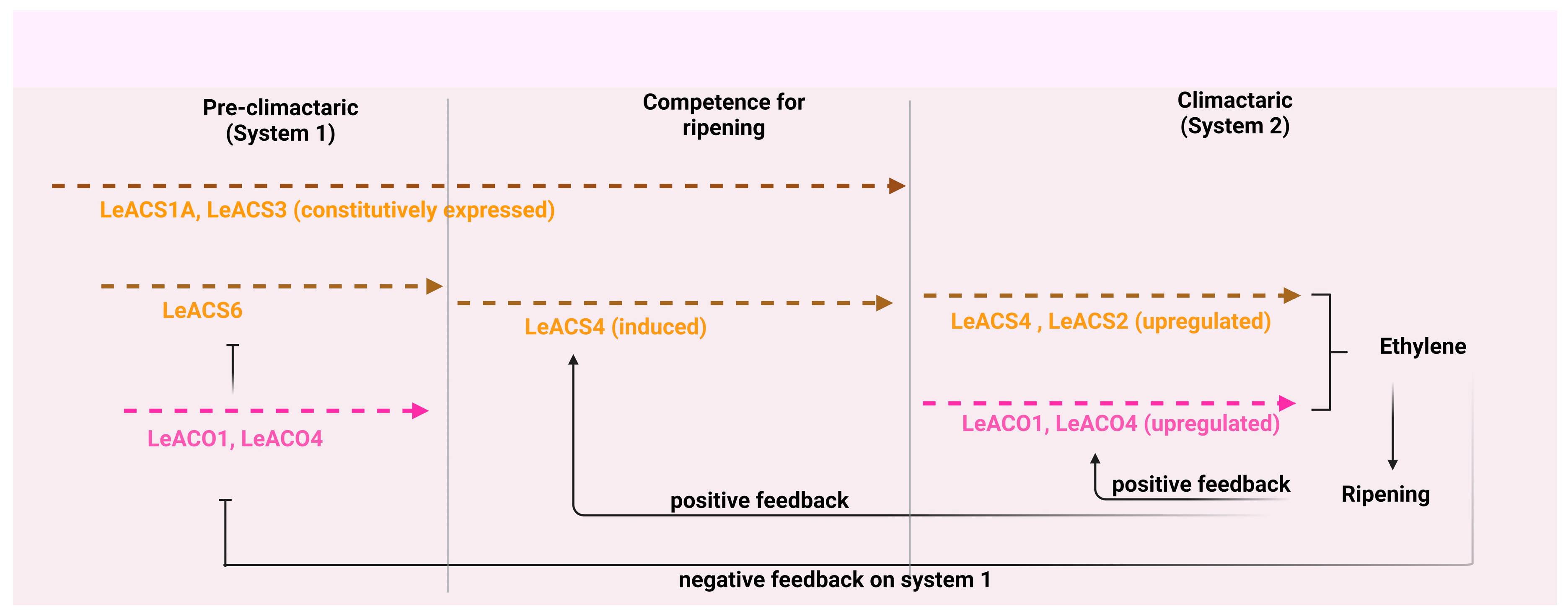
4.4. Fruit ripening
4.5. Chloroplast development
4.6. Photosynthesis
4.7. Senescence
4.8. Abscission
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chang, C.; Wang, B.; Shi, L.; Li, Y.; Duo, L.; Zhang, W. Alleviation of salinity stress-induced inhibition of seed germination in cucumber (Cucumis sativus L.) by ethylene and glutamate. J Plant Physiol. 2010, 167, 1152–1156. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.R.; Iqbal, N.; Masood, A.; Per, T.S.; Khan, N.A. Salicylic acid alleviates adverse effects of heat stress on photosynthesis through changes in proline production and ethylene formation. Plant Signal. Behav. 2013, 8, e26374. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: a master regulator of salinity stress tolerance in plants. Biomol. 2020, 10, 959. [Google Scholar] [CrossRef]
- Adams, D.O.; Yang, S. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Nat. Acad. Sci. 1979, 76, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Kende, H. Ethylene biosynthesis. Annu. Rev. Plant Biol. 1993, 44, 283–307. [Google Scholar] [CrossRef]
- Houben, M.; Van de Poel, B. 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO): the enzyme that makes the plant hormone ethylene. Front. Plant Sci. 2019, 10, 695. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Chang, K.N.; Yazaki, J.; Ecker, J.R. Interplay between ethylene, ETP1/ ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009, 23, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Zhang, C.; Ji, Y.; Zhao, Q.; He, W.; An, F.; Guo, H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012, 22, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, L.; Qi, B.; Zhao, B.; Ko, E.E.; Riggan, N.D.; Qiao, H. EIN2 mediates direct regulation of histone acetylation in the ethylene response. Proc. Nat. Acad. Sci. 2017, 114, 10274–10279. [Google Scholar] [CrossRef]
- Barry, C.S.; Llop-Tous, M.I.; Grierson, D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 2000, 123, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Kitagawa, M.; Ihashi, N.; Yabe, K.; Kimbara, J.; Yasuda, J.; Ito, H.; Inakuma, T.; Hiroi, S.; Kasumi, T. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 2008, 55, 212–223. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Quan, R.; Wang, X.C.; Huang, R. Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol. 2009, 150, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Harkey, A.F.; Yoon, G.M.; Seo, D.H.; DeLong, A.; Muday, G.K. Light modulates ethylene synthesis, signaling, and downstream transcriptional networks to control plant development. Front. Plant Sci. 2019, 10, 1094. [Google Scholar] [CrossRef] [PubMed]
- Pattyn, J.; Vaughan-Hirsch, J.; Van de Poel, B. The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 2021, 229, 770–782. [Google Scholar] [CrossRef]
- Tan, S.T.; Xue, H.W. Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep. 2014, 9, 1692–1702. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.H.; Yoon, G.M. Light-induced stabilization of ACS contributes to hypocotyl elongation during the dark-to-light transition in Arabidopsis seedlings. The Plant J. 2019, 98, 898–911. [Google Scholar] [CrossRef]
- Skottke, K.R.; Yoon, G.M.; Kieber, J.J.; DeLong, A. Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 2011, 7, e1001370. [Google Scholar] [CrossRef]
- Gautam, H.; Sehar, Z.; Khan, N.A. Ethylene: A gaseous signaling molecule with diverse roles. In The Plant Hormone Ethylene; Nafees A. Khan, Antonio Ferrante, Sergi Munné-Bosch, Ed.; Academic Press, 2023; pp. 1–13. [Google Scholar] [CrossRef]
- Khan, N.A.; Khan, M.I.R.; Ferrante, A.; Poor, P. Ethylene: a key regulatory molecule in plants. Front. Plant Sci. 2017, 8, 1782. [Google Scholar] [CrossRef] [PubMed]
- Millenaar, F.F.; Cox, M.C.; van Berkel, Y.E.D.J.; Welschen, R.A.; Pierik, R.; Voesenek, L.A.; Peeters, A.J. Ethylene-induced differential growth of petioles in Arabidopsis. Analyzing natural variation, response kinetics, and regulation. Plant Physiol. 2005, 137, 998–1008. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A.; Mir, M.R.; Nazar, R.; Singh, S. The application of ethephon (an ethylene releaser) increases growth, photosynthesis and nitrogen accumulation in mustard (Brassica juncea L.) under high nitrogen levels. Plant Biol. 2008, 10, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Nazar, R.; Syeed, S.; Masood, A.; Khan, N.A. Exogenously-sourced ethylene increases stomatal conductance, photosynthesis, and growth under optimal and deficient nitrogen fertilization in mustard. J. Exp. Bot. 2011, 62, 4955–4963. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yin, Y.; Jiang, W.; Peng, D.; Yang, D.; Cui, Y.; Wang, Z. Severe water deficit-induced ethylene production decreases photosynthesis and photochemical efficiency in flag leaves of wheat. Photosynthetica 2014, 52, 341–350. [Google Scholar] [CrossRef]
- Dai, H.; Fu, M.; Yang, X.; Chen, Q. Ethylene inhibited sprouting of potato tubers by influencing the carbohydrate metabolism pathway. J. Food Sci. Tech. 2016, 53, 3166–3174. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, V.L.; Pereira, A.M.; Pereira, A.S.; Silva, V.F.; Costa, L.C.; Bastos, C.E.; Ribeiro, D.M.; Caldana, C.; Sulpice, R.; Nunes-Nesi, A.; et al. Physiological and metabolic bases of increased growth in the tomato ethylene-insensitive mutant Never ripe: extending ethylene signaling functions. Plant Cell Rep. 2021, 40, 1377–1393. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yang, M.; Wang, J.; Jiang, C.Z.; Wang, Q. Application of exogenous ethylene inhibits postharvest peel browning of ‘Huangguan’pear. Front. Plant Sci. 2017, 7, 2029. [Google Scholar] [CrossRef]
- Zhang, X.; Li, B.; Zhang, X.; Wang, C.; Zhang, Z.; Sun, P. Exogenous application of ethephon regulates flower abscission, shoot growth, and secondary metabolites in Camellia sinensis. Sci. Hortic. 2022, 304, 111333. [Google Scholar] [CrossRef]
- Lyu, J.; Wu, Y.; Jin, X.; Tang, Z.; Liao, W.; Dawuda, M.M.; Calderón-Urrea, A. Proteomic analysis reveals key proteins involved in ethylene-induced adventitious root development in cucumber (Cucumis sativus L.). PeerJ 2021, 9, e10887. [Google Scholar] [CrossRef]
- Shinozaki, Y.; Hao, S.; Kojima, M.; Sakakibara, H.; Ozeki-Iida, Y.; Zheng, Y.; Ariizumi, T. Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J. 2015, 83, 237–251. [Google Scholar] [CrossRef]
- Raz, V.; Koornneef, M. Cell division activity during apical hook development. Plant Physiol. 2001, 125, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Provost, C.M.; Turner, S.R. Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling. PLoS Genet. 2012, 8, e1002997. [Google Scholar] [CrossRef]
- Ruzicka, K.; Ljung, K.; Vanneste, S.; Podhorská, R.; Beeckman, T.; Friml, J.; Benková, E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 2007, 19, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Martínez, O.; Pernas, M.; Carol, R.J.; Dolan, L. Ethylene modulates stem cell division in the Arabidopsis thaliana root. Sci. 2007, 317, 507–510. [Google Scholar] [CrossRef]
- Thomann, A.; Lechner, E.; Hansen, M.; Dumbliauskas, E.; Parmentier, Y.; Kieber, J.; Scheres, B.; Genschik, P. Arabidopsis CULLIN3 genes regulate primary root growth and patterning by ethylene-dependent and -independent mechanisms. PLoS Genet. 2009, 5, e1000328. [Google Scholar] [CrossRef] [PubMed]
- Zluhan-Martínez, E.; López-Ruíz, B.A.; García-Gómez, M.L.; García-Ponce, B.; de la Paz Sánchez, M.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. Integrative roles of phytohormones on cell proliferation, elongation and differentiation in the Arabidopsis thaliana primary root. Front. Plant Sci. 2021, 12, 659155. [Google Scholar] [CrossRef] [PubMed]
- Street, I.H.; Aman, S.; Zubo, Y.; Ramzan, A.; Wang, X.; Shakeel, S.N.; Kieber, J.J.; Schaller, G.E. Ethylene inhibits cell proliferation of the Arabidopsis root meristem. Plant Physiol. 2015, 169, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Skirycz, A.; Claeys, H.; De Bodt, S.; Oikawa, A.; Shinoda, S.; Andriankaja, M.; Maleux, K.; Eloy, N.B.; Coppens, F.; Yoo, S.D.; et al. Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. The Plant Cell 2011, 23, 1876–1888. [Google Scholar] [CrossRef] [PubMed]
- Marsch-Martinez, N.; Greco, R.; Becker, J.D.; Dixit, S.; Bergervoet, J.H.; Karaba, A.; de Folter, S.; Pereira, A. BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol. Biol. 2006, 62, 825–843. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Van den Broeck, L.; Inzé, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Claeys, H.; De Bodt, S.; Inzé, D. Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 2014, 19, 231–239. [Google Scholar] [CrossRef]
- Polko, J.K.; van Rooij, J.A.; Vanneste, S.; Pierik, R.; Ammerlaan, A.M.; Vergeer-van Eijk, M.H.; McLoughlin, F.; Gühl, K.; Van Isterdael, G.; Voesenek, L.A.; et al. Ethylene-mediated regulation of A2-type CYCLINs modulates hyponastic growth in Arabidopsis. Plant Physiol. 2015, 169, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Van de Poel, B.; Smet, D.; Van Der Straeten, D. Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol. 2015, 169, 61–72. [Google Scholar] [CrossRef]
- Polko, J.K.; van Zanten, M.; van Rooij, J.A.; Marée, A.F.; Voesenek, L.A.; Peeters, A.J.; Pierik, R. Ethylene-induced differential petiole growth in Arabidopsis thaliana involves local microtubule reorientation and cell expansion. New Phytol. 2012, 193, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.P.; Woeste, K.E.; Theologis, A.; Kieber, J.J. Recessive and dominant mutations in the ethylene biosynthetic gene ACS 5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. PNAS. 1998, 95, 4766–4771. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Hall, B.P.; Gao, Z.; Schaller, G.E. A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 2007, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Liu, G.; Xiao, J. The Arabidopsis EIN2 restricts organ growth by retarding cell expansion. Plant Signal. Behav. 2015, 10, e1017169. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.I.; Wang, X.; Thibault, D.M.; Kim, H.J.; Bombyk, M.M.; Binder, B.M.; Shakeel, S.N.; Schaller, G.E. The ARGOS gene family functions in a negative feedback loop to desensitize plants to ethylene. BMC Plant Biol. 2015, 15, 1–14. [Google Scholar] [CrossRef]
- Shi, J.; Habben, J.E.; Archibald, R.L.; Drummond, B.J.; Chamberlin, M.A.; Williams, R.W.; Lafitte, H.R.; Weers, B.P. Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiol. 2015, 169, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Dodd, I.C.; Davies, W.J.; Wilkinson, S. Ethylene limits abscisic acid- or soil drying-induced stomatal closure in aged wheat leaves. Plant Cell Environ. 2013, 36, 1850–1859. [Google Scholar] [CrossRef]
- Fiorani, F.; Bogemann, G.M.; Visser, E.J.; Lambers, H.; Voesenek, L.A. Ethylene emission and responsiveness to applied ethylene vary among Poa species that inherently differ in leaf elongation rates. Plant Physiol. 2002, 129, 1382–1390. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Reid, D.M. The role of endogenous ethylene in the expansion of Helianthus annuus leaves. Can. J. Bot. 1997, 75, 501–508. [Google Scholar] [CrossRef]
- Khan, N.A. The influence of exogenous ethylene on growth and photosynthesis of mustard (Brassica juncea) following defoliation. Sci. Hortic. 2005, 105, 499–505. [Google Scholar] [CrossRef]
- Tholen, D.; Voesenek, L.A.C.J.; Poorter, H. Ethylene insensitivity does not increase leaf area or relative growth rate in Arabidopsis, Nicotiana tabacum, and Petunia x hybrida. Plant Physiol. 2004, 134, 1803–1812. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene role in plant growth, development and senescence. Interaction with other phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.J.; Pannell, J.R. Sex determination: separate sexes are a double turnoff in melons. Curr. Biol. 2016, 26, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Yin, T.; Quinn, J.A. A Mechanistic Model of a Single Hormone Regulating Both Sexes in Flowering Plants. Bulletin of the Torrey Botanical Club 1992, 119, 431–441. [Google Scholar] [CrossRef]
- Zhang, H.; Li, S.; Yang, L.; Cai, G.; Chen, H.; Gao, D.; Lin, T.; Cui, Q.; Wang, D.; Li, Z.; et al. Gain-of-function of the 1-aminocyclopropane-1-carboxylate synthase gene ACS1G induces female flower development in cucumber gynoecy. The Plant Cell 2021, 33, 306–321. [Google Scholar] [CrossRef]
- Chen, H.; Sun, J.; Li, S.; Cui, Q.; Zhang, H.; Xin, F.; Wang, H.; Lin, T.; Gao, D.; Wang, S.; et al. An ACC oxidase gene essential for cucumber carpel development. Mol. Plant. 2016, 9, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Trebitsh, T.; Staub, J.E.; O'Neill, S.D. Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the female (F) locus that enhances female sex expression in cucumber. Plant Physiol. 1997, 113, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Ning, Q.; Jian, Y.; Du, Y.; Li, Y.; Shen, X.; Jia, H.; Zhang, Z. An ethylene biosynthesis enzyme controls quantitative variation in maize ear length and kernel yield. Nat. Commun. 2021, 12, 5832. [Google Scholar] [CrossRef]
- García, A.; Aguado, E.; Garrido, D.; Martínez, C.; Jamilena, M. Two androecious mutations reveal the crucial role of ethylene receptors in the initiation of female flower development in Cucurbita pepo. The Plant J. 2020, 103, 1548–1560. [Google Scholar] [CrossRef]
- Yu, D.; Li, X.; Li, Y.; Ali, F.; Li, F.; Wang, Z. Dynamic roles and intricate mechanisms of ethylene in epidermal hair development in Arabidopsis and cotton. New Phytol. 2022, 234, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Dolan, L. The role of ethylene in root hair growth in Arabidopsis. J. Plant. Nutr. Soil Sci. 2001, 164, 141–145. [Google Scholar] [CrossRef]
- Li, M.; Zhu, Y.; Li, S.; Zhang, W.; Yin, C.; Lin, Y. Regulation of phytohormones on the growth and development of plant root hair. Front. Plant Sci. 2022, 13, 865302. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, P.; Li, B.; Li, P.; Wen, X.; An, F.; Gong, Y.; Xin, Y.; Zhu, Z.; Wang, Y.; et al. Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. PNAS. 2017, 114, 13834–13839. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Tao, R.; Feng, Y.; Xiao, Z.; Zhang, D.; Peng, Y.; Wen, X.; Wang, Y.; Guo, H. EIN3 and RSL4 interfere with an MYB–bHLH–WD40 complex to mediate ethylene-induced ectopic root hair formation in Arabidopsis. PNAS. 2021, 118, e2110004118. [Google Scholar] [CrossRef]
- Xiao, F.; Gong, Q.; Zhao, S.; Lin, H.; Zhou, H. MYB30 and ETHYLENE INSENSITIVE3 antagonistically modulate root hair growth in Arabidopsis. Plant J. 2021, 106, 480–492. [Google Scholar] [CrossRef]
- Yan, A.; Wu, M.; Zhao, Y.; Zhang, A.; Liu, B.; Schiefelbein, J.; Gan, Y. Involvement of C2H2 zinc finger proteins in the regulation of epidermal cell fate determination in Arabidopsis. J. Integr. Plant Biol. 2014, 56, 1112–1117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Huang, L.; Yan, A.; Liu, Y.; Liu, B.; Yu, C.; Zhang, A.; Schiefelbein, J.; Gan, Y. Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J. Exp. Bot. 2016, 67, 6363–6372. [Google Scholar] [CrossRef] [PubMed]
- Mangano, S.; Denita-Juarez, S.P.; Choi, H.S.; Marzol, E.; Hwang, Y.; Ranocha, P.; Velasquez, S.M.; Borassi, C.; Barberini, M.L.; Aptekmann, A.A.; et al. Molecular link between auxin and ROS-mediated polar growth. PANS. 2017, 114, 5289–5294. [Google Scholar] [CrossRef]
- Takahashi, H.; Kawahara, A.; Inoue, Y. Ethylene promotes the induction by auxin of the cortical microtubule randomization required for low-pH-induced root hair initiation in lettuce (Lactuca sativa L.) seedlings. Plant Cell Physiol. 2003, 44, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Bruex, A.; Kainkaryam, R.M.; Wieckowski, Y.; Kang, Y.H.; Bernhardt, C.; Xia, Y.; Zheng, X.; Wang, J.Y.; Lee, M.M.; Benfey, P.; et al. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 2012, 8, 1002446. [Google Scholar] [CrossRef]
- Pitts, R.J.; Cernac, A.; Estelle, M. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 1998, 16, 553–560. [Google Scholar] [CrossRef]
- Strader, L.C.; Chen, G.L.; Bartel, B. Ethylene directs auxin to control root cell expansion. Plant J. 2010, 64, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Muday, G.K.; Rahman, A.; Binder, B.M. Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 2012, 17, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Ge, Y.; Chen, L.; Yan, F.; Cai, L.; Zhao, H.; Lei, D.; Jiang, J.; Wang, M.; Tao, Y. SAV4 is required for ethylene-induced root hair growth through stabilizing PIN2 auxin transporter in Arabidopsis. New Phytol. 2022, 234, 1735–1752. [Google Scholar] [CrossRef]
- Liu, Y.; Tang, M.; Liu, M.; Su, D.; Chen, J.; Gao, Y.; Li, Z. The molecular regulation of ethylene in fruit ripening. Small Methods 2020, 4, 1900485. [Google Scholar] [CrossRef]
- Liu, M.; Pirrello, J.; Chervin, C.; Roustan, J.P.; Bouzayen, M. Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol. 2015, 169, 2380–2390. [Google Scholar] [CrossRef]
- Nakatsuka, A.; Murachi, S.; Okunishi, H.; Shiomi, S.; Nakano, R.; Kubo, Y.; Inaba, A. Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant physiol. 1998, 118, 1295–1305. [Google Scholar] [CrossRef]
- Symons, G.M.; Chua, Y.J.; Ross, J.J.; Quittenden, L.J.; Davies, N.W.; Reid, J.B. Hormonal changes during non-climacteric ripening in strawberry. J. Exp. Bot. 2012, 63, 4741–4750. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.K.; Motyka, V.; Pokorna, E.; Dobrev, P.I.; Lacek, J.; Shao, J.; Lewers, K.S.; Mattoo, A.K. Comprehensive profiling of endogenous phytohormones and expression analysis of 1-aminocyclopropane-1-carboxylic acid synthase gene family during fruit development and ripening in octoploid strawberry (Fragaria× ananassa). Plant Physiol. Biochem. 2003, 196, 186–196. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.F.; Wei, W.; Fan, Z.Q.; Deng, W.; Li, Z.G.; Bouzayen, M.; Pirrello, J.; Lu, W.J.; Chen, J.Y. MaXB3 modulates MaNAC2, MaACS1, and MaACO1 stability to repress ethylene biosynthesis during banana fruit ripening. Plant Physiol. 2020, 184, 1153–1171. [Google Scholar] [CrossRef] [PubMed]
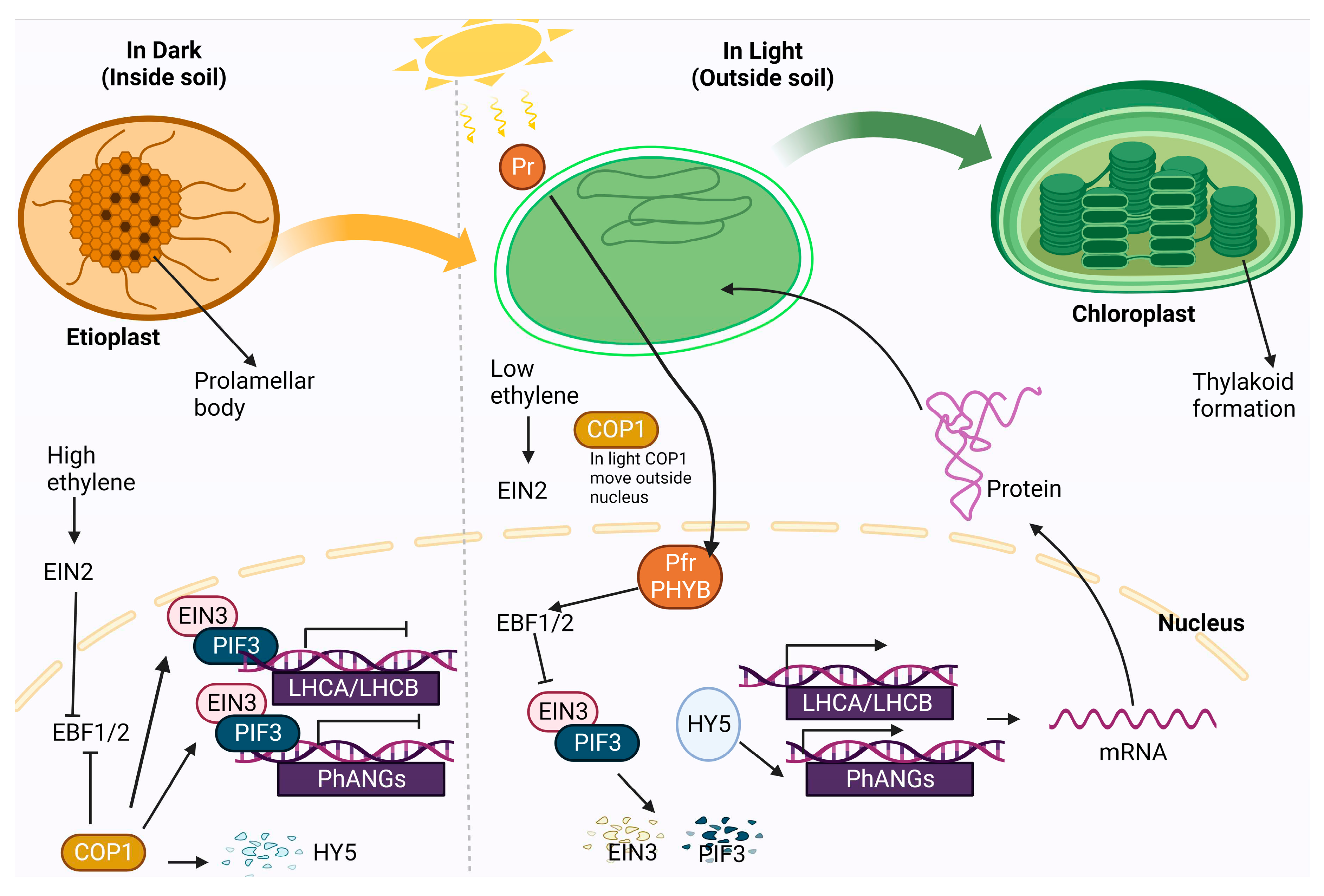
- Cackett, L.; Luginbuehl, L.H.; Schreier, T.B.; Lopez-Juez, E.; Hibberd, J.M. Chloroplast development in green plant tissues: the interplay between light, hormone, and transcriptional regulation. New Phytol. 2022, 233, 2000–2016. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Su, Y.S.; Lagarias, J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef]
- Leivar, P.; Monte, E. PIFs: systems integrators in plant development. The Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Liu, R.; Xue, C.; Shen, X.; Wei, N.; Deng, X.W.; Zhong, S. Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol. 2016, 26, 139–149. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Zhao, Q.; Ji, Y.; Li, W.; Jiang, Z.; Yu, X.; Zhang, C.; Han, Y.; He, W.; Liu, Y.; et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. The Plant Cell 2010, 22, 2384–2401. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Verdeja, T.; Vuorijoki, L.; Strand, A. Emerging from the darkness: interplay between light and plastid signaling during chloroplast biogenesis. Physiol. Plant. 2020, 169, 397–406. [Google Scholar] [CrossRef]
- Zhong, S.; Zhao, M.; Shi, T.; Shi, H.; An, F.; Zhao, Q.; Guo, H. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. PNAS. 2009, 106, 21431–21436. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Shi, H.; Xue, C.; Wei, N.; Guo, H.; Deng, X.W. Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. PNAS. 2014, 111, 3913–3920. [Google Scholar] [CrossRef]
- Liu, X.; Liu, R.; Li, Y.; Shen, X.; Zhong, S.; Shi, H. EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. The Plant Cell 2017, 29, 3051–3067. [Google Scholar] [CrossRef]
- Desikan, R.; Last, K.; Harrett-Williams, R.; Tagliavia, C.; Harter, K.; Hooley, R.; Hancock, J.T.; Neill, S.J. Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 2006, 47, 907–916. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sano, T.; Tamaoki, M.; Nakajima, N.; Kondo, N.; Hasezawa, S. Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 2005, 138, 2337–2343. [Google Scholar] [CrossRef] [PubMed]
- Azoulay-Shemer, T.; Schulze, S.; Nissan-Roda, D.; Bosmans, K.; Shapira, O.; Weckwerth, P.; Zamora, O.; Yarmolinsky, D.; Trainin, T.; Kollist, H.; et al. A role for ethylene signaling and biosynthesis in regulating and accelerating CO2-and abscisic acid-mediated stomatal movements in Arabidopsis. New Phytol. 2023, 238, 2460–2475. [Google Scholar] [CrossRef]
- Arve, L.E.; Torre, S. Ethylene is involved in high air humidity promoted stomatal opening of tomato (Lycopersicon esculentum) leaves. Funct. Plant Biol. 2015, 42, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Ceusters, J.; Van de Poel, B. Ethylene exerts species-specific and age-dependent control of photosynthesis. Plant physiol. 2018, 176, 2601–2612. [Google Scholar] [CrossRef]
- Tholen, D.; Pons, T.L.; Voesenek, L.A.C.J.; Poorter, H. Ethylene insensitivity results in down-regulation of Rubisco expression and photosynthetic capacity in tobacco. Plant Physiol. 2007, 144, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Tholen, D.; Pons, T.L.; Voesenek, L.A.C.J.; Poorter, H. The role of ethylene perception in the control of photosynthesis. Plant Signal. Behav. 2008, 3, 108–109. [Google Scholar] [CrossRef]
- Grbic, V.; Bleecker, A.B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995, 8, 595–602. [Google Scholar] [CrossRef]
- Monteiro, C.C.; Carvalho, R.F.; Gratão, P.L.; Carvalho, G.; Tezotto, T.; Medici, L.O.; Peres, L.E.P.; Azevedo, R.A. Biochemical responses of the ethylene insensitive Never ripe tomato mutant subjected to cadmium and sodium stresses. Environ. Exp. Bot. 2011, 71, 306–320. [Google Scholar] [CrossRef]
- Chen, Z.; Gallie, D.R. Ethylene regulates energy-dependent non-photochemical quenching in Arabidopsis through repression of the xanthophyll cycle. PLoS One 2015, 10, e0144209. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.D.; Cho, Y.H.; Yoo, S.D. Phytohormone ethylene-responsive Arabidopsis organ growth under light is in the fine regulation of Photosystem II deficiency-inducible AKIN10 expression. Sci. Rep. 2017, 7, 2767. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.D.; Hanson, P.J.; Gunderson, C.A. Assessing the influence of exogenous ethylene on electron transport and fluorescence quenching in leaves of Glycine max. Environ. Exp. Bot. 1992, 32, 449–455. [Google Scholar] [CrossRef]
- Xie, X.L.; Xia, X.J.; Kuang, S.; Zhang, X.L.; Yin, X.R.; Yu, J.Q.; Chen, K.S. A novel ethylene responsive factor CitERF13 plays a role in photosynthesis regulation. Plant Sci. 2017, 256, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Nazar, R.; Khan, M.I.R.; Khan, N.A. Variation in photosynthesis and growth of mustard cultivars: role of ethylene sensitivity. Sci. Hortic. 2012, 135, 1–6. [Google Scholar] [CrossRef]
- Khan, N.A. An evaluation of the effects of exogenous ethephon, an ethylene releasing compound, on photosynthesis of mustard (Brassica juncea) cultivars that differ in photosynthetic capacity. BMC Plant Biol. 2004, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef]
- van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flugge, U.I.; Kunze, R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant physiol. 2006, 141, 776–792. [Google Scholar] [CrossRef] [PubMed]
- Tsuchisaka, A.; Yu, G.; Jin, H.; Alonso, J.M.; Ecker, J.R.; Zhang, X.; Gao, S.; Theologis, A. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genet. 2009, 183, 979–1003. [Google Scholar] [CrossRef] [PubMed]
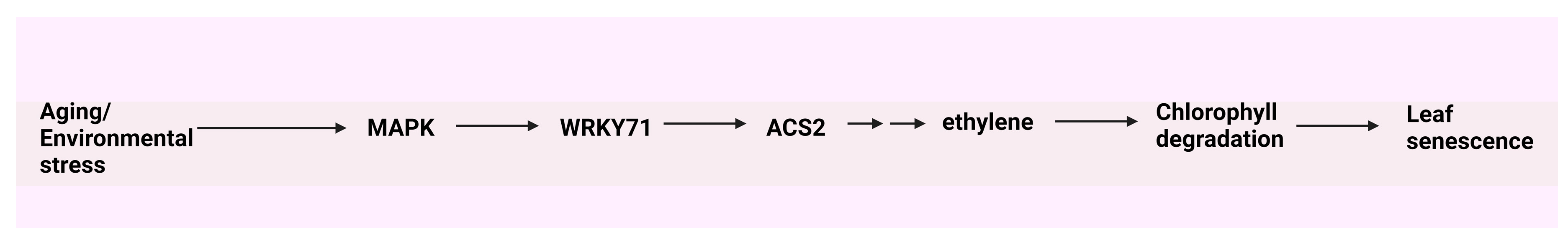
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Sci. 2009, 323, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef]
- Yu, Y.; Qi, Y.; Xu, J.; Dai, X.; Chen, J.; Dong, C.H.; Xiang, F. Arabidopsis WRKY71 regulates ethylene-mediated leaf senescence by directly activating EIN2, ORE1 and ACS2 genes. The Plant J. 2021, 107, 1819–1836. [Google Scholar] [CrossRef] [PubMed]
- Dar, R.A.; Nisar, S.; Tahir, I. Ethylene: a key player in ethylene sensitive flower senescence: a review. Sci. Hortic. 2021, 290, 110491. [Google Scholar] [CrossRef]
- Ten Have, A.; Woltering, E.J. Ethylene biosynthetic genes are differentially expressed during carnation (Dianthus caryophyllus L.) flower senescence. Plant Mol. Biol. 1997, 34, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Satoh, S. Ethylene Production and Petal Wilting during Senescence of Cut Carnation (Dianthus caryophyllus) Flowers and Prolonging Their Vase Life by Genetic Transformation. J. Japan. Soc. Hort. Sci. 2011, 80, 127–135. [Google Scholar] [CrossRef]
- Shibuya, K.; Yoshioka, T.; Hashiba, T.; Satoh, S. Role of the gynoecium in natural senescence of carnation (Dianthus caryophyllus L.) flowers. J. Exp. Bot. 2000, 51, 2067–2073. [Google Scholar] [CrossRef] [PubMed]
- Norikoshi, R.; Niki, T.; Ichimura, K. Differential regulation of two 1-aminocyclopropane-1-carboxylate oxidase (ACO) genes, including the additionally cloned DcACO2, during senescence in carnation flowers. Postharvest Biol. Technol. 2022, 183, 111752. [Google Scholar] [CrossRef]
- Tang, X.; Gomes, A.M.T.R.; Bhatia, A.; Woodson, W.R. Pistil specifc and ethylene-regulated expression of 1-aminocyclopropene-1-carboxylate oxidase genes on petunia flower. Plant Cell 1994, 6, 1227–1239. [Google Scholar] [CrossRef]
- Llop-Tous, I.; Barry, C.S.; Grierson, D. Regulation of ethylene biosynthesis in response to pollination in tomato flowers. Plant Physiol. 2000, 123, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Li, Y.; Tan, H.; Yang, F.; Ma, N.; Gao, J. Expression of ethylene biosynthetic and receptor genes in rose floral tissues during ethylene-enhanced flower opening. J. Exp. Bot. 2008, 59, 2161–2169. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Woodson, W.R. Differential expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in carnation. Plant Physiol. 1999, 119, 755–764. [Google Scholar] [CrossRef]
- Okamoto, M.; Niki, T.; Azuma, M.; Shibuya, K.; Ichimura, K. Expression of ethylene biosynthesis genes in the gynoecium and receptacle associated with sepal abscission during senescence in Delphinium grandiflorum. Plant Growth Regul. 2022, 97, 593–609. [Google Scholar] [CrossRef]
- Tanase, K.; Onozaki, T. Regulation of ethylene-and senescence-related genes in pot carnation flowers during flower senescence. J. Hortic. 2016, 85, 254–263. [Google Scholar] [CrossRef]
- In, B.C.; Ha, S.T.; Lee, Y.S.; Lim, J.H. Relationships between the longevity, water relations, ethylene sensitivity, and gene expression of cut roses. Postharvest Biol. Technol. 2017, 131, 74–83. [Google Scholar] [CrossRef]
- Ji, X.; Wang, M.; Xu, Z.; Wang, K.; Sun, D.; Niu, L. PlMYB308 regulates flower senescence by modulating ethylene biosynthesis in Herbaceous Peony. Front. Plant Sci. 2022, 13, 872442. [Google Scholar] [CrossRef]
- Patharkar, O.R.; Walker, J.C. Advances in abscission signaling. J. Exp. Bot. 2018, 69, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Meir, S.; Philosoph-Hadas, S.; Riov, J.; Tucker, M. L.; Patterson, S.E.; Roberts, J.A. Re-evaluation of the ethylene-dependent and-independent pathways in the regulation of floral and organ abscission. J. Exp. Bot. 2019, 70, 1461–1467. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.M. Ethylene and abscission. Physiol. Plant. 1997, 100, 567–576. [Google Scholar] [CrossRef]
- Botton, A.; Ruperti, B. The yes and no of the ethylene involvement in abscission. Plants 2019, 8, 187. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Zhang, L.; Yang, X.; Zhong, G. Profiling gene expression in citrus fruit calyx abscission zone (AZ-C) treated with ethylene. Mol. Genet. Genom. 2015, 290, 1991–2006. [Google Scholar] [CrossRef]
- Hartmond, U.; Yuan, R.; Burns, J.K.; Grant, A.; Kender, W.J. Citrus fruit abscission induced by methyl-jasmonate. J. Am. Soc. Hortic. Sci. 2000, 125, 547–552. [Google Scholar] [CrossRef]
- Merelo, P.; Agustí, J.; Ventimilla, D.; Talón, M.; Tadeo, F.R. Vesicular trafficking in abscission zone cells during ethylene-promoted fruit abscission in citrus. Acta Hortic. 2019, 1230, 41–48. [Google Scholar] [CrossRef]
- Dal Cin, V.; Barbaro, E.; Danesin, M.; Murayama, H.; Velasco, R.; Ramina, A. Fruitlet abscission: a cDNA-AFLP approach to study genes differentially expressed during shedding of immature fruits reveals the involvement of a putative auxin hydrogen symporter in apple (Malus domestica L. Borkh). Gene 2009, 442, 26–36. [Google Scholar] [CrossRef]
- Li, J.; Zhu, H.; Yuan, R. ; Profiling the expression of genes related to ethylene biosynthesis, ethylene perception, and cell wall degradation during fruit abscission and fruit ripening in apple. J. Am. Soc. Hortic. Sci. 2010, 135, 391–401. [Google Scholar] [CrossRef]
- Woltering, E.J.; van Doorn, W.C. Role of ethylene in senescence of petals: morphological and taxonomical relationships. J. Exp. Bot. 1998, 208, 1605–1616. [Google Scholar] [CrossRef]
- Zhao, M.; Li, C.; Ma, X.; Xia, R.; Chen, J.; Liu, X.; Ying, P.; Peng, M.; Wang, J.; Shi, C.L.; et al. KNOX protein KNAT1 regulates fruitlet abscission in litchi by repressing ethylene biosynthetic genes. J.Exp. Bot. 2020, 71, 4069–4082. [Google Scholar] [CrossRef]
| Plant species | Ethylene concentration | Response | Reference |
| Arabidopsis thaliana | 1-5 µL L-1 ethylene | Strong hyponastic response of leaf | [20] |
| Brassica juncea | 400 µL L-1 ethephon | Net photosynthesis and stomatal conductance decreases | [21] |
| Brassica juncea | 200 µL L-1 ethephon | Net photosynthesis and stomatal conductance increases | [22] |
| Triticum aestivum | 50mM ethephon | No effect on photosynthesis and stomatal conductance increases | [23] |
| Solanum tuberosum | 199.3 µL L-1 ethylene | inhibited the sprouting of potato tubers by influencing their carbohydrate metabolism. | [24] |
| Solanum lycopersicum | 1 mM ethephon | Reduced growth due to alteration in cell wall composition (accumulation of cellulose and lignin) and reduction in photosynthesis | [25] |
| Pyrus bretschneideri | 5 μL L-1 ethylene | Ethylene effectively inhibited postharvest peel browning by enhancing the activity of antioxidant enzymes such as CAT, APX, and SOD, while reducing Polyphenol oxidase activity. Additionally, it led to lower rates of electrolyte leakage and respiration in the peel. | [26] |
| Camellia sinensis | 3000mg L-1 ethephon | Induced abscission of flower and flower bud, enhances shoot length and caffeine content | [27] |
| Cucumis sativus | 0.5 Μm ethrel | Adventitious root formation | [28] |
| Solanum lycopersicum | 0,0.2, 1 and 2mM ACC | Ethylene suppresses the process of fruit set by affecting the metabolism of gibberellin | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





