1. Introduction
Lucky bamboo (
Dracaena sanderiana hort. ex Mast. =
Dracaena braunii) is a domestic species that is one of the most popular ornamental houseplants in China [
1,
2]. Lucky bamboo belongs to an entirely different taxonomic order from true bamboos, although the stems have a similar appearance [
3]. The history of the lucky bamboo plant goes back to Chinese culture for more than 4000 years [
4], because they are believed to bring good luck and fortune into life. While the lucky bamboo plants are easy to care for and grows well under diverse indoor conditions, they still inevitably encounter several diseases worldwide, including leaf spots caused by
Alternaria alternata in Egypt [
5], leaf blight caused by
Phytophthora nicotianae in Brazil [
6], stem rot caused by
Aspergillus niger [
7] and
Fusarium solani [
8] in Iran, stem and root rot caused by
Fusarium proliferatum in Iraq [
9], leaf chlorosis caused by pepper mild mottle virus (PMMoV) in Korea [
10], and antharacnoses caused by
Colletotrichum dracaenophilum in Bulgaria [
11], Iran [
12], USA [
13], Egypt [
3], and Brazil [
14], respectively, as well as
Colletotrichum karstii [
15] and
Colletotrichum truncatum [
16] in China.
In March 2022, typical anthracnose symptoms were observed on D. sanderiana plants grown in a greenery retail store located in Nanjing, China (32°6′27.281′’N,118°50′24.169′’E). About 40% of the lucky bamboo plants for sale were affected. Ornamentals are expected to have an impeccable appearance, and anthracnose poses a threat to the ornamental value of lucky bamboo. Therefore, the present study aims to (i) isolate the pathogen causing anthracnose, (ii) identify the causal agent using morphological characterization and molecular analysis, and (iii) assess its pathogenicity.
2. Results
2.1. Isolation and Morphological Characteristics of Isolates
Disease symptoms on stems initially appeared as large, sunken, black lesions, covered with numerous black acervuli. Orange-to-pink spore masses were visible within the senescent and dead plants. Symptomatic stem tissues gradually became necrotic and turned soft, and several leaves wilted and eventually abscised from the plants (
Figure 1).
Thirty-two fungal isolates with a notable colony morphology for the
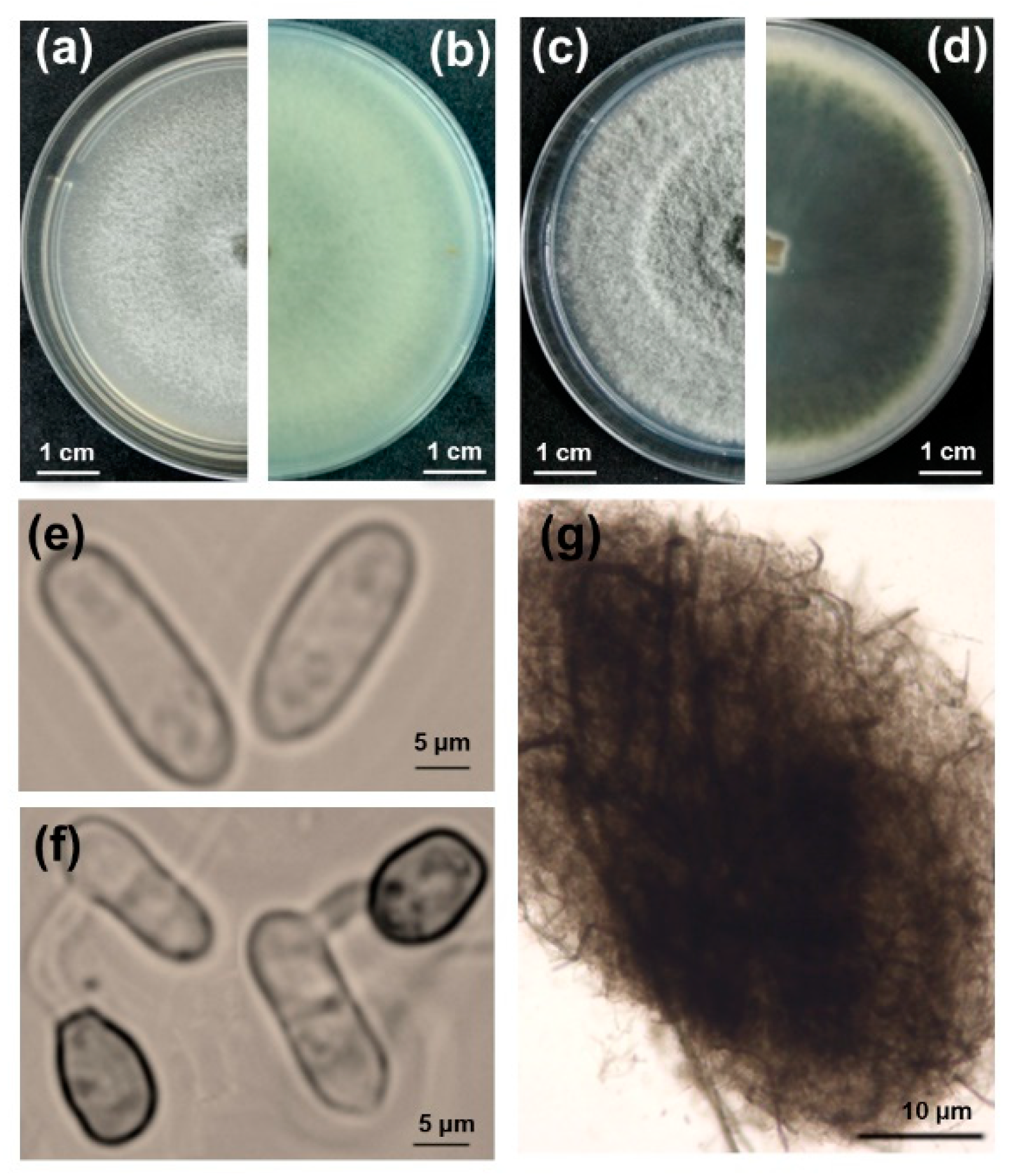
Colletotrichum taxon were obtained from five distinct symptomatic lucky bamboo plants. Follwing that, pure cultures of the 32 isolates were obtained by single-spore isolation. Colonies grown on V8 agar plates in the dark initially produced white, thin, and flat mycelia and eventually became pale gray in the center with concentric rings (
Figure 2a,b). Colonies on potato dextrose agar (PDA) had fluffy mycelia, pale white margins with concentric rings, and an olive-green center, which gradually turned dark green with age and extended to the margins after seven days of incubation in the dark. The reverse side of the colonies completely turned dark green, and the production of crystals in PDA agar was observed when the colonies were incubated at 25°C with a 12-hour photoperiod for nine days (
Figure 2c,d). Microscopically, conidia were hyaline, unicellular, and cylindrical with rounded ends, measuring 10.8 to 18.5 × 2.8 to 6.9 μm (
Figure 2e). Acervuli were circular to elliptical with no setae (
Figure 2g). Appressoria were formed at the tip of the germ tube and were brown and ovate to obovate, measuring 6 to 10 × 4.5 to 7.5 μm (
Figure 2f).
2.2. Molecular characterization
Fragments of the 573 bp internal transcribed spacer (ITS) region, 282 bp actin (ACT), and 277 bp glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were respectively amplified from the genomic DNA of all 32 single-spore isolates. Multiple sequence alignments by ClustalW revealed that the ITS, ACT and GAPDH sequences of 32 isolates shared 100% sequence similarity with each other. One representative single-spore isolate, named FGZ-1, was deposited in the China Center of Industrial Culture Collection (CICC) under number CICC 41715. The Basic Local Alignment Search Tool (BLAST) search in GenBank revealed that the obtained sequences of FGZ-1 (GenBank accession nos. MH752444.1-ITS, MH757114.1-ACT, and MH757113.1-GAPDH) shared 99-100% similarity with related sequences of C. gloeosporioides in Genbank (strain Sour8-accession no. KX227593.1, strain BWH1-accession no. KF712382.1, and strain TAX-1-accession no. HM575314.1, respectively).
2.3. Phylogenetic analysis
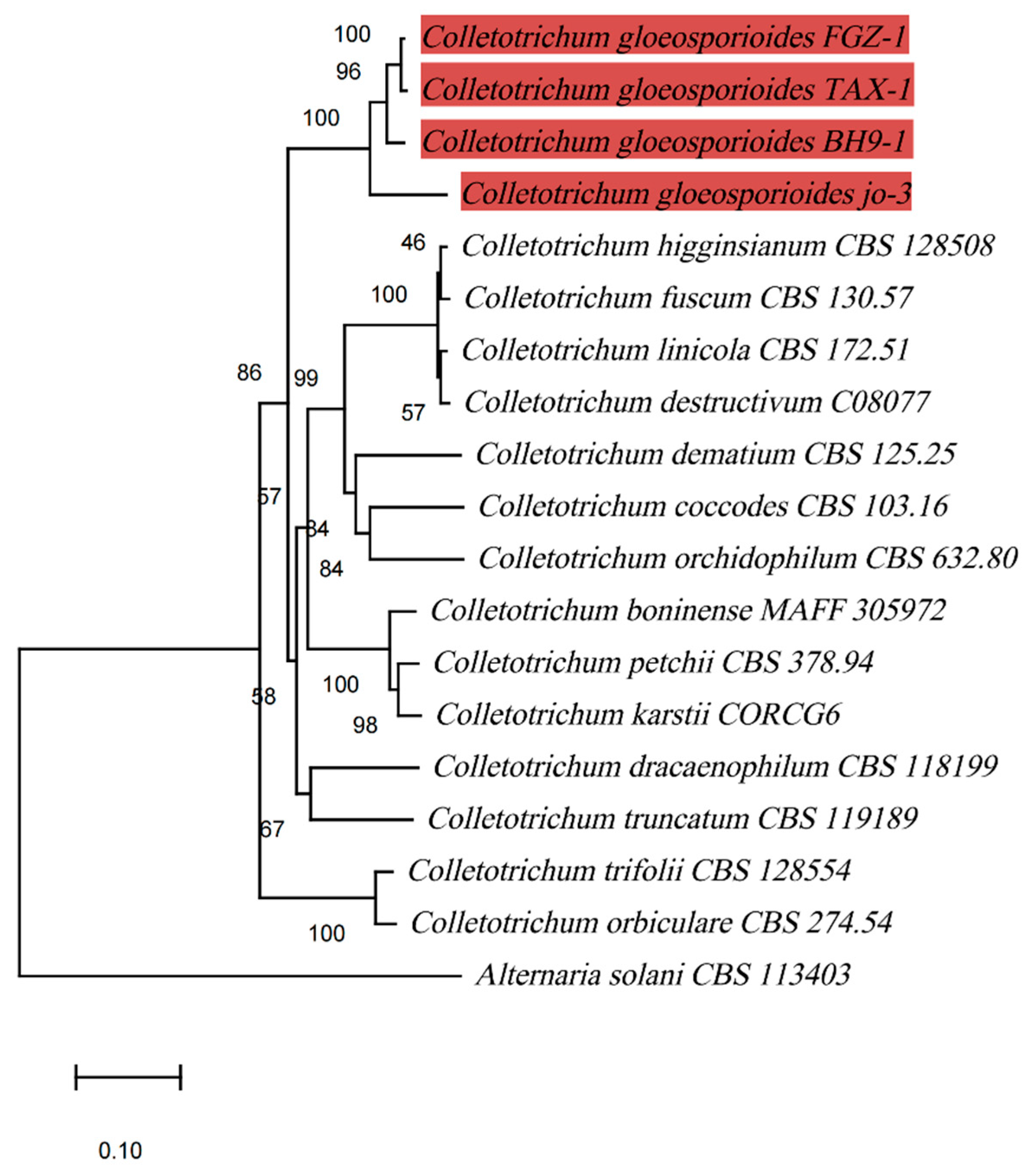
In the phylogenetic tree established on the basis of the concatenated
ITS,
ACT and
GAPDH sequences, isolate FGZ-1 was placed within a monophyletic clade that comprised three reference isolates of
Colletotrichum gloeosporioides with a robust bootstrap and formed a separate clade distinct from other
Colletotrichum species (
Figure 3). Isolate FGZ-1 was separated from
C. dracaenophilum, which was reported to commonly cause anthracnose on lucky bamboo in Bulgaria, Iran, USA, Egypt, and Brazil [
3,
11,
12,
13,
14]. In addition to
C. dracaenophilum, there have been new reports of another three species of
Colletotrichum associated with anthracnose of lucky bamboo
, namely,
C. petchii [
17],
C. karstii [
15], and
C. truncatum [
16], which were also clearly separated from
C. gloeosporioides isolate FGZ-1 (
Figure 3).
2.4. Pathogenicity Tests
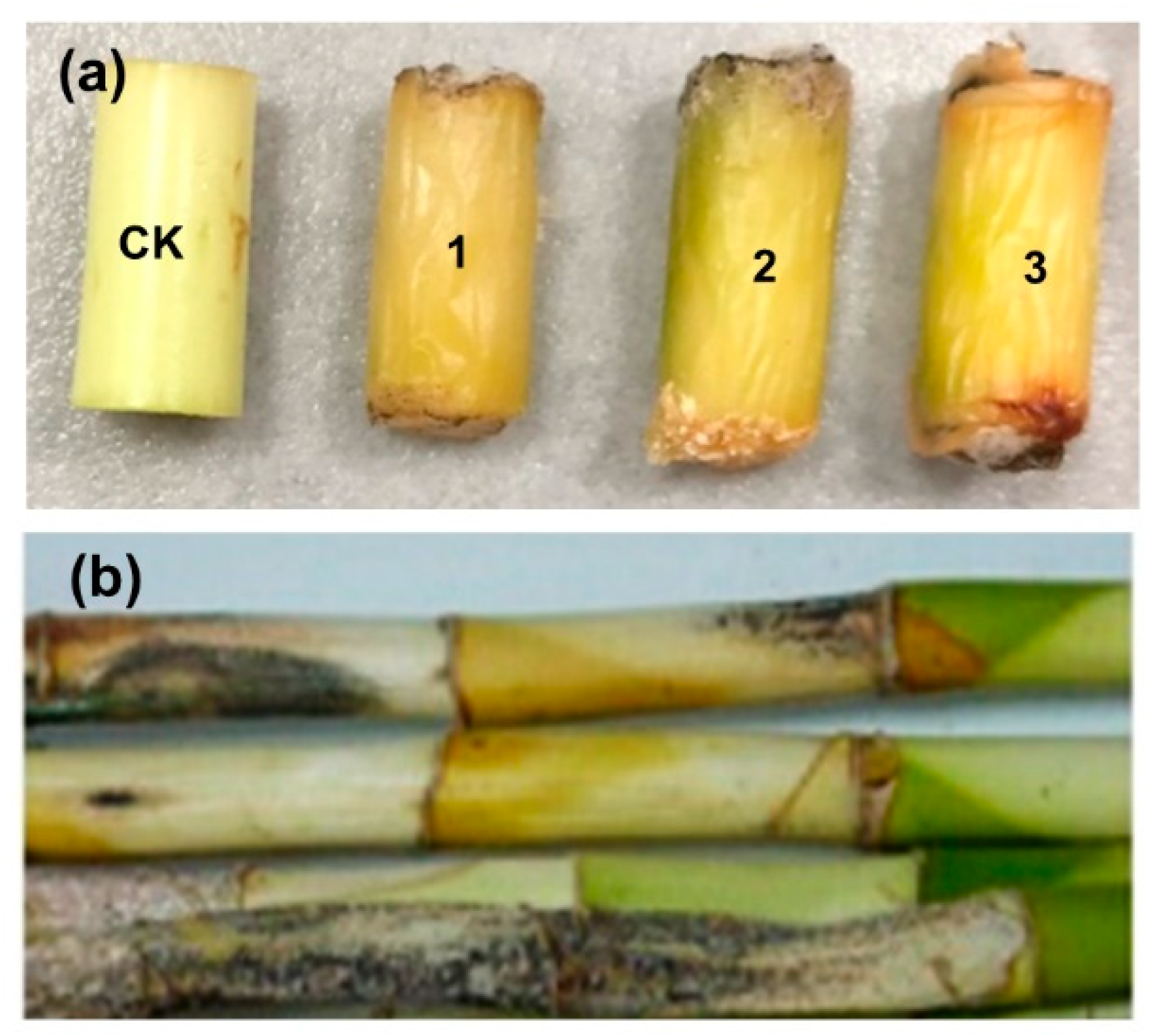
Symptoms of black spots and stem rot on lucky bamboo segments were observed three weeks after being artificially inoculated with mycelial plugs of isolate FGZ-1, and the disease severity reached 100%, while the controls with pure agar plugs remained symptomless (
Figure 4a). Similar disease symptoms were observed on the whole one-year-old plants after eight weeks of inoculation (
Figure 4b). All of the inoculated stem segments and whole plants showed typical anthracnose symptoms similar to those of naturally diseased lucky bamboo plants (
Figure 1). Koch’s postulates were fulfilled by the reisolation of fungal colonies from all symptomatic stems and plants but not from controls, and these isolates had the same colony morphology, microscopic morphology, and molecular characteristics as described above. Similar results were obtained from the three repetitions of the experiment.
3. Discussion
Based on the above morphological characterization, multilocus phylogenetic analysis, and pathogenicity tests, isolates were identified as
Colletotrichum gloeosporioides (Penz.) Penz. & Sacc. [
18] and found to be the causal agent of anthracnose of lucky bamboo in China. On the other hand,
C. gloeosporioides is one of the most common
Colletotrichum fungal pathogens, infecting many economically important plants throughout the world, including citrus, yam, papaya, avocado, coffee, eggplant, loquat, sweet pepper, and tomato [
19]. So far,
C. dracaenophilum is the most widely reported
Colletotrichum species that can cause anthracnose disease on
D. sanderiana plants [
3,
11,
12,
13,
14]. In the United States, Sharma, et al. [
13] previously reported that an isolate of the
C. gloeosporioides species complex caused anthracnose disease in lucky bamboo but was less pathogenic than isolates of
C. dracaenophilum. Moreover, in China, Liu, et al. [
16] and Li, et al. [
15], respectively, provided the first reports that lucky bamboo anthracnose is caused by
C. truncatum and
C. karstii. Nevertheless, to the best of our knowledge, the occurrence of anthracnose symptoms caused by
C. gloeosporioides was observed for the first time on
D. sanderiana in China. The report of
C. gloeosporioides on lucky bamboo in China widens the range of
Colletotrichum species involved in the anthracnose disease of this popular ornamental houseplant. The occurrence and spread of
C. gloeosporioides could be a new potential threat to lucky bamboo production in China. Further investigation is needed on the development of molecular diagnostic techniques and control measures for the anthracnose disease.
4. Materials and Methods
4.1. Sampling and Isolation
Infected stem samples displaying typical anthracnose symptoms were collected from five independent lucky bamboo plants. Stem tissues containing acervuli and spore masses were surface sterilized in a 1% sodium hypochlorite solution for 2 minutes, and rinsed three to four times with sterilized distilled water. The stem tissues were then aseptically cut into small pieces (approximately 0.5 cm long), plated onto potato dextrose agar (PDA) containing rifampicin (100 µg/ml) and streptomycin sulfate (100 µg/ml), and incubated at 25 °C in the dark. After 3 days, the emerging fungal colonies that grew from the stem tissues were sub-cultured on PDA and incubated at 25 °C in the dark. Pure cultures of the isolates were obtained using a single-spore isolation method described by Fei
, et al. [
20].
4.2. DNA Extraction, PCR Amplification and Sequencing
Mycelia of isolates were obtained by inoculating the mycelial plugs into 100 ml of liquid potato dextrose broth (PDB) in a 250-ml baffle flask, culturing them at 25°C with shaking for at least 3 days, and harvesting them by filtration [
21]. Total genomic DNA was extracted from the mycelium using the DNeasy Plant Mini Kit (Qiagen, USA) according to the manufacturer’s protocols. The amount and purity of extracted DNA were determined by a NanoDrop ND-3300 fluorospectrometer (Thermo-Fisher Scientific, USA). DNA integrity was visualized by electrophoresis on 1% (w/v) agarose gels that were stained with ethidium bromide (0.1 mg/l) and viewed under transmitted ultraviolet light.
The internal transcribed spacer (ITS) region of ribosomal DNA (rDNA) was amplified using universal primers ITS1 and ITS4 [
22]. The
actin (
ACT) gene was amplified using primer pairs ACT-512F and ACT-783R [
23]. The
glyceraldehyde-3-phosphate dehydrogenase (
GAPDH) gene was amplified using primers GDF1 and GDR1 [
24]. PCR amplification was carried out in 50-μl reaction mixtures containing 25 μl of 2
PrimeSTAR Max Premix (Takara, Japan), 0.3 μM each primer (GenScript Corporation, China), and 20 ng of genomic DNA. All reactions were performed under the following thermal conditions: 30 cycles of 98°C for 10 s, 55°C for 5 s, and 72°C for 5 s using a PTC2000 PCR instrument (MJ Research, USA). Following PCR amplification, the quality and concentration of PCR products were verified on 1% agarose gels stained with ethidium bromide (0.1 mg/l), viewed with transmitted ultraviolet light, and the sizes of PCR products were determined against Marker II (Tiangen, China). The bands of expected size were excised from the agarose gel, purified by the Agarose Gel DNA Extraction Kit from Takara, and subsequently ligated into the pMDTM19-T vector (Takara, Japan) at 4 °C overnight. Ligated DNA was transformed into
Escherichia coli DH5α cells (Tiangen, China) as described previously [
25]. Positive clones were selected by PCR amplification of inserts using the M13 forward primer and the reverse primers of the corresponding targets. The colony PCRs were carried out directly with a suspension of transformed bacteria in ultrapure water. Clones containing the bands of interest were selected according to the expected PCR product sizes. Plasmid DNA was isolated from the positive clones using the plasmid DNA purification kit (Promega Corporation, USA) according to the manufacturer’s instructions. The inserts were sequenced bi-directionally using universal primers M13-F/R. Sequencing was performed by the GenScript Corporation (Nanjing, China).
4.3. Sequence Alignment and Phylogenetic Analysis
Comparative analysis of the inserted fragments (
ITS,
ACT, and
GAPDH) of all the isolates was performed with ClustalW in BioEdit version 7.0.90 (
www.mbio.ncsu.edu/BioEdit/bioedit.html). Sequences of selected isolates were subsequently deposited in GenBank with accession numbers. The
ITS,
ACT, and
GAPDH sequences obtained in this study, together with the sequences of 16 strains representing closely related species of
Colletotrichum retrieved from GenBank (
Table 1), were aligned using MAFFT version 7 [
26] (
http://mafft.cbrc.jp/alignment/server/). For each strain, the sequences of
ITS,
ACT, and
GAPDH were optimized to the same length and concatenated in the SATé-II v.2.2.7 (Simultaneous Alignment and Tree Estimation) high-throughput alignment platform (
http://phylo.bio.ku.edu/software/sate/sate.html) [
27]. Phylogenetic analyses of the concatenated sequence data of
ITS,
ACT, and
GAPDH were performed by the Maximum Composite Likelihood (ML) method using the substitution model of the Tamura-Nei model [
28], with sequence distances calculated in MEGA version X [
29].
Alternaria solani CBS 113403 was included as an outgroup in the phylogenetic analyses. Confidence intervals were estimated using bootstrap analysis with 1,000 replications. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
4.4. Morphology Assay
Pure culture FGZ-1 was grown on potato dextrose agar (PDA) (200 g of potato, 20 g of dextrose, 15 g of agar powder, and 1,000 ml of distilled water) or V8 juice medium (100 ml of V8 tomato juice, 1 g of CaCO
3, and 15 g of agar per 1000 ml of distilled water) [
30] and incubated at 25±2°C with a 12-h photoperiod for analyzing the colony morphology [
31]. The colony morphology and color were recorded on day nine. The surface of the PDA cultures was flooded with sterilized distilled water and gently scraped to prepare spore suspension. The conidial suspension was then filtered through two layers of Miracloth (Calbiochem, San Diego, CA) to remove mycelia and then placed at 25°C in darkness on the hydrophobic surface of a GelBond membrane (Lonza) to allow germination and appressoria formation. The detailed fungal structures (conidia, germ tube, appressorium, and acervuli) of the obtained isolates were examined under a Zeiss Axio Scope. A1 microscope (equipped with a high-resolution Carl Zeiss AxioCam MRc5x microscopy camera), and at least 30 units per structure were measured.
4.5. Pathogenicity assay
One fungal isolate (FGZ-1) was used as inoculum. For pathogenicity assessment, healthy one-year-old lucky bamboo plants were bought from a flower market in Nanjing, China, planted individually in containers with water, and placed in a greenhouse at day/night temperatures of 26°/23°C under a 16-h light/8-h dark photoperiod for five days. The stems were cut into cylindrical segments (approximately 0.5 cm in width
2 cm in height) and surface sterilized with 70% ethanol, and the top and bottom bases were inoculated with mycelial plugs (5 mm diameter from 7-day-old PDA cultures). Sterile PDA plugs were inoculated as a control. Inoculation sites were subsequently covered with Parafilm strips to prevent dehydration and to hold the mycelial plugs in position. In each inoculation procedure, there were three stem segments inoculated per treatment, and the pathogenicity test was repeated three times. All treated stem segments were incubated at 25°C in a moist chamber (relative humidity >90%) with a 12-h photoperiod and observed for symptom appearance every three days after inoculation. For the pathogenicity assay on live plants, the conidial suspension was prepared as mentioned above and adjusted to 1
10
6 conidia/ml using a hemocytometer. Four plants per treatment were spray- inoculated with 10 ml of conidial suspension, sealed with plastic wrap to prevent desiccation following the method described by Xiong, et al. [
30], and placed back in a greenhouse. Simultaneously, sterile distilled water was used as a control. All experiments were repeated three times. Fungi were reisolated from any resulting lesions, and the morphological and cultural characteristics of the isolated fungi were compared with those of the original fungus.
Author Contributions
Conceptualization, Q.X. and Y.Z.; methodology, Q.X., Y.Z., X.W., W.W., Q.Z., and J.L.; analysis of results, Q.X., Y.Z., Y.Q. and X.W.; resources, Q.X., W.W., Q.Z., and J.L.; writing—original draft preparation, Q.X. and Y.Z.; writing—review and editing, Q.X. and X.W.; supervision, Q.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China under Grant no. 31600512; China Postdoctoral Science Foundation (2021M691605); Postdoctoral Science Foundation of Jiangsu Province (2021K641C); Postgraduate Research & Practice Innovation Program of Jiangsu Province (SJCX23_0350); Students Practice Innovation and Training Program of Nanjing Forestry University under Grant nos. 2022NFUSPITP0364 and 202310298135Y; and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Acknowledgments
The authors are grateful to Kai Tao from Oregon Health and Science University for the English revision of the text.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abdel-Rahman, T.F.M.; Abdel-Megeed, A.; Salem, M.Z.M. Characterization and control of Rhizoctonia solani affecting lucky bamboo (Dracaena sanderiana hort. ex. Mast.) using some bioagents. Sci. Rep. 2023, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Junaid, A.; Mujib, A.; Sharma, M. Cell and tissue culture of Dracaena sanderiana Sander ex Mast: a review. Hamdard Medicus 2009, 52, 31–35. [Google Scholar]
- Morsy, A.A.; Elshahawy, I.E. Anthracnose of lucky bamboo Dracaena sanderiana caused by the fungus Colletotrichum dracaenophilum in Egypt. J. Adv. Res. 2016, 7, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Chongtham, N.; Bisht, M.S. Bamboo shoot: superfood for nutrition, health and medicine; CRC Press: 2020.
- Hilal, A.; El-Argawy, E.; Korany, A.E.; Fekry, T. Chemical and Biological Control of Dracaena marginata Leaf Spots in Northern Egypt. Int. J. Agric. Biol. 2016, 18, 1201–1212. [Google Scholar] [CrossRef]
- Dos Santos, M.; Luz, E.; de Souza, J. First record of Phytophthora nicotianae causing leaf blight on Dracaena sanderiana. New Dis. Rep. 2011, 24, 28–28. [Google Scholar] [CrossRef]
- Abbasi, M.; Aliabadi, F. First Report of Stem Rot of Dracaena Caused by Aspergillus niger in Iran. Plant Heal. Prog. 2008, 9. [Google Scholar] [CrossRef]
- Abedi-Tizaki, M.; Zafari, D.; Sadeghi, J. First report of Fusarium solani causing stem rot of Dracaena in Iran. J. Plant Prot. Res. 2016, 56, 100–103. [Google Scholar] [CrossRef]
- Lahuf, A. First report of Fusarium proliferatum causing stem and root rot on lucky bamboo (Dracaena braunii) in Iraq. Hell. Plant Prot. J. 2019, 12, 1–5. [Google Scholar] [CrossRef]
- Kim, S.-W.; Jeong, Y.; Yang, K.-Y.; Jeong, R.-D. First report of natural infection of Dracaena braunii by pepper mild mottle virus in Korea. J. Plant Pathol. 2022, 104, 1579–1579. [Google Scholar] [CrossRef]
- Bobev, S.G.; Castlebury, L.A.; Rossman, A.Y. First Report of Colletotrichum dracaenophilum on Dracaena sanderiana in Bulgaria. Plant Dis. 2008, 92, 173–173. [Google Scholar] [CrossRef] [PubMed]
- Komaki, A.M.; Aghapour, B.; Aghajani, M.A. First report of Colletotrichum dracaenophilum on Dracaena sanderiana. Rostaniha 2012, 13, 111–112. [Google Scholar] [CrossRef]
- Sharma, K.; Merritt, J.L.; Palmateer, A.; Goss, E.; Smith, M.; Schubert, T.; Johnson, R.S.; van Bruggen, A.H. Isolation, Characterization, and Management of Colletotrichum spp. Causing Anthracnose on Lucky Bamboo (Dracaena sanderiana). HortScience 2014, 49, 453–459. [Google Scholar] [CrossRef]
- Macedo, D.M.; Barreto, R.W. Colletotrichum dracaenophilum causes anthracnose on Dracaena braunii in Brazil. Australas. Plant Dis. Notes 2016, 11, 1–3. [Google Scholar] [CrossRef]
- Li, Y.L.; Yan, Z.B.; Wang, Y.H.; Lin, Q.K.; Wang, S.B.; Zhou, Z. First Report of Colletotrichum karstii Causing Anthracnose on Lotus Bamboo (Dracaena sanderiana) in China. Plant Dis. 2018, 102, 2641–2641. [Google Scholar] [CrossRef]
- Liu, Y.L.; Lu, J.N.; Zhou, Y.H. First Report of Colletotrichum truncatum Causing Anthracnose of Lucky Bamboo in Zhanjiang, China. Plant Dis. 2019, 103, 2947–2948. [Google Scholar] [CrossRef]
- Damm, U.; Sato, T.; Alizadeh, A.; Groenewald, J.Z.; Crous, P.W. The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Studies in mycology 2019, 92, 1–46. [Google Scholar]
- Agostini, J.P.; Timmer, L.W.; Mitchell, D. Morphological and pathological characteristics of strains of Colletotrichum gloeosporioides from citrus. Phytopathology 1992, 82, 1377–1382. [Google Scholar] [CrossRef]
- Farr, D.; Rossman, A. Fungal Databases, US National Fungus Collections, ARS, USDA. Retrieved January 16, 2018. 2018.
- Fei, L.-W.; Lu, W.-B.; Xu, X.-Z.; Yan, F.-C.; Zhang, L.-W.; Liu, J.-T.; Bai, Y.-J.; Li, Z.-Y.; Zhao, W.-S.; Yang, J.; et al. A rapid approach for isolating a single fungal spore from rice blast diseased leaves. J. Integr. Agric. 2019, 18, 1415–1418. [Google Scholar] [CrossRef]
- Xiong, Q.; Xu, J.; Zhao, Y.; Wang, K. CtPMK1, a mitogen-activated-protein kinase gene, is required for conidiation, appressorium formation, and pathogenicity ofColletotrichum truncatumon soybean. Ann. Appl. Biol. 2015, 167, 63–74. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 553–556. [Google Scholar] [CrossRef]
- Guerber, J.C.; Liu, B.; Correll, J.C.; Johnston, P.R. Characterization of Diversity in Colletotrichum acutatum sensu lato by Sequence Analysis of Two Gene Introns, mtDNA and Intron RFLPs, and Mating Compatibility. Mycologia 2003, 95, 872–95. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning; Cold spring harbor laboratory press New York: 1989; Volume 2.
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
- Liu, K.; Warnow, T.J.; Holder, M.T.; Nelesen, S.M.; Yu, J.; Stamatakis, A.P.; Linder, C.R. SATé-II: Very Fast and Accurate Simultaneous Estimation of Multiple Sequence Alignments and Phylogenetic Trees. Syst. Biol. 2011, 61, 90–90. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhang, L.; Waletich, J.; Zhang, L.; Zhang, C.; Zheng, X.; Qian, Y.; Zhang, Z.; Wang, Y.; Cheng, Q. Characterization of the Papain-Like Protease p29 of the Hypovirus CHV1-CN280 in Its Natural Host Fungus Cryphonectria parasitica and Nonhost Fungus Magnaporthe oryzae. Phytopathology® 2019, 109, 736–747. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Z.; Cai, L.; Hyde, K.; Yu, Z.; McKenzie, E. Colletotrichum anthracnose of Amaryllidaceae. Fungal Divers. 2009, 39, 123–146. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).