Submitted:
18 September 2023
Posted:
19 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial strains and media
2.2. Biofilm culture conditions
2.3. Scanning electron microscopy
2.4. Calcofuor white staining
2.5. Biofilm enzymatic degradation assay
2.6. Antimicrobial susceptibility
3. Results
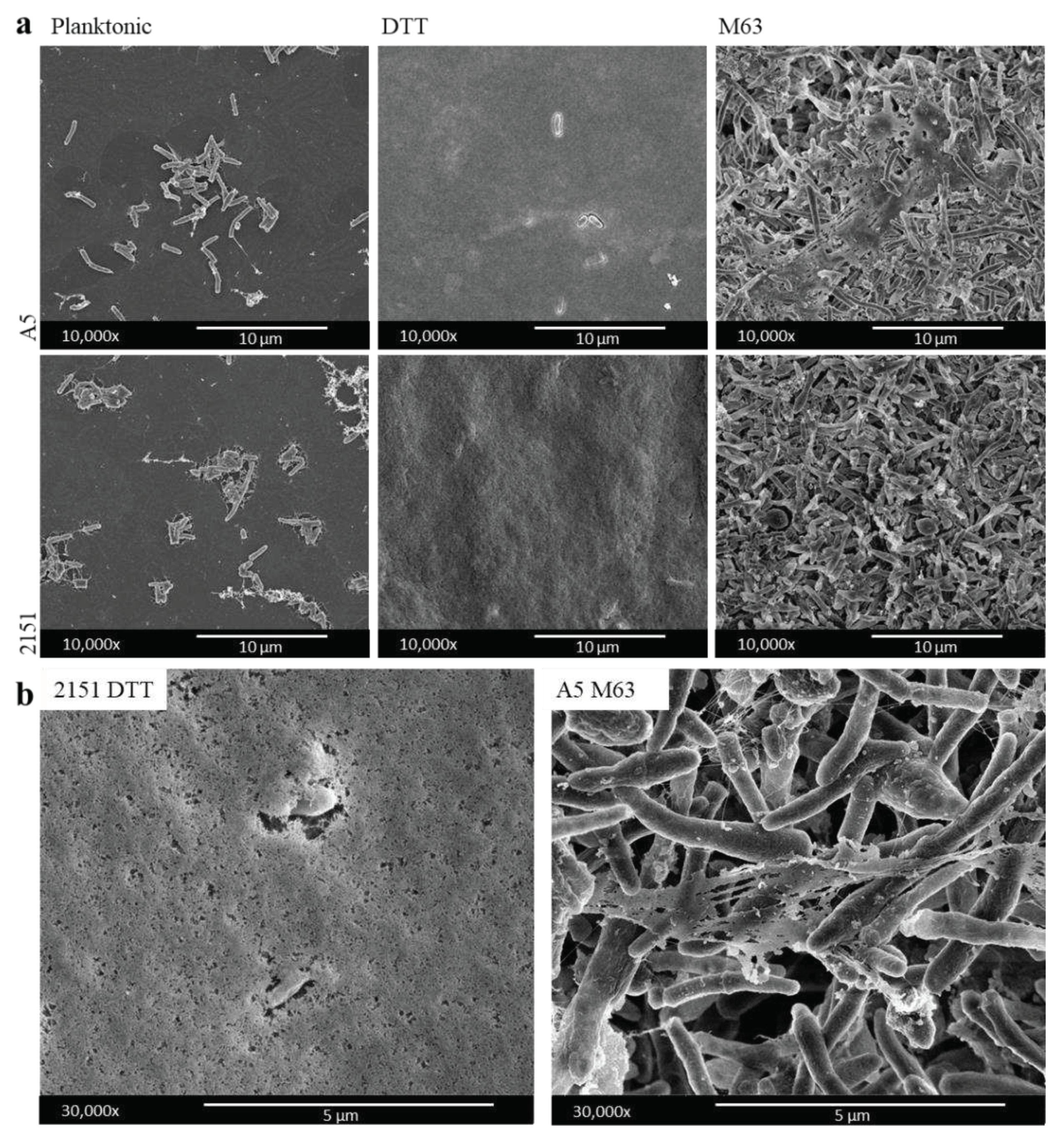
3.1. Biofilm models display differences in ultrastructure cell
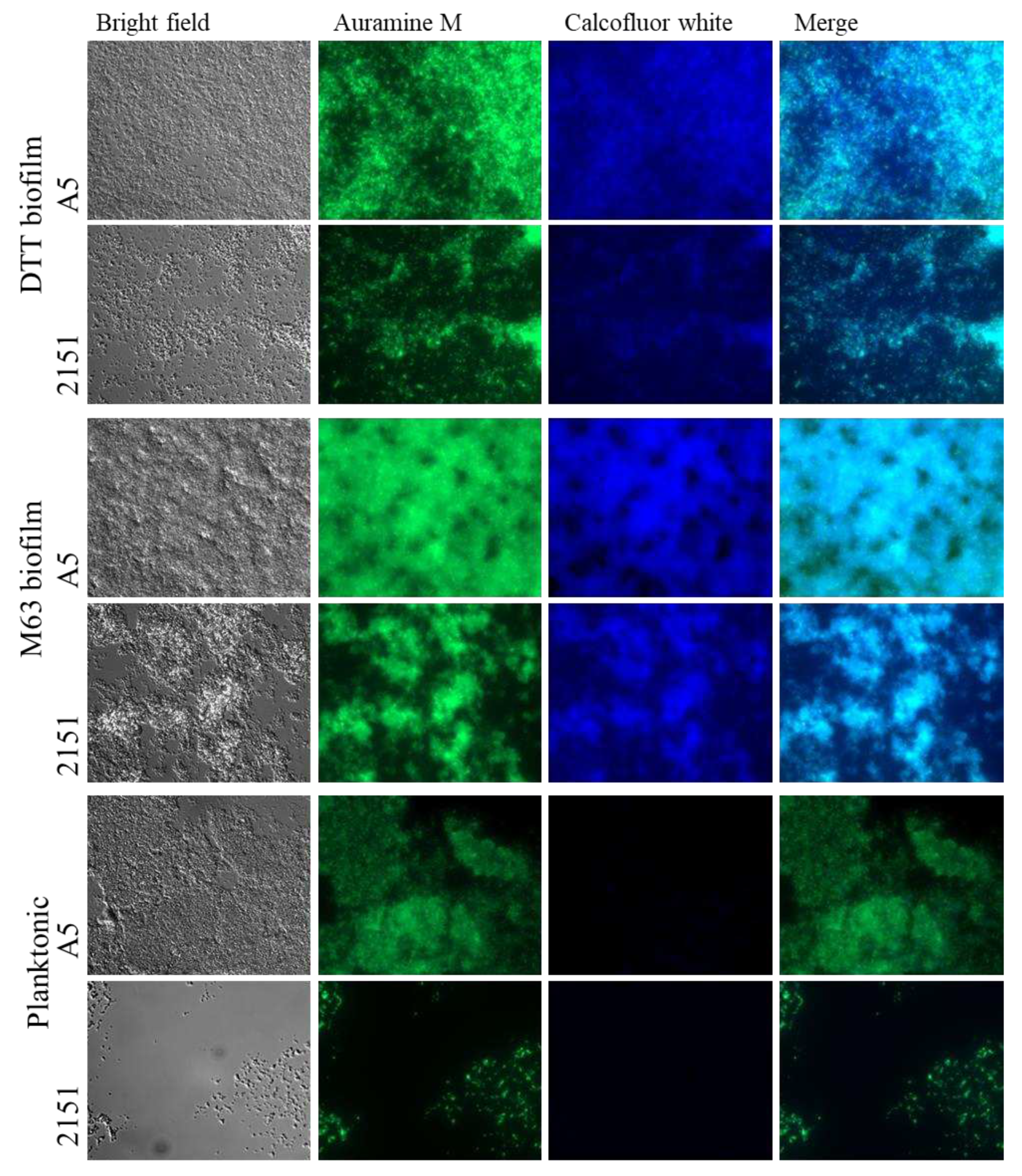
3.2. Calcofluor white staining suggests the presence of cellulose in biofilm ECM in both models
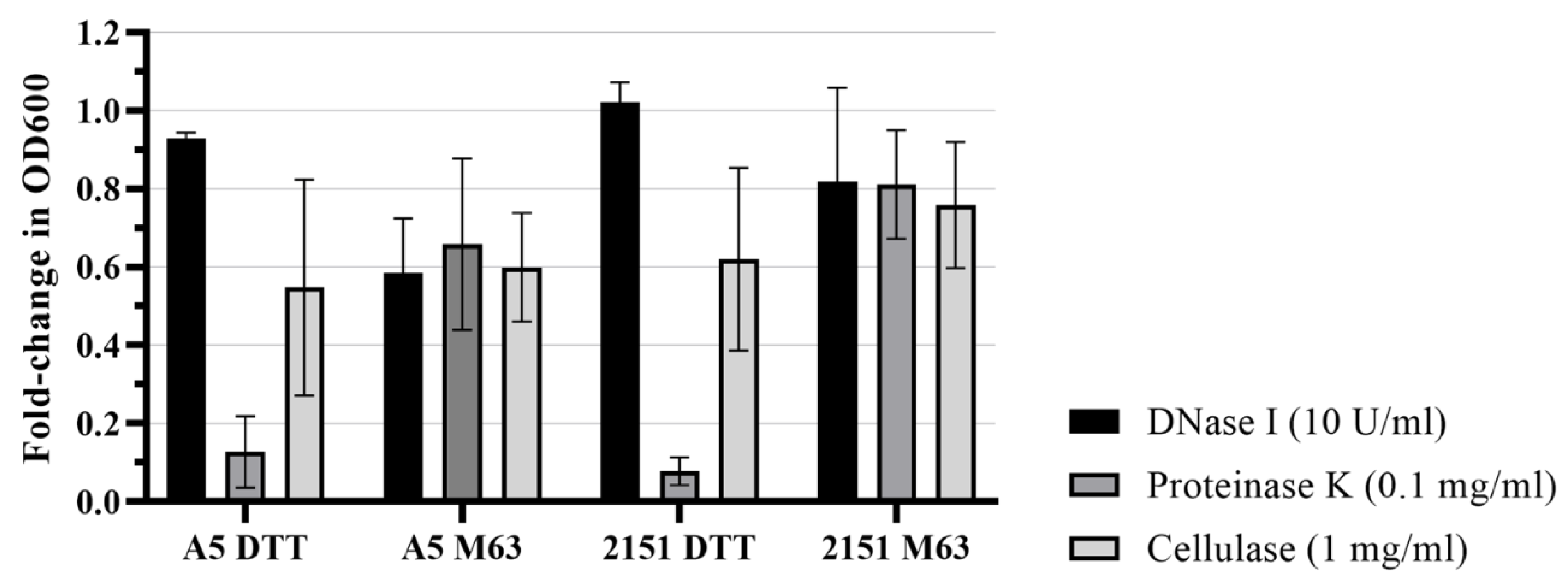
3.3. Biofilm models differ in structurally important ECM molecules
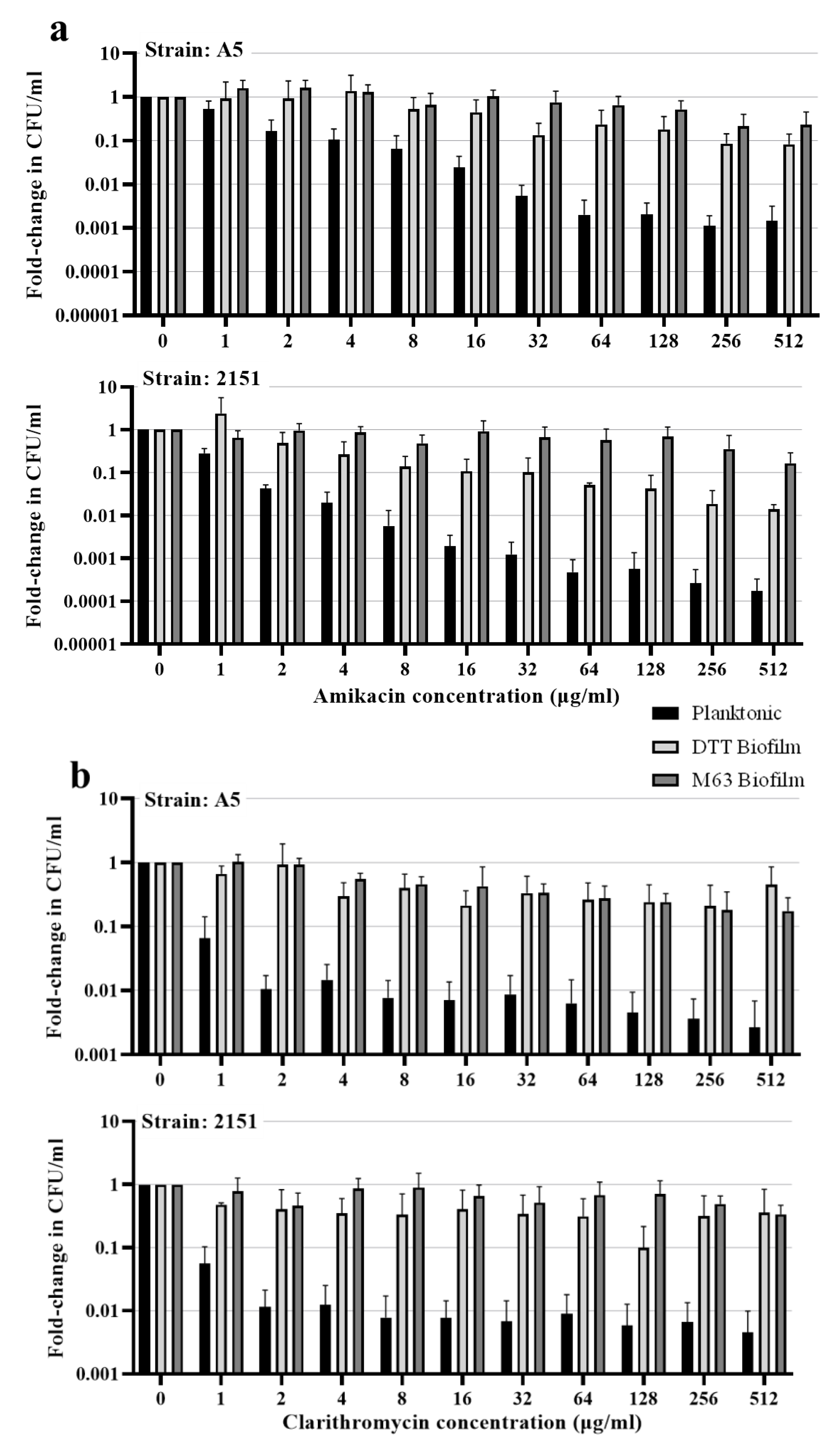
3.4. Biofilm resident M. avium is resistant to killing by antimicrobial drugs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chalmers, J.D.; Aksamit, T.; Carvalho, A.C.C.; Rendon, A.; Franco, I. Non-Tuberculous Mycobacterial Pulmonary Infections. Pulmonology, 2018, 24, 120–131. [Google Scholar] [CrossRef]
- Hoefsloot, W.; Van Ingen, J.; Andrejak, C.; Ängeby, K.; Bauriaud, R.; Bemer, P.; Beylis, N.; Boeree, M.J.; Cacho, J.; Chihota, V.; Chimara, E.; Churchyard, G.; Cias, R.; Daza, R.; Daley, C.L.; Dekhuijzen, P.N.R.; Domingo, D.; Drobniewski, F.; Esteban, J.; Fauville-Dufaux, M.; Folkvardsen, D.B.; Gibbons, N.; Gómez-Mampaso, E.; Gonzalez, R.; Hoffmann, H.; Hsueh, P.R.; Indra, A.; Jagielski, T.; Jamieson, F.; Jankovic, M.; Jong, E.; Keane, J.; Koh, W.J.; Lange, B.; Leao, S.; Macedo, R.; Mannsåker, T.; Marras, T.K.; Maugein, J.; Milburn, H.J.; Mlinkó, T.; Morcillo, N.; Morimoto, K.; Papaventsis, D.; Palenque, E.; Paez-Peña, M.; Piersimoni, C.; Polanová, M.; Rastogi, N.; Richter, E.; Ruiz-Serrano, M.J.; Silva, A.; Da Silva, M.P.; Simsek, H.; Van Soolingen, D.; Szabó, N.; Thomson, R.; Fernandez, T.T.; Tortoli, E.; Totten, S.E.; Tyrrell, G.; Vasankari, T.; Villar, M.; Walkiewicz, R.; Winthrop, K.L.; Wagner, D. The Geographic Diversity of Nontuberculous Mycobacteria Isolated from Pulmonary Samples: An NTM-NET Collaborative Study. Eur. Respir. J. 2013, 42, 1604–1613. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Upadhyay, V. Epidemiology, Diagnosis & Treatment of Non-Tuberculous Mycobacterial Diseases. Indian Journal of Medical Research, 2020, 152, 185–226. [Google Scholar] [CrossRef]
- Prevots, D.R.; Marras, T.K. Epidemiology of Human Pulmonary Infection with Nontuberculous Mycobacteria a Review. Clinics in Chest Medicine, 2015, 36, 13–34. [Google Scholar] [CrossRef] [PubMed]
- Falkinham, III, J.O. Surrounded by Mycobacteria: Nontuberculous Mycobacteria in the Human Environment. J. Appl. Microbiol., 2009, 107, 356–367. [CrossRef] [PubMed]
- Tzou, C.L.; Dirac, M.A.; Becker, A.L.; Beck, N.K.; Weigel, K.M.; Meschke, J.S.; Cangelosi, G.A. Association between Mycobacterium Avium Complex Pulmonary Disease and Mycobacteria in Home Water and Soil a Case-Control Study. Ann. Am. Thorac. Soc., 2020, 17, 57–62. [Google Scholar] [CrossRef]
- Falkinham, J.O.; Iseman, M.D.; de Haas, P.; van Soolingen, D. Mycobacterium Avium in a Shower Linked to Pulmonary Disease. J. Water Health, 2008, 6, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Mangione, E.J.; Huitt, G.; Lenaway, D.; Beebe, J.; Bailey, A.; Figoski, M.; Rau, M.P.; Albrecht, K.D.; Yakrus, M.A. Nontuberculous Mycobacterial Disease Following Hot Tub Exposure. Emerg. Infect. Dis., 2001, 7, 1039–1042. [Google Scholar] [CrossRef]
- Nishiuchi, Y.; Maekura, R.; Kitada, S.; Tamaru, A.; Taguri, T.; Kira, Y.; Hiraga, T.; Hirotani, A.; Yoshimura, K.; Miki, M.; Ito, M. The Recovery of Mycobacterium Avium-Intracellulare Complex (MAC) from the Residential Bathrooms of Patients with Pulmonary MAC. Clin. Infect. Dis., 2007, 45, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, L.A.; Falkinham, J.O. Aerosolization of Mycobacterium Avium and Mycobacterium Abscessus from a Household Ultrasonic Humidifier. J. Med. Microbiol., 2018, 67, 1491–1495. [Google Scholar] [CrossRef]
- Falkinham, J. Mycobacterium Avium Complex: Adherence as a Way of Life. AIMS Microbiol., 2018, 4, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Norton, C.D.; LeChevallier, M.W.; Falkinham, J.O. Survival of Mycobacterium Avium in a Model Distribution System. Water Res., 2004, 38, 1457–1466. [Google Scholar] [CrossRef]
- Steed, K.A.; Falkinham, J.O. Effect of Growth in Biofilms on Chlorine Susceptibility of Mycobacterium Avium and Mycobacterium Intracellulare. Appl. Environ. Microbiol., 2006, 72, 4007–4011. [Google Scholar] [CrossRef]
- Carter, G.; Wu, M.; Drummond, D.C.; Bermudez, L.E. Characterization of Biofilm Formation by Clinical Isolates of Mycobacterium Avium. J. Med. Microbiol., 2003, 52, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Mcnabe, M.; Tennant, R.; Danelishvili, L.; Young, L.; Bermudez, L.E. Mycobacterium Avium Ssp. Hominissuis Biofilm Is Composed of Distinct Phenotypes and Influenced by the Presence of Antimicrobials. Clin. Microbiol. Infect., 2011, 17, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Bajeli, S.; Kaushal, D.; Radotra, B.D.; Kumar, A. Biofilm Formation in the Lung Contributes to Virulence and Drug Tolerance of Mycobacterium Tuberculosis. Nat. Commun., 2021, 12. [Google Scholar] [CrossRef]
- Chakraborty, P.; Kumar, A. The Extracellular Matrix of Mycobacterial Biofilms: Could We Shorten the Treatment of Mycobacterial Infections? Microbial Cell, 2019, 6, 105–122. [Google Scholar] [CrossRef]
- Freeman, R.; Geier, H.; Weigel, K.M.; Do, J.; Ford, T.E.; Cangelosi, G.A. Roles for Cell Wall Glycopeptidolipid in Surface Adherence and Planktonic Dispersal of Mycobacterium Avium. Appl. Environ. Microbiol., 2006, 72, 7554–7558. [Google Scholar] [CrossRef]
- Recht, J.; Kolter, R. Glycopeptidolipid Acetylation Affects Sliding Motility and Biofilm Formation in Mycobacterium Smegmatis. J. Bacteriol., 2001, 183, 5718–5724. [Google Scholar] [CrossRef]
- Van Wyk, N.; Navarro, D.; Blaise, M.; Berrin, J.-G.; Henrissat, B.; Drancourt, M.; Kremer, L. Characterization of a Mycobacterial Cellulase and Its Impact on Biofilm-and Drug-Induced Cellulose Production. Glycobiology, 2017, 27, 392–399. [Google Scholar] [CrossRef]
- Trivedi, A.; Mavi, P.S.; Bhatt, D.; Kumar, A. Thiol Reductive Stress Induces Cellulose-Anchored Biofilm Formation in Mycobacterium Tuberculosis. Nat. Commun., 2016, 7. [Google Scholar] [CrossRef]
- Mavi, P.S.; Singh, S.; Kumar, A. Media Component Bovine Serum Albumin Facilitates the Formation of Mycobacterial Biofilms in Response to Reductive Stress. BMC Microbiol., 2023, 23, 111. [Google Scholar] [CrossRef] [PubMed]
- Rose, S.J.; Babrak, L.M.; Bermudez, L.E. Mycobacterium Avium Possesses Extracellular DNA That Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics. PLoS One, 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Nishiuchi, Y. Ultrastructure of the Mycobacterium Avium Subsp. Hominissuis Biofilm. Microbes Environ., 2021, 36, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dziadek, J.; Rutherford, S.A.; Madiraju, M. V.; Atkinson, M.A.L.; Rajagopalan, M. Conditional Expression of Mycobacterium Smegmatis FtsZ, an Essential Cell Division Gene. Microbiology, 2003, 149, 1593–1603. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Carette, X.; Potluri, L.P.; Sharp, J.D.; Xu, R.; Prisic, S.; Husson, R.N. Investigating Essential Gene Function in Mycobacterium Tuberculosis Using an Efficient CRISPR Interference System. Nucleic Acids Res., 2016, 44. [Google Scholar] [CrossRef]
- Hett, E.C.; Rubin, E.J. Bacterial Growth and Cell Division: A Mycobacterial Perspective. Microbiol. Mol. Biol. Rev., 2008, 72, 126–156. [Google Scholar] [CrossRef]
- Rose, S.J.; Bermudez, L.E. Identification of Bicarbonate as a Trigger and Genes Involved with Extracellular DNA Export in Mycobacterial Biofilms. MBio, 2016, 7. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y. Bacterial Cellulose Biosynthesis: Diversity of Operons, Subunits, Products, and Functions. Trends in Microbiology, 2015, 23, 545–557. [Google Scholar] [CrossRef]
- Dumitrache, A.D.; Wolfaardt, G.; Allen, G.; Liss, S.N.; Lynd, L.R. Form and Function of Clostridium Thermocellum Biofilms. Appl. Environ. Microbiol., 2013, 79, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Falkinham, J.O. Growth in Catheter Biofilms and Antibiotic Resistance of Mycobacterium Avium. J. Med. Microbiol., 2007, 56, 250–254. [Google Scholar] [CrossRef]
- Ojha, A.; Anand, M.; Bhatt, A.; Kremer, L.; Jacobs, W.R.; Hatfull, G.F. GroEL1: A Dedicated Chaperone Involved in Mycolic Acid Biosynthesis during Biofilm Formation in Mycobacteria. Cell, 2005, 123, 861–873. [Google Scholar] [CrossRef]
- Ojha, A.; Hatfull, G.F. The Role of Iron in Mycobacterium Smegmatis Biofilm Formation: The Exochelin Siderophore Is Essential in Limiting Iron Conditions for Biofilm Formation but Not for Planktonic Growth. Mol. Microbiol., 2007, 66, 468–483. [Google Scholar] [CrossRef]
- Ojha, A.K.; Baughn, A.D.; Sambandan, D.; Hsu, T.; Trivelli, X.; Guerardel, Y.; Alahari, A.; Kremer, L.; Jacobs, W.R.; Hatfull, G.F. Growth of Mycobacterium Tuberculosis Biofilms Containing Free Mycolic Acids and Harbouring Drug-Tolerant Bacteria. Mol. Microbiol., 2008, 69, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Ojha, A.K.; Trivelli, X.; Guerardel, Y.; Kremer, L.; Hatfull, G.F. Enzymatic Hydrolysis of Trehalose Dimycolate Releases Free Mycolic Acids during Mycobacterial Growth in Biofilms. J. Biol. Chem., 2010, 285, 17380–17389. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Danelishvili, L.; Wu, M.; MacNab, M.; Bermudez, L.E. Mycobacterium Avium Genes Associated with the Ability to Form a Biofilm. Appl. Environ. Microbiol., 2006, 72, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Maya-Hoyos, M.; Leguizamón, J.; Mariño-Ramírez, L.; Soto, C.Y. Sliding Motility, Biofilm Formation, and Glycopeptidolipid Production in Mycobacterium Colombiense Strains. Biomed Res. Int., 2015, 2015, 419549–10. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Sweet, L. The Mycobacterial Glycopeptidolipids: Structure, Function, and Their Role in Pathogenesis. Glycobiology, 2008, 18, 832–841. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



