Submitted:
18 September 2023
Posted:
20 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. The proposed method
-
Initialization Step
- (a)
- Set as the number of chromosomes.
- (b)
- Set the maximum number of allowed generations.
- (c)
- Initialize randomly the chromosomes in in S.
- (d)
- Set as the selection rate of the algorithm, with .
- (e)
- Set as the mutation rate, with .
- (f)
- Set iter=0.
-
Fitness calculation Step
- (a)
-
Fordo
- i
- Calculate the fitness of chromosome .
- (b)
- EndFor
-
Genetic operations step
- (a)
- Selection procedure. The chromosomes are sorted according to their fitness values. The chromosomes with the lowest fitness values are transferred intact to the next generation. The remain chromosomes are substituted by offspings created in the crossover procedure. During the selection process for each offspring two parents are selected from the population using the tournament selection.
- (b)
-
Crossover procedure: For every pair of selected parents two additional chromosomes and are produced using the following equations:The value is a randomly selected number with [56].
- (c)
- Mutation procedure: For each element of every chromosome, a random number is drawn. The corresponding element is altered randomly if .
-
Termination Check Step
- (a)
- Set
- (b)
- If or the proposed stopping rule of Tsoulos [57] is hold, then goto Local Search Step, else goto b.
- Local Search Step. Apply a local search procedure to chromosome of the population with the lowest fitness value and report the obtained minimum. In the current work the BFGS variant of Powell [58] was used as a local search procedure.
2.1. Proposed initialization Distribution
| Algorithm 1: The k-Means algorithm. |
|
2.2. Chromosome rejection rule
| Algorithm 2: Chromosome rejection rule |
|
2.3. The proposed sampling procedure
- Take random samples from the objective function using uniform distribution
- Calculate the k centers of the points using the k-means algorithm provided in algorithm 1.
- Remove from the set of centers C, points that are closed to each other.
- Return the set of centers C as the set of chromosomes.
3. Experiments
3.1. Test functions
- Bf1 (Bohachevsky 1) function:
- Bf2 (Bohachevsky 2) function:with .
- Branin function: with .
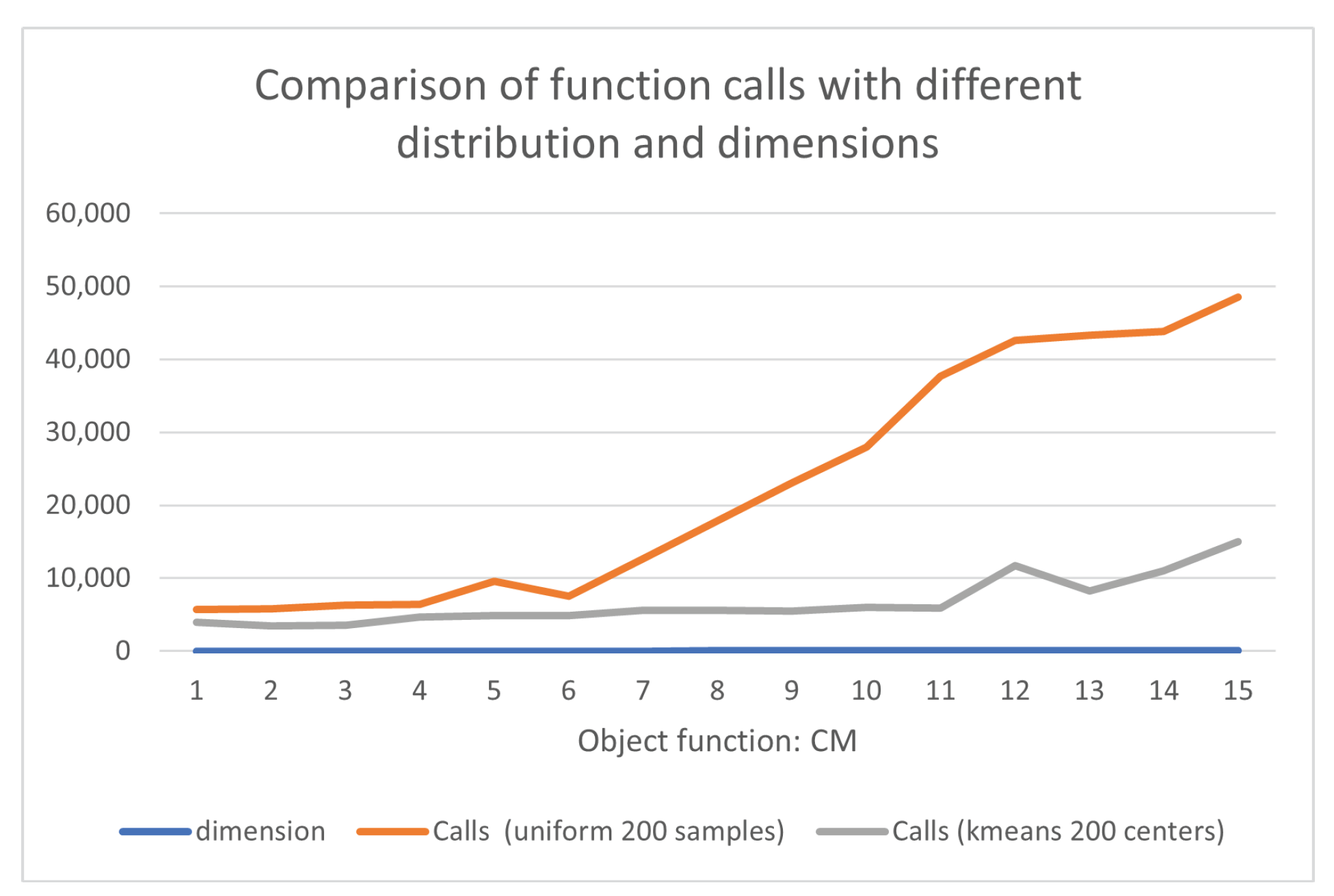
- CM function:where . In the conducted experiments the value was used.
- Camel function:
- Easom function:with
-
Exponential function, defined as:The values were used in the executed experiments.
- Griewank2 function:
- Griewank10 function. The function is given by the equationwith .
- Gkls function. , is a function with w local minima, described in [72] with and n a positive integer between 2 and 100. The values and were used in the conducted experiments.
-
Goldstein and Price functionWith .
- Hansen function: , .
- Hartman 3 function:with and and
- Hartman 6 function:with and and
-
Potential function. The molecular conformation corresponding to the global minimum of the energy of N atoms interacting via the Lennard-Jones potential[73] is used a test function here and it is defined by:The values were used in the conducted experiments.
- Rastrigin function.
-
Rosenbrockfunction.The values were used in the conducted experiments.
- Shekel 5function.
- Shekel 7 function.
- Shekel 10 function.
-
Sinusoidal function:The values of and was used in the conducted experiments.
-
Test2N function:The function has in the specified range and in our experiments we used .
- Test30N function:with , with local minima in the search space. For our experiments we used .
3.2. Experimental results
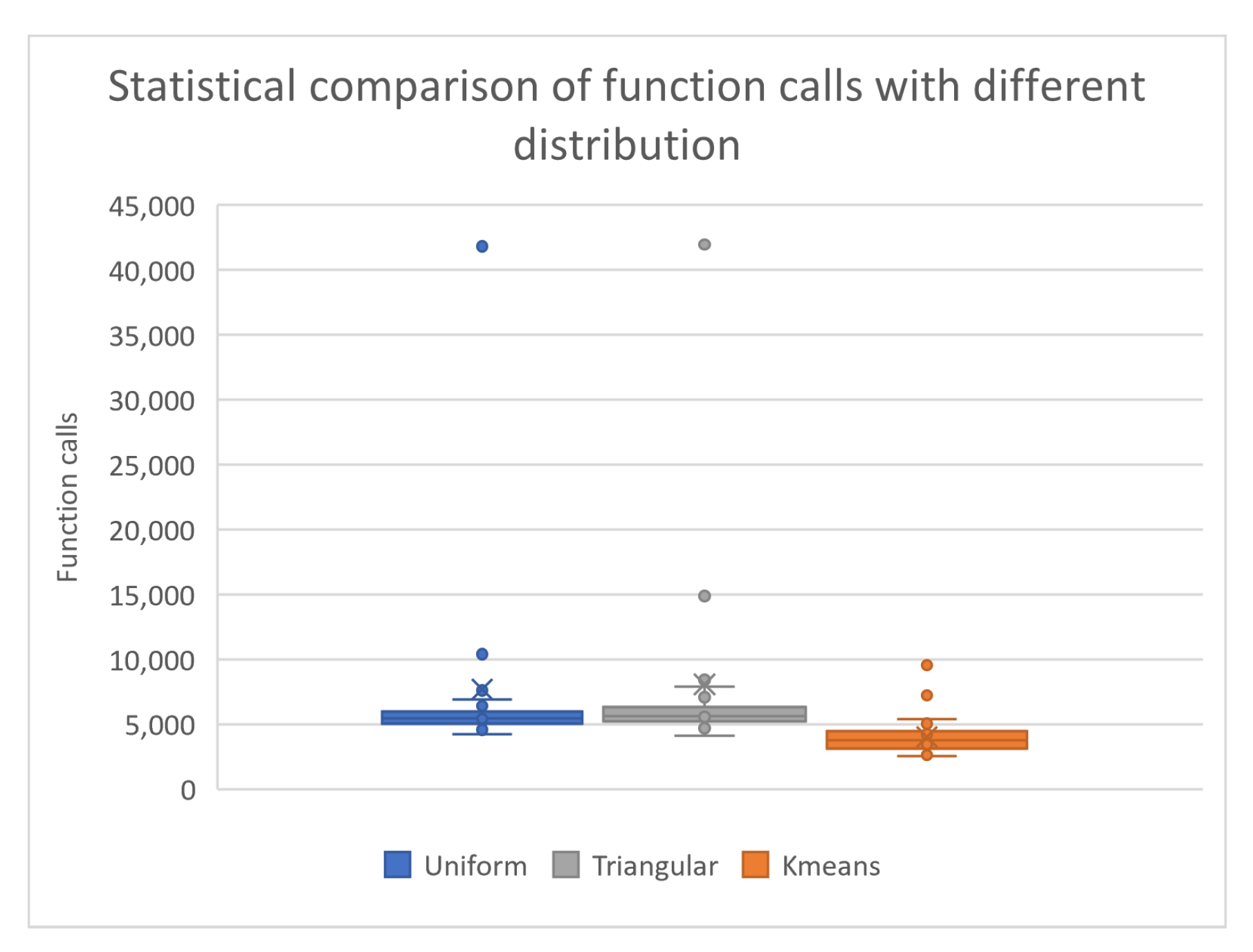
- The column UNIFORM indicates the incorporation of uniform sampling in the genetic algorithm. In this case, randomly selected chromosomes using uniform sampling are used in the genetic algorithm.
- The column TRIANGULAR defines the usage of triangular distribution for the initial samples of the genetic algorithm. For this case, randomly selected chromosomes with triangular distribution are used in the genetic algorithm.
- The column KMEANS denotes the application of k - means sampling as proposed here in the genetic algorithm. In this case, randomly selected points were sampled from the objective function and k centers were produced using the k - means algorithm. In order to have a reliable comparison with the above distributions, the number of centers equals the number of randomly generated chromosomes .
- The numbers in cells represent the average number of function calls required to obtain the global minimum. The fraction in parentheses denotes the percentage where the global minimum was successfully discovered. If this fraction is absent, then the global minimum was successfully discovered in all runs.
- In every table, an additional line was added under the name TOTAL, representing the total number of function calls and, in parentheses, the average success rate in finding the global minimum.
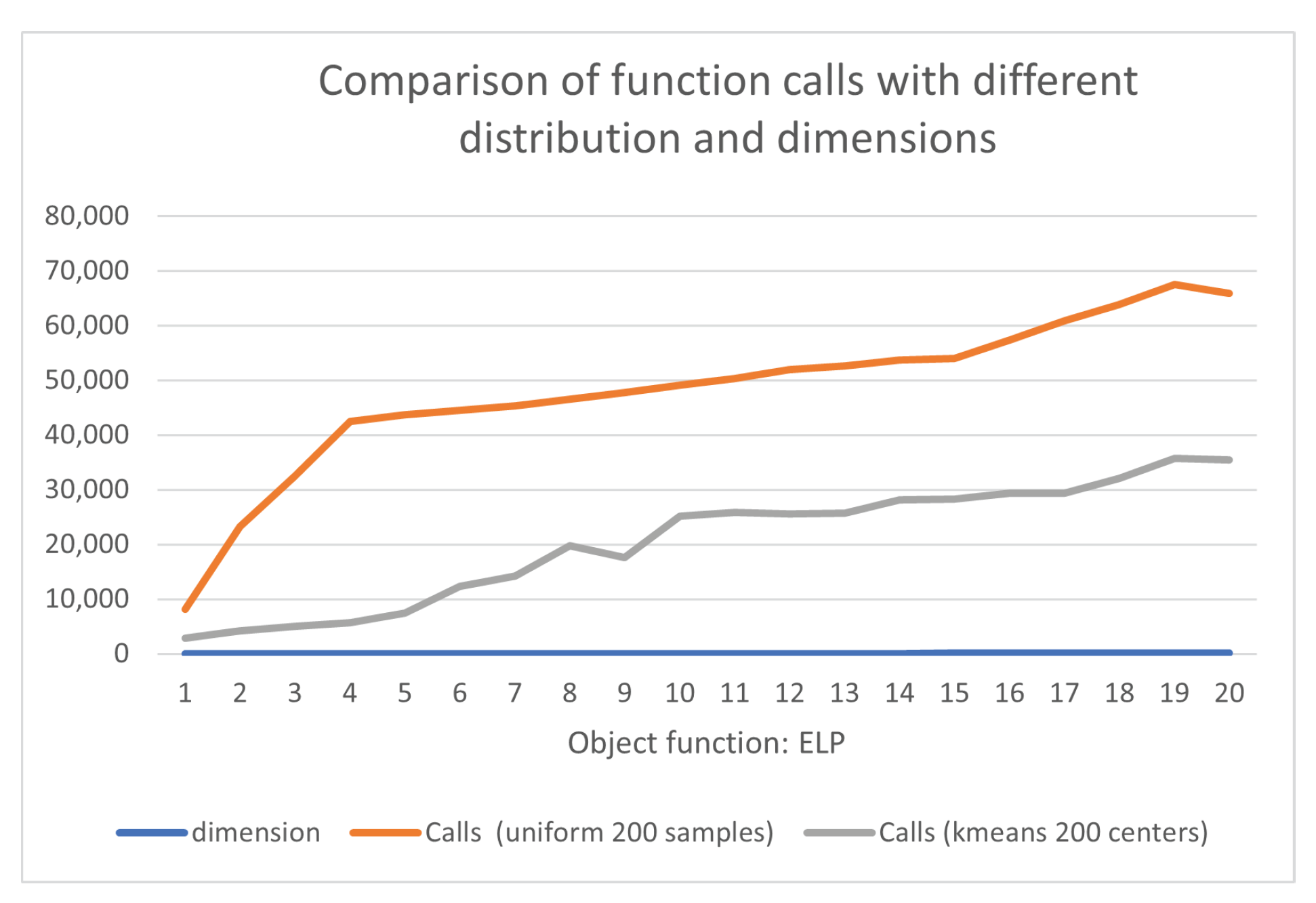
- High Conditioned Elliptic function, defined as
- Cm function, defined as
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- L. Yang, D. Robin, F. Sannibale, C. Steier, W. Wan, Global optimization of an accelerator lattice using multiobjective genetic algorithms, Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 609, pp. 50-57, 2009. [CrossRef]
- E. Iuliano, Global optimization of benchmark aerodynamic cases using physics-based surrogate models, Aerospace Science and Technology 67, pp.273-286, 2017. [CrossRef]
- Q. Duan, S. Sorooshian, V. Gupta, Effective and efficient global optimization for conceptual rainfall-runoff models, Water Resources Research 28, pp. 1015-1031 , 1992. [CrossRef]
- S. Heiles, R. L. Johnston, Global optimization of clusters using electronic structure methods, Int. J. Quantum Chem. 113, pp. 2091– 2109, 2013. [CrossRef]
- W.H. Shin, J.K. Kim, D.S. Kim, C. Seok, GalaxyDock2: Protein–ligand docking using beta-complex and global optimization, J. Comput. Chem. 34, pp. 2647– 2656, 2013.
- A. Liwo, J. Lee, D.R. Ripoll, J. Pillardy, H. A. Scheraga, Protein structure prediction by global optimization of a potential energy function, Biophysics 96, pp. 5482-5485, 1999. [CrossRef]
- Zwe-Lee Gaing, Particle swarm optimization to solving the economic dispatch considering the generator constraints, IEEE Transactions on 18 Power Systems, pp. 1187-1195, 2003.
- C. D. Maranas, I. P. Androulakis, C. A. Floudas, A. J. Berger, J. M. Mulvey, Solving long-term financial planning problems via global optimization, Journal of Economic Dynamics and Control 21, pp. 1405-1425, 1997. [CrossRef]
- Eva K. Lee, Large-Scale Optimization-Based Classification Models in Medicine and Biology, Annals of Biomedical Engineering 35, pp 1095-1109, 2007. [CrossRef]
- Y. Cherruault, Global optimization in biology and medicine, Mathematical and Computer Modelling 20, pp. 119-132, 1994. [CrossRef]
- M.A. Wolfe, Interval methods for global optimization, Applied Mathematics and Computation 75, pp. 179-206, 1996.
- T. Csendes and D. Ratz, Subdivision Direction Selection in Interval Methods for Global Optimization, SIAM J. Numer. Anal. 34, pp. 922–938, 1997. [CrossRef]
- W. L. Price, Global optimization by controlled random search, Journal of Optimization Theory and Applications 40, pp. 333-348, 1983.
- Ivan Krivy, Josef Tvrdik, The controlled random search algorithm in optimizing regression models, Computational Statistics & Data Analysis 20, pp. 229-234, 1995. [CrossRef]
- M.M. Ali, A. Torn, and S. Viitanen, A Numerical Comparison of Some Modified Controlled Random Search Algorithms, Journal of Global Optimization 11,pp. 377–385,1997. [CrossRef]
- S. Kirkpatrick, CD Gelatt, , MP Vecchi, Optimization by simulated annealing, Science 220, pp. 671-680, 1983.
- L. Ingber, Very fast simulated re-annealing, Mathematical and Computer Modelling 12, pp. 967-973, 1989.
- R.W. Eglese, Simulated annealing: A tool for operational research, Simulated annealing: A tool for operational research 46, pp. 271-281, 1990. [CrossRef]
- R. Storn, K. Price, Differential Evolution - A Simple and Efficient Heuristic for Global Optimization over Continuous Spaces, Journal of Global Optimization 11, pp. 341-359, 1997. [CrossRef]
- J. Liu, J. Lampinen, A Fuzzy Adaptive Differential Evolution Algorithm. Soft Comput 9, pp.448–462, 2005.
- J. Kennedy and R. Eberhart, Particle swarm optimization, Proceedings of ICNN’95 - International Conference on Neural Networks, 1995, pp. 1942-1948 vol.4. [CrossRef]
- Riccardo Poli, James Kennedy kennedy, Tim Blackwell, Particle swarm optimization An Overview, Swarm Intelligence 1, pp 33-57, 2007.
- Ioan Cristian Trelea, The particle swarm optimization algorithm: convergence analysis and parameter selection, Information Processing Letters 85, pp. 317-325, 2003. [CrossRef]
- M. Dorigo, M. Birattari and T. Stutzle, Ant colony optimization, IEEE Computational Intelligence Magazine 1, pp. 28-39, 2006.
- K. Socha, M. Dorigo, Ant colony optimization for continuous domains, European Journal of Operational Research 185, pp. 1155-1173, 2008. [CrossRef]
- M. Perez, F. Almeida and J. M. Moreno-Vega, Genetic algorithm with multistart search for the p-Hub median problem, Proceedings. 24th EUROMICRO Conference (Cat. No.98EX204), Vasteras, Sweden, 1998, pp. 702-707 vol.2.
- H. C. B. d. Oliveira, G. C. Vasconcelos and G. B. Alvarenga, A Multi-Start Simulated Annealing Algorithm for the Vehicle Routing Problem with Time Windows, 2006 Ninth Brazilian Symposium on Neural Networks (SBRN’06), Ribeirao Preto, Brazil, 2006, pp. 137-142.
- B. Liu, L. Wang, Y.H. Jin, F. Tang, D.X. Huang, Improved particle swarm optimization combined with chaos, Chaos Solitons and Fractals 25, pp. 1261-1271, 2005. [CrossRef]
- X.H. Shi, Y.C. Liang, H.P. Lee, C. Lu, L.M. Wang, An improved GA and a novel PSO-GA based hybrid algorithm, Information Processing Letters 93, pp. 255-261, 2005. [CrossRef]
- Harish Garg, A hybrid PSO-GA algorithm for constrained optimization problems, Applied Mathematics and Computation 274, pp. 292-305, 2016. [CrossRef]
- J. Larson and S.M. Wild, Asynchronously parallel optimization solver for finding multiple minima, Mathematical Programming Computation 10, pp. 303-332, 2018. [CrossRef]
- H.P.J. Bolton, J.F. Schutte, A.A. Groenwold, Multiple Parallel Local Searches in Global Optimization. In: Dongarra J., Kacsuk P., Podhorszki N. (eds) Recent Advances in Parallel Virtual Machine and Message Passing Interface. EuroPVM/MPI 2000. Lecture Notes in Computer Science, vol 1908. Springer, Berlin, Heidelberg, 2000.
- R. Kamil, S. Reiji, An Efficient GPU Implementation of a Multi-Start TSP Solver for Large Problem Instances, Proceedings of the 14th Annual Conference Companion on Genetic and Evolutionary Computation, pp. 1441-1442, 2012.
- Van Luong T., Melab N., Talbi EG. (2011) GPU-Based Multi-start Local Search Algorithms. In: Coello C.A.C. (eds) Learning and Intelligent Optimization. LION 2011. Lecture Notes in Computer Science, vol 6683. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-25566-3_24. [CrossRef]
- J.H. Holland, Genetic algorithms. Scientific american 267, pp. 66-73, 1992.
- J. Stender, Parallel Genetic Algorithms:Theory & Applications. Edition: IOS Press, 1993.
- D. Goldberg, Genetic Algorithms in Search, Optimization and Machine Learning, Addison-Wesley Publishing Company, Reading, Massachussets, 1989.
- Z. Michaelewicz, Genetic Algorithms + Data Structures = Evolution Programs. Springer - Verlag, Berlin, 1996. [CrossRef]
- S. Ansari, K. Alnajjar, M. Saad, S. Abdallah, A. Moursy, Automatic Digital Modulation Recognition Based on Genetic-Algorithm-Optimized Machine Learning Models, IEEE Access 10, pp. 50265-50277, 2022. [CrossRef]
- Y. Ji, S. Liu, M. Zhou, Z. Zhao, X. Guo, L. Qi, L., A machine learning and genetic algorithm-based method for predicting width deviation of hot-rolled strip in steel production systems. Information Sciences 589, pp. 360-375, 2022.
- Y. Hervis Santana, R. Martinez Alonso, G. Guillen Nieto, L. Martens, W. Joseph, D. Plets, Indoor genetic algorithm-based 5G network planning using a machine learning model for path loss estimation. Applied Sciences 12, 3923, 2022. [CrossRef]
- X. Liu, D. Jiang, B. Tao, G. Jiang, Y. Sun, J. Kong, B. Chen, Genetic algorithm-based trajectory optimization for digital twin robots. Frontiers in Bioengineering and Biotechnology 9, 793782, 2022. [CrossRef]
- K. Nonoyama, Z. Liu, T. Fujiwara, M.M. Alam, T. Nishi, Energy-efficient robot configuration and motion planning using genetic algorithm and particle swarm optimization. Energies 15, 2074, 2022. [CrossRef]
- K. Liu, B. Deng, Q. Shen, J. Yang, Y. Li, Optimization based on genetic algorithms on energy conservation potential of a high speed SI engine fueled with butanol–gasoline blends, Energy Reports 8, pp. 69-80, 2022. [CrossRef]
- G. Zhou, Z. Zhu, S. Luo, Location optimization of electric vehicle charging stations: Based on cost model and genetic algorithm, Energy 247, 123437, 2022. [CrossRef]
- Q. Chen, X. Hu, Design of intelligent control system for agricultural greenhouses based on adaptive improved genetic algorithm for multi-energy supply system, Energy Reports 8, pp. 12126-12138, 2022. [CrossRef]
- D. Min, Z. Song, H. Chen, T. Wang, T. Zhang, Genetic algorithm optimized neural network based fuel cell hybrid electric vehicle energy management strategy under start-stop condition, Applied Energy 306, 118036, 2022. [CrossRef]
- R.I. Doewes, R. Nair, T. Sharma, Diagnosis of COVID-19 through blood sample using ensemble genetic algorithms and machine learning classifier, World Journal of Engineering 19, pp. 175-182, 2022. [CrossRef]
- S. Choudhury, M. Rana, A. Chakraborty, S. Majumder, S. Roy, A. RoyChowdhury, S. Datta, Design of patient specific basal dental implant using Finite Element method and Artificial Neural Network technique. Journal of Engineering in Medicine 236, pp. 1375-1387, 2022. [CrossRef]
- M.I. El-Anwar, M.M. El-Zawahry, A three dimensional finite element study on dental implant design, Journal of Genetic Engineering and Biotechnology 9, pp. 77-82, 2011. [CrossRef]
- Zheng, Q. & Zhong, J. (2022). Design of Automatic Pronunciation Error Correction System for Cochlear Implant Based on Genetic Algorithm. ICMMIA: Application of Intelligent Systems in Multi-modal Information Analytics pp 1041–1047.
- O. Brahim, B. Hamid, N. Mohammed, Optimal design of inductive coupled coils for biomedical implants using metaheuristic techniques. E3S Web Conf.: Volume 351, 2022. [CrossRef]
- E. Tokgoz, M.A. Carro, Applications of Artificial Intelligence, Machine Learning, and Deep Learning on Facial Plastic Surgeries. Springer: Cosmetic and Reconstructive Facial Plastic Surgery pp 281–306, 2023.
- B. Wang, J.F. Gomez-Aguilar, Z. Sabir, M.A.Z. Raja, W. Xia, H. Jahanshahi, M.O. Alassafi, F. Alsaadi, Surgery Using The Capability Of Morlet Wavelet Artificial Neural Networks, Fractals30, 2240147, 2023.
- M. Ahmed, R. Seraj, S.M.S. Islam, The k-means algorithm: A comprehensive survey and performance evaluation, Electronics 9, 1295, 2020. [CrossRef]
- P. Kaelo, M.M. Ali, Integrated crossover rules in real coded genetic algorithms, European Journal of Operational Research 176, pp. 60-76, 2007. [CrossRef]
- I.G. Tsoulos, Modifications of real code genetic algorithm for global optimization, Applied Mathematics and Computation 203, pp. 598-607, 2008. [CrossRef]
- M.J.D Powell, A Tolerant Algorithm for Linearly Constrained Optimization Calculations, Mathematical Programming 45, pp. 547-566, 1989. [CrossRef]
- J. Kennedy, R. Eberhart, Particle swarm optimization, In: Proceedings of IEEE International Conference on Neural Networks pp. 1942-1948, 1995.
- H.G. Beyer, H.P. Schwefel, Evolution strategies–A comprehensive introduction, Natural Computing 1, pp. 3-52, 2002.
- Y. LeCun, Y. Bengio, G. Hinton, Deep learning, Nature 521, pp. 436-444, 2015.
- J.H. Holland, Adaptation in Natural and Artificial Systems, University of Michigan Press, 1975.
- D. Whitley, The GENITOR algorithm and selection pressure: Why rank-based allocation of reproductive trials is best, In Proceedings of the Third International Conference on Genetic Algorithms, pp. 116-121, 1994.
- A.E. Eiben, J.E. Smith, Introduction to Evolutionary Computing, Springer, 2015.
- S. Lloyd, Least squares quantization in PCM, IEEE Transactions on Information Theory 28, pp. 129–137, 1982.
- J.B. MacQueen, Some Methods for classification and Analysis of Multivariate Observations. Proceedings of 5th Berkeley Symposium on Mathematical Statistics and Probability. Vol. 1. University of California Press. pp. 281–297. MR 0214227. Zbl 0214.46201, 1967.
- A.K. Jain, M.N. Murty, P.J. Flynn, Data clustering: A review, ACM Computing Surveys 31, pp. 264-323, 1999.
- C.M. Bishop, Pattern recognition and machine learning, Springer, 2006.
- T. Hastie, R. Tibshirani, J. Friedman, The elements of statistical learning: Data mining, inference, and prediction, Springer, 2009.
- M. Montaz Ali, Charoenchai Khompatraporn, Zelda B. Zabinsky, A Numerical Evaluation of Several Stochastic Algorithms on Selected Continuous Global Optimization Test Problems, Journal of Global Optimization 31, pp 635-672, 2005. [CrossRef]
- C.A. Floudas, P.M. Pardalos, C. Adjiman, W. Esposoto, Z. Gümüs, S. Harding, J. Klepeis, C. Meyer, C. Schweiger, Handbook of Test Problems in Local and Global Optimization, Kluwer Academic Publishers, Dordrecht, 1999. [CrossRef]
- M. Gaviano, D.E. Ksasov, D. Lera, Y.D. Sergeyev, Software for generation of classes of test functions with known local and global minima for global optimization, ACM Trans. Math. Softw. 29, pp. 469-480, 2003.
- J.E. Lennard-Jones, On the Determination of Molecular Fields, Proc. R. Soc. Lond. A 106, pp. 463–477, 1924.
- W. Gropp, E. Lusk, N. Doss, A. Skjellum, A high-performance, portable implementation of the MPI message passing interface standard, Parallel Computing 22, pp. 789-828, 1996.
- R. Chandra, L. Dagum, D. Kohr, D. Maydan,J. McDonald and R. Menon, Parallel Programming in OpenMP, Morgan Kaufmann Publishers Inc., 2001.
| PARAMETER | MEANING | VALUE |
|---|---|---|
| Number of chromosomes | 200 | |
| Initial samples for k-means | 2000 | |
| k | Number of centers in k-means | 200 |
| Maximum number of allowed generations | 200 | |
| Selection rate | 0.9 | |
| Mutation rate | 0.05 | |
| Small value used in comparisons |
| PROBLEM | UNIFORM | TRIANGULAR | KMEANS |
|---|---|---|---|
| BF1 | 5731 | 5934 | 4478 |
| BF2 | 5648(0.97) | 5893 | 4512 |
| BRANIN | 4680 | 4835 | 4627 |
| CM4 | 5801 | 5985 | 4431 |
| CAMEL | 4965 | 5099 | 4824 |
| EASOM | 5657 | 7089 | 4303 |
| EXP4 | 4934 | 4958 | 4539 |
| EXP8 | 5021 | 5187 | 4689 |
| EXP16 | 5063 | 5246 | 4874 |
| EXP32 | 5044 | 5244 | 5016 |
| GKLS250 | 4518 | 4710 | 4525 |
| GKLS350 | 4650 | 4833 | 4637 |
| GOLDSTEIN | 8099 | 8537 | 7906 |
| GRIEWANK2 | 5500(0.97) | 5699(0.97) | 4324 |
| GRIEWANK10 | 6388(0.70) | 7482(0.63) | 4559 |
| HANSEN | 5681(0.93) | 6329 | 6357 |
| HARTMAN3 | 4950 | 5157 | 4998 |
| HARTMAN6 | 5288 | 5486 | 5258 |
| POTENTIAL3 | 5587 | 5806 | 5604 |
| POTENTIAL5 | 7335 | 7824 | 7450 |
| RASTRIGIN | 5703 | 5848 | 4481 |
| ROSENBROCK4 | 4241 | 4441 | 4241 |
| ROSENBROCK8 | 41802 | 41965 | 4523 |
| ROSENBROCK16 | 42196 | 42431 | 4962 |
| SHEKEL5 | 5488(0.97) | 5193(0.97) | 5232(0.97) |
| SHEKEL7 | 5384 | 5711(0.97) | 5695(0.97) |
| SHEKEL10 | 6360 | 5989 | 6396 |
| TEST2N4 | 5000 | 5179 | 5047 |
| TEST2N5 | 5306 | 5309 | 5039 |
| TEST2N6 | 5245 | 5492 | 5107 |
| TEST2N7 | 5282(0.93) | 5583 | 5216 |
| SINU4 | 4844 | 5046 | 4899 |
| SINU8 | 5368 | 5503 | 5509 |
| SINU16 | 6919 | 5583 | 5977 |
| TEST30N3 | 7215 | 8115 | 5270 |
| TEST30N4 | 7073 | 7455 | 6712 |
| Total | 273966(0.98) | 282176(0.985) | 186217(0.998) |
| dimension | Calls (uniform 200 samples) | Calls (kmeans 200 centers) |
|---|---|---|
| 5 | 15637 | 4332 |
| 10 | 24690 | 4486 |
| 15 | 39791 | 4743 |
| 20 | 42976 | 5194 |
| 25 | 43617 | 7152 |
| 30 | 44502 | 6914 |
| 35 | 45252 | 15065 |
| 40 | 46567 | 13952 |
| 45 | 47640 | 15193 |
| 50 | 49393 | 22535 |
| 55 | 50062 | 23692 |
| 60 | 52293 | 25570 |
| 65 | 52546 | 25678 |
| 70 | 53346 | 28153 |
| 75 | 54110 | 28328 |
| 80 | 57209 | 29320 |
| 85 | 60970 | 29371 |
| 90 | 65319 | 32121 |
| 95 | 68097 | 35721 |
| 100 | 66803 | 35396 |
| TOTAL | 980820 | 392916 |
| dimension | Calls (uniform 200 samples) | Calls (kmeans 200 centers) |
|---|---|---|
| 2 | 5665 | 4718 |
| 4 | 6212 | 4431 |
| 6 | 7980 | 4390 |
| 8 | 9917 | 4449 |
| 10 | 12076(0.97) | 4481 |
| 12 | 14672 | 4565 |
| 14 | 18708(0.87) | 4685 |
| 16 | 23251(0.77) | 4687 |
| 18 | 24624(0.77) | 4766 |
| 20 | 30153(0.80) | 4848 |
| 22 | 35851(0.77) | 15246(0.97) |
| 24 | 43677(0.93) | 7865(0.93) |
| 26 | 41492(0.77) | 5627 |
| 28 | 38017(0.73) | 10566(0.97) |
| 30 | 47538(0.83) | 24803(0.90) |
| TOTAL | 359833(0.84) | 110127(0.98) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





