Submitted:
19 September 2023
Posted:
20 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
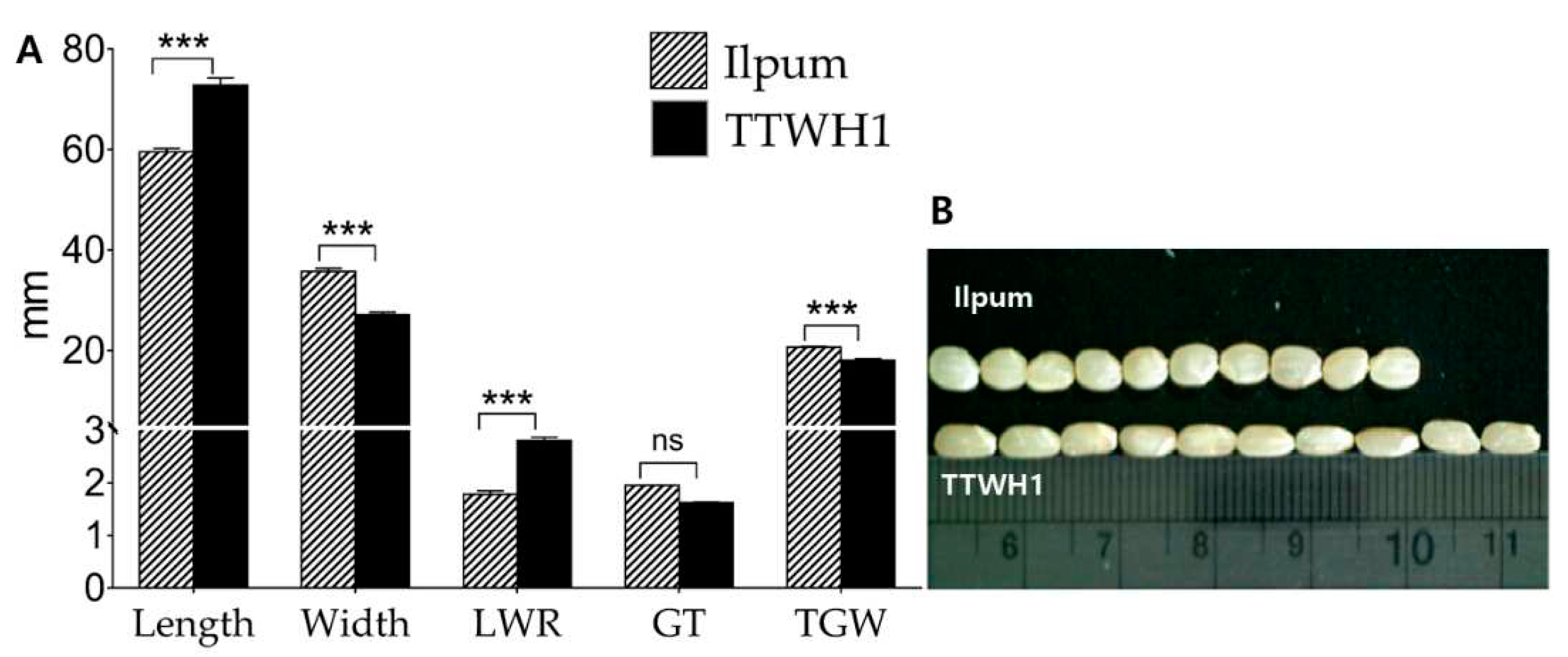
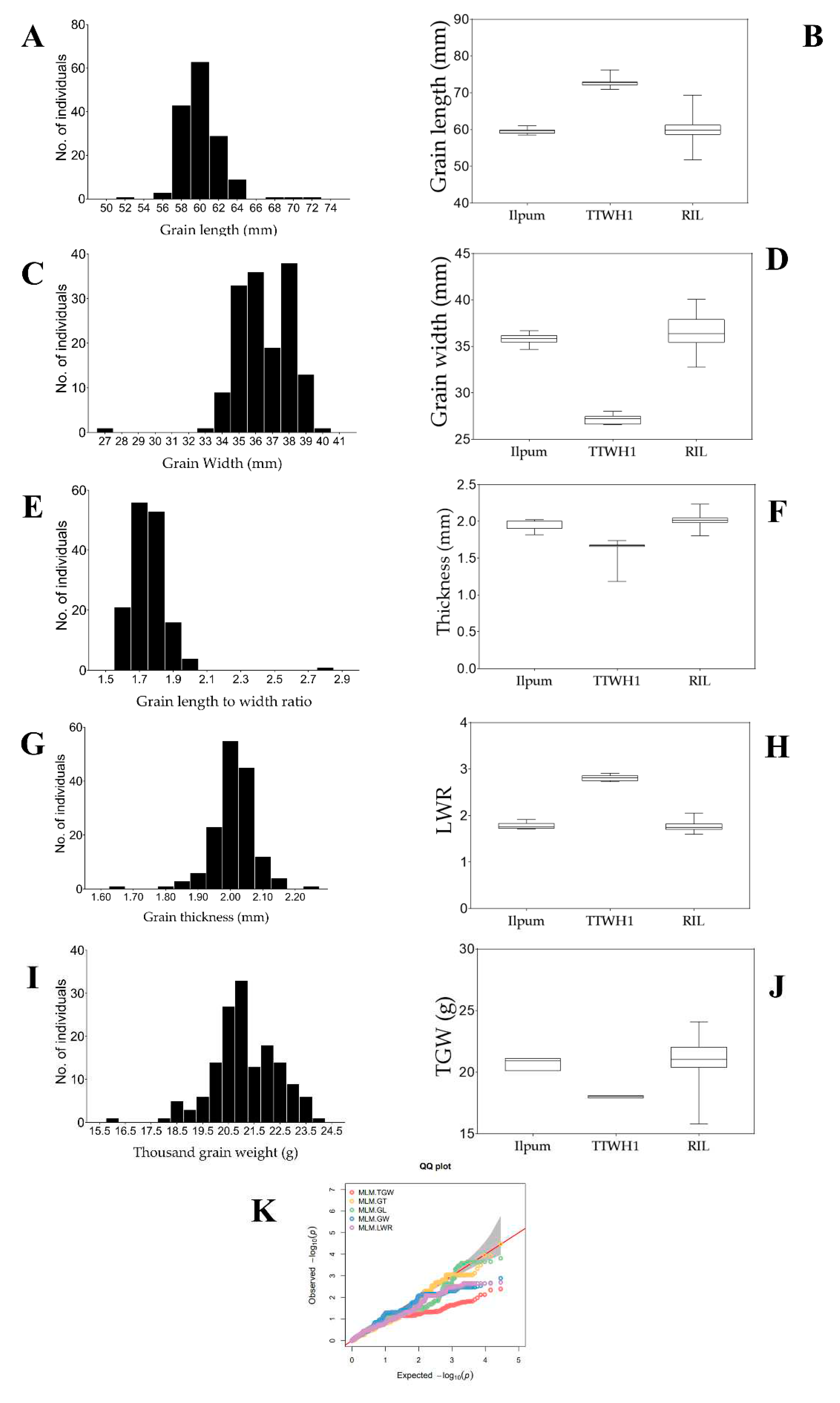
2.1. Diverging Grain Phenotypes between Parental Lines and RILs
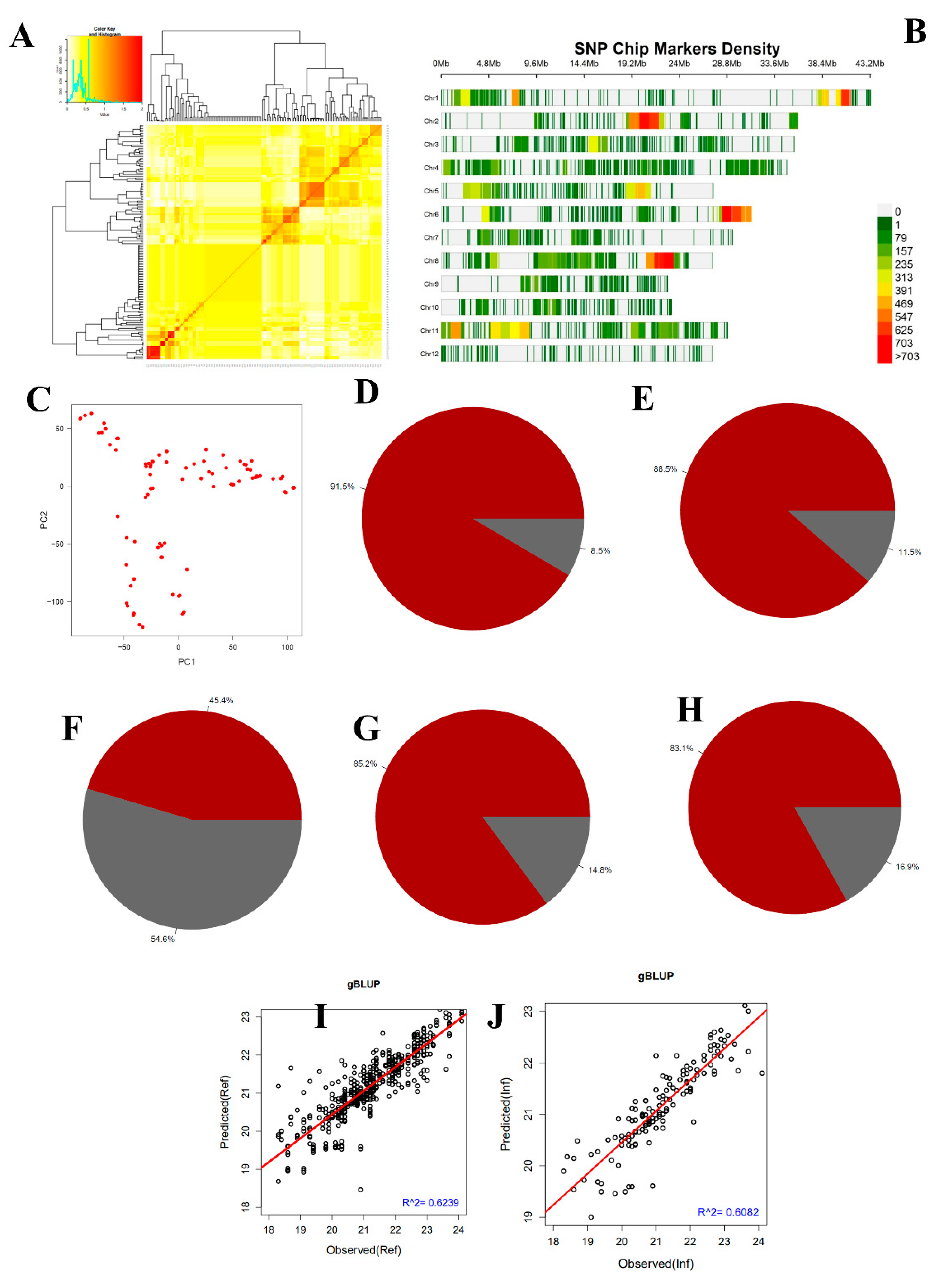
2.2. Relatedness, Correlation, Heritability, and Genomic Selection
2.3. Identified QTLs for Grain Size and Shape in Rice through GWAS with Multiple GAPIT Models
2.4. Putative Candidate Genes Harbored by Grain Traits-Related QTLs
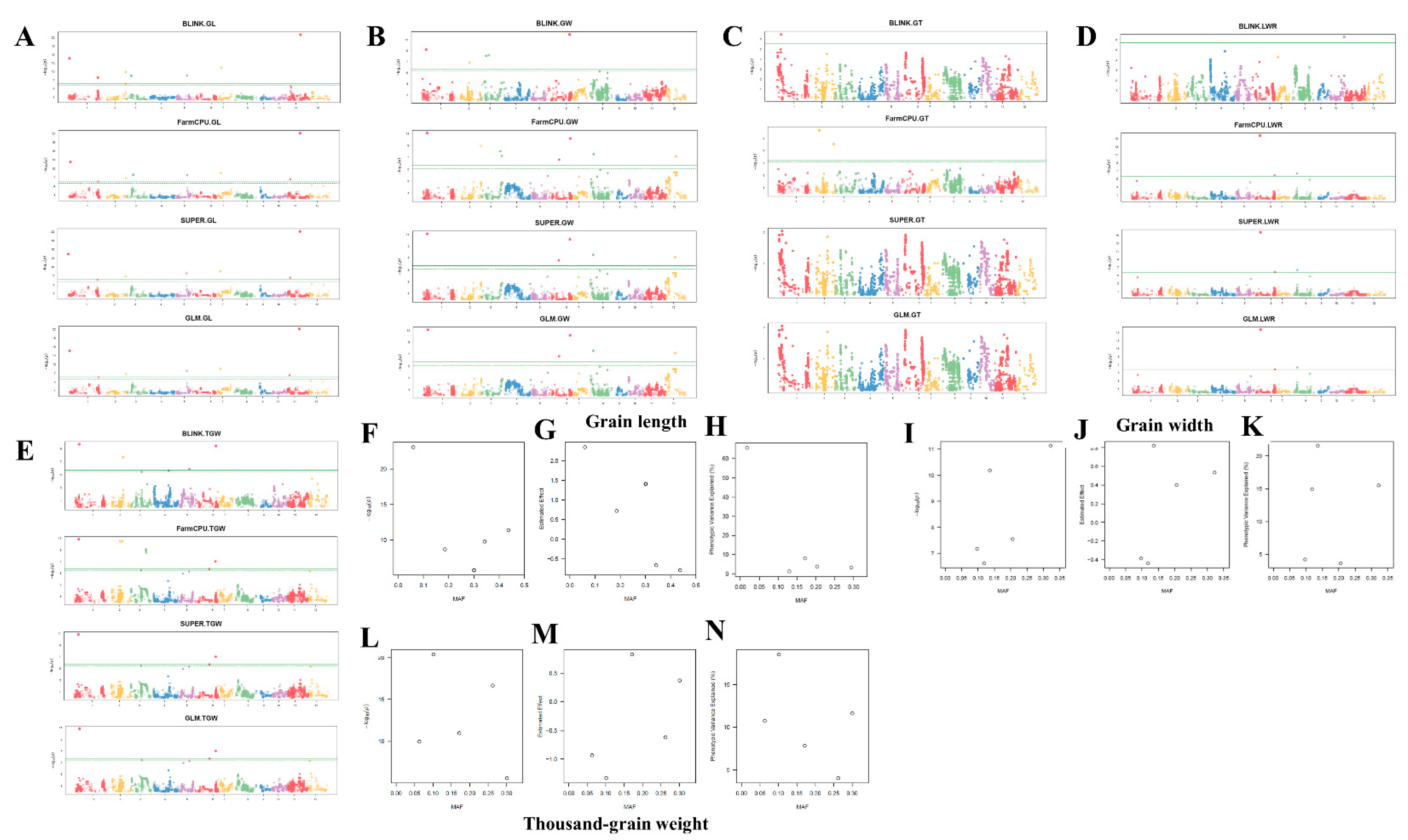
| Traits/QTLs | SNP markers | Chr | Position (bp) | PVE (%) | GWAS-GAPIT Models | Allele | |||
|---|---|---|---|---|---|---|---|---|---|
| Grain length | |||||||||
| qGL1-1BFSG | AX-95918134 | 1 | 3820526 | 72.5 | BLINK | FarmCPU | SUPER | GLM | TTWH1 |
| qGL11-1 BFSG | AX-274862201 | 11 | 16356105 | 31.9 | BLINK | FarmCPU | SUPER | GLM | Ilpum |
| qGL2-1BFSG | AX-115751685 | 2 | 35100558 | 22.9 | BLINK | FarmCPU | SUPER | GLM | TTWH1 |
| qGL3-1BF | AX-154437636 | 3 | 2253428 | 5.5 | BLINK | FarmCPU | - | - | Ilpum |
| qGL2-2BF | AX-154880023 | 2 | 35133528 | 3.1 | BLINK | FarmCPU | - | - | TTWH1 |
| qGL7-1BFSG | AX-153903748 | 7 | 6690089 | 2.6 | BLINK | FarmCPU | SUPER | GLM | TTWH1 |
| qGL1-2BF | AX-279584700 | 1 | 41489588 | 1.7 | BLINK | FarmCPU | - | - | TTWH1 |
| qGL5BFSG | AX-282746698 | 5 | 16169537 | 1.4 | BLINK | FarmCPU | - | - | Ilpum |
| qGL11-2FSG | AX-115751092 | 11 | 2823622 | 14.0 | - | FarmCPU | SUPER | GLM | Ilpum |
| qGL3-2Farm | AX-154551783 | 3 | 8242439 | 8.0 | - | FarmCPU | - | - | TTWH1 |
| Grain Width | |||||||||
| qGW1-1BFSG | AX-273945773 | 1 | 5623288 | 18.9 | BLINK | FarmCPU | SUPER | GLM | Ilpum |
| qGW2-1Blink | AX-279699609 | 2 | 10805604 | 14.9 | BLINK | - | - | - | TTWH1 |
| qGW1-2BF | AX-115791785 | 1 | 43103625 | 8.8 | BLINK | FarmCPU | - | - | TTWH1 |
| qGW1-3Blink | AX-281116133 | 1 | 20864932 | 6.9 | BLINK | - | - | - | Ilpum |
| qGW6-1BF | AX-115737727 | 6 | 28484619 | 6.2 | BLINK | FarmCPU | - | - | Ilpum |
| qGW3-1Blink | AX-154073979 | 3 | 7895651 | 4.8 | BLINK | - | - | - | Ilpum |
| qGW3-2Blink | AX-115811160 | 3 | 14888685 | 4.1 | BLINK | - | - | - | Ilpum |
| qGW6-2FSG | AX-273990782 | 6 | 13986482 | 14.9 | - | FarmCPU | SUPER | GLM | TTWH1 |
| qGW1-4Farm | AX-280898927 | 1 | 2483022 | 9.4 | - | FarmCPU | - | - | TTWH1 |
| qGW12FSG | AX-284265976 | 12 | 13013702 | 4.2 | - | FarmCPU | SUPER | GLM | TTWH1 |
| qGW8 FSG | AX-115796459 | 8 | 3875546 | 3.7 | - | FarmCPU | SUPER | GLM | Ilpum |
| qGW2-2Farm | AX-279994820 | 2 | 35461009 | 3.5 | BLINK | FarmCPU | - | - | TTWH1 |
| qGW2-3 Farm | AX-154042022 | 2 | 24704256 | 3.4 | BLINK | FarmCPU | - | - | Ilpum |
| qGW3-3 Farm | AX-154797543 | 3 | 2973374 | 1.2 | BLINK | FarmCPU | - | - | Ilpum |
| Grain thickness | |||||||||
| qGT1Blink | AX-279261704 | 1 | 18023142 | 74.9 | BLINK | - | - | - | TTWH1 |
| qGT2-1Farm | AX-154787777 | 2 | 2118477 | 54.9 | - | FarmCPU | - | - | TTWH1 |
| qGT2-2Farm | AX-154913392 | 2 | 25105471 | 5.3 | - | FarmCPU | - | - | Ilpum |
| Length-to-Width Ratio | |||||||||
| qLWR10Blink | AX-115835839 | 10 | 22038978 | 26.5 | BLINK | - | - | - | Ilpum |
| qLWR2BFSG | AX-274833045 | 2 | 10000097 | 15.2 | BLINK | FarmCPU | SUPER | GLM | TTWH1 |
| qLWR1-1BF | AX-154960834 | 1 | 1595394 | 13.5 | BLINK | FarmCPU | - | - | TTWH1 |
| qLWR1-2Blink | AX-115737888 | 1 | 600441 | 10.7 | BLINK | - | - | - | TTWH1 |
| qLWR3Blink | AX-154834762 | 3 | 8098398 | 6.9 | BLINK | - | - | - | TTWH1 |
| qLWR6-1FSG | AX-115851421 | 6 | 10178858 | 30.5 | - | FarmCPU | SUPER | GLM | Ilpum |
| qLWR6-2FSG | AX-155522120 | 6 | 30842264 | 9.4 | - | FarmCPU | SUPER | GLM | TTWH1 |
| qLWR8FSG | AX-154176130 | 8 | 5398451 | 5.9 | - | FarmCPU | SUPER | GLM | TTWH1 |
| Thousand grain weight | |||||||||
| qTGW6BFSG | AX-115737727 | 6 | 28484619 | 32.8 | BLINK | FarmCPU | SUPER | GLM | Ilpum |
| qTGW2-1BF | AX-279699609 | 2 | 10805604 | 18. 6 | BLINK | FarmCPU | - | - | TTWH1 |
| qTGW3-2Farm | AX-123153600 | 3 | 7887961 | 13.9 | - | FarmCPU | - | - | Ilpum |
| qTGW1-1Blink | AX-154298059 | 1 | 5644298 | 11.6 | BLINK | - | - | - | Ilpum |
| qTGW3-1BF | AX-154471576 | 3 | 15332432 | 10.7 | BLINK | FarmCPU | - | - | TTWH1 |
| qTGW2-2BF | AX-154096541 | 2 | 10773042 | 7.8 | BLINK | FarmCPU | - | - | Ilpum |
| qTGW1-2BFSG | AX-154333920 | 1 | 5860250 | 4.9 | BLINK | FarmCPU | SUPER | GLM | Ilpum |
| qTGW1-3BF | AX-154810092 | 1 | 42931550 | 4.03 | BLINK | FarmCPU | - | - | TTWH1 |
| No. | japonica/indica | Description | Biological process | Molecular function | Cellular component | CDS japonica vs indica/Similar Report |
|---|---|---|---|---|---|---|
| qGL1-1BFSG | Chr1:3804000..3883000 | |||||
| 1 | Os01g07870 | ATP-bindinc cassette (ABC) transporter family protein, Peroxidase 56 | Transport | Hydrolase activity, transporter activity | Extracellular region, integral component of membrane, vacuole | -; [46] |
| 2 | Os01g07880 | OsbZIP01/OsRE1, Transcription factor HY5, putative, expressed | Post-embryonic development, signal transduction, secondary metabolism | Sequence-specific DNA binding transcription factor activity | Nucleus | -; [47] |
| 4 | Os01g07910/ BGIOSGA002284 | NADH-cytochrome b5 reductase, putative | Response to stress, response to abiotic stimulus | Binding, catalytic activity | Cell wall, mitochondrion | 100% similar |
| 5 | Os01g07920 | Prolyl 4-hydroxylase, putative | Protein modification process | Binding, catalytic activity | Golgi apparatus, vacuole, membrane | -; - |
| 6 | Os01g07930/ BGIOSGA002287 | Zinc finger C-x8-C-x5-C-x3-H (CCCH)-domain containing protein family, transcription factor | Biosynthetic process | Sequence-specific DNA binding transcription factor activity | - | 100% similar |
| 7 | Os01g07940/ BGIOSGA002282 | AGC_PVPK_like_kin82y.3 - ACG kinases include homologs to PKA, PKG and PKC | Reproduction, post-embryonic development, embryo development, protein modification process | Nucleotide binding, kinase activity | - | Deletion in japonica (126–131 bp); indica (924–932, 1201–3 bp), and SNPs |
| 8 | Os01g07950 | OsGrx_S15.2 - glutaredoxin subgroup II | Cellular homeostasis | Binding | Mitochondrion | -; - |
| 9 | Os01g07960 | Acyl-protein thioesterase, Similar to Biostress-resistance-related protein | - | Hydrolase activity | - | - |
| 10 | Os01g07980 | Ankyrin, putative, expressed. SGT1, suppressor of G2 allele of SKP1; Provisional | - | Binding | - | -; [48] |
| 11 | Os01g08000 | Fibronectin type 3 and ankyrin repeat domains 1 protein | - | Protein binding | - | -; - |
| 12 | Os01g08020/ BGIOSGA002278 | Boron transporter protein, Bicarbonate transporter, eukaryotic domain containing protein. | Anion transport, | Borate efflux transmembrane transporter activity; inorganic anion exchanger activity | Integral component of membrane | Deletion in japonica (1–174 bp) |
| qGW1-1BFSG | Chr1:5623500..5684500 | |||||
| 13 | Os01g10550 | DEFL35 - Defensin-like DEFL family | - | |||
| 14 | Os01g10580/ BGIOSGA002958 | B-box (BBx) zinc finger family protein, transcription factor | Post-embryonic development, cellular component organization, secondary metabolic process, response to abiotic stimulus | Sequence-specific DNA binding transcription factor activity | Nucleoplasm | Deletion in indica (1–184; 197; 241; 841–846) |
| 15 | Os01g10590/ BGIOSGA002959 | OsFTL8 FT-Like8 homologous to Flowering Locus T gene | Flower development, reproduction, post-embryonic development, response to abiotic stimulus | Protein binding, lipid binding | Nucleus, cytoplasm | Deletion in japonica (161–194 bp) |
| 16 | Os01g10600 | OsNIP1;2 encoding Aquaporin protein, putative, expressed | Transport | Transporter activity | Membrane, plasma membrane | -; [49] |
| 17 | Os01g10610/ BGIOSGA002172 | BRI1-EMS-SUPPRESSOR1/ BRASSINOZANOL RESISTANT 1 (BES1/BZR1); transcriptional repressor family protein. | Brassinosteroids signaling- | - | - | Deletion in indica (1–93 bp) and SNPs (831:G/T, 836: T/C, 879: C/T); [50,51] |
| qGT1-1Blink | Chr1:17993000..18054000 | |||||
| 18 | Os01g32780/ BGIOSGA001545 | Universal stress protein domain-containing protein, UspA domain containing protein | Response to stress, response to molecule of fungal origin | - | - | 100% similar; [52] |
| 19 | Os01g32800/ BGIOSGA001543 | Proteasome subunit, putative, expressed. PCI domain, also known as PINT motif (Proteasome, Int-6, Nip-1, and TRIP-15). | Protein metabolic process | Protein binding | Nucleus, intracellular, cytosol, proteasome complex | Deletion in indica (972–1004 bp) |
| 20 | Os01g32870 | Heat shock protein DnaJ, Similar to Chaperone protein dnaJ 15 (Protein ALTERED RESPONSE TO GRAVITY) (AtARG1) (AtJ15) (AtDjB15). | Protein metabolic process, response to abiotic stimulus, protein binding tropism | - | - | -; - |
| 21 | Os01g32880 | AP-3 complex subunit delta, Armadillo-type fold domain containing protein | Intra-Golgi vesicle-mediated transport, protein storage vacuole organization | Transporter activity, protein binding | Membrane, cytoplasm, Golgi apparatus | -; - |
| 22 | Os01g32920/ BGIOSGA003627 | ZOS1-08 - C2H2 zinc finger protein, expressed, Transcription factor | Biosynthetic process | Sequence-specific DNA binding transcription factor activity | Intracellular | SNP689: T/C |
| 23 | Os01g32930/ BGIOSGA003628 | SGT1-specific (SGS) domain-containing protein | Embryo development, reproduction, post-embryonic development, protein binding, signal transduction, protein metabolic process, response to biotic stimulus | - | Cytosol | Deletion in indica (1–12, 270, 275–294 bp), japoica (479–484), SNPs (272: T/C, 319: C/A, 447: C/T, 504–505: GG/AC) |
| qGT2-1Farm | Chr2:2088000..2151000 | |||||
| 24 | Os02g04630 | Sodium/calcium exchanger protein, putative, expressed. The Ca2+:Cation Antiporter (CaCA) Family (TC 2.A.19) proteins | Transport | Transporter activity | Cell, vacuole, membrane | -; - |
| 25 | Os02g04640/ BGIOSGA007162 | PHOSPHATE STARVATION RESPONSE 3 (OsPHR3), Myb-like DNA-binding domain containing protein, , transcription factor | Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | Sequence-specific DNA binding transcription factor activity | - | 100% similar; [53] |
| 26 | Os02g04650/ BGIOSGA007161 | Activator of 90 kDa heat shock protein ATPase homolog | Catabolic process | Enzyme regulator activity, protein binding | - | 100% similar; - |
| 27 | Os02g04660 | Arginine N-methyltransferase 5 | Response to abiotic stimulus, protein modification process | Transferase activity | Cytosol | -; - |
| 28 | Os02g04670/ BGIOSGA007498 | Glucan endo-1,3-beta-glucosidase precursor | Carbohydrate metabolic process | Binding, hydrolase activity | Plasma membrane, membrane | Deletion in japonica (44–52 bp) |
| 29 | Os02g04680/ BGIOSGA007499 | Squamosa promoter-binding-like protein 3 (OsSPL3) - SBP-box gene family member, Transcription factor | Flower development, multicellular organismal development | Sequence-specific DNA binding transcription factor activity | Nucleus | 100% similar; [54] |
| 30 | Os02g04690 | Cycloartenol synthase | Multicellular organismal development, cellular component organization, lipid metabolic process | Catalytic activity | Vacuole | -; - |
| 31 | Os02g04700 | tRNA synthetases class II domain-containing protein | Translation | Catalytic activity, nucleic acid binding | Cytosol, cytoplasm | -; - |
| 32 | Os02g04710 | Cycloartenol synthase | Multicellular organismal development, cellular component organization, lipid metabolic process | Catalytic activity | Vacuole | -; - |
| 33 | Os02g04725 | Dolichol phosphate-mannose biosynthesis regulatory protein | Macromolecule biosynthetic process | - | Cell, integral component of endoplasmic reticulum membrane | -; - |
| qLWR2BFSG | Chr2:9970000..10030000 | |||||
| 34 | Os02g17350/ BGIOSGA007951 | VPS-27, Hrs, and STAM (VHS) and GGA and Tom1 (GAT) domain-containing protein | Transport | Transporter activity | Golgi apparatus, plasma membrane | 100% similar |
| 35 | Os02g17360/ BGIOSGA006711 | Restorer of fertility gene, Rf, pentatricopeptide repeat (PPR) repeat domain-containing protein | Mitochondrial cytoplasmic male sterility (CMS) | Nuclease activity | Plastid, mitochondrion | deletion in indica (1–84 bp); [55] |
| 36 | Os02g17380 | Fertility restorer 2 (Rf2), Mitochondrial glycine-rich protein, Fertility restoration in LD-CMS | - | - | - | -; [56] |
| 37 | Os02g17390/ BGIOSGA007953 | ABNORMAL INFLORENSCENCE MERISTEM 1(MFP/AIM1); 3-hydroxyacyl-CoA dehydrogenase | Flower development, multicellular organismal development, post-embryonic development, lipid metabolic process | Catalytic activity | Plastid, cell wall, peroxisome | 100% similar; [45] |
| 38 | Os02g17400/ BGIOSGA006709 | Leucine-rich repeat protein | Signal transduction, response to biotic stimulus, response to stress | - | Cell wall | Deletion in indica (96–101 bp) |
| 39 | Os02g17460 | Tesmin/TSO1-like CXC domain-containing protein; transcrption factor | Biosynthetic process | Sequence-specific DNA binding transcription factor activity | - | -; - |
| qTGW6BFSG | Chr6:28484608..28484625 | |||||
| 40 | Os06g46910/ BGIOSGA023481 | ZOS6-07 - C2H2 zinc finger transcription factor, expressed | Biosynthetic process | Sequence-specific DNA binding transcription factor activity | Intracellular | (SNP329: A/C; SNP445: T/C; SNP676: A/G; SNP1318: G/A); [57] |
| 41 | Os06g46920 | Dihydroflavonol-4-reductase, NAD(P)-binding domain containing protein | Fatty acid catabolic process, gibberellin (GA) biosynthesis process, Seed dormancy process, GA-mediated signaling pathway | Cinnamyl-alcohol dehydrogenase activity, coenzyme binding, nucleotide binding, catalytic activity | - | -; - |
| 42 | Os06g46930/ BGIOSGA020659 | 50S ribosomal protein L24, chloroplast precursor (CL24) | Pastid translation | Structural constituent of ribosome | Ribosome, plastid, large ribosomal subunit, chloroplast stroma | (SNP51: T/G; SNP282: A/G) |
| 43 | Os06g46940 | Os6bglu25 - beta-glucosidase homologue, similar to Os3bglu6, expressed | Carbohydrate metabolic process | Hydrolase activity, binding | Cell wall | -; [58] |
| 44 | Os06g46950/ BGIOSGA023482 | carotenoid cleavage dioxygenase 1(OsCCD1), EF-hand calcium (Ca2+)-binding protein familyexpressed | Anatomical structure morphogenesis, cellular component organization, cell differentiation, multicellular organismal development | Calcium ion binding | - | 100% similar; [59,60] |
| 45 | Os06g46995 | Armadillo/beta-catenin repeat family protein, putative, expressed | - | Protein binding | - | -; - |
| 46 | Os06g47000/ BGIOSGA020655 | External NADH-ubiquinone oxidoreductase 1, mitochondrial precursor, putative, expressed | Metabolic process | Catalytic activity | Membrane, mitochondrion | 100% similar; - |
3. Discussion
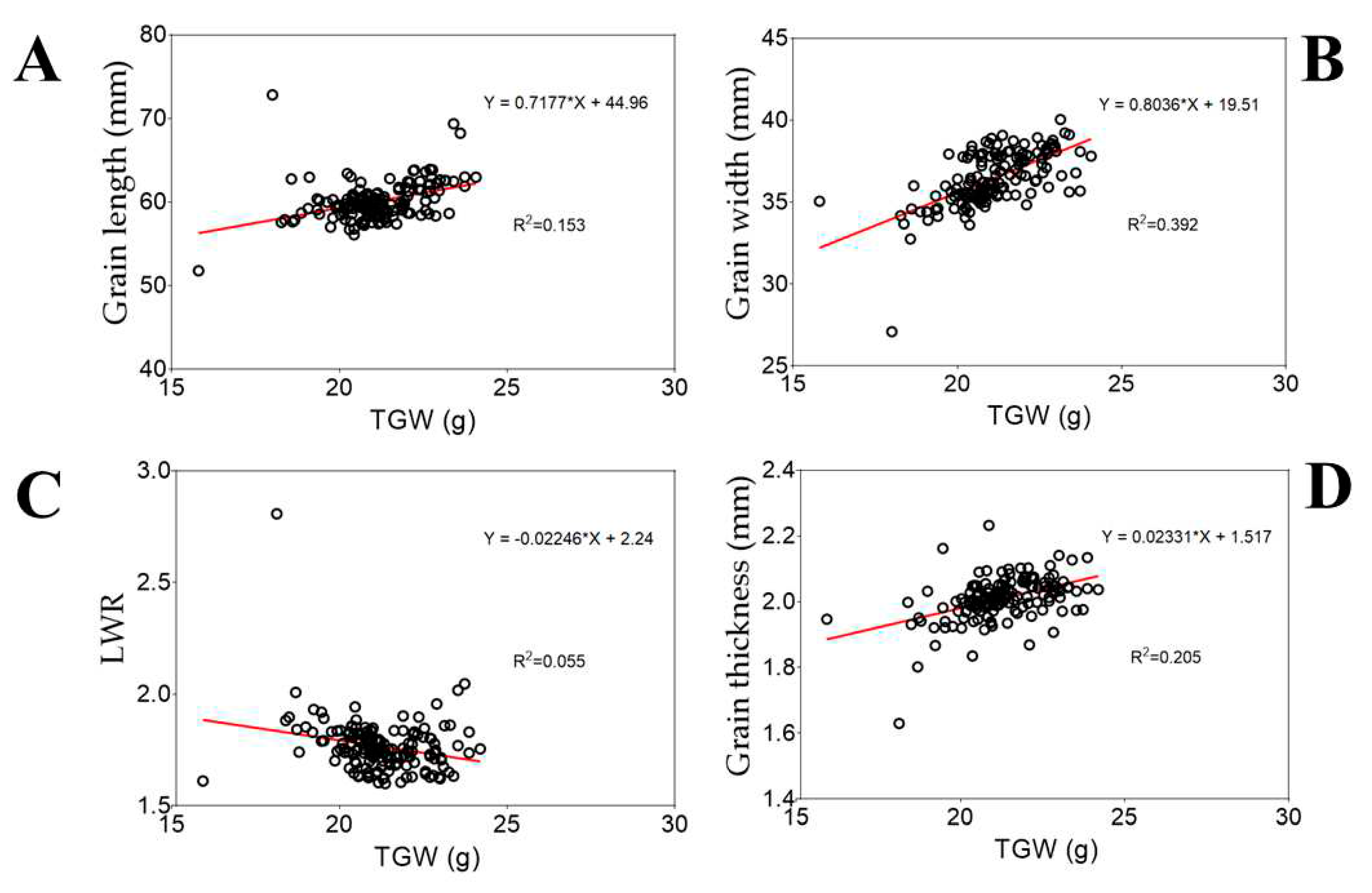
3.1. Grain Length, Width, and Thickness are Closely Related to Thousand Grain Weight but Not Length-to-Width Ratio
3.2. Genomic Estimated Breeding Value of RILs Population and Traits Heritability
3.3. The qGL1-1BFSG QTL Harbors Genes Involved in Post-Embryonic Development and Reproduction
3.4. The Grain Width, Thickness, and LWR-Associated QTLs qGW1-1BSFG, qGT1Blink, qGT2-1Farm and qLWR2BSFG Carry Genes Involved in Flower Development, Post-Embryonic Development and Reproduction
3.5. The Grain Weight-Related QTL qTGW6BFSG Harbors Genes Associated with Anatomical Structure Morphogenesis, Cell Differentiation, and Carbohydrate Metabolism
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campbell, M.T.; Bandillo, N.; Al Shiblawi, F.R.A.; Sharma, S.; Liu, K.; Du, Q.; Schmitz, A.J.; Zhang, C.; Véry, A.-A.; Lorenz, A.J.; et al. Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PLOS Genet. 2017, 13, e1006823. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-W.; Kabange, N.R.; Phyo, Z.; Park, S.-Y.; Lee, S.-M.; Lee, J.-Y.; Shin, D.; Cho, J.H.; Park, D.-S.; Ko, J.-M.; et al. Combined Linkage Mapping and Genome-Wide Association Study Identified QTLs Associated with Grain Shape and Weight in Rice (Oryza sativa L.). Agronomy 2020, 10, 1532. [Google Scholar] [CrossRef]
- Aljumaili, S.J.; Rafii, M.Y.; Latif, M.A.; Sakimin, S.Z.; Arolu, I.W.; Miah, G. Genetic Diversity of Aromatic Rice Germplasm Revealed By SSR Markers. BioMed Res. Int. 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Adjah, K.L.; Asante, M.D.; Toure, A.; Aziadekey, M.; Amoako-Andoh, F.O.; Frei, M.; Diallo, Y.; Agboka, K. Improvement of Rice Production under Drought Conditions in West Africa: Application of QTLs in Breeding for Drought Resistance. Rice Sci. 2022, 29, 512–521. [Google Scholar] [CrossRef]
- Desa, U.J.U.D.; economic, U.N.D.o.; org, s.a.u. World population projected to reach 9.8 billion in 2050, and 11.2 billion in 2100. 2017.
- Bazrkar-Khatibani, L.; Fakheri, B.-A.; Hosseini-Chaleshtori, M.; Mahender, A.; Mahdinejad, N.; Ali, J.J.I.J.o.G. Genetic mapping and validation of quantitative trait loci (QTL) for the grain appearance and quality traits in rice (Oryza sativa L.) by using recombinant inbred line (RIL) population. 2019, 2019.
- Sharma, N.; Khanna, R.J.R.a.i.g.c.r. Rice grain quality: current developments and future prospects. 2019, 5772, 89367.
- Feng, F.; Li, Y.; Qin, X.; Liao, Y.; Siddique, K.H.M. Changes in Rice Grain Quality of Indica and Japonica Type Varieties Released in China from 2000 to 2014. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Yuan, L.; Denning, G.; Mew, T.J.D.G. , Mew TW, editors. Hybrid rice breeding for super high yield. 1998, 10–12. [Google Scholar]
- Dixit, S.; Singh, A.; Kumar, A. Rice Breeding for High Grain Yield under Drought: A Strategic Solution to a Complex Problem. Int. J. Agron. 2014, 2014, 1–15. [Google Scholar] [CrossRef]
- Goto, A.; Sasahara, H.; Shigemune, A.; Miura, K. Hokuriku 193: A New High-yielding Indica Rice Cultivar Bred in Japan. Jpn. Agric. Res. Quarterly: JARQ 2009, 43, 13–18. [Google Scholar] [CrossRef]
- He, P.; Li, S.G.; Qian, Q.; Ma, Y.Q.; Li, J.Z.; Wang, W.M.; Chen, Y.; Zhu, L.H. Genetic analysis of rice grain quality. Theor. Appl. Genet. 1999, 98, 502–508. [Google Scholar] [CrossRef]
- Juliano, B.; Duff, B.J.r.g.m.; quality issues. Los Banos, L., IRRI. Rice grain quality as an emerging priority in National rice breeding programmes. 1991, 55-64.
- Hori, K.; Yamamoto, T.; Yano, M. Genetic dissection of agronomically important traits in closely related temperate japonica rice cultivars. Breed. Sci. 2017, 67, 427–434. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Duan, P.; Li, Y. Control of grain size in rice. Plant Reprod. 2018, 31, 237–251. [Google Scholar] [CrossRef]
- Zhao, D.-S.; Li, Q.-F.; Zhang, C.-Q.; Zhang, C.; Yang, Q.-Q.; Pan, L.-X.; Ren, X.-Y.; Lu, J.; Gu, M.-H.; Liu, Q.-Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Mahender, A.; Anandan, A.; Pradhan, S.K.; Pandit, E. Rice grain nutritional traits and their enhancement using relevant genes and QTLs through advanced approaches. SpringerPlus 2016, 5, 1–18. [Google Scholar] [CrossRef]
- Nagata, K.; Ando, T.; Nonoue, Y.; Mizubayashi, T.; Kitazawa, N.; Shomura, A.; Matsubara, K.; Ono, N.; Mizobuchi, R.; Shibaya, T.; et al. Advanced backcross QTL analysis reveals complicated genetic control of rice grain shape in a japonica × indica cross. Breed. Sci. 2015, 65, 308–318. [Google Scholar] [CrossRef]
- Yoshida, S.; Ikegami, M.; Kuze, J.; Sawada, K.; Hashimoto, Z.; Ishii, T.; Nakamura, C.; Kamijima, O. QTL Analysis for Plant and Grain Characters of Sake-brewing Rice Using a Doubled Haploid Population. Breed. Sci. 2002, 52, 309–317. [Google Scholar] [CrossRef]
- Yao, G.-X.; Li, J.-J.; Zhang, Q.; Hu, G.-L.; Chen, C.; Tang, B.; Zhang, H.-L.; Li, Z.-C. Mapping QTLs for Grain Weight and Shape Using Four Sister Near Isogenic Lines in Rice (Oryza sativa L.). Acta Agron. Sin. 2010, 36, 1310–1317. [Google Scholar] [CrossRef]
- Yong-Zhong, X.; Yi-Fang, T.; Cai-Guo, X.; Jin-Ping, H.; Xin-Li, S.J.J.o.I.P.B. Mapping quantitative trait loci for grain appearance traits of rice using a recombinant inbred line population. 2001, 43, 840.
- Rabiei, B.; Valizadeh, M.; Ghareyazie, B.; Moghaddam, M.; Ali, A. Identification of QTLs for rice grain size and shape of Iranian cultivars using SSR markers. Euphytica 2004, 137, 325–332. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, H.; Gu, Y.; Xia, D.; Wu, B.; Gao, G.; Zhang, Q.; He, Y. Mapping and verification of grain shape QTLs based on high-throughput SNP markers in rice. Mol. Breed. 2019, 39, 42. [Google Scholar] [CrossRef]
- Jian-long, X.; Qing-zhong, X.; Li-jun, L.; Zhi-kang, L.J.C.J.o.R.S. Genetic dissection of grain weight and its related traits in rice (Oryza sativa L. ). 2002, 16, 6. [Google Scholar]
- Li, P.; Chen, Y.-H.; Lu, J.; Zhang, C.-Q.; Liu, Q.-Q.; Li, Q.-F. Genes and Their Molecular Functions Determining Seed Structure, Components, and Quality of Rice. Rice 2022, 15, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. 2010, 107, 19579–19584. [Google Scholar] [CrossRef]
- Song, X.-J.; Huang, W.; Shi, M.; Zhu, M.-Z.; Lin, H.-X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- Huang, K.; Wang, D.; Duan, P.; Zhang, B.; Xu, R.; Li, N.; Li, Y.J.T.P.J. WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. 2017, 91, 849-860.
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef]
- Duan, P.; Ni, S.; Wang, J.; Zhang, B.; Xu, R.; Wang, Y.; Chen, H.; Zhu, X.; Li, Y.J.N.p. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. 2015, 2, 1-5.
- Qi, P.; Lin, Y.-S.; Song, X.-J.; Shen, J.-B.; Huang, W.; Shan, J.-X.; Zhu, M.-Z.; Jiang, L.; Gao, J.-P.; Lin, H.-X.J.C.r. The novel quantitative trait locus GL3. 1 controls rice grain size and yield by regulating Cyclin-T1; 3. 2012, 22, 1666–1680. [Google Scholar]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. 2012, 109, 21534–21539. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, J.; Zheng, X.; Wu, F.; Lin, Q.; Heng, Y.; Tian, P.; Cheng, Z.; Yu, X.; Zhou, K.; et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef]
- Hu, Z.; Lu, S.-J.; Wang, M.-J.; He, H.; Sun, L.; Wang, H.; Liu, X.-H.; Jiang, L.; Sun, J.-L.; Xin, X.J.M.P. A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. 2018, 11, 736-749.
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.-I.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef]
- Shi, C.; Dong, N.; Guo, T.; Ye, W.; Shan, J.; Lin, H. A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 2020, 103, 1174–1188. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.J.R.G. , Genetics; Breeding. Gene Network of Grain Size and Number in Rice. 2018, 191–206. [Google Scholar]
- Li, N.; Xu, R.; Li, Y. Molecular Networks of Seed Size Control in Plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Li, J.J.A.r.o.g. Molecular genetic dissection of quantitative trait loci regulating rice grain size. 2014, 48, 99-118.
- Xu, L.; Zhao, H.; Ruan, W.; Deng, M.; Wang, F.; Peng, J.; Luo, J.; Chen, Z.; Yi, K. ABNORMAL INFLORESCENCE MERISTEM1 Functions in Salicylic Acid Biosynthesis to Maintain Proper Reactive Oxygen Species Levels for Root Meristem Activity in Rice. Plant Cell 2017, 29, 560–574. [Google Scholar] [CrossRef]
- Li, H.; Liu, Y.; Qin, H.; Lin, X.; Tang, D.; Wu, Z.; Luo, W.; Shen, Y.; Dong, F.; Wang, Y.; et al. A rice chloroplast-localized ABC transporter ARG1 modulates cobalt and nickel homeostasis and contributes to photosynthetic capacity. New Phytol. 2020, 228, 163–178. [Google Scholar] [CrossRef]
- Chai, J.; Zhu, S.; Li, C.; Wang, C.; Cai, M.; Zheng, X.; Zhou, L.; Zhang, H.; Sheng, P.; Wu, M.; et al. OsRE1 interacts with OsRIP1 to regulate rice heading date by finely modulating Ehd1 expression. Plant Biotechnol. J. 2020, 19, 300–310. [Google Scholar] [CrossRef]
- Zhang, X.; Li, D.; Zhang, H.; Wang, X.; Zheng, Z.; Song, F. Molecular characterization of rice OsBIANK1, encoding a plasma membrane-anchored ankyrin repeat protein, and its inducible expression in defense responses. Mol. Biol. Rep. 2009, 37, 653–660. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, S.; Li, C.; Hu, T.; Hou, S.; Bai, Q.; Ji, X.; Xu, F.; Guo, C.; Huang, M.; et al. The plasma membrane-localized OsNIP1;2 mediates internal aluminum detoxification in rice. Front. Plant Sci. 2022, 13, 970270. [Google Scholar] [CrossRef]
- Yang, S.; Yuan, D.; Zhang, Y.; Sun, Q.; Xuan, Y.H. BZR1 Regulates Brassinosteroid-Mediated Activation of AMT1;2 in Rice. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, X.; Zhu, W.; Lin, H.; Chen, X.; Li, Y.; Ye, W.; Yin, Z. Molecular Traits and Functional Exploration of BES1 Gene Family in Plants. Int. J. Mol. Sci. 2022, 23, 4242. [Google Scholar] [CrossRef]
- Sauter, M.; Rzewuski, G.; Marwedel, T.; Lorbiecke, R. The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 2002, 53, 2325–2331. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Ruan, W.; Li, C.; Huang, F.; Zeng, M.; Liu, Y.; Yu, Y.; Ding, X.; Wu, Y.; Wu, Z.; et al. Integrative Comparison of the Role of the PHOSPHATE RESPONSE1 Subfamily in Phosphate Signaling and Homeostasis in Rice. Plant Physiol. 2015, 168, 1762–1776. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Zhou, H.-Z.; Wu, Y.; Zhang, H.; Lin, J.; Jiang, X.; He, Q.; Zhu, J.; Li, Y.; Yu, H.; et al. OsSPL3, an SBP-Domain Protein, Regulates Crown Root Development in Rice. Plant Cell 2019, 31, 1257–1275. [Google Scholar] [CrossRef]
- Jiang, H.; Lu, Q.; Qiu, S.; Yu, H.; Wang, Z.; Yu, Z.; Lu, Y.; Wang, L.; Xia, F.; Wu, Y.; et al. Fujian cytoplasmic male sterility and the fertility restorer gene OsRf19 provide a promising breeding system for hybrid rice. 2022, 119. [CrossRef]
- Itabashi, E.; Iwata, N.; Fujii, S.; Kazama, T.; Toriyama, K.J.T.p.j. The fertility restorer gene, Rf2, for Lead Rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein. 2011, 65, 359-367.
- Agarwal, P.; Arora, R.; Ray, S.; Singh, A.K.; Singh, V.P.; Takatsuji, H.; Kapoor, S.; Tyagi, A.K. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 2007, 65, 467–485. [Google Scholar] [CrossRef]
- Ren, R.; Li, D.; Zhen, C.; Chen, D.; Chen, X. Specific roles of Os4BGlu10, Os6BGlu24, and Os9BGlu33 in seed germination, root elongation, and drought tolerance in rice. Planta 2019, 249, 1851–1861. [Google Scholar] [CrossRef] [PubMed]
- Ilg, A.; Yu, Q.; Schaub, P.; Beyer, P.; Al-Babili, S. Overexpression of the rice carotenoid cleavage dioxygenase 1 gene in Golden Rice endosperm suggests apocarotenoids as substrates in planta. Planta 2010, 232, 691–699. [Google Scholar] [CrossRef]
- Jing, P.; Zou, J.; Kong, L.; Hu, S.; Wang, B.; Yang, J.; Xie, G. OsCCD1, a novel small calcium-binding protein with one EF-hand motif, positively regulates osmotic and salt tolerance in rice. Plant Sci. 2016, 247, 104–114. [Google Scholar] [CrossRef]
- Senthilnathan, S.J.A.a.S. Usefulness of correlation analysis. 2019.
- Li, R.; Li, M.; Ashraf, U.; Liu, S.; Zhang, J. Exploring the Relationships Between Yield and Yield-Related Traits for Rice Varieties Released in China From 1978 to 2017. Front. Plant Sci. 2019, 10, 543. [Google Scholar] [CrossRef]
- Xie, Q.; Mayes, S.; Sparkes, D.L. Carpel size, grain filling, and morphology determine individual grain weight in wheat. J. Exp. Bot. 2015, 66, 6715–6730. [Google Scholar] [CrossRef]
- Huang, R.; Jiang, L.; Zheng, J.; Wang, T.; Wang, H.; Huang, Y.; Hong, Z. Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci. 2013, 18, 218–226. [Google Scholar] [CrossRef]
- Yang, W.; Peng, S.; Dionisio-Sese, M.L.; Laza, R.C.; Visperas, R.M. Grain filling duration, a crucial determinant of genotypic variation of grain yield in field-grown tropical irrigated rice. Field Crop. Res. 2008, 105, 221–227. [Google Scholar] [CrossRef]
- Du, Z.; Huang, Z.; Li, J.; Bao, J.; Tu, H.; Zeng, C.; Wu, Z.; Fu, H.; Xu, J.; Zhou, D.; et al. qTGW12a, a naturally varying QTL, regulates grain weight in rice. Theor. Appl. Genet. 2021, 134, 2767–2776. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Ahamed, K.U.; Rahman, S. Yield and yield contributing attributes of rice (Oryza Sativa L.) under different planting dates in boro season. Asian-Australasian J. Biosci. Biotechnol. 2018, 3, 162–169. [Google Scholar] [CrossRef]
- Bhat, J.A.; Ali, S.; Salgotra, R.K.; Mir, Z.A.; Dutta, S.; Jadon, V.; Tyagi, A.; Mushtaq, M.; Jain, N.; Singh, P.K.; et al. Genomic Selection in the Era of Next Generation Sequencing for Complex Traits in Plant Breeding. Front. Genet. 2016, 7, 221. [Google Scholar] [CrossRef]
- E Meuwissen, T.H.; Hayes, B.J.; E Goddard, M. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Chen, J.; Liu, K.; Zha, W.; Zhou, L.; Li, M.; Xu, H.; Li, P.; Chen, Z.; Yang, G.; Chen, P.; et al. Identification and verification of grain shape QTLs by SNP array in rice. PLOS ONE 2021, 16, e0260133. [Google Scholar] [CrossRef]
- Aslam, K.; Naveed, S.A.; Sabar, M.; Shabir, G.; Shah, S.M.; Khan, A.R.; Shah, M.M.; Fiaz, S.; Xu, J.; Arif, M. Identification of QTLs for rice grain size and weight by high-throughput SNP markers in the IR64 x Sadri population. Front. Genet. 2022, 13, 955347. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Chu, L.; Zhang, Y.; Bian, Y.; Xiao, J.; Xu, D. HY5: A Pivotal Regulator of Light-Dependent Development in Higher Plants. Front. Plant Sci. 2022, 12, 800989. [Google Scholar] [CrossRef] [PubMed]
- Jonassen, E.M.; Sévin, D.C.; Lillo, C. The bZIP transcription factors HY5 and HYH are positive regulators of the main nitrate reductase gene in Arabidopsis leaves, NIA2, but negative regulators of the nitrate uptake gene NRT1.1. J. Plant Physiol. 2009, 166, 2071–2076. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhang, H.; Zhang, H.; Deng, X.W.; Wei, N. HY5 regulates nitrite reductase 1 (NIR1) and ammonium transporter1;2 (AMT1;2) in Arabidopsis seedlings. Plant Sci. 2015, 238, 330–339. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Yanagisawa, S. Light signalling-induced regulation of nutrient acquisition and utilisation in plants. Semin. Cell Dev. Biol. 2018, 83, 123–132. [Google Scholar] [CrossRef]
- Yang, Y.; Liang, T.; Zhang, L.; Shao, K.; Gu, X.; Shang, R.; Shi, N.; Li, X.; Zhang, P.; Liu, H. UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat. Plants 2018, 4, 98–107. [Google Scholar] [CrossRef]
- Chen, X.; Yao, Q.; Gao, X.; Jiang, C.; Harberd, N.P.; Fu, X. Shoot-to-Root Mobile Transcription Factor HY5 Coordinates Plant Carbon and Nitrogen Acquisition. Curr. Biol. 2016, 26, 640–646. [Google Scholar] [CrossRef]
- Zhang, X.; Huai, J.; Shang, F.; Xu, G.; Tang, W.; Jing, Y.; Lin, R. A PIF1/PIF3-HY5-BBX23 Transcription Factor Cascade Affects Photomorphogenesis. Plant Physiol. 2017, 174, 2487–2500. [Google Scholar] [CrossRef]
- Bursch, K.; Toledo-Ortiz, G.; Pireyre, M.; Lohr, M.; Braatz, C.; Johansson, H. Identification of BBX proteins as rate-limiting cofactors of HY5. Nat. Plants 2020, 6, 921–928. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F.J.T.P.C. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. 2013, 25, 1243-1257.
- Heng, Y.; Lin, F.; Jiang, Y.; Ding, M.; Yan, T.; Lan, H.; Zhou, H.; Zhao, X.; Xu, D.; Deng, X.W.J.P.P. B-Box containing proteins BBX30 and BBX31, acting downstream of HY5, negatively regulate photomorphogenesis in Arabidopsis. 2019, 180, 497-508.
- Li, C.; Qi, L.; Zhang, S.; Dong, X.; Jing, Y.; Cheng, J.; Feng, Z.; Peng, J.; Li, H.; Zhou, Y.; et al. Mutual upregulation of HY5 and TZP in mediating phytochrome A signaling. Plant Cell 2021, 34, 633–654. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Guo, Y.; Wu, C.; Yang, G.; Li, Y.; Zheng, C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom. 2008, 9, 44–44. [Google Scholar] [CrossRef] [PubMed]
- Han, G.L.; Qiao, Z.Q.; Li, Y.X.; Wang, C.F.; Wang, B.S. The Roles of CCCH Zinc-Finger Proteins in Plant Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 8327. [Google Scholar] [CrossRef]
- Chen, F.; Liu, H.-L.; Wang, K.; Gao, Y.-M.; Wu, M.; Xiang, Y. Identification of CCCH Zinc Finger Proteins Family in Moso Bamboo (Phyllostachys edulis), and PeC3H74 Confers Drought Tolerance to Transgenic Plants. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Li, J.; Mahajan, A.; Tsai, M.-D. Ankyrin Repeat: A Unique Motif Mediating Protein−Protein Interactions. Biochemistry 2006, 45, 15168–15178. [Google Scholar] [CrossRef]
- Gu, H.; Zhang, K.; Chen, J.; Gull, S.; Chen, C.; Hou, Y.; Li, X.; Miao, J.; Zhou, Y.; Liang, G. OsFTL4, an FT-like Gene, Regulates Flowering Time and Drought Tolerance in Rice (Oryza sativa L.). Rice 2022, 15, 1–15. [Google Scholar] [CrossRef]
- Bai, B.; Lu, N.; Li, Y.; Guo, S.; Yin, H.; He, Y.; Sun, W.; Li, W.; Xie, X. OsBBX14 promotes photomorphogenesis in rice by activating OsHY5L1 expression under blue light conditions. Plant Sci. 2019, 284, 192–202. [Google Scholar] [CrossRef]
- Lei, L.; Cao, L.; Ding, G.; Zhou, J.; Luo, Y.; Bai, L.; Xia, T.; Chen, L.; Wang, J.; Liu, K.; et al. OsBBX11 on qSTS4 links to salt tolerance at the seeding stage in Oryza sativa L. ssp. Japonica. Front. Plant Sci. 2023, 14, 1139961. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The Rice B-Box Zinc Finger Gene Family: Genomic Identification, Characterization, Expression Profiling and Diurnal Analysis. PLOS ONE 2012, 7, e48242. [Google Scholar] [CrossRef]
- He, Y.; Chen, S.; Liu, K.; Chen, Y.; Cheng, Y.; Zeng, P.; Zhu, P.; Xie, T.; Chen, S.; Zhang, H.J.P.B.J. OsHIPL1, a hedgehog-interacting protein-like 1 protein, increases seed vigour in rice. 2022, 20, 1346-1362.
- Shi, H.; Li, X.; Lv, M.; Li, J.J.I.J.o.M.S. BES1/BZR1 family transcription factors regulate plant development via brassinosteroid-dependent and independent pathways. 2022, 23, 10149.
- Tong, H.; Liu, L.; Jin, Y.; Du, L.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C.J.T.P.C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. 2012, 24, 2562-2577.
- Verma, L.; Bhadouria, J.; Bhunia, R.K.; Singh, S.; Panchal, P.; Bhatia, C.; Eastmond, P.J.; Giri, J. Monogalactosyl diacylglycerol synthase 3 affects phosphate utilization and acquisition in rice. J. Exp. Bot. 2022, 73, 5033–5051. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Łyskowski, A.; Jaremko, .; Jaremko, M. Genetic and Molecular Factors Determining Grain Weight in Rice. Front. Plant Sci. 2021, 12, 605799. [CrossRef]
- Li, G.; Tang, J.; Zheng, J.; Chu, C. Exploration of rice yield potential: Decoding agronomic and physiological traits. Crop. J. 2021, 9, 577–589. [Google Scholar] [CrossRef]
- Zhang, Z.; Todhunter, R.J.; Buckler, E.S.; Van Vleck, L.D. Technical note: Use of marker-based relationships with multiple-trait derivative-free restricted maximal likelihood. J. Anim. Sci. 2007, 85, 881–885. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Zhou, Y.; Summers, R.J.G. BLINK: a package for the next level of genome-wide association studies with both individuals and markers Meng Huang. 2018, 8, 1-12.
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLOS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Proceedings of the Nucleic acids symposium series; 1999; pp. 95–98. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





