Introduction
Influenza viruses belong to the Orthomyxoviridae family [
1], and together with the Amnoonviridae family [
2], they constitute the order Articulavirales [
3]. Members of this family possess a segmented negative-sense single-stranded RNA genome. The number of genome segments varies depending on the genus: Orthomyxoviridae has 6–8 segments, while Amnoonviridae has 10 segments. Influenza viruses are classified into four genera: Alphainfluenzavirus, Betainfluenzavirus, Gammainfluenzavirus, and Deltainfluenzavirus. Each genus has a single ratified species: influenza A virus (IAV), influenza B virus (IBV), influenza C virus (ICV), and influenza D virus (IDV), respectively [
4]. The Orthomyxoviridae family includes not only influenza viruses but also other orthomyxoviruses categorized into different genera such as Thogotovirus, Isavirus, Mykissvirus, Sardinovirus, and Quarajavirus [
1,
4,
5,
6]. Most recently, it has been proposed to reclassify the genus Quarajavirus as the family Quaranjaviridae. Additionally, a novel and divergent family, tentatively named Cnidenomoviridae, has been discovered in Cnidaria (including corals) and is now included in the order Articulavirales, bringing the total number of families in this order to four [
7].
IAV exhibits the highest genetic variability and the broadest host range among the four types of influenza viruses. IAVs are further divided into subtypes based on the antigenicity of two envelope glycoproteins: rod-shaped haemagglutinin (HA) and mushroom-shaped neuraminidase (NA). To date, 16 HA types and nine NA types of IAV have been described [
8], and all of these subtypes, which have a worldwide distribution, are considered to have originated from the silent reservoir in wild aquatic birds [
9]. However, two novel influenza A-like virus subtypes, designated “H17N10” and “H18N11,” have only been detected in bats from Central and South America [
10,
11]. Moreover, the recent identification of a highly divergent influenza virus associated with the sturgeon virus suggests that fish may have been one of the earliest hosts of influenza viruses [
7].
Influenza viruses infect many wild birds and mammals, including humans, adapting to new hosts and forming several lineages that are specific to humans, horses, swine, dogs, and other animals [
12]. Thus, depending on their origin host, IAVs can be classified as avian influenza viruses (AIVs), swine influenza viruses (SIVs), or other animal influenza viruses. In human populations, seasonal epidemics and occasional pandemics are caused by mutations in the genomes of human influenza viruses. The IAV subtypes that circulate in bird populations can mutate into the highly pathogenic avian influenza (HPAI) phenotype, which causes severe disease in poultry and results in high mortality rates. These mutations frequently occur in the AIV H5 and H7 subtypes [
13]. Conversely, viruses that cause mild diseases in poultry are referred to as low pathogenic avian influenza (LPAI) viruses. Since 2022, outbreaks of HPAI H5N1 viruses have been detected first in Europe, then in North America [
14], and subsequently in Latin America in commercial poultry, backyard poultry, wild birds, and mammals, including humans [
15,
16]. This is an unprecedented occurrence. The current HPAI epidemic persists, with 38 poultry-related outbreaks and 152 incidents involving non-poultry avian and mammalian species reported globally, except Oceania, according to the World Animal Health Information System situation report covering the 14 July to 24 August 2023 period [
17]. Approximately 230,000 poultry birds perished or were culled during this time. Outbreaks are anticipated to decline, particularly among poultry, although sporadic non-poultry avian incidents continue, such as the recent sizable mortality event in Norway's Troms Og Finnmark region, where 12,000 black-legged kittiwake birds succumbed to the virus. The ongoing circulation of the virus in all global regions, except Oceania, remains a significant concern, encompassing both poultry and other animal species [
17]. This review provides a summary of the latest developments in the evolution of different IAV subtypes in birds and mammals, including humans, in Chile, with particular emphasis on the current status of HPAI H5N1 viruses (
Table S1).
Influenza A virus subtypes in commercial poultry
AIV H7N3
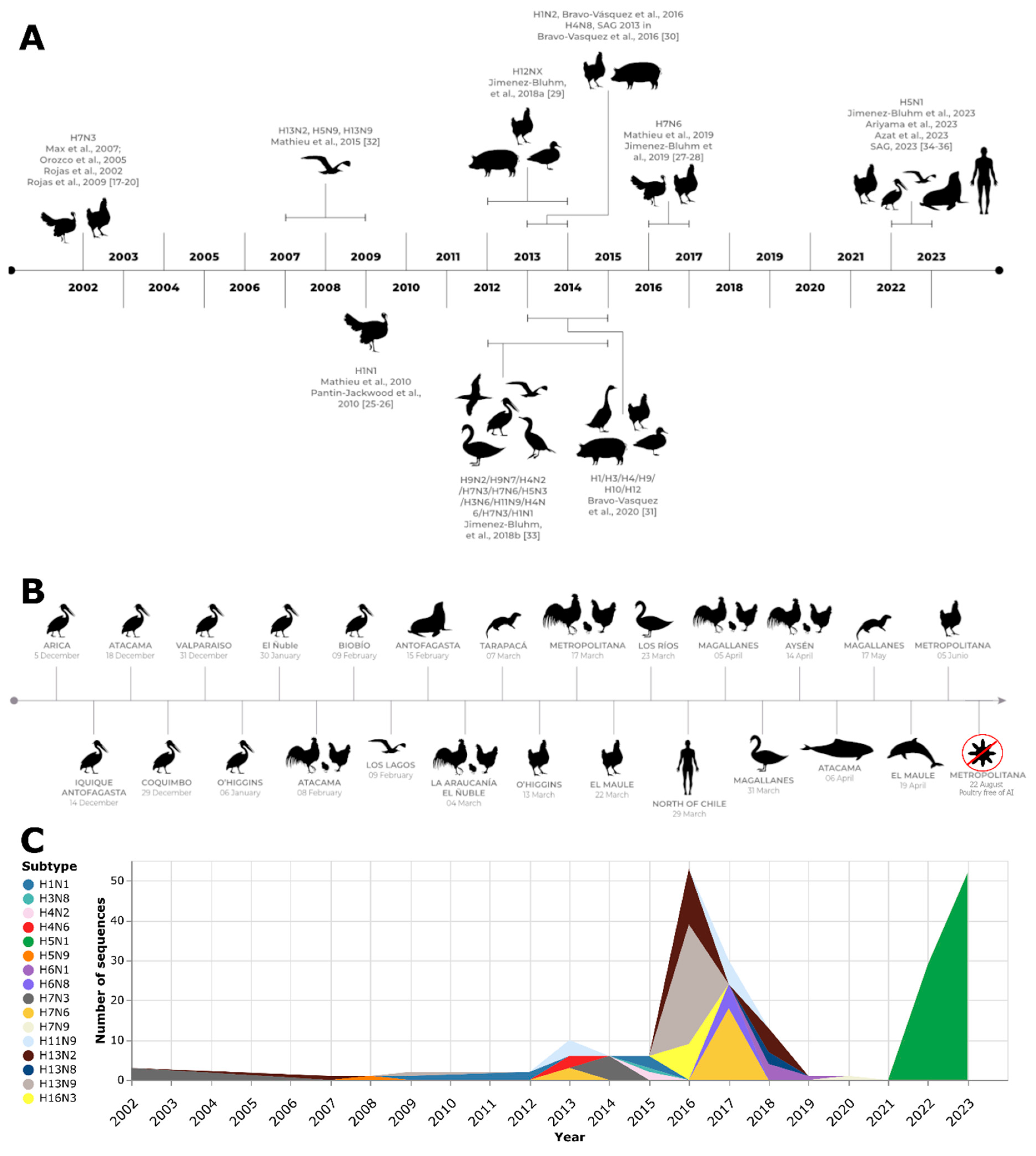
In May 2002, an HPAI virus outbreak occurred in Chile, marking the first occurrence of such a virus in the country [
18] (
Figure 1). This outbreak was attributed to the H7N3 subtype and classified as lineage A/chicken/Chile/184240-4322/2002(H7N3), which had evolved from a previous LPAI virus. The epicenter of the outbreak was a broiler breeder farm located in the densely populated poultry region of San Antonio, V Region, Chile. The farm encompassed 27 poultry sheds housing birds ranging from 1 to 79 weeks of age, as well as a hatchery [
19]. Between April and May 2002, the farm experienced a clinical disease characterized by a relatively low mortality rate, a slight decline in egg production, and cases of salpingo-peritonitis. In certain cases, mortality was so sudden that no clinical signs were observable. The necropsy revealed cyanotic combs and wattles, as well as petechial hemorrhages in various organs such as the muscles, heart, pancreas, and legs. Additionally, subcutaneous edema was present. Subsequent surveillance activities identified a second outbreak occurring one week later at a turkey breeding farm owned by the same company. This farm, situated 4 km from the initial outbreak, consisted of eight pens that housed turkeys ranging in age from 6 to 59 weeks, alongside a hatchery. The infection was confined to only 25% of the sheds. Clinical signs predominantly affected the upper respiratory tract and were followed by a sudden increase in mortality. Cloacal swabs were collected, and an HPAI virus was isolated from two samples from this farm, which were identical to those obtained from the index case [
19].
In response to these two outbreaks, Chilean authorities implemented an extensive and rigorous sanitation program to curb the spread of the disease and eradicate it as swiftly as possible [
19]. Furthermore, a comprehensive set of measures has been employed to control the disease. These measures include establishing surveillance zones, implementing nationwide control strategies, enforcing quarantine measures, conducting depopulation procedures, intensifying surveillance, controlling movement, and enhancing biosecurity. Thorough cleaning, disinfection, and sentinel monitoring were conducted in addition to zoning procedures, which facilitated the safe exportation of unaffected poultry products [
19].
Although positive serologic results for H5N2 were detected in other poultry farms, no clinical signs were observed, and the virus was not isolated. Further investigation revealed that the positive serology stemmed from a contaminated batch of an imported vaccine for the inclusion body hepatitis virus [
21,
22].
The initial identification of LPAI H7N3 was later followed by the detection of HPAI H7N3, which was accompanied by increased mortality rates. Detailed analysis of the viral genomes has yielded valuable insights, revealing minimal genetic differences between the low and highly pathogenic strains, except for a significant alteration in the cleavage site of the HA protein. The LPAI H7N3 virus exhibited a cleavage site that is similar to that of other LPAI H7 viruses. In contrast, the HPAI H7N3 isolates displayed a 30-nucleotide insertion at this specific site. This insertion likely resulted from recombination events between the HA and nucleoprotein genes of the LPAI H7N3, which led to an increase in pathogenicity. A comparative analysis of the full sequences of the eight gene segments confirmed that the Chilean viruses have a distinct nature compared to other AIVs, forming a unique clade that is exclusive to South America. These findings highlight the critical importance of continuously monitoring and surveillance AIVs. These viruses can undergo mutations that significantly increase their pathogenicity [
13]. In addition, identifying a distinct viral clade that is specific to South America underscores the necessity for tailored understanding and control strategies to effectively manage AIV outbreaks in the region [
23].
During a comprehensive survey of wild waterfowl in Bolivia in 2001, researchers isolated a strain of AIV H7N3 designated A/Cinnamon Teal/Bolivia/4537/01 from a Cinnamon Teal (Anas cyanoptera) specimen. Phylogenetic analysis revealed interesting insights into the genetic characteristics of this viral strain. The Bolivian isolate’s NA and matrix (M) genes shared the highest sequence similarity with North American AIV isolates found in wild birds, indicating a potential genetic link. Surprisingly, the nonstructural (NS) gene displayed a closer relationship to an equine influenza virus, suggesting a possible cross-species transmission event. Additionally, the remaining genes showed the greatest resemblance to isolates obtained from the HPAI H7N3 outbreak in commercial poultry in Chile in May 2002 [
19], indicating a potential connection between these events. Further investigations into the characteristics of the A/Cinnamon Teal/Bolivia/4537/01 strain revealed that the cleavage site of the HA protein, as well as pathogenesis studies conducted in chickens, indicated that it is a low-pathogenicity AIV. Interestingly, the infective dose required to cause infection in chickens was approximately 105 times higher than that required for turkeys, suggesting potential variations in host specificity within this H7N3 AIV strain. These findings highlight the complex nature of AIVs, their genetic diversity, and their ability to infect different host species. The identification of genetic similarities between AIV isolates from wild birds in North America, equine influenza viruses, and outbreaks in Chilean commercial poultry further emphasizes the importance of ongoing surveillance and comprehension of AIVs in both wild and domestic bird populations [
24].
Pandemic Influenza A (H1N1) 2009 (H1N1pdm09)
The pandemic Influenza A (H1N1) 2009 (H1N1pdm09) virus, which caused the fourth influenza pandemic [
25], emerged as a triple reassortant virus in pigs [
26]. It was first recognized in Canada in May 2009, where it caused respiratory disease in pigs, and in Chile in June 2009, where it led to a significant decrease in egg production in turkeys [
27]. During this outbreak, there was a decrease in egg production and shell quality among turkey flocks on two farms (A and B) located in the Valparaiso Region of Chile. The suspicion of AIV led to the collection of blood samples from the affected turkeys on August 14, 2009, for serological testing [
28]. The agar gel immunodiffusion (AGID) assay detected IAV antibodies in 140 out of 227 turkeys sampled, with a higher proportion of positive cases in Farm A (80%) compared to Farm B (32%) [
28]. Consequently, the Chilean Agricultural and Livestock Service (SAG) implemented control measures, including quarantine, intensified biosecurity measures, epidemiologic investigations, and postmortem examinations [
28]. Sampling for the detection of viral RNA was conducted on August 16, 2009, on affected flocks, surrounding premises, and neighboring turkey farms. Two days later, the results showed that the infection was only present in the breeder turkeys from the initially affected flocks of Farms A and B. The real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR) identified RNA corresponding to the IAV M gene but not the H5 or H7 genes. Viral RNA was detected in some cloacal and tracheal swabs but not in the homogenates of turkey embryo lungs and tracheas. This indicates that there was no vertical transmission. Necropsies revealed specific lesions in some birds, while others were recovering. Fecal samples collected from wild birds near Farm A tested negative for the influenza virus. Subtyping tests identified the virus subtype as H1N1. On August 19, 2009, SAG authorities coordinated with the Chilean Public Health Institute (ISP) to isolate the virus and sequence its genome. The viral sequences exhibited nearly identical characteristics to the novel Influenza A (H1N1) pdm09 virus [
28], suggesting reverse zoonosis, which is the transmission of the Influenza A (H1N1) pdm09 virus from humans to birds. The virus was classified as belonging to the A/turkey/Chile/28317-6504-3/2009(H1N1) lineage. Follow-up testing indicated that the virus was eliminated from turkeys within 2–4 weeks. During September and October, SAG implemented an RT-qPCR assay to detect the N1 gene of the Influenza A (H1N1) pdm09 virus as the flock’s egg production gradually recovered. The most recent evidence of infection was obtained on August 31, 2009, indicating that the virus had been successfully eradicated from the turkeys [
28].
AIV H7N6
In December 2016, a turkey farm located in the Valparaiso Region of Chile was infected with LPAI H7N6. Two weeks later, another turkey farm located 70 km north of the first site was also infected. The animals presented a variety of gross lesions, including bursitis, catarrhal to mucopurulent caseous sinusitis, tracheitis, caseous/purulent airsacculitis, polyserositis, pericarditis/hydropericardium, congestion, pulmonary edema, mucopurulent to caseous pneumonia, localized subcutaneous emphysema, mild splenomegaly, pancreatitis, petechiae, and ecchymosis focused on the epicardium and coronary fat, fibrin-purulent pericarditis, pulmonary congestion/mucopurulent to caseous pneumonia, and pleuritis. On January 28, 2017, a backyard poultry farm also tested positive for AIV through the AGID test. However, the PCR test result was negative. Various measures, such as depopulation, zoning, animal movement control, and active surveillance, were implemented to contain the outbreak. The 2016–2017 Chilean LPAI outbreak in turkeys involved a group of viruses that belonged to a monophyletic clade. These viruses share the closest common ancestor with two other viruses, A/yellow_billed_pintail/Chile/10/2014(H7N3) and A/yellow_billed_teal/Chile/9/2013(H7N6), which were collected from wild water birds in the same area. The A/turkey/Chile/2017(H7N6) LPAI virus is part of a native South American lineage. Phylogenetic analysis revealed a close relationship between the A/turkey/Chile/2017(H7N6) LPAI viruses and AIV found in wild aquatic birds in Chile and Bolivia, as well as the A/chicken/Chile 2002 virus, which caused the HPAI H7N3 outbreak in Chile. Based on the HA phylogeny, these South American AIVs formed a distinct cluster [
29,
30].
IAV subtypes in backyard poultry and swine
IAV H12NX
The study by Jimenez-Bluhm et al., 2018 [
31] presents compelling evidence supporting the exposure of poultry in backyard productive systems (BPS) in central Chile to IAV. Between 2012 and 2014, this surveillance study employed a comprehensive analysis of serological and molecular data, which revealed a significant presence and circulation of IAV within backyard populations. Utilizing RT-qPCR, IAV RNA was successfully detected in poultry during three out of four sampling seasons, which further confirms the existence of the virus. Additionally, seroconversion, a confirmatory diagnosis through serology, was observed in 60% of the sampled counties among farm animals, including poultry and swine. These findings collectively highlight the widespread dissemination of IAV among the animal populations studied in central Chile. Although attempts to isolate IAVs from positive samples were unsuccessful, a complete H12 HA sequence was obtained from a cloacal swab of a domestic Muscovy duck (Cairina moschata), and the virus was classified as lineage A/Muscovy duck/Chile/3/2013(H12). The closest sequence comparison revealed a 94% identity with an H12 virus obtained from Alberta in 2003 (A/pintail/Alberta/49/2003) [
31].
SIV H1N2
Bravo-Vasquez et al., 2016 [
32] conducted a study to assess the exposure of backyard poultry and swine in the BPS of the El Yali wetland ecosystem to IAV. The study aimed to determine seropositivity rates and identify specific strains of IAV. During the initial sampling period, 31 BPS were analyzed, and it was found that 42% of them were seropositive for IAV (with a 95% confidence interval ranging from 22% to 49%). During the sampling phase in 2014, which involved 40 BPS, the seropositivity rate increased to 60% (with a 95% confidence interval ranging from 43% to 72%). All nine BPS exhibited seropositivity in both sampling periods. The RT-qPCR assay revealed that 27% of the BPS (with a 95% confidence interval ranging from 14% to 39%) tested positive for the IAV M gene. The cycle threshold (Ct) values of these positive samples ranged from 33.69 to 37.82. However, even though these values were below the RT-qPCR Ct cutoff of 38, they were still relatively high for subtyping or viral isolation. In addition, eight farms (which account for 73% of the RT-qPCR-positive farms) tested positive for IAV through the enzyme-linked immunosorbent assay (ELISA) test. Notably, one BPS showed simultaneous positivity for IAV in multiple species, including poultry, swine, and geese. On this farm, an SIV was successfully identified and subtyped as H1N2 [
32].
IAV H1/H3/H4/H9/H10/H12
Bravo-Vasquez et al., 2020 [
33] described the different risk factors associated with contracting the influenza virus in backyard animal production systems. Through their studies, researchers have identified various subtypes of the avian IAV, including H1, H3, H4, H9, H10, and H12. In terms of seroprevalence at the BPS level, the study found rates of 34.7% (95% CI: 23.1%–46.2%), 19.7% (95% CI: 9.9%–30.6%), and 11.7% (95% CI: 7.2%–16.4%) for the Metropolitan, Valparaiso, and LGB O’Higgins regions, respectively. Regarding prevalence at the BPS level, it was 4.2% (95% CI: 0.0%–8.8%), 8.2% (95% CI: 0.8%–14.0%), and 9.2% (95% CI: 4.8%–13.1%) for the corresponding regions. All of this data was collected from a total of 329 samples obtained from the respective regions of Central Chile [
33].
Avian influenza virus subtypes in wild birds
AIVs H13N2, H5N9, and H13N9
During the period between 2007 and 2009, there were reports of dead birds found along the coastlines of the northern regions (Arica and Atacama) and the central region (Valparaíso) of Chile [
34]. The deceased birds were sampled as part of the surveillance program conducted by the SAG to detect IAVs. Using RT-qPCR, four infected birds were identified, and strains of AIV subtypes A/seagull/Chile/SAG14259/2007(H13N2), A/wild bird/Chile/1805/2008(H5N9), and A/seagull/Chile/5775/2009(H13N9) were isolated. Sequence analysis of gene segments from all of these Chilean strains revealed a closer similarity to AIV strains identified in aquatic birds from North America’s western, central, and eastern regions than to other South American strains found along the Pacific and Atlantic coasts. This suggests that the distribution of Chilean AIV strains may be associated with migratory flyways based on geographical regions. Additionally, these findings also indicate that the distribution of strains was independent in both the Chilean and South American contexts. The observed homogeneity and similarity to North American lineages are also evident in the A/black-bellied whistling duck/Colombia/1/2011(H5N2) strain [
34].
AIVs H9N2, H9N7, H4N2, H7N3, H7N6, H5N3, H3N6, H11N9, H4N6, H7N3, and H1N1
In a study conducted by Jiménez-Bluhm et al., 2018 [
35] in 2012–2015, a comprehensive analysis of fresh wild bird feces in Central and Northern Chile revealed significant findings. A total of 4,036 fecal samples were collected from various sampling sites, out of which 115 samples tested positive for the IAV M gene using RT-qPCR. The prevalence of the influenza virus varies depending on the season and site of collection. A significant increase in positivity was observed during the summer/fall seasons compared to the winter/spring seasons (p=0.007), indicating a seasonal variation in the presence of the virus. Additionally, it was also noted that three sites, which were heavily sampled, accounted for over half of the total sampling effort and yielded 73% of the positive samples. Among the identified bird species, yellow-billed pintails (Anas georgica) and yellow-billed teals (Anas flavrostris) from the Anseriformes order exhibit the highest prevalence of AIV infection and display the greatest AIV strain diversity. Various other species were identified as hosts, such as the Chiloé wigeon (Anas sibilatrix), mallards (Anas platyrhynchos), red-fronted coot (Fulica rufifrons), oystercatchers (Haematopus), gulls (Larus), black-necked stilts (Himantopus mexicanus), gray plovers (Pluvialis squatarola), and whimbrels (Numenius phaeopus). Full genomic sequences were obtained from 16 isolates, while partial sequences were obtained from 20 positive swab samples. The identified AIV strains exhibited diverse combinations of HA and NA subtypes, including H1, H3, H4, H5, H6, H7, H8, H9, H11, H13, N1, N2, N3, N6, and N9. These strains showed unique gene combinations that resembled viruses isolated from both North and South America [
35].
Highly pathogenic avian influenza H5N1 in birds, non-human mammals, and human
In early December 2022, an increase in mortality was observed among wild birds, primarily pelicans, along the north coast of Chile. These birds were found either dead or in critical condition. SAG collected a total of 1,368 samples from both domestic birds (n=1080) and wild birds (n=288) to detect HPAI and conduct an epidemiological investigation. As of December 22, 2022, a total of 578 RT-qPCR reactions and 790 AGID tests had been performed on the collected samples. The VetMAX-Gold AIV Detection Kit (Applied Biosystems™) was used for the RT-qPCR analysis, with a specific focus on targeting the AIV M gene. Various sample types were examined, including tissues (n=13; 15% positivity), tracheal swabs (n=248; 17% positivity), cloacal swabs (n=314; 15% positivity), and oral swabs (n=3; 33% positivity). Notably, no positive AIV cases were detected in the tested poultry samples. Among the species affected by AIV, pelicans (Pelecanus thagus) were the most frequently infected, accounting for 54% (n=50) of the cases. They were followed by vultures (Cathartes aura) and Peruvian boobies (Sula variegata), which indicate their susceptibility to AIV infection. Out of the 10 samples that were sequenced, the average coverage depth was determined to be 33.381x, suggesting that the sequencing was performed at a sufficient level. Among these samples, nine out of 10 complete genomes were successfully obtained. All the sequenced samples were identified as belonging to the H5N1 subtype and the 2.3.4.4b H5 clade. Comparisons with sequences available on the GISAID database revealed that A/Peru/LIM-003/2022 and A/Peru/LAM-002/2022 (GISAID Isolate IDs EPI_ISL_16249730 and EPI_ISL_16249681) exhibited the closest genetic similarity in terms of the HA gene. Similar results were also observed when analyzing the phylogenies of the NA and internal genes, further supporting the genetic relatedness of these samples to previously identified strains. The analysis conducted provides support for the introduction of the HPAI H5N1 clade 2.3.4.4b into the Americas via the Atlantic Flyway. Furthermore, it suggests that the virus has subsequently spread to other migratory bird routes within the region. This finding indicates the potential for significant dissemination of the HPAI H5N1 clade 2.3.4.4b strain among bird populations across different migratory pathways in the Americas [
36].
In their recent study, Jimenez-Bluhm et al., 2023 [
37] conducted extensive AIV surveillance in Chile since 2015, focusing on wild birds and their potential interactions with domestic poultry and humans. To enhance their monitoring efforts, they intensified surveillance in the Lluta River estuary by conducting biweekly sampling starting in August 2022. During August and September of 2022, the prevalence of AIV remained below 1%, as measured by M gene-specific qRT-PCR. However, there was a notable increase in November (2.6%) and December (11.7%), which coincided with the arrival of migratory birds from the Northern Hemisphere. Among the 69 AIV-positive environmental fecal samples collected in November and December (out of a total of 2023 samples), seven samples (10.1%) were identified as belonging to the A/H5 subtype of the HA protein. One sample was obtained in late November, while the remaining six were collected in December. Using Cytochrome Oxidase I speciation, the researchers determined that the A/H5-positive samples originated from various bird species, including the Peruvian pelican, Franklin’s gull, gray gull, elegant tern, and black skimmer. Three of the samples underwent full genome sequencing, resulting in the generation of two complete genomes from the gray gull and black skimmer. However, the Peruvian pelican sample only yielded a partial genome due to a low viral load. A comparison of the A/H5 hemagglutinin genes showed genetic similarity between the Chilean samples and the A/H5N1 viruses detected in both Peruvian pelicans and chickens during the same period. It is believed that these strains, which were found in both Peru and Chile, originated from A/H5N1 viruses that caused widespread outbreaks in poultry and wild birds in North America. Nucleotide and amino acid similarity analyses revealed a high degree of identity among the Chilean A/H5N1 HA sequences, with significant similarity to the A/H5N1 clade 2.3.4.4b candidate vaccine strain, A/Astrakhan/3212/2020.
In their study, Azat et al., 2023 [
38] conducted a spatiotemporal cluster analysis of H5N1 cases in both wild and domestic birds in Chile. The data used in the analysis was reported by SAG to the World Animal Health Information System (WAHIS) and covered the period from December 9, 2022, to March 3, 2023. The analysis identified 14 statistically significant clusters of H5N1 outbreaks, indicating a progression of the epidemic wave from the north to the south of Chile. These clusters were dispersed throughout the country, varying in size from two to 19 birds that tested positive for H5N1 in each cluster. Eight clusters had a radius of less than 1 km, while the remaining six clusters had radii ranging from 4 to 29 km. Notably, four clusters were located in the central zone of Chile, near densely populated areas, while one cluster (#12) was identified in the northern city of Tocopilla, where the first human case of H5N1 occurred at a later time. The study also revealed a robust linear correlation between distance and time since the first outbreak, suggesting a gradual, wave-like dissemination of H5N1. This relationship was further supported by the absence of spatial autocorrelation in the data, as indicated by the non-significant Global Moran’s I index. Additionally, the presence of H5N1 outbreaks in birds was found to be correlated with various ecological and human-related factors, including bird diversity, human activity, precipitation during the wettest month, minimum temperature during the coldest month, and mean diurnal temperature range. On the other hand, the presence of H5N1 was negatively correlated with the distance to the nearest urban center, precipitation seasonality, and isothermality. No significant associations were found between the presence of H5N1 and the annual mean temperature, temperature seasonality, precipitation variables, distance to the closest SAG office, human total population, or density. These findings highlight the complex interplay between ecological and human factors in the distribution and spread of HPAI H5N1 in Chile.
AIV H5N1 in wild birds
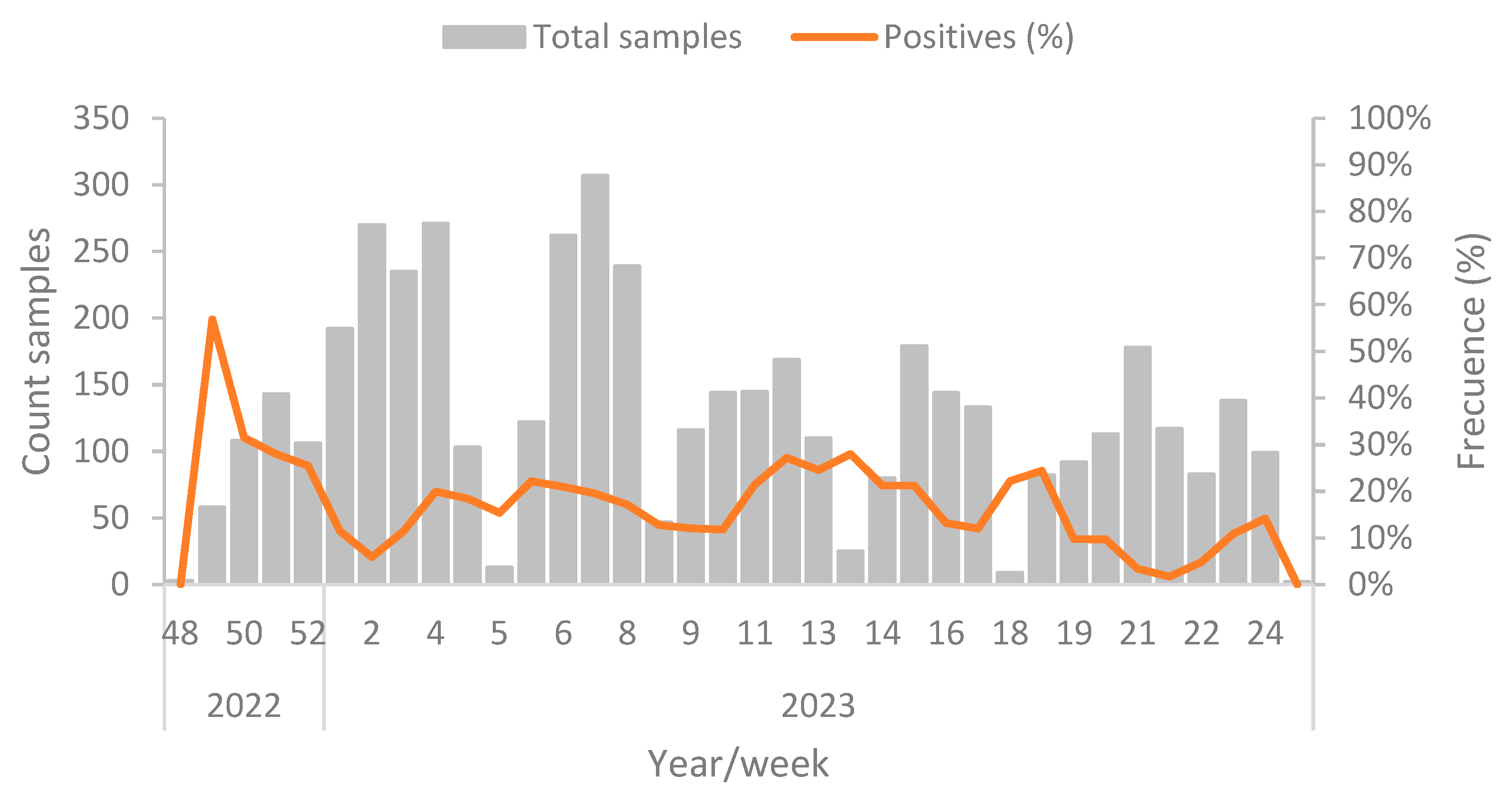
From December 1, 2022, to June 19, 2023, a total of 4,637 samples from wild birds originating from the entire territory of Chile have been analyzed [
39]. Of this total, 778 samples have tested AIV-positive using RT-qPCR that targets the M gene, resulting in a cumulative prevalence rate of 16.77% during the specified period. The highest number of positive samples was recorded during week 49 of 2022, and the positivity rate has remained consistent throughout the entire period, with an average prevalence of 17.0%. There is no apparent correlation between prevalence and the number of samples analyzed, as shown in
Figure 2.
Figure 2.
Positive prevalence of H5N1 cases in wild birds by epidemiological week. All samples were sequenced and categorized as the H5N1 subtype [
39]. The timing of sampling during the migration period could explain the higher prevalence of positive cases observed in the northern and central regions of the country (
Figure 3).
Figure 2.
Positive prevalence of H5N1 cases in wild birds by epidemiological week. All samples were sequenced and categorized as the H5N1 subtype [
39]. The timing of sampling during the migration period could explain the higher prevalence of positive cases observed in the northern and central regions of the country (
Figure 3).
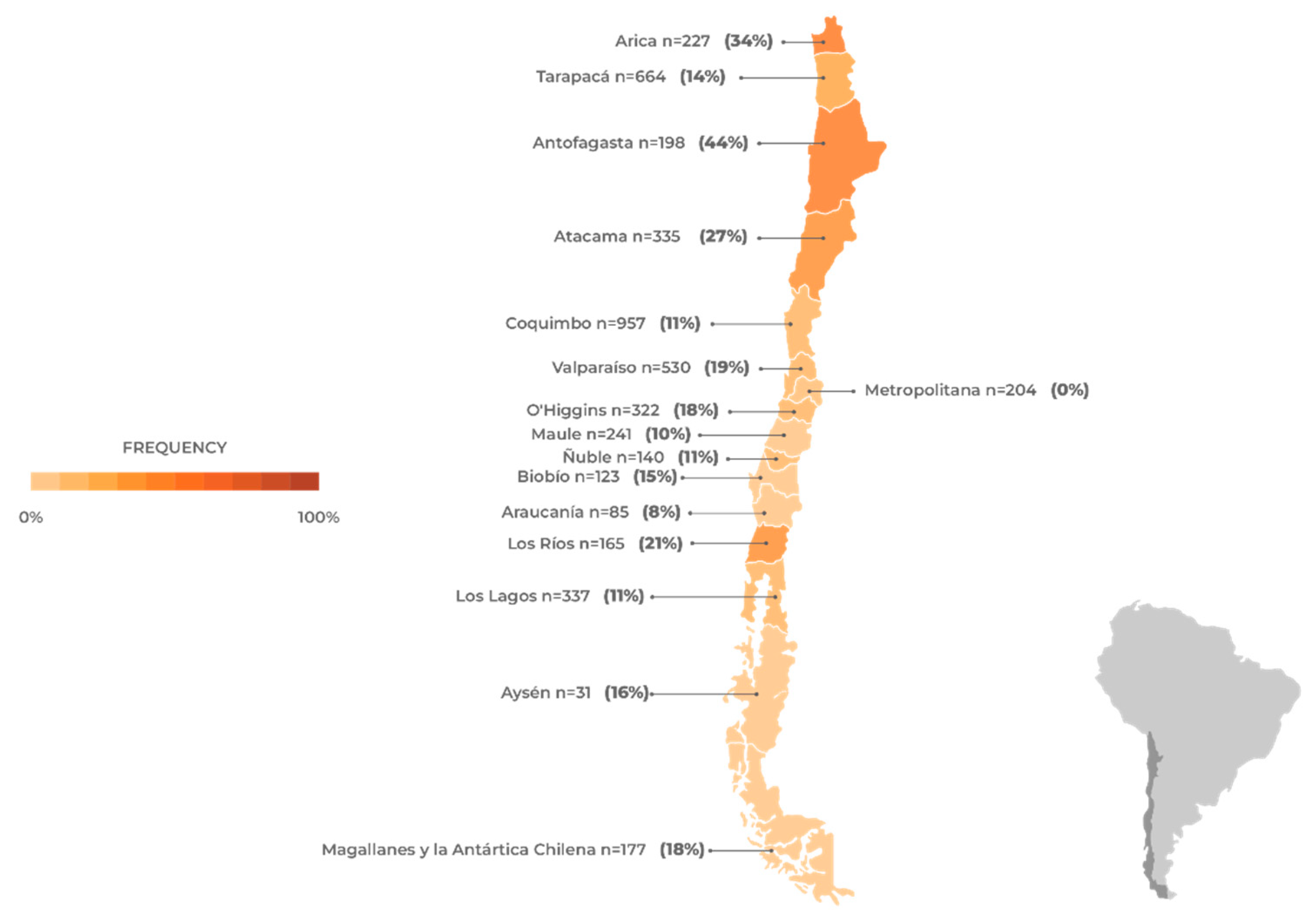
Figure 3.
Avian influenza (H5N1) frequency in wild birds in Chile. The number of samples and the frequency of positives are shown by region based on data obtained from the official surveillance program of the Chilean Agricultural and Livestock Service [
39] between December 5th, 2022, and June 19th, 2023. The number of samples and frequency of positives for the Humboldt Penguin (Spheniscus humboldti) and Magellanic Penguin (Spheniscus magellanicus) analyzed in the official surveillance program of the National Fisheries and Aquaculture Service [
40] are also included.
Figure 3.
Avian influenza (H5N1) frequency in wild birds in Chile. The number of samples and the frequency of positives are shown by region based on data obtained from the official surveillance program of the Chilean Agricultural and Livestock Service [
39] between December 5th, 2022, and June 19th, 2023. The number of samples and frequency of positives for the Humboldt Penguin (Spheniscus humboldti) and Magellanic Penguin (Spheniscus magellanicus) analyzed in the official surveillance program of the National Fisheries and Aquaculture Service [
40] are also included.
The avian species that showed the highest positivity rates are linked to coastal waterways, and a significant number of them are migratory (
Figure 4). Many of these species exhibit local or intercontinental migratory habits, which could explain the observed correlation of genotypes in the Northern Hemisphere.
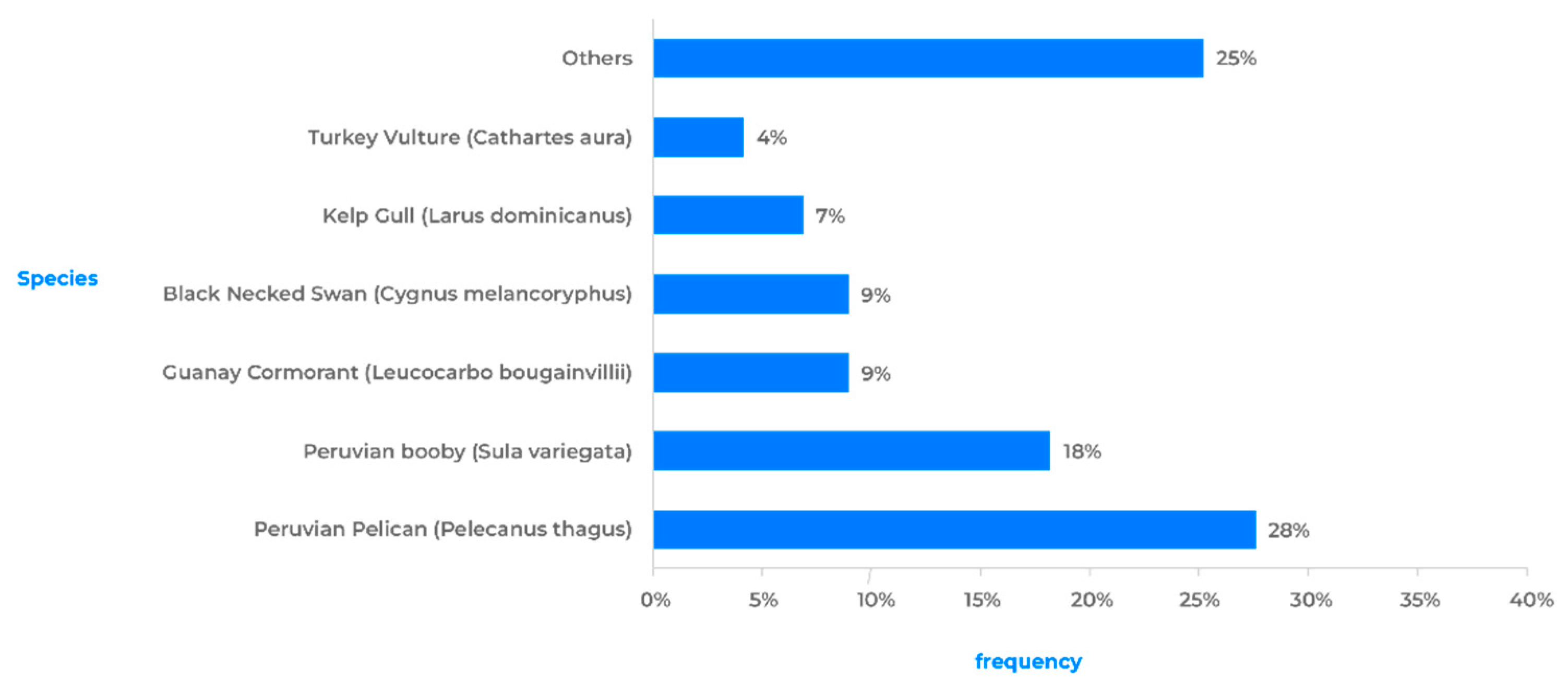
Among the orders of birds with a higher prevalence of positive species, the species belonging to the orders Pelecaniformes (pelicans, herons, and ibises), Suliformes (cormorants and boobies), and Charadriiformes (gulls, terns, and sandpipers) stand out (
Table 1). It has been observed that 75% of all positive avian cases are represented by a limited group of wild bird species. The Humboldt’s Pelican (Pelecanus thagus, Pelecaniformes) has shown the highest susceptibility, accounting for 28% of all positive cases. It is followed by the Peruvian Booby (Sula variegata, Suliformes) and Guanay Cormorant (Leucocarbo bougainvillea, Suliformes), which account for 18% and 9% of positive cases, respectively. Although these three species do not exhibit long-distance migratory behavior, their distribution ranges span a significant portion of the Pacific coast (Chile, Peru, Ecuador, and Colombia), and they are frequently observed together in large flocks feeding near the coast [
41].
On the other hand, specimens of the Kelp Gull (Larus dominicanus, Charadriiformes) and Black-necked Swan (Cygnus melancoryphus, Anseriformes) have a high frequency of positive cases, each accounting for 9% and 7% respectively. Among the long-distance migratory birds (USA-Chile), the presence of positive cases was detected in individuals of Franklin's Gull (Leucophaeus pipixcan) and Sanderling (Calidris alba). Finally, although less frequently, positive cases have been detected in groups of raptors such as the Black Vulture (Coragyps atratus), Peregrine Falcon (Falco peregrinus), and Chilean Hawk (Parabuteo unicinctus) [
39].
AIV H5N1 in marine mammals
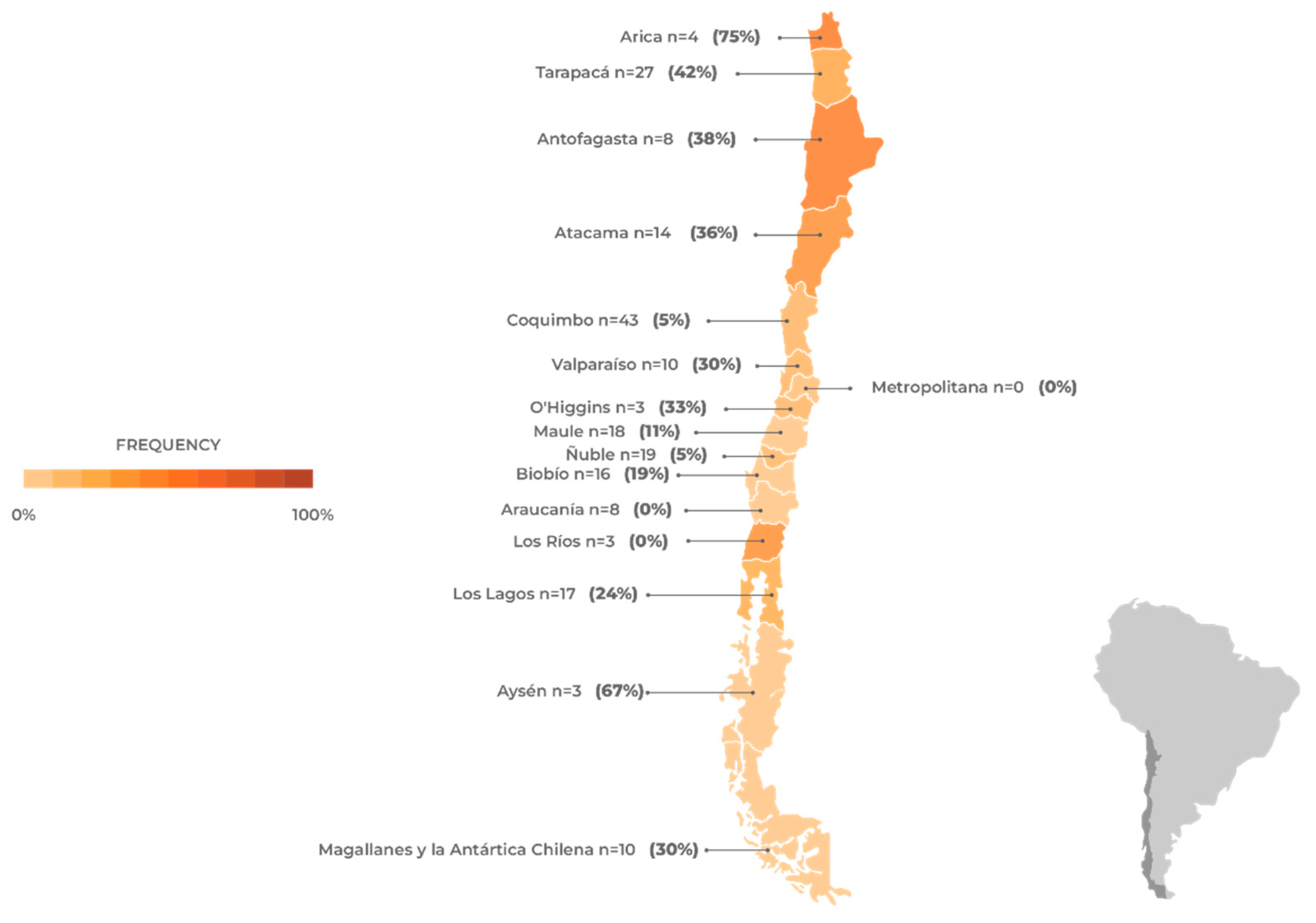
Since the first case of HPAI H5N1 in wild birds, there has been a constant occurrence of the virus affecting different species of marine mammals and birds across various regions of Chile (
Table 2). Therefore, there is currently a state of high alert due to reports of HPAI H5N1 in marine mammals, which coincides with the appearance of H5N1 in wild birds. The data presented in
Figure 5 correspond to the total number of animals sampled and the percentages of animals that tested positive for AIV H5N1, categorized by region in Chile. All data were collected by SERNAPESCA [
40]. Based on this data, the regions of Tarapacá, Coquimbo, Maule, and Los Lagos had the highest number of cases sampled. However, it is worth noting that the virus has the highest prevalence in the regions of Antofagasta, Atacama, and Biobío, with a prevalence rate exceeding 20%. This information provides an overview of the distribution of AIV H5N1 cases across different regions of Chile, highlighting both the quantity of sampled cases and the virus’s prevalence in each region.
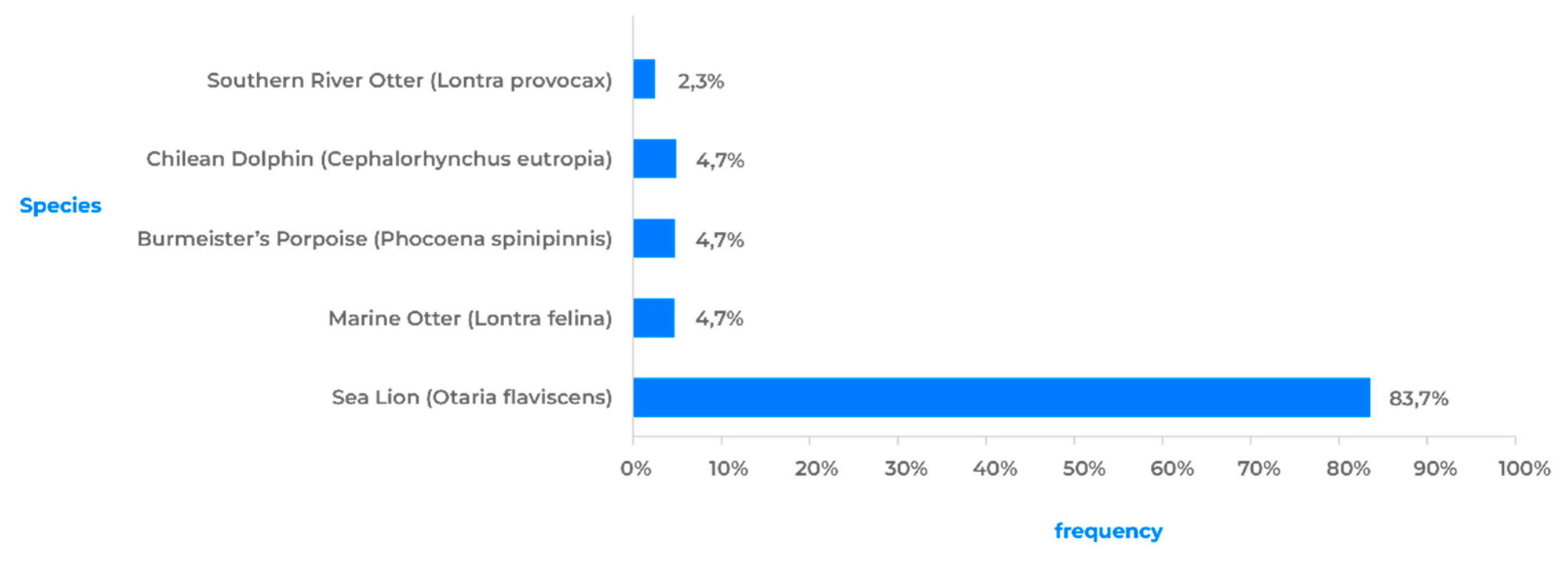
On February 15, 2023, a sea lion (Otaria flavescens) in the Antofagasta Region (
Figure 6) became the first marine mammal in Chile to test positive for AIV H5N1. This is significant because sea lions are present in a large part of Chile’s continental territory and can travel over 200 km in search of food, making them highly susceptible to infection. To date, 32 out of over 144 sea lions that were sampled in Chile have been found positive for AIV H5N1. Another species of marine mammal is the Chungungo (Lontra felina), a carnivorous mammal found from Peru to Argentina. Recently, two cases of AIV H5N1 have been detected in Chungungos, in the northern regions of Chile. One endemic species of a marine mammal is the Chilean dolphin (Cephalorhynchus eutropia), which has been found in two locations in the Maule and Ñuble regions. Finally, the spiny porpoise (Phocoena spinipinnis) is a small cetacean found in the coastal waters of South America. Positive cases of this species have been reported in the Antofagasta and Atacama regions.
The transmission of IAVs between different animal species is a crucial factor in their evolution and ecology. The continued transmission of these viruses from bird species to marine mammal species may play an important role in the development of new viral strains [
42]. In addition, following interspecies transmission of IAVs, other factors, such as coinfection with other pathogens or predisposing environmental conditions, can enhance the virus’s pathogenicity for marine and terrestrial mammals.
The potential for marine mammals to host or carry zoonotic pathogens, such as HPAI H5N1, is a topic that requires further investigation. Moreover, the significant migration of marine mammals along the Chilean continental coast may increase the likelihood of direct contact with humans, thereby playing a key role in the spread of the H5N1 virus.
AIV H5N1 in humans
On March 29, 2023, the Chilean Ministry of Health reported to the World Health Organization (WHO) the first confirmed case of human infection with the AIV H5 virus in the Antofagasta Region [
43]. This case represents the first documented occurrence of human infection with AIV H5 in Chile and the third case in South America [
16]. The patient from Chile is a 53-year-old man who lives in the Antofagasta region. On March 13, 2023, he presented symptoms of a cough and sore throat. There was no reported history of comorbidities or recent travel. As his health deteriorated, he was admitted to the Antofagasta Hospital on March 21, 2023. A nasopharyngeal swab was collected as
part of regular monitoring for severe acute respiratory infections (SARIs). Subsequently, on March 27th, a bronchoalveolar sample was collected and tested positive for a non-sub-typeable IAV using RT-qPCR. The sample was sent to the Institute of Public Health (ISP), where it was confirmed to be positive for AIV H5 [
16].
A recent study analyzed whole-genome H5N1 virus sequences from 77 birds, 8 marine mammals, and the human case [
44]. Remarkably, two mutations in the PB2 segment (Q591K and D701N) associated with mammalian hosts adaptation were identified in Northern Chile during March 2023. D701N was described in four samples, one sanderling, two sea lions, and the human case. Two viruses with D701N also included the Q591K mutation, these are the human case and one sea lion. Notably, these two substitutions showed a frequency of 0.2% for D701N and 0% for Q591K in H5N1 virus sequences collected globally excluding Chile and Peru, since January 01, 2021. In Chile, although these four PB2 H5N1 virus sequences form a cluster it remains unclear whether the D701N mutation was transmitted from sea lion, birds, or if these correspond to independent de novo emergence and there has been no transmission of D701N between hosts. This study also revealed that the D701N substitution was present in 52.9 - 70.9% of sequence reads, signifying the coexistence of both genotypes within the host. Castillo et al., 2023 also confirmed the human case as subtyped in the clade 2.3.4.4b [
45]. Other zoonotic mutations had been described in other mammals [
46].
Perspectives on AIV H5N1 in Chile
A review of the current status of HPAI H5N1 and the evolution of IAV in Chile shows that over the last two decades, different outbreaks in both wild and domestic bird species have been genetically related to viruses in North America. This supports the hypothesis of an introduction through the Atlantic Flyway, rather than local dissemination from South America. Moreover, it further emphasizes the relevance of utilizing biosecurity measures and genomic surveillance approaches to properly control virus dissemination.
The first outbreak of avian flu in Chile was detected in poultry in 2002. The isolated virus belongs to the H7N3 subtype and showed relevant sequence identity with a Bolivian isolate from 2001 that is related to North American isolates. Later in 2009, an outbreak on turkey farms took place. This time, the identified subtype was H1N1, and its sequence identity highlighted its significant relatedness to the pandemic H1N1 strain. Between 2012 and 2014, a surveillance study of the BPS in central Chile determined the circulation of the virus in backyard populations. Then, in 2017, a second turkey farm outbreak was studied. The identified subtype is H7N6, and its sequence analysis revealed significant relatedness to isolates from wild aquatic birds in previous years, underscoring the importance of genomic surveillance not only in farms and BPS but also in wild birds.
From 2007 to 2009, a surveillance study of wild aquatic birds identified the H5N9, H13N2, and H13N9 subtypes. Notably, these isolates showed a closer genetic identity to North American viruses than to South American strains. This finding further emphasizes the critical role of migratory flyways in determining the genetic diversity of the virus in Chile. Then, from 2012 to 2015, a surveillance study of wild birds exhibited a high variability of subtypes and host species. The study revealed that the season and site of the collection are relevant factors in virus prevalence.
HPAI H5N1 viruses belonging to clade 2.3.4.4b and originating from the A/goose/Guangdong/1/1996 lineage pose a serious threat to public health and food security due to their zoonotic potential and poultry economic losses worldwide [
47]. Recently, H5N1 viruses from this clade have been spreading widely among wild birds in Europe [
48], Asia [
49], and Africa [
50]. Normally, sporadic outbreaks occur in the autumn and subside by the following spring. However, since 2022, outbreaks have continued to occur during the summer in Europe and North America [
51,
52]. Thus, in early 2022, the HPAI H5N1 clade 2.3.4.4b was detected in North America, most likely introduced by migratory birds from Europe [
53], and starting in late 2022, it spread throughout South America [
54]. The first case of this disease in Chile was detected on December 7, 2022, in a
Pelecanus thagus specimen [
36]. In the subsequent months, fatal AIV H5N1 infections were documented in various avian species, marine mammals, and one human case [
16,
39,
40]. Phylogenetic analysis of the H5N1 sequences indicates that the recent avian sequences from Chile form a subcluster with Peruvian avian sequences sampled in late November 2022 [
36]. This suggests a common origin and adds to the massive mortality observed in over 3000 sea lions (
Otaria flavescens) in Peru [
15]. The species has also shown fatal cases in Chile [
40]. Moreover, these Chilean-Peruvian subcluster sequences are located within a larger cluster that encompasses sequences from Ecuador, Mexico, and the USA [
36]. These sequences were mostly sampled in late November 2022 in Ecuador and mid-October 2022 in Mexico and the USA, strengthening the notion that the Atlantic migratory route was the source of the introduction of the AIV H5N1 2.3.4.4b clade to the Americas.
Chile’s first human case of H5N1 was genetically analyzed and identified as belonging to the 2.3.4.4b clade [
16]. Furthermore, its sequence was determined to be 99% identical to H5N1 virus samples found in wild birds in Chile. However, there is currently no evidence of human-to-human transmission of the circulating virus. The importance of implementing biosecurity measures and genomic surveillance approaches is growing. Therefore, conducting additional studies to characterize this virus should be a priority.
Conclusions
In conclusion, this review summarizes recent updates on the evolution of the different IAV subtypes in birds and mammals including humans, in Chile. IAV shows a wide diversity of HA/NA subtypes and host species in Chile. The HPAI H5N1 viruses first detected in North America in early 2022 after their introduction from Europe, were subsequently detected in Latin America, including Chile in commercial poultry, backyard poultry, wild birds, and in mammals, including one human case. The distribution and spread of AIV H5N1 in Chile indicated a complex interplay between ecological and human factors in that it was negatively correlated with distance to the closest urban center and precipitation and temperature seasonality. It is evident that HPAI H5N1 in Chile was introduced via the Atlantic migratory flyways, which have been shown as important pathways leading to the AIV diversity in Chile as opposed to local transmission from other countries in South America. The presence of these viruses in Chile underscores the need for increased biosecurity on poultry farms and continuous genomic surveillance approaches to understand and control AIVs in both wild and domestic bird populations in Chile, which appear to form a distinct cluster to the South American AIVs.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org. Table S1: List of Influenza A virus isolates from avian hosts in Chile.
Author Contributions
Conceptualization M.G. and F.K.; investigation, data curation, writing—original draft preparation M.G., M.O., D.C., J.P.; writing—review and editing M.G., M.O., M.K. and F.K.
Funding
This work was supported by Centro de Investigaciones Biológicas Aplicadas (CIBA), Puerto Montt 5480000, Chile. The funder had no role in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- McCauley JW, Hongo S, Kaverin NV, Kochs G, Lamb RA, Matrosovich MN, Perez DR, Palese P, Presti RM, Rimstad E, et al. International Committee on the Taxonomy of Viruses: 2019; Negative Sense RNA Viruses: Orthomyxoviridae. Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/negative-sense-rna-viruses-2011/w/negrna_viruses/209/Orthomyxoviridae (accessed on 5 June 2023).
- Turnbull OMH, Ortiz-Baez AS, Eden JS, Shi M, Williamson JE, Gaston TF, et al. Meta-Transcriptomic Identification of Divergent Amnoonviridae in Fish. Vol. 12, Viruses. MDPI AG; 2020. p. 1254. [CrossRef]
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Vol. 2020, Database. Oxford University Press (OUP); 2020. [CrossRef]
- Parry R, Wille M, Turnbull O, Geoghegan J, Holmes E. Divergent Influenza-Like Viruses of Amphibians and Fish Support an Ancient Evolutionary Association. Vol. 12, Viruses. MDPI AG; 2020. p. 1042. [CrossRef]
- García-Sastre, A. 2020. 2021.022M.A.v1.Orthomyxoviridae_1ngen_1nsp_Mykiss.zip. Available online: https://ictv.global/filebrowser/download/5269 (accessed on 2 June 2023).
- Mohr PG, Godwin SE, Morrison RN, Carson J, Crane MStJ, Moody NJG. 2020. 2021.023M.R.Orthomyxoviridae_1ngen_1nsp_Sardino.zip. 2021. Available online: https://ictv.global/filebrowser/download/5365 (accessed on 2 June 2023).
- Petrone ME, Parry R, Mifsud JCO, Van Brussel K, Vorhees I, Richards ZT, et al. Evidence for an aquatic origin of influenza virus and the order Articulavirales. Cold Spring Harbor Laboratory; 2023. [CrossRef]
- Wille M, Holmes EC. The Ecology and Evolution of Influenza Viruses. Vol. 10, Cold Spring Harbor Perspectives in Medicine. Cold Spring Harbor Laboratory; 2019. p. a038489. [CrossRef]
- Short KR, Richard M, Verhagen JH, van Riel D, Schrauwen EJA, van den Brand JMA, et al. One health, multiple challenges: The inter-species transmission of influenza A virus. Vol. 1, One Health. Elsevier BV; 2015. p. 1–13. [CrossRef]
- Yang W, Schountz T, Ma W. Bat Influenza Viruses: Current Status and Perspective. Vol. 13, Viruses. MDPI AG; 2021. p. 547. [CrossRef]
- Uribe M, Rodríguez-Posada ME, Ramirez-Nieto GC. Molecular Evidence of Orthomyxovirus Presence in Colombian Neotropical Bats. Vol. 13, Frontiers in Microbiology. Frontiers Media SA; 2022. [CrossRef]
- Kessler S, Harder T, Schwemmle M, Ciminski K. Influenza A Viruses and Zoonotic Events—Are We Creating Our Own Reservoirs?. Vol. 13, Viruses. MDPI AG; 2021. p. 2250. [CrossRef]
- Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus ADME, Fouchier RAM. Global Patterns of Influenza A Virus in Wild Birds. Vol. 312, Science. American Association for the Advancement of Science (AAAS); 2006. p. 384–8. [CrossRef]
- Kandeil A, Patton C, Jones JC, Jeevan T, Harrington WN, Trifkovic S, et al. Rapid evolution of A(H5N1) influenza viruses after intercontinental spread to North America. Vol. 14, Nature Communications. Springer Science and Business Media LLC; 2023. [CrossRef]
- Gamarra-Toledo V, Plaza PI, Gutiérrez R, Inga-Diaz G, Saravia-Guevara P, Pereyra-Meza O, et al. Mass Mortality of Marine Mammals Associated to Highly Pathogenic Influenza Virus (H5N1) in South America. Cold Spring Harbor Laboratory; 2023. [CrossRef]
- Centers for Disease Control and Prevention. Human Infection with highly pathogenic avian influenza A(H5N1) virus in Chile. 2023. Available online: https://www.cdc.gov/flu/avianflu/spotlights/2022-2023/chile-first-case-h5n1-addendum.htm (accessed on 3 May 2023).
- World Animal Health Information System. HIGH PATHOGENICITY AVIAN INFLUENZA (HPAI) – SITUATION REPORT. 2023. Available online: https://www.woah.org/en/document/high-pathogenicity-avian-influenza-hpai-situation-report-47/.
- Orozco, C. The SPS Agreement and crisis management: The Chile–EU avian influenza experience. Managing the Challenges of WTO Participation. Cambridge University Press; 2005. p. 150–66. [CrossRef]
- Max V, Herrera J, Moreira R, Rojas H. Avian Influenza in Chile: A Successful Experience. Vol. 51, Avian Diseases. American Association of Avian Pathologists (AAAP); 2007. p. 363–5. [CrossRef]
- Nucleotide. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information, 2004. Available online: https://www.ncbi.nlm.nih.gov/nuccore (accessed on 17 May 2023).
- Rojas H, Moreira R, Avalos P, Capua I, Marangon S. Avian influenza in poultry in Chile. Vet Rec. 2002 Aug 10;151(6):188.
- Rojas Olavarría, H., Moreira Zúñiga, R., & Mundial, B. (2009). Influenza aviar en chile 2002: una sinopsis. Available online: https://www2.sag.gob.cl/pecuaria/bvo/bvo_6_numero_especial_oct_2006/articulos/sinopsis_IA_2002.pdf (accessed on 2 June 2023).
- Suarez DL, Senne DA, Banks J, Brown IH, Essen SC, Lee CW, et al. Recombination Resulting in Virulence Shift in Avian Influenza Outbreak, Chile. Vol. 10, Emerging Infectious Diseases. Centers for Disease Control and Prevention (CDC); 2004. p. 693–9. [CrossRef]
- Spackman E, McCracken KG, Winker K, Swayne DE. H7N3 Avian Influenza Virus Found in a South American Wild Duck Is Related to the Chilean 2002 Poultry Outbreak, Contains Genes from Equine and North American Wild Bird Lineages, and Is Adapted to Domestic Turkeys. Vol. 80, Journal of Virology. American Society for Microbiology; 2006. p. 7760–4. [CrossRef]
- Kibenge, FSB. A One Health approach to mitigate the impact of influenza A virus (IAV) reverse zoonosis is by vaccinating humans and susceptible farmed and pet animals. American Journal of Veterinary Research, June 2023. [CrossRef]
- Trifonov V, Khiabanian H, Rabadan R. Geographic dependence, surveillance, and origins of the 2009 influenza A (H1N1) virus. N Engl J Med 2009;361:115-119. [CrossRef]
- Pantin-Jackwood M, Wasilenko JL, Spackman E, Suarez DL, Swayne DE. Susceptibility of turkeys to pandemic-H1N1 virus by reproductive tract insemination. Virol J 2010;7: 27. [CrossRef]
- Mathieu C, Moreno V, Retamal P, Gonzalez A, Rivera A, Fuller J, et al. Pandemic (H1N1) 2009 in Breeding Turkeys, Valparaiso, Chile. Vol. 16, Emerging Infectious Diseases. Centers for Disease Control and Prevention (CDC); 2010. p. 709–11. [CrossRef]
- Mathieu C, Gonzalez A, Garcia A, Johow M, Badia C, Jara C, et al. H7N6 low pathogenic avian influenza outbreak in commercial turkey farms in Chile caused by a native South American Lineage. Vol. 68, Transboundary and Emerging Diseases. Hindawi Limited; 2019. p. 2–12. [CrossRef]
- Jimenez-Bluhm P, Bravo-Vasquez N, Torchetti MK, Killian ML, Livingston B, Herrera J, et al. Low pathogenic avian influenza (H7N6) virus causing an outbreak in commercial Turkey farms in Chile. Vol. 8, Emerging Microbes & Infections. Informa UK Limited; 2019. p. 479–85. [CrossRef]
- Jimenez-Bluhm P, Di Pillo F, Bahl J, Osorio J, Schultz-Cherry S, Hamilton-West C. Circulation of influenza in backyard productive systems in central Chile and evidence of spillover from wild birds. Vol. 153, Preventive Veterinary Medicine. Elsevier BV; 2018. p. 1–6. [CrossRef]
- Bravo-Vasquez N, Di Pillo F, Lazo A, Jiménez-Bluhm P, Schultz-Cherry S, Hamilton-West C. Presence of influenza viruses in backyard poultry and swine in El Yali wetland, Chile. Vol. 134, Preventive Veterinary Medicine. Elsevier BV; 2016. p. 211–5. [CrossRef]
- Bravo-Vasquez N, Baumberger C, Jimenez-Bluhm P, Di Pillo F, Lazo A, Sanhueza J, et al. Risk factors and spatial relative risk assessment for influenza A virus in poultry and swine in backyard production systems of central Chile. Vol. 6, Veterinary Medicine and Science. Wiley; 2020. p. 518–26. [CrossRef]
- Mathieu C, Moreno V, Pedersen J, Jeria J, Agredo M, Gutiérrez C, et al. Avian Influenza in wild birds from Chile, 2007–2009. Vol. 199, Virus Research. Elsevier BV; 2015. p. 42–5. [CrossRef]
- Jiménez-Bluhm P, Karlsson EA, Freiden P, Sharp B, Di Pillo F, Osorio JE, et al. Wild birds in Chile Harbor diverse avian influenza A viruses. Vol. 7, Emerging Microbes & Infections. Informa UK Limited; 2018. p. 1–4. [CrossRef]
- Ariyama N, Pardo-Roa C, Muñoz G, Aguayo C, Ávila C, Mathieu C, et al. Emergence and rapid dissemination of highly pathogenic avian influenza virus H5N1 clade 2.3.4.4b in wild birds, Chile. 2023. [CrossRef]
- Jimenez-Bluhm P, Siegers JY, Tan S, Sharp B, Freiden P, Johow M, et al. Detection and Phylogenetic Analysis of Highly Pathogenic A/H5N1 Avian Influenza Clade 2.3.4.4b Virus in Chile, 2022. Emerging Microbes & Infections. Informa UK Limited; 2023. [CrossRef]
- Azat C, Alvarado-Rybak M, Aguilera JF, Benavides JA. Spatio-temporal dynamics and drivers of Highly Pathogenic Avian Influenza H5N1 in Chile. Cold Spring Harbor Laboratory; 2023. [CrossRef]
- SAG. Servicio Agrícola y Ganadero, Ministerio de Agricultura de Chile. Plataforma de influenza aviar. 2023. Available online: https://www.sag.gob.cl/ia (accessed on 5 June 2023).
- SERNAPESCA. Servicio Nacional de Pesca. Plataforma de influenza aviar. 2023. Available online: http://www.sernapesca.cl/influenza-aviar (accessed on 5 June 2023).
- eBird. eBird: An online database of bird distribution and abundance. eBird, Cornell Lab of Ornithology, Ithaca, New York, 2021. Available online: http://www.ebird.org (accessed on 30 May 2023).
- Reperant LA, Kuiken T, Osterhaus ADME. Adaptive pathways of zoonotic influenza viruses: From exposure to establishment in humans. Vol. 30, Vaccine. Elsevier BV; 2012. p. 4419–34. [CrossRef]
- World Health Organization (WHO): Human Infection caused by Avian Influenza A (H5N1) - Chile. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON461 (accessed on 5 June 2023).
- Pardo-Roa, C., Nelson, M. I., Ariyama, N., Aguayo, C., Almonacid, L. I., Munoz, G., Navarro, C., Avila, C., Ulloa, M., Reyes, R., Luppichini, E. F., Mathieu, C., Vergara, R., González, Á., González, C. G., Araya, H., Fernández, J., Fasce, R., Johow, M., … Neira, V. Cross-species transmission and PB2 mammalian adaptations of highly pathogenic avian influenza A/H5N1 viruses in Chile. Cold Spring Harbor Laboratory. 2023. [CrossRef]
- Castillo, A., Fasce, R., Parra, B., Andrade, W., Covarrubias, P., Hueche, A., Campano, C., Tambley, C., Rojas, M., Araya, M., Hernández, F., Bustos, P., & Fernández, J. The first reported case of human infection with H5N1 avian Influenza A virus in Chile. In Journal of Travel Medicine. Oxford University Press (OUP), 2023. [CrossRef]
- Vreman, S., Kik, M., Germeraad, E., Heutink, R., Harders, F., Spierenburg, M., Engelsma, M., Rijks, J., van den Brand, J., & Beerens, N. Zoonotic Mutation of Highly Pathogenic Avian Influenza H5N1 Virus Identified in the Brain of Multiple Wild Carnivore Species. In Pathogens (Vol. 12, Issue 2, p. 168). MDPI AG, 2023. [CrossRef]
- Chmielewski R, Swayne DE. Avian Influenza: Public Health and Food Safety Concerns. Vol. 2, Annual Review of Food Science and Technology. Annual Reviews; 2011. p. 37–57. [CrossRef]
- European Food Safety Authority, European Centre for Disease Prevention and Control, European Union Reference Laboratory for Avian Influenza, Adlhoch C, Fusaro A, Gonzales JL, et al. Avian influenza overview December 2022 – March 2023. EFS2. 2023 Mar;21(3). Available online: https://data.europa.eu/doi/10.2903/j.efsa.2023.7917.
- Sagong M, Lee Y, Song S, Cha RM, Lee E, Kang Y, et al. Emergence of clade 2.3.4.4b novel reassortant H5N1 high pathogenicity avian influenza virus in South Korea during late 2021. Vol. 69, Transboundary and Emerging Diseases. Hindawi Limited; 2022. [CrossRef]
- El-Shesheny R, Moatasim Y, Mahmoud SH, Song Y, El Taweel A, Gomaa M, et al. Highly Pathogenic Avian Influenza A(H5N1) Virus Clade 2.3.4.4b in Wild Birds and Live Bird Markets, Egypt. Vol. 12, Pathogens. MDPI AG; 2022. p. 36. [CrossRef]
- European Food Safety Authority, European Centre for Disease Prevention and Control, European Union Reference Laboratory for Avian Influenza, Adlhoch C, Fusaro A, Gonzales JL, et al. Avian influenza overview June – September 2022. EFS2. 2022 Oct;20(10). Available online: https://data.europa.eu/doi/10.2903/j.efsa.2022.7597.
- Harvey JA, Mullinax JM, Runge MC, Prosser DJ. The changing dynamics of highly pathogenic avian influenza H5N1: Next steps for management & science in North America. Vol. 282, Biological Conservation. Elsevier BV; 2023. p. 110041. [CrossRef]
- Günther A, Krone O, Svansson V, Pohlmann A, King J, Hallgrimsson GT, et al. Iceland as Stepping Stone for Spread of Highly Pathogenic Avian Influenza Virus between Europe and North America. Vol. 28, Emerging Infectious Diseases. Centers for Disease Control and Prevention (CDC); 2022. [CrossRef]
- Leguia M, Garcia-Glaessner A, Muñoz-Saavedra B, Juarez D, Barrera P, Calvo-Mac C, et al. Highly pathogenic avian influenza A (H5N1) in marine mammals and seabirds in Peru. Cold Spring Harbor Laboratory; 2023. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).