1. Introduction
Medical imaging plays a pivotal role in many medical diagnostics tasks. In order to obtain an accurate diagnosis, both high quality image acquisition and interpretation are required [
1]. Acquisition technology has significantly improved over the past decades, with available scanners that enable higher resolution and shortened acquisition times [
2,
3,
4,
5]. Furthermore, the image interpretation process has undergone major advancements during the last decade, due to deep learning methods and accessibility to high performance computational resources. Deep learning has been shown to yield state-of-the-art performance in terms of accuracy and computational speed [
7,
8,
9,
10]. The combination of these technological advancements thus has the potential to revolutionize medical imaging by playing a key role as a support tool in the radiological workflow [
11]. The U.S. Food and Drug Administration (FDA) has approved few diagnostic procedures utilizing deep learning and artificial intelligence (AI) technologies [
12,
13,
14]. Nonetheless, three major challenges remain for deep diagnostic models, namely: (1) lack of accessibility to interpreted medical images and data, which is required for the training process; (2) limited reliability and robustness to noise or other unknown perturbations in the data acquisition process [
15,
16]; and limited generalization to data sets characterized by distributions that differ from that of the training data [
18,
19]. These challenges stand at the core of the bottleneck that currently exists between theory and practice in terms of incorporating this technology in actual clinical settings.
Typically, the performance of a machine learning (ML) algorithm for a given task is assessed by metrics such as accuracy, specificity, and sensitivity. However, especially when considering clinical applicability of such algorithms, robustness is an additional important performance that should be considered. ‘Robustness’ is a general term, which may be referred to in different contexts, such as robustness to deterministic input variations (e.g., contrast) [
17], out-of-distribution (OOD) inputs [
18], random input perturbations or noise [
20], adversarial attacks [
15,
21,
22,
23,
24], and more. The underlying approach at the core of robustness assessment, which is common to these different contexts, is to examine the extent of the variation that may be applied upon the algorithm’s input whilst obtaining valid predictions at the output. In any case, evaluating the robustness of a deep learning model aimed for clinical tasks should be addressed in addition to the accuracy and specificity metrics, and according to the application at hand.
One straightforward approach to increase the robustness of deep neural networks (DNN) to unknown perturbations is data augmentation, where the network is trained for a larger data set and/or a broadened set of data distributions. However, this approach is basically focused on the desired output of the network according to the required task [
25]; it does not introduce any new information that may affect the learning process of the network to resemble a human perspective that is derived from the specific problem or task at hand. An approach to address this issue that has been discussed in recent years is the inclusion of physics information or prior knowledge into the learning process or the network design [
25,
26,
27]. The assumption at the base of inclusion of prior domain-related knowledge is that it may shift the learning mechanism such as to produce improved stabilization of the network [
26]
. In addition, it has been shown that the inclusion of prior physics knowledge may improve the generalization capacity of the network. Furthermore, it has the important advantage of supporting training on small amounts of data or in cases where clinical ground truth (GT) is un-available [
26]
.
An additional important issue is the choice of network architecture, which may significantly affect the network’s performance. In [
28], the problem of neural architecture search in the context of physics-based learning has been addressed, and a scheme for automatically finding an optimal architecture was proposed in a general formulation that isn’t limited to the medical case. However, the performance of this approach has yet to be demonstrated for medical applications.
A comprehensive survey of works for which medical knowledge domain is integrated into deep learning techniques is given in [
25], categorizing the studies according to their medical tasks, namely: disease diagnosis, lesion/organ abnormality detection and lesion/organ segmentation. An additional central review paper is [
29], which includes an overview of deep learning methods incorporating physics information for MRI reconstruction tasks. In [
29], the physics-driven approaches are classified into the principal mechanisms of inclusion of the physics-information, namely: physics-based loss functions, Plug-and-Play (PnP) methods, generative models and unrolled networks.
In this paper, we survey recent works that utilize the approach of inclusion of prior knowledge into the learning process for various medical imaging modalities, tasks and applications and discuss how this improves the training mechanism and performance of the deep neural models. We follow the classification method as proposed in [
29], and discuss the common thread of the physics-inclusion mechanisms for various imaging modalities, such as Diffusion-Weighted MRI, Ultrasound imaging, PET-CT and more. It is important to note that our goal in this review is not to cover all the research papers within the scope of physics-based deep learning; we mainly focus on recent works, the majority of which from the previous five or six years, and discuss their general common principles in the context of the different physics-inclusion mechanism. To conclude this survey, we outline current challenges and future research directions within this context.
2. Physics-Driven Loss Terms
In [
30], the problem of producing in-silico patient-specific cardiac mechanical models is discussed. This problem, which may be solved by computational heavy finite element models (FEMs), is addressed here by a deep neural network approach that aims to replace the FEM solution. A cost term that includes the effect of the heart microstructure and its contraction is applied in order to avoid the need to compute input-output pairs for supervised network training. This enables the simulation of full cardiac cycles whilst replacing heavy and time-consuming finite element model computations. The minimization of this cost function is practically the solution to the momentum balance equations, which model the dynamics of the heart microstructure. In addition, the last layer of the network constraints the solution to belong to a prescribed linear subspace that represents left ventricular dynamics. The inclusion of the physics information as part of the loss function and the allowed set of solutions enables to bypass heavy computations whilst providing the network with a generalization capacity for individualized in-silico models.
In [
31], an unsupervised physics-informed DNN is proposed for the problem of the Intra-Voxel Incoherent Motion (IVIM) model parameters’ estimation for Diffusion-Weighted MRI. The forward model equation is incorporated into the loss function of the network, thus enabling the iterative learning of the IVIM parameters, which serve as useful biomarkers for various clinical applications. The training process is applied for synthetically generated data that is produced according to the forward model equations, and this enables unsupervised learning without requiring neither clinical data nor its corresponding ground truth (GT) parameters maps.
A similar approach is applied in [
26] for the ophthalmology medical domain, where the bio-optical parameters of the intraocular lens should be accurately evaluated for cataract surgeries. To achieve that, a DNN that incorporates the optical ray propagation model in the eye is trained on synthetically generated bio-metric data. A physics-induced loss term includes the expressions for the eye’ optical system and its minimization yields the required intraocular lens parameters. The proposed training is shown to be superior to systems with a standard training approach as well as to the state-of-the-art of intraocular calculation on five biometric datasets. It is thus concluded that the inclusion of physics knowledge into the network enables the generalization of the network model for a wide range of eye biometric data, without requiring big clinical data sets.
In [
32], a novel method for strain reconstruction maps in ultrasound elastography imaging is proposed, based on the incorporation of implicit physics information that relates the RF data, the tissue displacement and the predicted elastography map in a causal manner. The inclusion of the implicit tissue displacement prior (termed as “privileged information”) is enabled by applying a strategy to generate triplets of training data, privileged information and training labels based on numerical biomechanics and ultrasound physics simulations. A physics-driven loss term based on the implicit privileged information is applied as feedback information to an intermediate layer of the network’s architecture. In other words, the physics-based loss term is utilized in combination with a tailored architecture to produce the predicted tissue strain map. In addition, the physics-driven approach is applied to produce lacking implicit tissue displacement datasets to be used in the training process. The performance of this method is demonstrated for simulated data, phantom data and clinical liver and breast imaging data.
For under-sampled MRI reconstruction, where fast signal acquisition is enabled by sampling in the k-space (the transform domain), several works have shown that applying a loss term that enforces consistency in the k-space domain may yield improved quality for higher sampling rates [
33,
34]. In [
35], a few low-frequency terms from the k-space are included in addition to the consistency enforcement, such that the overall structure of the MRI image is preserved and anomaly location uncertainty in the image domain is avoided. Similarly, in [
18] a transform-based consistency was applied to generate standard-dose PET images from their low-dose counterparts and corresponding multi-contrast MRI images. For that, the sinogram-based physics of the PET imaging system and the uncertainty in the DNN output through the per-voxel heteroscedasticity were utilized in combination, resulting in improved robustness of the network to out-of-distribution (OOD) acquisitions. It is noted in [
18] that in the context of PET imaging, OOD inputs may be attributed to variations in different factors, such as photon counts, reconstruction protocol, hardware, and more.
3. Unrolled Networks
Network unrolling (or unfolding) was originally proposed in [
36], motivated by the need to connect iterative algorithms to deep neural networks and specifically to improve the computational efficiency of sparse coding. In the general context of iterative algorithms, the core idea of network unrolling is to map each iteration of the algorithm into a single network layer; these layers are concatenated to form the network’s architecture. The input’s propagation through the network is thus equivalent to executing the algorithm’s iterations for a finite number of times that is the number of layers. The network may be trained using backpropagation and its obtained parameters transfer to the algorithm parameters, such as the model and regularization coefficients. Thus, the unrolled network may be viewed as a parameters’ optimization algorithm that is directly related to the original optimization problem; in this manner, unrolled networks reflect the knowledge domain and improve the lack of interpretability that is common in other architectures [
19].
In [
37] the problem of parameters’ estimation for the Neurite Orientation Dispersion and Density Imaging (NODDI) model in Diffusion MRI (dMRI) is addressed. The NODDI bio-physical model has been widely used for tissue microstructure characterization of white matter in the brain [
38]. In [
37], an unfolded network for the NODDI’s parameters optimization has been proposed and compared to a previous multi-layer perceptron (MLP) proposed in [
39]. Results for clinical brain scans demonstrated the superior performance of the unrolled network compared to the previously proposed MLP in terms of computational speed and accuracy. It is discussed that while the MLP is a generic feed-forward network, the new unrolled network structure is derived from the iterative update of the solver of the NODDI model equations, thus feeding each layer not merely from the previous later, but also from a “short-cut” input signal that is the input of the first layer. In this manner, the unrolling technique applies information from the physics of the knowledge domain, which is shown to significantly improve the network’s performance.
A separate and significant group of works in the context of computational MRI discusses the task of image reconstruction from k-space sampling with sub-Nyquist rate, termed also compressive sensing (CS). In CS, the compressibility of MR images is utilized to obtain MRI reconstruction from sub-sampled k-space measurements, in a manner that enables faster image acquisition rates, which are clinically applicable. To that end, a regularized least squares problem that consists of a sparse linear transform needs to be solved. One method to solve this problem is the alternating direction method of multipliers (ADMM) [
40], which may be solved by an unrolling deep neural network as described in [
41]. In [
41], each layer of the proposed ADMM-Net architecture corresponds to an iteration in the ADMM algorithm. Results on brain and chest MR images demonstrate the high performance of ADMM-Net in terms of accuracy and computational speed. In [
42]
a more general formulation of ADMM-Net is proposed, by extending the sparse transform expression; it is shown that this extension enables to achieve optimal image recovery for different imaging tasks with different types of images.
In [
43], an accelerated MRI reconstruction method that embeds total variation (TV) regularization into an unrolled deep neural network is proposed. The network is designed to learn a complete reconstruction process for complex-valued multichannel MR data. It is trained on complete clinical musculoskeletal images that are retrospectively sub-sampled with different acceleration rates. The proposed TV-embedding unrolled network is shown to preserve important spatial features not presented in the training data for a wide range of pathologies, whilst achieving high reconstruction speed.
In [
44], a novel recurrent neural network architecture for the reconstruction of k-space under-sampled cardiac MR images is proposed. In addition to embedding the iterative nature of the traditional fast MR optimization algorithms, the proposed scheme captures the temporal dependencies of the images. This novel architecture is demonstrated to be able to learn the temporal dependencies and the iterative reconstruction process with only a small number of parameters, whilst producing superior reconstruction quality and computational speed compared to previous methods.
An additional work in which an unrolled network architecture for MRI reconstruction is proposed is [
45], where the key significance lies in the reduction of the number of network parameters. Experiments on clinical brain images demonstrate the improvement in the robustness of the training process and the improved quality of the obtained reconstructions.
In [
46], a new method for clutter suppression in contrast enhanced ultrasound (US) imaging is proposed, which is based on robust Principal Component Analysis (PCA) and unfolded deep neural networks. Specifically, the optimization required for the solution of the robust PCA is applied by using an unfolded network. In this manner, the contrast enhanced US image is separated from the outliers’ image. The method is trained in a supervised manner on sets of separated enhanced US images from both in-vivo rats’ brain scans and simulated data. Experimental results on simulated and in-vivo images demonstrate the improved performance of the proposed method in terms of image quality (high contrast-to-noise ratios) as well as reduced computational speed.
In [
47], an unrolled network architecture is proposed for solving the inverse problem of CT reconstruction. The method, which is based on the Primal-Dual Hybrid Gradient optimization algorithm, consists CNNs in both the reconstruction and data space. The network is trained on simulated data and accounts for the physical modeling of the problem by incorporating a non-linear forward operator, which relates the signal to the observed data. Results on simulated low-dose CT and human phantoms demonstrate the high performance of the method in terms of reconstruction quality.
4. Generative Models
An important class of physics-induced networks for image reconstruction relies on the concept of Deep Image Prior (DIP) originally proposed in [
48,
49]. In DIP, an implicit prior is enforced in order to constrain the solution space. More specifically, a generator CNN reconstructs an image from a random latent vector. Notably, rather than training a CNN on a large dataset of example images and ground truths, the generator network is fitted to a single degraded image. In other words, the CNN weights are utilized as a parametrization of the restored image; these weights are randomly initialized and optimized to fit a given degraded image as well as a task-dependent observation model [
48]. The application of DIP has been studied for MRI reconstruction in [
50], for which the observation model encapsulated the physics of the problem and was determined as the k-space transform (i.e., applying a Fourier transform followed by a sampling operator). The generator network’s weights were not trained based on a training dataset; they were updated based on a single under-sampled k-space data. The network’s performance was evaluated for clinical knee and brain MR images, and its superiority in terms of reconstruction accuracy as well as its generalization capacity was demonstrated compared to previous state-of-the-art reconstruction methods.
A different group of works is based on generative adversarial networks (GANs), in which a generative CNN is trained in a supervised manner that consists of two sub-networks: the generator model, which is trained to generate new examples (or images), and the discriminator model, whose goal is to classify examples as either real (i.e., taken from the image domain) or fake (i.e., generated). These two models are trained together in an adversarial fashion, until the discriminator model is fooled at a prevalence rate meaning that the generator model is sufficiently plausible. In [
51], the concept of GAN was utilized for CS in MRI. To generate high-quality MR images, a mixture of cost functions was applied such that the generator was trained to remove the aliasing artifacts whilst preserving the texture details. The discriminator network was trained by applying a perceptual lost and high-quality MR images. Performance evaluation of this scheme (named GANCS) was applied on a contrast-enhanced MR dataset of pediatric patients. The generated images were rated by expert radiologists and were scored to be of higher quality in terms of preservation of fine texture details, compared with previous methods.
5. Plug-and-Play Methods
In Plug-and-Play (PnP) algorithms, the image de-noising problem is de-coupled from the forward model signal recovery [
52]. The image denoiser may be chosen from a variety of state-of-the-art denoisers. This approach may be utilized especially for problems in which the forward model may significantly change among different scans, as is the case for CS in MRI. Since the CNN-based denoisers are trained independently of the specific forward model of the problem, it may improve the generalization capacity of the trained network for different data sets. Specifically, for the case of CS in MRI, PnP algorithms may exploit diverse image structures for training. For example, in [
52] PnP methods were applied for MR knee and cardiac cines for image reconstruction from highly under-sampled data; the CNN denoisers were tailored specifically for each application, and the comparable performance of PnP methods with respect to previous CS deep-learning methods was shown even for significantly reduced training data.
In [
53], the problem of Diffusion-Weighted MRI (dMRI) reconstruction from under-sampled k-space and q-space data was addressed by learning the signal manifold corresponding to the measurement k- and q-space from bio-physical modelling, such that no in-vivo data was required. In addition, a PnP algorithm for the de-noising of the data was utilized. Results of human brain scans demonstrated the capacity of the proposed scheme to accurately recover dMRI from accelerated q- and k-space acquisitions.
In [
54], it is suggested to broaden the PnP approach by training the denoiser network on pairs of fully sampled images with their generated artifact contaminated images. In this manner, the denoiser is provided with additional model information regarding the resulting aliasing artifacts. In other words, the forward model is leveraged as a prior to improve the denoiser’s performance. The proposed method is validated on experimental
in-vivo liver data for different acceleration rates.
6. Conclusion and Future Challenges
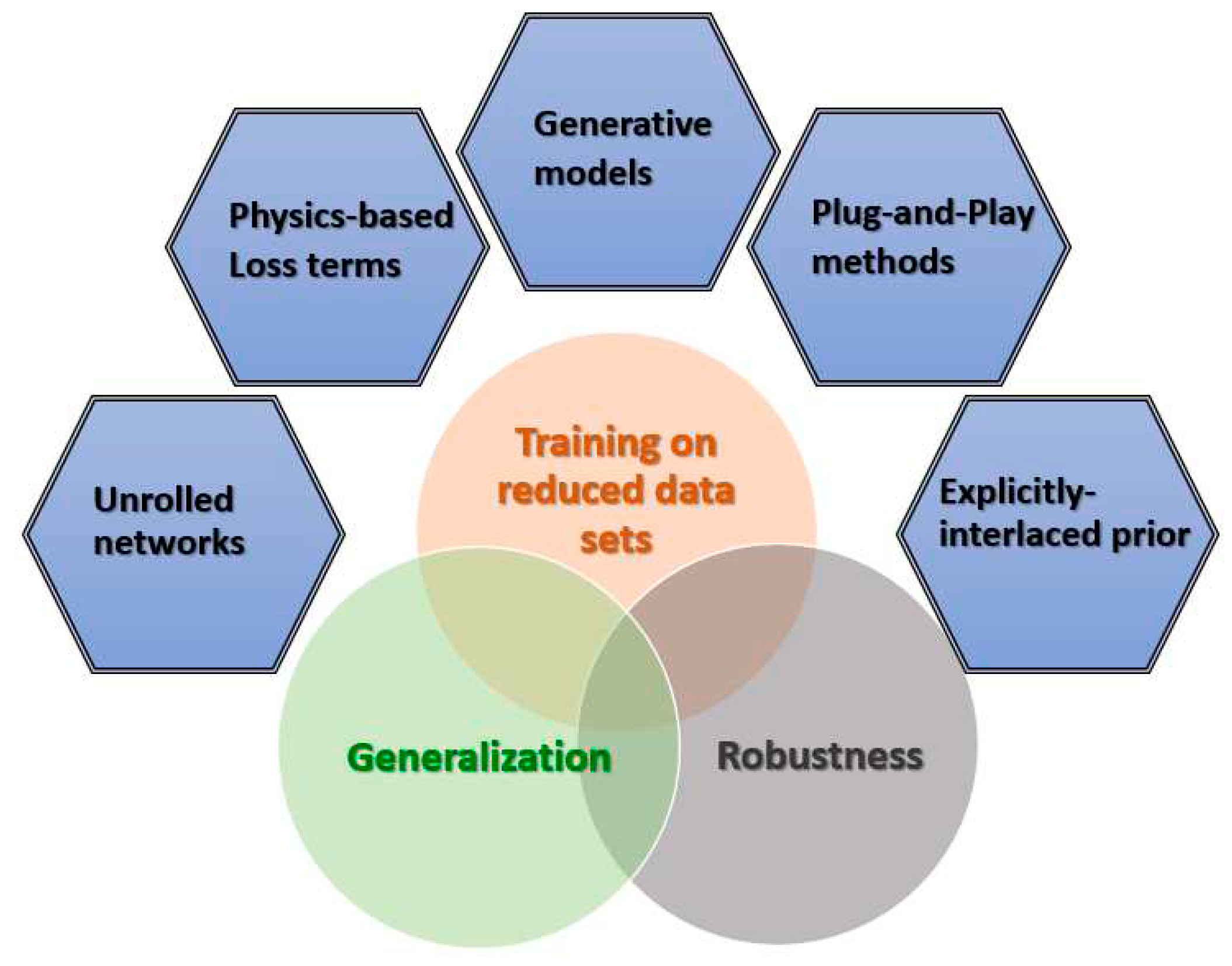
The unprecedented performance of DNN-based systems has been demonstrated over a wide range of medical imaging tasks and applications. Nonetheless, major challenges in terms of limited robustness, limited stability and generalization capacities, as well as small or unavailable training datasets, still exist and impede incorporation of such systems into daily clinical use. In this paper, we surveyed recent works aiming at addressing these issues. The underlying common thread of the reviewed works is the inclusion of a prior, specific-domain knowledge or information into either the DNNs’ architecture, learning process or both. We classified these works into four main categories, namely:
(1) physics-driven loss-based networks; (2) unrolled networks; (3) generative models (GANs); and (4) plug-and-play methods. The list of works is summarized in Table 1. For all of these categories, some sort of a known (or partially known) forward model of the problem is typically utilized in the training or learning process. Specifically, in unrolled networks, an iterative solver of an optimization problem directly translates into the network’s architecture as concatenated layers corresponding to the solver’s iterations.
The surveyed works span over a wide range of imaging modalities (MRI, US, X-Ray CT, PET-CT, OCT, Ultrasound Elastography), medical imaging tasks (classification, regression, image reconstruction) and applications in different clinical domains, such as ophthalmology, cardio-vascularity, neuroimaging, digestive system imaging, chest imaging and more. For the various discussed applications, the added value of the inclusion of the physics prior in terms of improved robustness and generalization, and/or the ability to train the networks over limited data sets, is demonstrated over simulated or clinical settings.
Physics-informed neural networks within the context of medical imaging is an important and exciting research field that is extensively progressing. Additional future research is required to address existing major challenges, such as the generalization capacity of the network models. In addition, methods to quantitatively assess the robustness and generalization capacities are yet to be developed and tested in order to obtain clinically applicable deep learning based systems.
An additional interesting research direction is the design of optimal network architectures. In many cases, currently existing network architectures and learning mechanisms may be improved by
explicitly incorporating additional knowledge-domain priors; for example, for the case where the forward model parameters that relate the network’s input to its output are at least partly known, their respective value may be explicitly introduced as additional inputs to the layers of the network. More generally, an important future challenge is to design improved architectures that better integrate the “engineering knowledge” of the problem and harness it for better regularized learning mechanisms. Such an approach, where the physical prior is explicitly interlaced into the network’s architecture, may be either applied as stand-alone or in combination with the previously mentioned methods, as depicted in
Figure 1; furthermore, it has the potential to produce more robust and generalizable models and henceforth advance their applicability to clinical practice.
Funding
Research authority, University of Haifa, Haifa, Israel.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Irrelevant.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Greenspan, H.; van Ginneken, B.; Summers, R.M. Guest Editorial Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Trans. Med Imaging 2016, 35, 1153–1159. [Google Scholar] [CrossRef]
- Matrone, G.; Ramalli, A.; Savoia, A.S.; Tortoli, P.; Magenes, G. High Frame-Rate, High Resolution Ultrasound Imaging With Multi-Line Transmission and Filtered-Delay Multiply And Sum Beamforming. IEEE Trans. Med Imaging 2016, 36, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Gholipour, A.; A Estroff, J.; Warfield, S.K. Robust Super-Resolution Volume Reconstruction From Slice Acquisitions: Application to Fetal Brain MRI. IEEE Trans. Med Imaging 2010, 29, 1739–1758. [Google Scholar] [CrossRef] [PubMed]
- Baran, J.M. and Webster, J.G., 2009, September. Design of low-cost portable ultrasound systems. In 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society (pp. 792-795). IEEE.
- Kim, G.-D.; Yoon, C.; Kye, S.-B.; Lee, Y.; Kang, J.; Yoo, Y.; Song, T.-K. A single FPGA-based portable ultrasound imaging system for point-of-care applications. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2012, 59, 1386–1394. [Google Scholar] [CrossRef]
- Ghafoorian, M.; Karssemeijer, N.; Heskes, T.; van Uder, I.W.M.; de Leeuw, F.E.; Marchiori, E.; van Ginneken, B.; Platel, B. Non-uniform patch sampling with deep convolutional neural networks for white matter hyperintensity segmentation. 2016. [Google Scholar] [CrossRef]
- Zhou, S. , Nie, D. , Adeli, E., Gao, Y., Wang, L., Yin, J. and Shen, D., Granada, Spain, 16-20 September 2018, Proceedings, Part IV 11 (pp. 488-496). Springer International Publishing., 2018. Fine-grained segmentation using hierarchical dilated neural networks. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference. [Google Scholar]
- Zhang, L.; Wang, M.; Liu, M.; Zhang, D. A Survey on Deep Learning for Neuroimaging-Based Brain Disorder Analysis. Front. Neurosci. 2020, 14, 779. [Google Scholar] [CrossRef]
- Wang, M.; Lian, C.; Yao, D.; Zhang, D.; Liu, M.; Shen, D. Spatial-Temporal Dependency Modeling and Network Hub Detection for Functional MRI Analysis via Convolutional-Recurrent Network. IEEE Trans. Biomed. Eng. 2019, 67, 2241–2252. [Google Scholar] [CrossRef]
- Jie, B.; Liu, M.; Lian, C.; Shi, F.; Shen, D. Designing weighted correlation kernels in convolutional neural networks for functional connectivity based brain disease diagnosis. Med Image Anal. 2020, 63, 101709. [Google Scholar] [CrossRef]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.W.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: a systematic review and meta-analysis. npj Digit. Med. 2021, 4, 1–23. [Google Scholar] [CrossRef]
- Finlayson, S.G.; Chung, H.W.; Kohane, I.S.; Beam, A.L. Adversarial attacks against medical deep learning systems. arXiv 2018, arXiv:1804.05296. [Google Scholar]
- Topol, E.J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Benjamens, S.; Dhunnoo, P.; Meskó, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. npj Digit. Med. 2020, 3, 1–8. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, T.; Li, Z.; Liu, M.; Zhang, D. Towards evaluating the robustness of deep diagnostic models by adversarial attack. Med Image Anal. 2021, 69, 101977. [Google Scholar] [CrossRef]
- Springenberg, M.; Frommholz, A.; Wenzel, M.; Weicken, E.; Ma, J.; Strodthoff, N. From modern CNNs to vision transformers: Assessing the performance, robustness, and classification strategies of deep learning models in histopathology. Med Image Anal. 2023, 87, 102809. [Google Scholar] [CrossRef] [PubMed]
- Billot, B.; Greve, D.N.; Puonti, O.; Thielscher, A.; Van Leemput, K.; Fischl, B.; Dalca, A.V.; Iglesias, J.E. SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining. Med Image Anal. 2023, 86, 102789–102789. [Google Scholar] [CrossRef]
- Sudarshan, V.P.; Upadhyay, U.; Egan, G.F.; Chen, Z.; Awate, S.P. Towards lower-dose PET using physics-based uncertainty-aware multimodal learning with robustness to out-of-distribution data. Med Image Anal. 2021, 73, 102187. [Google Scholar] [CrossRef] [PubMed]
- Monga, V.; Li, Y.; Eldar, Y.C. Algorithm Unrolling: Interpretable, Efficient Deep Learning for Signal and Image Processing. IEEE Signal Process. Mag. 2021, 38, 18–44. [Google Scholar] [CrossRef]
- Barbieri, S.; Gurney-Champion, O.J.; Klaassen, R.; Thoeny, H.C. Deep learning how to fit an intravoxel incoherent motion model to diffusion-weighted MRI. Magn. Reson. Med. 2020, 83, 312–321. [Google Scholar] [CrossRef]
- Bortsova, G.; González-Gonzalo, C.; Wetstein, S.C.; Dubost, F.; Katramados, I.; Hogeweg, L.; Liefers, B.; van Ginneken, B.; Pluim, J.P.; Veta, M.; et al. Adversarial attack vulnerability of medical image analysis systems: Unexplored factors. Med Image Anal. 2021, 73, 102141. [Google Scholar] [CrossRef]
- Szegedy, C.; Zaremba, W.; Sutskever, I.; Bruna, J.; Erhan, D.; Goodfellow, I.; Fergus, R. Intriguing properties of neural networks. arXiv 2013, arXiv:1312.6199. [Google Scholar]
- Ortiz-Jimenez, G.; Modas, A.; Moosavi-Dezfooli, S.-M.; Frossard, P. Optimism in the Face of Adversity: Understanding and Improving Deep Learning Through Adversarial Robustness. Proc. IEEE 2021, 109, 635–659. [Google Scholar] [CrossRef]
- Carlini, N. and Wagner, D., 2017, May. Towards evaluating the robustness of neural networks. In 2017 IEEE symposium on security and privacy (sp) (pp. 39-57).
- Xie, X.; Niu, J.; Liu, X.; Chen, Z.; Tang, S.; Yu, S. A survey on incorporating domain knowledge into deep learning for medical image analysis. Med Image Anal. 2021, 69, 101985. [Google Scholar] [CrossRef]
- Burwinkel, H.; Matz, H.; Saur, S.; Hauger, C.; Trost, M.; Hirnschall, N.; Findl, O.; Navab, N.; Ahmadi, S.-A. Physics-aware learning and domain-specific loss design in ophthalmology. Med Image Anal. 2021, 76, 102314. [Google Scholar] [CrossRef]
- Lucas, A.; Iliadis, M.; Molina, R.; Katsaggelos, A.K. Using Deep Neural Networks for Inverse Problems in Imaging: Beyond Analytical Methods. IEEE Signal Process. Mag. 2018, 35, 20–36. [Google Scholar] [CrossRef]
- Ba, Y.; Zhao, G.; Kadambi, A. Blending diverse physical priors with neural networks. arXiv 2019, arXiv:1910.00201. [Google Scholar]
- Hammernik, K.; Kustner, T.; Yaman, B.; Huang, Z.; Rueckert, D.; Knoll, F.; Akcakaya, M. Physics-Driven Deep Learning for Computational Magnetic Resonance Imaging: Combining physics and machine learning for improved medical imaging. IEEE Signal Process. Mag. 2023, 40, 98–114. [Google Scholar] [CrossRef] [PubMed]
- Buoso, S.; Joyce, T.; Kozerke, S. Personalising left-ventricular biophysical models of the heart using parametric physics-informed neural networks. Med. Image Anal. 2021, 71, 102066. [Google Scholar] [CrossRef]
- Kaandorp, M.P.T.; Barbieri, S.; Klaassen, R.; van Laarhoven, H.W.M.; Crezee, H.; While, P.T.; Nederveen, A.J.; Gurney-Champion, O.J. Improved unsupervised physics-informed deep learning for intravoxel incoherent motion modeling and evaluation in pancreatic cancer patients. Magn. Reson. Med. 2021, 86, 2250–2265. [Google Scholar] [CrossRef]
- Gao, Z.; Wu, S.; Liu, Z.; Luo, J.; Zhang, H.; Gong, M.; Li, S. Learning the implicit strain reconstruction in ultrasound elastography using privileged information. Med Image Anal. 2019, 58, 101534. [Google Scholar] [CrossRef]
- Schlemper, J.; Caballero, J.; Hajnal, J.V.; Price, A.N.; Rueckert, D. A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction. IEEE Trans. Med. Imaging 2017, 37, 491–503. [Google Scholar] [CrossRef]
- Yang, G.; Yu, S.; Dong, H.; Slabaugh, G.; Dragotti, P.L.; Ye, X.; Liu, F.; Arridge, S.; Keegan, J.; Guo, Y.; et al. DAGAN: Deep De-Aliasing Generative Adversarial Networks for Fast Compressed Sensing MRI Reconstruction. IEEE Trans. Med Imaging 2017, 37, 1310–1321. [Google Scholar] [CrossRef]
- Hyun, C.M.; Kim, H.P.; Lee, S.M.; Lee, S.; Seo, J.K. Deep learning for under-sampled MRI reconstruction. Phys. Med. Biol. 2018, 63, 135007. [Google Scholar] [CrossRef] [PubMed]
- Gregor, K. and LeCun, Y. , 2010, June. Learning fast approximations of sparse coding. In Proceedings of the 27th international conference on international conference on machine learning (pp. 399-406).
- Ye, C. Tissue microstructure estimation using a deep network inspired by a dictionary-based framework. Med Image Anal. 2017, 42, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K.; Hori, M.; Aoki, S. NODDI in clinical research. J. Neurosci. Methods 2020, 346, 108908. [Google Scholar] [CrossRef]
- Golkov, V.; Dosovitskiy, A.; Sperl, J.I.; Menzel, M.I.; Czisch, M.; Samann, P.; Brox, T.; Cremers, D. q-Space Deep Learning: Twelve-Fold Shorter and Model-Free Diffusion MRI Scans. IEEE Trans. Med Imaging 2016, 35, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y.; Yin, W. A Fast Alternating Direction Method for TVL1-L2 Signal Reconstruction From Partial Fourier Data. IEEE J. Sel. Top. Signal Process. 2010, 4, 288–297. [Google Scholar] [CrossRef]
- Sun, J.; Li, H.; Xu, Z. Deep ADMM-Net for compressive sensing MRI. Adv. Neural Inf. Process. Syst. 2016, 29. [Google Scholar]
- Yang, Y.; Sun, J.; Li, H.; Xu, Z. ADMM-CSNet: A Deep Learning Approach for Image Compressive Sensing. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 42, 521–538. [Google Scholar] [CrossRef]
- Hammernik, K.; Klatzer, T.; Kobler, E.; Recht, M.P.; Sodickson, D.K.; Pock, T.; Knoll, F. Learning a variational network for reconstruction of accelerated MRI data. Magn. Reson. Med. 2017, 79, 3055–3071. [Google Scholar] [CrossRef]
- Qin, C.; Schlemper, J.; Caballero, J.; Price, A.N.; Hajnal, J.V.; Rueckert, D. Convolutional Recurrent Neural Networks for Dynamic MR Image Reconstruction. IEEE Trans. Med Imaging 2018, 38, 280–290. [Google Scholar] [CrossRef]
- Aggarwal, H.K.; Mani, M.P.; Jacob, M. MoDL: Model-Based Deep Learning Architecture for Inverse Problems. IEEE Trans. Med Imaging 2018, 38, 394–405. [Google Scholar] [CrossRef]
- Solomon, O.; Cohen, R.; Zhang, Y.; Yang, Y.; He, Q.; Luo, J.; van Sloun, R.J.G.; Eldar, Y.C. Deep Unfolded Robust PCA With Application to Clutter Suppression in Ultrasound. IEEE Trans. Med Imaging 2019, 39, 1051–1063. [Google Scholar] [CrossRef]
- Adler, J.; Öktem, O. Learned primal-dual reconstruction. IEEE Trans. Med. Imaging 2018, 37, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Ulyanov, D. , Vedaldi, A. and Lempitsky, V., 2018. Deep image prior. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 9446-9454). [Google Scholar]
- Golts, A. , Freedman, D. and Elad, M., 2018. Deep energy: Using energy functions for unsupervised training of dnns.
- Pour Yazdanpanah, A.; Afacan, O.; Warfield, S.K. Non-learning based deep parallel MRI reconstruction (NLDpMRI). 10949, 1094. [Google Scholar] [CrossRef]
- Mardani, M.; Gong, E.; Cheng, J.Y.; Vasanawala, S.S.; Zaharchuk, G.; Xing, L.; Pauly, J.M. Deep Generative Adversarial Neural Networks for Compressive Sensing MRI. IEEE Trans. Med Imaging 2018, 38, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Bouman, C.A.; Buzzard, G.T.; Chan, S.; Liu, S.; Reehorst, E.T.; Schniter, P. Plug-and-Play Methods for Magnetic Resonance Imaging: Using Denoisers for Image Recovery. IEEE Signal Process. Mag. 2020, 37, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Mani, M.; Magnotta, V.A.; Jacob, M. qModeL: A plug-and-play model-based reconstruction for highly accelerated multi-shot diffusion MRI using learned priors. Magnetic resonance in medicine 2021, 86, 835–851. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, Y.; Eldeniz, C.; Gan, W.; An, H.; Kamilov, U.S. RARE: Image Reconstruction Using Deep Priors Learned Without Groundtruth. IEEE J. Sel. Top. Signal Process. 2020, 14, 1088–1099. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).