Submitted:
15 September 2023
Posted:
19 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
| Primers | Forward | Reverse |
| hVEGF | 5’-GTCGGGCCTCCGAAACCATG-3′ | 5’-CCTGGTGAGAGATCTGGTTC-3′ |
| hBcl2 | 5′-CTGGAGAGTGCTGAAGATTGATG-3′ | 5′-CAATCACGCGGAACACTTGATTC-3′ |
| hMcl1 | 5’-GATCAGTATATACACTTCAG-3′ | 5’-CAGGTGCAGCCTGTACTTGTC-3′ |
| h-actin | 5’-GCCCTGGACTTCGAGCAAGA-3′ | 5’-TGCCAGGGTACATGGTGGTG-3′ |
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Faderl, S.; Talpaz, M.; Estrov, Z.; O'Brien, S.; Kurzrock, R.; Kantarjian, H.M. The biology of chronic myeloid leukemia. N Engl J Med 1999, 341, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Ottmann, O.G.; Hoelzer, D. The ABL tyrosine kinase inhibitor STI571 (Glivec) in Philadelphia positive acute lymphoblastic leukemia - promises, pitfalls and possibilities. Hematol J 2002, 3, 2–6. [Google Scholar] [CrossRef]
- Radich, J.P.; Dai, H.; Mao, M.; Oehler, V.; Schelter, J.; Druker, B.; Sawyers, C.; Shah, N.; Stock, W.; Willman, C.L.; et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A 2006, 103, 2794–2799. [Google Scholar] [CrossRef] [PubMed]
- Talpaz, M.; Silver, R.T.; Druker, B.J.; Goldman, J.M.; Gambacorti-Passerini, C.; Guilhot, F.; Schiffer, C.A.; Fischer, T.; Deininger, M.W.; Lennard, A.L.; et al. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: Results of a phase 2 study. Blood 2002, 99, 1928–1937. [Google Scholar] [CrossRef] [PubMed]
- Gambacorti-Passerini, C.B.; Gunby, R.H.; Piazza, R.; Galietta, A.; Rostagno, R.; Scapozza, L. Molecular mechanisms of resistance to imatinib in Philadelphia-chromosome-positive leukaemias. Lancet Oncol 2003, 4, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Meads, M.B.; Hazlehurst, L.A.; Dalton, W.S. The bone marrow microenvironment as a tumor sanctuary and contributor to drug resistance. Clin Cancer Res 2008, 14, 2519–2526. [Google Scholar] [CrossRef]
- Katz, B.Z.; Polliack, A. Cancer microenvironment, extracellular matrix, and adhesion molecules: The bitter taste of sugars in chronic lymphocytic leukemia. Leuk Lymphoma 2011, 52, 1619–1620. [Google Scholar] [CrossRef]
- Witz, I.P. The tumor microenvironment: The making of a paradigm. Cancer Microenviron 2009, 2 Suppl 1, 9–17. [Google Scholar] [CrossRef]
- Tesfai, Y.; Ford, J.; Carter, K.W.; Firth, M.J.; O'Leary, R.A.; Gottardo, N.G.; Cole, C.; Kees, U.R. Interactions between acute lymphoblastic leukemia and bone marrow stromal cells influence response to therapy. Leuk Res 2012, 36, 299–306. [Google Scholar] [CrossRef]
- Zhang, B.; Groffen, J.; Heisterkamp, N. Increased resistance to a farnesyltransferase inhibitor by N-cadherin expression in Bcr/Abl-P190 lymphoblastic leukemia cells. Leukemia 2007, 21, 1189–1197. [Google Scholar] [CrossRef]
- Jensen, P.O.; Mortensen, B.T.; Hodgkiss, R.J.; Iversen, P.O.; Christensen, I.J.; Helledie, N.; Larsen, J.K. Increased cellular hypoxia and reduced proliferation of both normal and leukaemic cells during progression of acute myeloid leukaemia in rats. Cell Prolif 2000, 33, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Wilson, W.R. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer 2004, 4, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Minakata, K.; Takahashi, F.; Nara, T.; Hashimoto, M.; Tajima, K.; Murakami, A.; Nurwidya, F.; Yae, S.; Koizumi, F.; Moriyama, H.; et al. Hypoxia induces gefitinib resistance in non-small-cell lung cancer with both mutant and wild-type epidermal growth factor receptors. Cancer Sci 2012, 103, 1946–1954. [Google Scholar] [CrossRef] [PubMed]
- Bruick, R.K. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci U S A 2000, 97, 9082–9087. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Chen, X.; Huang, R.; Zeng, H.; Gong, J.; Meng, W.; Lu, Y.; Zhao, F.; Wang, L.; Zhou, Q. BCL-xL is a target gene regulated by hypoxia-inducible factor-1{alpha}. J Biol Chem 2009, 284, 10004–10012. [Google Scholar] [CrossRef] [PubMed]
- Erler, J.T.; Cawthorne, C.J.; Williams, K.J.; Koritzinsky, M.; Wouters, B.G.; Wilson, C.; Miller, C.; Demonacos, C.; Stratford, I.J.; Dive, C. Hypoxia-mediated down-regulation of Bid and Bax in tumors occurs via hypoxia-inducible factor 1-dependent and -independent mechanisms and contributes to drug resistance. Mol Cell Biol 2004, 24, 2875–2889. [Google Scholar] [CrossRef]
- Kim, J.Y.; Ahn, H.J.; Ryu, J.H.; Suk, K.; Park, J.H. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1alpha. J Exp Med 2004, 199, 113–124. [Google Scholar] [CrossRef]
- Sowter, H.M.; Ratcliffe, P.J.; Watson, P.; Greenberg, A.H.; Harris, A.L. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res 2001, 61, 6669–6673. [Google Scholar]
- Comerford, K.M.; Wallace, T.J.; Karhausen, J.; Louis, N.A.; Montalto, M.C.; Colgan, S.P. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Res 2002, 62, 3387–3394. [Google Scholar]
- Walmsley, S.R.; Print, C.; Farahi, N.; Peyssonnaux, C.; Johnson, R.S.; Cramer, T.; Sobolewski, A.; Condliffe, A.M.; Cowburn, A.S.; Johnson, N.; et al. Hypoxia-induced neutrophil survival is mediated by HIF-1alpha-dependent NF-kappaB activity. J Exp Med 2005, 201, 105–115. [Google Scholar] [CrossRef]
- Dengler, V.L.; Galbraith, M.; Espinosa, J.M. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol 2014, 49, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Murakami, A.; Takahashi, F.; Nurwidya, F.; Kobayashi, I.; Minakata, K.; Hashimoto, M.; Nara, T.; Kato, M.; Tajima, K.; Shimada, N.; et al. Hypoxia increases gefitinib-resistant lung cancer stem cells through the activation of insulin-like growth factor 1 receptor. PLoS ONE 2014, 9, e86459. [Google Scholar] [CrossRef] [PubMed]
- Shain, K.H.; Yarde, D.N.; Meads, M.B.; Huang, M.; Jove, R.; Hazlehurst, L.A.; Dalton, W.S. Beta1 integrin adhesion enhances IL-6-mediated STAT3 signaling in myeloma cells: Implications for microenvironment influence on tumor survival and proliferation. Cancer Res 2009, 69, 1009–1015. [Google Scholar] [CrossRef]
- Nair, R.R.; Tolentino, J.H.; Argilagos, R.F.; Zhang, L.; Pinilla-Ibarz, J.; Hazlehurst, L.A. Potentiation of Nilotinib-mediated cell death in the context of the bone marrow microenvironment requires a promiscuous JAK inhibitor in CML. Leuk Res 2012, 36, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Traer, E.; Javidi-Sharifi, N.; Agarwal, A.; Dunlap, J.; English, I.; Martinez, J.; Tyner, J.W.; Wong, M.; Druker, B.J. Ponatinib overcomes FGF2-mediated resistance in CML patients without kinase domain mutations. Blood 2014, 123, 1516–1524. [Google Scholar] [CrossRef]
- Bewry, N.N.; Nair, R.R.; Emmons, M.F.; Boulware, D.; Pinilla-Ibarz, J.; Hazlehurst, L.A. Stat3 contributes to resistance toward BCR-ABL inhibitors in a bone marrow microenvironment model of drug resistance. Mol Cancer Ther 2008, 7, 3169–3175. [Google Scholar] [CrossRef]
- Okabe, S.; Tauchi, T.; Katagiri, S.; Tanaka, Y.; Ohyashiki, K. Combination of the ABL kinase inhibitor imatinib with the Janus kinase 2 inhibitor TG101348 for targeting residual BCR-ABL-positive cells. J Hematol Oncol 2014, 7, 37. [Google Scholar] [CrossRef]
- Ng, K.P.; Manjeri, A.; Lee, K.L.; Huang, W.; Tan, S.Y.; Chuah, C.T.; Poellinger, L.; Ong, S.T. Physiologic hypoxia promotes maintenance of CML stem cells despite effective BCR-ABL1 inhibition. Blood 2014, 123, 3316–3326. [Google Scholar] [CrossRef]
- Chen, W.; Qin, Y.; Liu, S. Cytokines, breast cancer stem cells (BCSCs) and chemoresistance. Clin Transl Med 2018, 7, 27. [Google Scholar] [CrossRef]
- Jiang, M.; He, G.; Wang, J.; Guo, X.; Zhao, Z.; Gao, J. Hypoxia induces inflammatory microenvironment by priming specific macrophage polarization and modifies LSC behaviour via VEGF-HIF1α signalling. Transl Pediatr 2021, 10, 1792–1804. [Google Scholar] [CrossRef]
- Kidan, N.; Khamaisie, H.; Ruimi, N.; Roitman, S.; Eshel, E.; Dally, N.; Ruthardt, M.; Mahajna, J. Ectopic Expression of Snail and Twist in Ph+ Leukemia Cells Upregulates CD44 Expression and Alters Their Differentiation Potential. J Cancer 2017, 8, 3952–3968. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Bartz, S.; Mao, M.; Li, L.; Kaelin, W.G., Jr. The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol Cell Biol 2007, 27, 2092–2102. [Google Scholar] [CrossRef] [PubMed]
- Stewart, S.A.; Dykxhoorn, D.M.; Palliser, D.; Mizuno, H.; Yu, E.Y.; An, D.S.; Sabatini, D.M.; Chen, I.S.; Hahn, W.C.; Sharp, P.A.; et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 2003, 9, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Najajreh, Y.; Khamaisie, H.; Ruimi, N.; Khatib, S.; Katzhendler, J.; Ruthardt, M.; Mahajna, J. Oleylamine-carbonyl-valinol inhibits auto-phosphorylation activity of native and T315I mutated Bcr-Abl, and exhibits selectivity towards oncogenic Bcr-Abl in SupB15 ALL cell lines. Mol Biol Rep 2013, 40, 2205–2213. [Google Scholar] [CrossRef]
- Gochman, E.; Mahajna, J.; Shenzer, P.; Dahan, A.; Blatt, A.; Elyakim, R.; Reznick, A.Z. The expression of iNOS and nitrotyrosine in colitis and colon cancer in humans. Acta Histochem 2012, 114, 827–835. [Google Scholar] [CrossRef]
- Mian, A.A.; Metodieva, A.; Badura, S.; Khateb, M.; Ruimi, N.; Najajreh, Y.; Ottmann, O.G.; Mahajna, J.; Ruthardt, M. Allosteric inhibition enhances the efficacy of ABL kinase inhibitors to target unmutated BCR-ABL and BCR-ABL-T315I. BMC Cancer 2012, 12, 411. [Google Scholar] [CrossRef]
- Koren Carmi, Y.; Khamaisi, H.; Adawi, R.; Noyman, E.; Gopas, J.; Mahajna, J. Secreted Soluble Factors from Tumor-Activated Mesenchymal Stromal Cells Confer Platinum Chemoresistance to Ovarian Cancer Cells. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Dotan, N.; Wasser, S.P.; Mahajna, J. Inhibition of the androgen receptor activity by Coprinus comatus substances. Nutr Cancer 2011, 63, 1316–1327. [Google Scholar] [CrossRef]
- Sahu, A.; Prabhash, K.; Noronha, V.; Joshi, A.; Desai, S. Crizotinib: A comprehensive review. South Asian J Cancer 2013, 2, 91–97. [Google Scholar] [CrossRef]
- Regev, O.; Kidan, N.; Nicola, M.; Khamisie, H.; Ruthardt, M.; Mahajna, J. Mesenchymal soluble factors confer imatinib drug resistance in chronic myelogenous leukemia cells. Arch Med Sci 2021, 17, 266–274. [Google Scholar] [CrossRef]
- Wu, D.; Yotnda, P. Induction and testing of hypoxia in cell culture. J Vis Exp 2011. [Google Scholar] [CrossRef]
- Selvendiran, K.; Bratasz, A.; Kuppusamy, M.L.; Tazi, M.F.; Rivera, B.K.; Kuppusamy, P. Hypoxia induces chemoresistance in ovarian cancer cells by activation of signal transducer and activator of transcription 3. Int J Cancer 2009, 125, 2198–2204. [Google Scholar] [CrossRef] [PubMed]
- Ostojic, A.; Vrhovac, R.; Verstovsek, S. Ruxolitinib: A new JAK1/2 inhibitor that offers promising options for treatment of myelofibrosis. Future Oncol 2011, 7, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Snell, C.E.; Turley, H.; McIntyre, A.; Li, D.; Masiero, M.; Schofield, C.J.; Gatter, K.C.; Harris, A.L.; Pezzella, F. Proline-hydroxylated hypoxia-inducible factor 1alpha (HIF-1alpha) upregulation in human tumours. PLoS ONE 2014, 9, e88955. [Google Scholar] [CrossRef] [PubMed]
- Hon, W.C.; Wilson, M.I.; Harlos, K.; Claridge, T.D.; Schofield, C.J.; Pugh, C.W.; Maxwell, P.H.; Ratcliffe, P.J.; Stuart, D.I.; Jones, E.Y. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature 2002, 417, 975–978. [Google Scholar] [CrossRef] [PubMed]
- Krishnamachary, B.; Penet, M.F.; Nimmagadda, S.; Mironchik, Y.; Raman, V.; Solaiyappan, M.; Semenza, G.L.; Pomper, M.G.; Bhujwalla, Z.M. Hypoxia regulates CD44 and its variant isoforms through HIF-1α in triple negative breast cancer. PLoS ONE 2012, 7, e44078. [Google Scholar] [CrossRef] [PubMed]
- Mabjeesh, N.J.; Escuin, D.; LaVallee, T.M.; Pribluda, V.S.; Swartz, G.M.; Johnson, M.S.; Willard, M.T.; Zhong, H.; Simons, J.W.; Giannakakou, P. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell 2003, 3, 363–375. [Google Scholar] [CrossRef]
- Frolova, O.; Samudio, I.; Benito, J.M.; Jacamo, R.; Kornblau, S.M.; Markovic, A.; Schober, W.; Lu, H.; Qiu, Y.H.; Buglio, D.; et al. Regulation of HIF-1alpha signaling and chemoresistance in acute lymphocytic leukemia under hypoxic conditions of the bone marrow microenvironment. Cancer Biol Ther 2012, 13, 858–870. [Google Scholar] [CrossRef]
- Mian, A.A.; Haberbosch, I.; Khamaisie, H.; Agbarya, A.; Pietsch, L.; Eshel, E.; Najib, D.; Chiriches, C.; Ottmann, O.G.; Hantschel, O.; et al. Crizotinib acts as ABL1 inhibitor combining ATP-binding with allosteric inhibition and is active against native BCR-ABL1 and its resistance and compound mutants BCR-ABL1(T315I) and BCR-ABL1(T315I-E255K). Ann Hematol 2021, 100, 2023–2029. [Google Scholar] [CrossRef]
- Albadari, N.; Deng, S.; Li, W. The transcriptional factors HIF-1 and HIF-2 and their novel inhibitors in cancer therapy. Expert Opinion on Drug Discovery 2019, 14, 667–682. [Google Scholar] [CrossRef]
- Kogita, A.; Togashi, Y.; Hayashi, H.; Sogabe, S.; Terashima, M.; De Velasco, M.A.; Sakai, K.; Fujita, Y.; Tomida, S.; Takeyama, Y.; et al. Hypoxia induces resistance to ALK inhibitors in the H3122 non-small cell lung cancer cell line with an ALK rearrangement via epithelial-mesenchymal transition. Int J Oncol 2014, 45, 1430–1436. [Google Scholar] [CrossRef] [PubMed]
- Adamski, J.; Price, A.; Dive, C.; Makin, G. Hypoxia-induced cytotoxic drug resistance in osteosarcoma is independent of HIF-1Alpha. PLoS ONE 2013, 8, e65304. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, Q.; Yu, T.; Sun, S.; Wang, W.; Liu, G. Hypoxia promotes drug resistance in osteosarcoma cells via activating AMP-activated protein kinase (AMPK) signaling. J Bone Oncol 2016, 5, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Petit, C.; Gouel, F.; Dubus, I.; Heuclin, C.; Roget, K.; Vannier, J.P. Hypoxia promotes chemoresistance in acute lymphoblastic leukemia cell lines by modulating death signaling pathways. BMC Cancer 2016, 16, 746. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Jiang, X.; Zhu, H.; Jiang, W.; Dong, X.; Qiao, H.; Sun, X.; Jiang, H. 2ME2 inhibits the activated hypoxia-inducible pathways by cabozantinib and enhances its efficacy against medullary thyroid carcinoma. Tumour Biol 2016, 37, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Azab, S.S.; Salama, S.A.; Hassan, M.H.; Khalifa, A.E.; El-Demerdash, E.; Fouad, H.; Al-Hendy, A.; Abdel-Naim, A.B. 2-Methoxyestradiol reverses doxorubicin resistance in human breast tumor xenograft. Cancer Chemother Pharmacol 2008, 62, 893–902. [Google Scholar] [CrossRef]
- Chauhan, D.; Catley, L.; Hideshima, T.; Li, G.; Leblanc, R.; Gupta, D.; Sattler, M.; Richardson, P.; Schlossman, R.L.; Podar, K.; et al. 2-Methoxyestradiol overcomes drug resistance in multiple myeloma cells. Blood 2002, 100, 2187–2194. [Google Scholar] [CrossRef]
- Lakhani, N.J.; Sarkar, M.A.; Venitz, J.; Figg, W.D. 2-Methoxyestradiol, a promising anticancer agent. Pharmacotherapy 2003, 23, 165–172. [Google Scholar] [CrossRef]
- Khamaisi, H.; Mahmoud, H.; Mahajna, J. 2-Hydroxyestradiol Overcomes Mesenchymal Stem Cells-Mediated Platinum Chemoresistance in Ovarian Cancer Cells in an ERK-Independent Fashion. Molecules 2022, 27. [Google Scholar] [CrossRef]
- Koren Carmi, Y.; Khamaisi, H.; Adawi, R.; Noyman, E.; Gopas, J.; Mahajna, J. Secreted Soluble Factors from Tumor-Activated Mesenchymal Stromal Cells Confer Platinum Chemoresistance to Ovarian Cancer Cells. International Journal of Molecular Sciences 2023, 24, 7730. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, H.; Shen, Y.; Zhou, X. Combined Effects of 2-Methoxyestradiol (Hypoxia-Inducible Factor 1alpha Inhibitor) and Dasatinib (A Second-Generation Tyrosine Kinase Inhibitor) on Chronic Myelocytic Leukemia Cells. J Immunol Res 2022, 2022, 6324326. [Google Scholar] [CrossRef] [PubMed]
- Van Veldhuizen, P.J.; Ray, G.; Banerjee, S.; Dhar, G.; Kambhampati, S.; Dhar, A.; Banerjee, S.K. 2-Methoxyestradiol modulates beta-catenin in prostate cancer cells: A possible mediator of 2-methoxyestradiol-induced inhibition of cell growth. Int J Cancer 2008, 122, 567–571. [Google Scholar] [CrossRef] [PubMed]
- LaVallee, T.M.; Zhan, X.H.; Johnson, M.S.; Herbstritt, C.J.; Swartz, G.; Williams, M.S.; Hembrough, W.A.; Green, S.J.; Pribluda, V.S. 2-methoxyestradiol up-regulates death receptor 5 and induces apoptosis through activation of the extrinsic pathway. Cancer Res 2003, 63, 468–475. [Google Scholar] [PubMed]
- Hernandez-Luna, M.A.; Rocha-Zavaleta, L.; Vega, M.I.; Huerta-Yepez, S. Hypoxia inducible factor-1alpha induces chemoresistance phenotype in non-Hodgkin lymphoma cell line via up-regulation of Bcl-xL. Leuk Lymphoma 2013, 54, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Sanchez, R.; Berzal, S.; Sanchez-Nino, M.D.; Neria, F.; Goncalves, S.; Calabia, O.; Tejedor, A.; Calzada, M.J.; Caramelo, C.; Deudero, J.J.; et al. AG490 promotes HIF-1alpha accumulation by inhibiting its hydroxylation. Curr Med Chem 2012, 19, 4014–4023. [Google Scholar] [CrossRef]
- Fernández-Sánchez, R.; Berzal, S.; Sánchez-Niño, M.D.; Neria, F.; Gonçalves, S.; Calabia, O.; Tejedor, A.; Calzada, M.J.; Caramelo, C.; Deudero, J.J.; et al. AG490 promotes HIF-1α accumulation by inhibiting its hydroxylation. Curr Med Chem 2012, 19, 4014–4023. [Google Scholar] [CrossRef]
- Jeong, C.H.; Lee, H.J.; Cha, J.H.; Kim, J.H.; Kim, K.R.; Kim, J.H.; Yoon, D.K.; Kim, K.W. Hypoxia-inducible factor-1 alpha inhibits self-renewal of mouse embryonic stem cells in Vitro via negative regulation of the leukemia inhibitory factor-STAT3 pathway. J Biol Chem 2007, 282, 13672–13679. [Google Scholar] [CrossRef]
- Regev, O.; Kidan, N.; Khamisie, H.; Mahajna, J. Crizotinib overcomes microenvironment mediated drug resistance in Ph positive leukemia. European Journal of Cancer 2016, 61, S92–S93. [Google Scholar] [CrossRef]
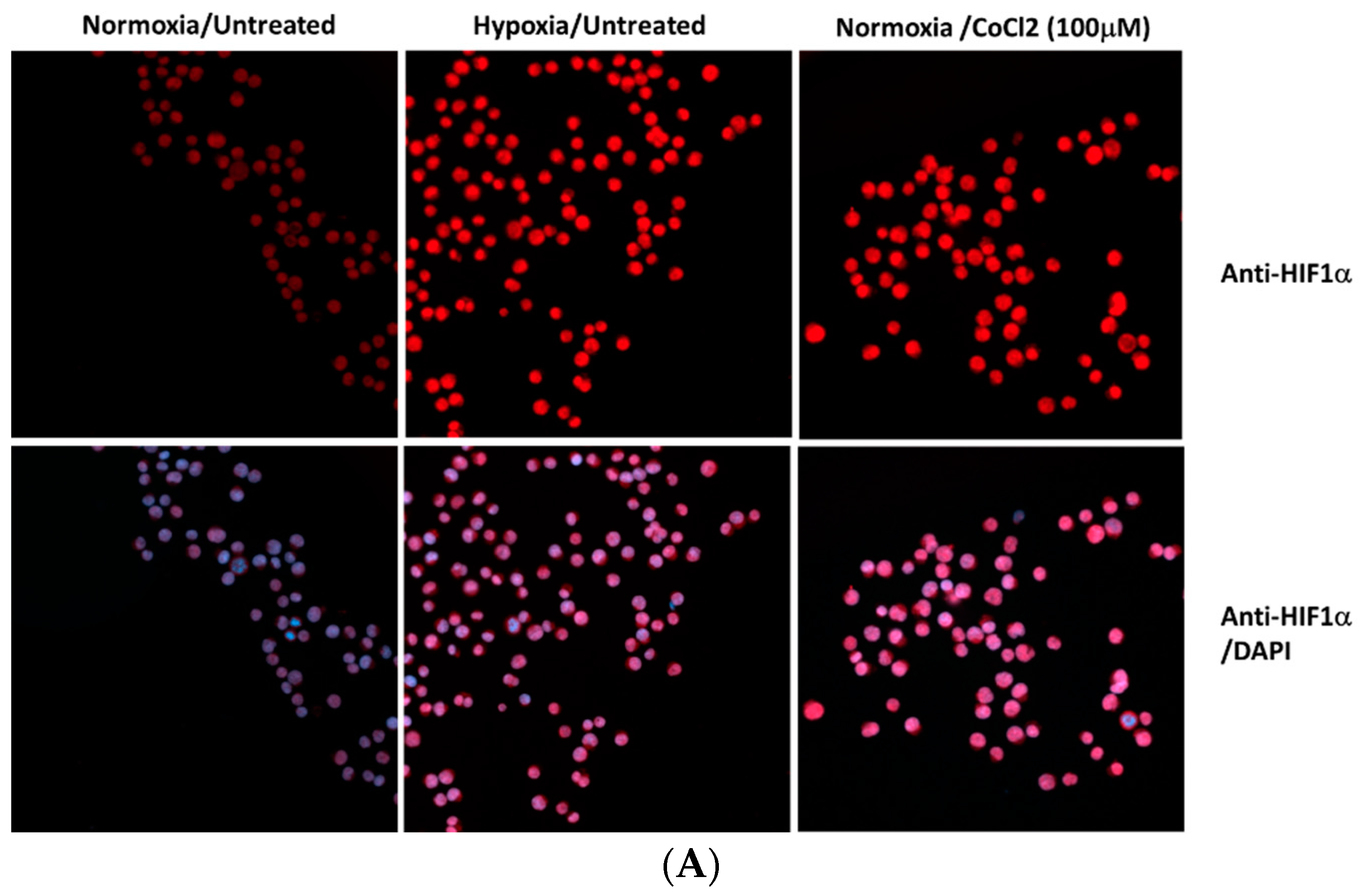
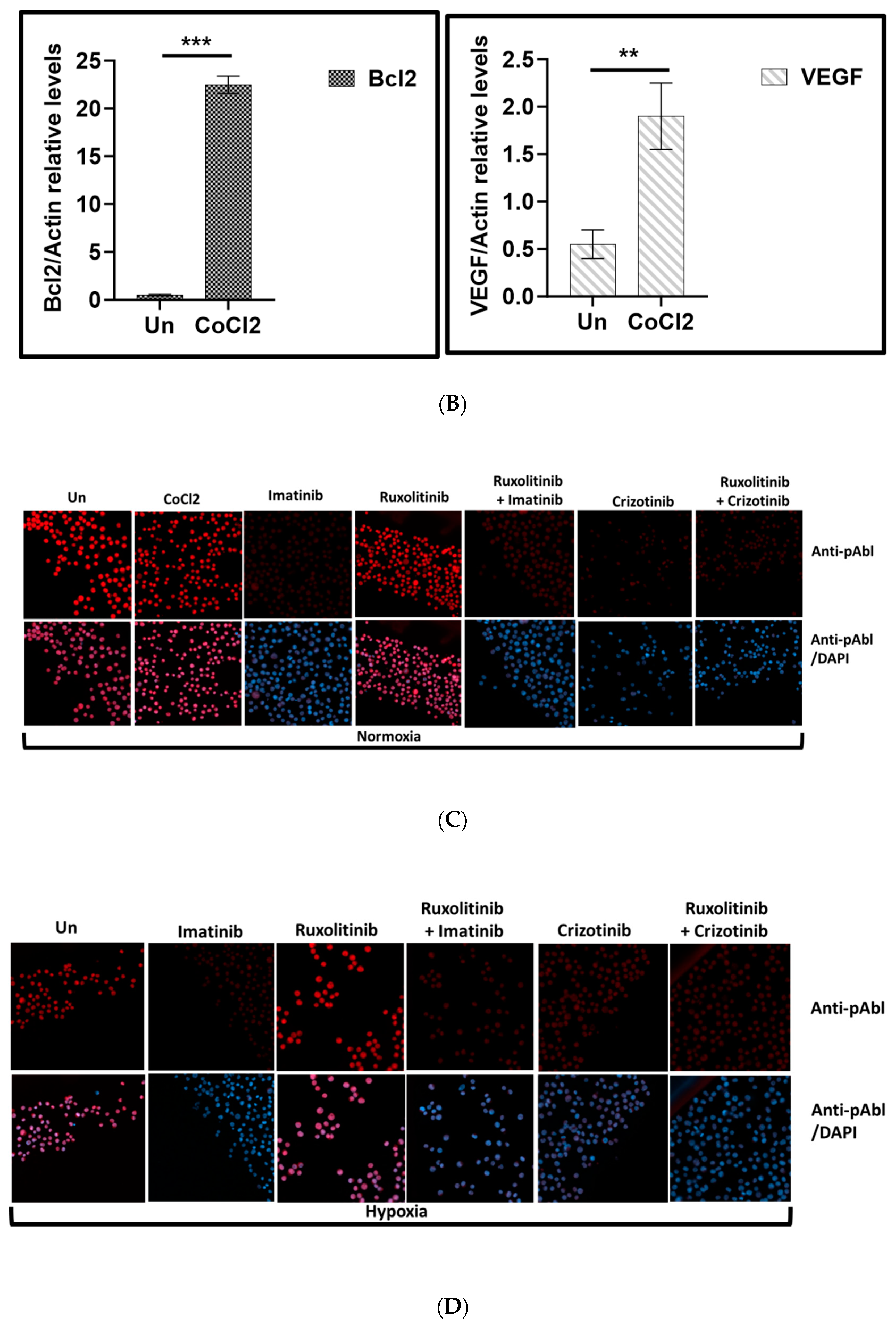
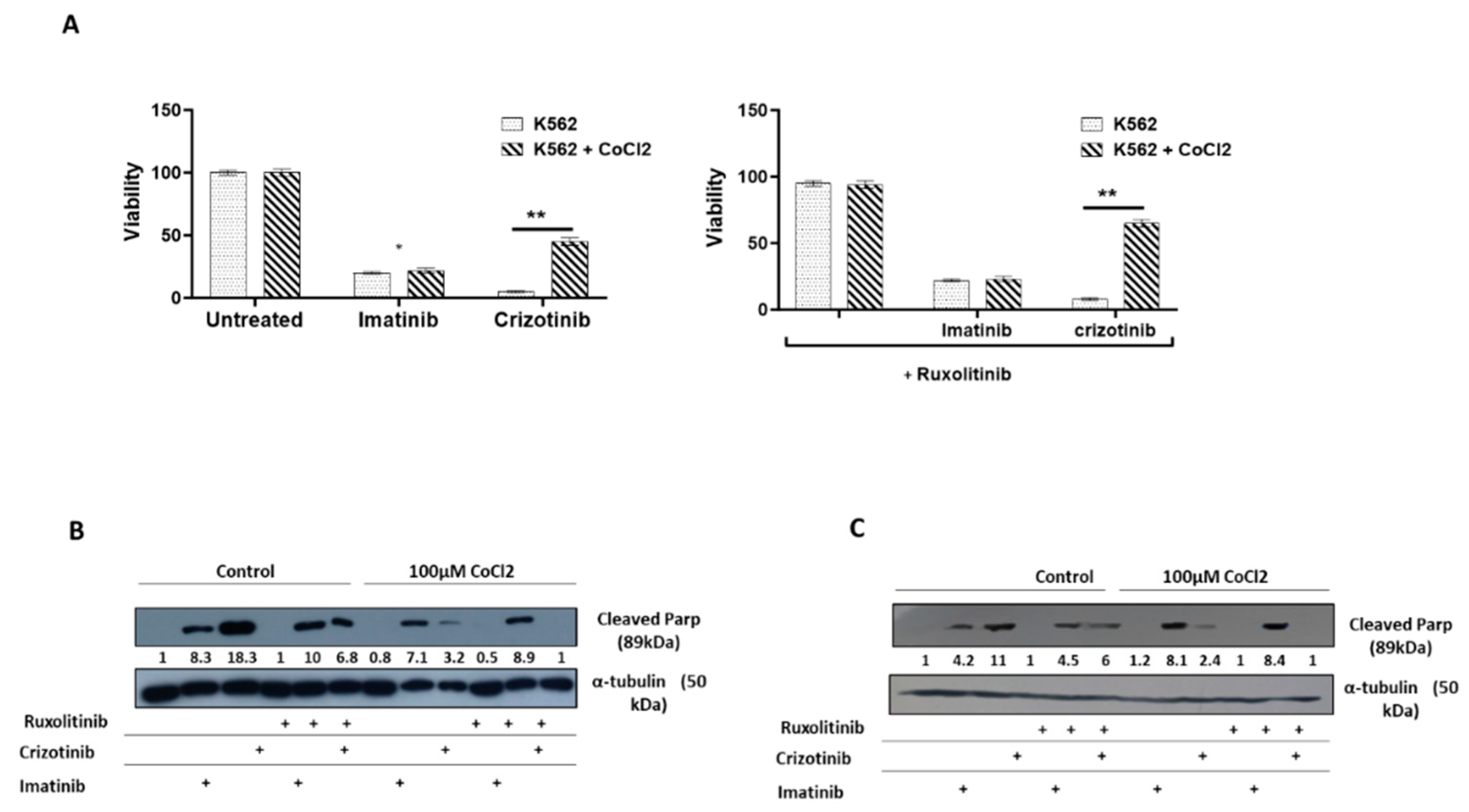
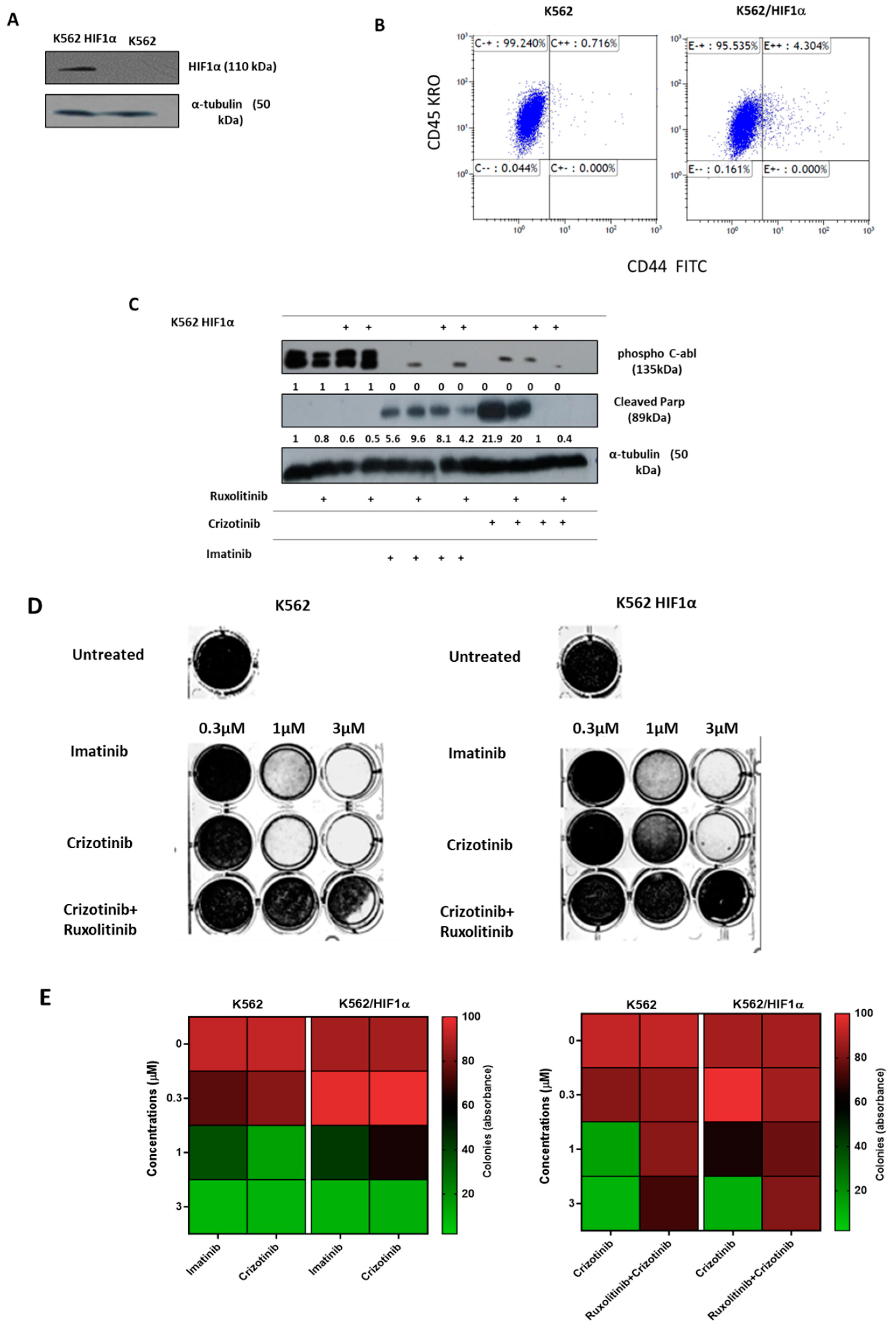

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





