1. Introduction
Colorectal cancer(CRC) has the third highest mortality rate in the world[
1]. Studies have pointed out that colorectal adenomatous polyp is one of the leading causes of CRC. Searching for and removing cancer precursor lesions, such as polyps, can significantly avoid the incidence of colorectal cancer. This is why it is so important that polyps in the colon are detected and treated at an early time.
Currently, the most effective measure to prevent CRC is to undergo regular colonoscopies and receive a resection for polyp removal[
2]. With modification of lifestyle and development of technology, the public’s willingness to undergo painless colonoscopy procedures has increased. Colonoscopy is a crucial medical procedure that helps doctors diagnose and treat various conditions related to the colon. During the procedure, the endoscopist carefully inserts a flexible endoscope into the rectum and navigates it through the entire length of the colon. This allows them to examine any abnormalities or growths in detail. However, colonoscopy is highly operator-dependence and subjectivity. It is both an expensive and consuming task. The detection of polyps is highly determined by the doctor’s experience and ability, which relates to patients’ risk of getting cancer[
3]. Due to polyps was detected manually by endoscopists, which resulted in high rate of missed detection. Early some classical segmentation methods[
4,
5,
6] was proposed to relieve the issue. However, these methods performed segmentation by extracting features such as size, shape, position, etc., which is difficult to segment diverse polyps accurately.
With emergence of Convolutional Neural Networks (CNNs), various imaging tasks rapidly apply the method. It is of great benefit to the segmentation of polyp. The methods based on CNN, such as FCN[
7,
8,
9], U-Net[
10,
11], PraNet[
12], HRENet[
13], PolypSeg[
14] and TGANet[
15], etc., have better results compared to the classical methods. However, there are still many problems, such as the higher missed rate for flat or diminutive polyps and multiple polyps.
In this paper, we evaluate segmentation SOTA methods on Kvasir-Sessile[
16] , Kvasir-SEG[
17] and CVC-ClinicDB[
18] dataset to supply comprehensive benckmark for the colonoscopy imgages. The most important contributions of the presented work can be summarised in four-fold:
We propose a novel deep neural network, called DoubleAANet, based on the backbone of ResNet50, which is an encoder-decoder architecture for segmentation of colonoscopic images. Our proposed method mines detail information and focuses more on multiple diminutive polyps for accurate polyp segmentation.
We design a simple yet effective auxiliary attention module (AAM) that can enhance high-level semantic features and learn more localized detailed features.
We develop a new area adaptive loss (AAL) function to further improve the segmentation performance. It works as a more efficient improvement to previous loss functions for addressing missed detection of multiple and diminutive polyps.
Extensive experiments on three variable size datasets and a cross dataset have demonstrated the efficient performance and generalization ability of DoubleAANet. Meanwhile, the study on robust estimation and ablation witnesses the model’s stability and the effectiveness of its key modules.
2. Related Works
Over the last decade, medical image segmentation became a hot area of research, with much effort going into the development of efficient methods and algorithms. In particular, most of the major work has concentrated on polyp segmentation. However, earlier methods can’t accurately segment various polyps by using manually created feature learning. More recently, CNNs-based methods have been demonstrated to outperform traditional segmentation algorithms. At the same time, the CNNs-based algorithms have received significant development, and have become the competitive methods for those participating in public challenges[
19,
20].
Long
et al. [
7] first solve image segmentation task by using fully convolutional networks (FCN), which is used to realize pixel-level classification in an image. Since then, the researchers have paid more attention to the convolutional neural network. U-Net[
10] was proposed in the same year as FCN, which has been extensively applied in medical image segmentation. With a relatively balanced U-shaped encoder-decoder architecture, It has developed into a basic network architecture for medical image segmentation. Badrinarayanan
et al. [
21] proposed a DL-based SegNet that was removed partly fully connected layers of encoder network. The trick makes SegNet dramatically lighter and more effective to train than many other newer network architectures. Deep layer aggregation[
22] was proposed to address the loss of marginal information and small targets, which resulted from up-sampling and down-sampling of deep network. Meanwhile, Zhou
et al. [
23] designed U-Net++ that optimize the architecture of original U-shaped. Guo
et al. [
24] used novel fully convolutional dilation neural network that produce the polyp occurrence confidence map (POCM). The polyps of image can obtain higher values of the POCM, which make its segmentation easier.
In recent years, more efficient CNNs backbone and additional modules were proposed, which have attained effective performance in terms of medical image segmentation. ResUNet[
25], ResUNet++[
26] and DoubleU-Net[
27] have stronger segmentation capability by using a U-shape structure combined with a more effective CNNs backbone. In addition to the optimization of backbone, DoubleU-Net also introduces Atrous Spatial Pyramid Pooling (ASPP)[
28] among the encoder block with the decoder block, which makes whole network faster and refines segmentation results. PraNet[
12] uses receptive field block (RFB)[
29] module to skip connection to attain multi-scale information. More recently, medical image segmentation have extensively applied multiple attention mechanisms, especially for segmentation of polyps which needs to focus on pixel-level localized and detailed information. PolypSeg[
14], ABC-Net[
30] and TGANet[
15] use different attention ways to improve performance. To enhance the feature representation capability, PolypSeg introduces the squeeze and excitation block (SEB)[
31]. TGANet uses three various attention modules,such as spatial attention module (SAM)[
32], channel attention module (CAM)[
32], and convolutional block attention module (CBAM)[
33] to obtain more precise edge cutting and higher accuracy.
The DoubleAANet we proposed uses more efficient ResNet50[
34] as the backbone. Meanwhile, we have also selectively added some additional modules to our network architecture, such as SCA-CNN proposed SAM and CAM; TGANet introduced feature enhancement module (FEM) and text guided attention. It has reached the outstanding performance of the current state-of-the-art.
3. Methods
3.1. Model Structure
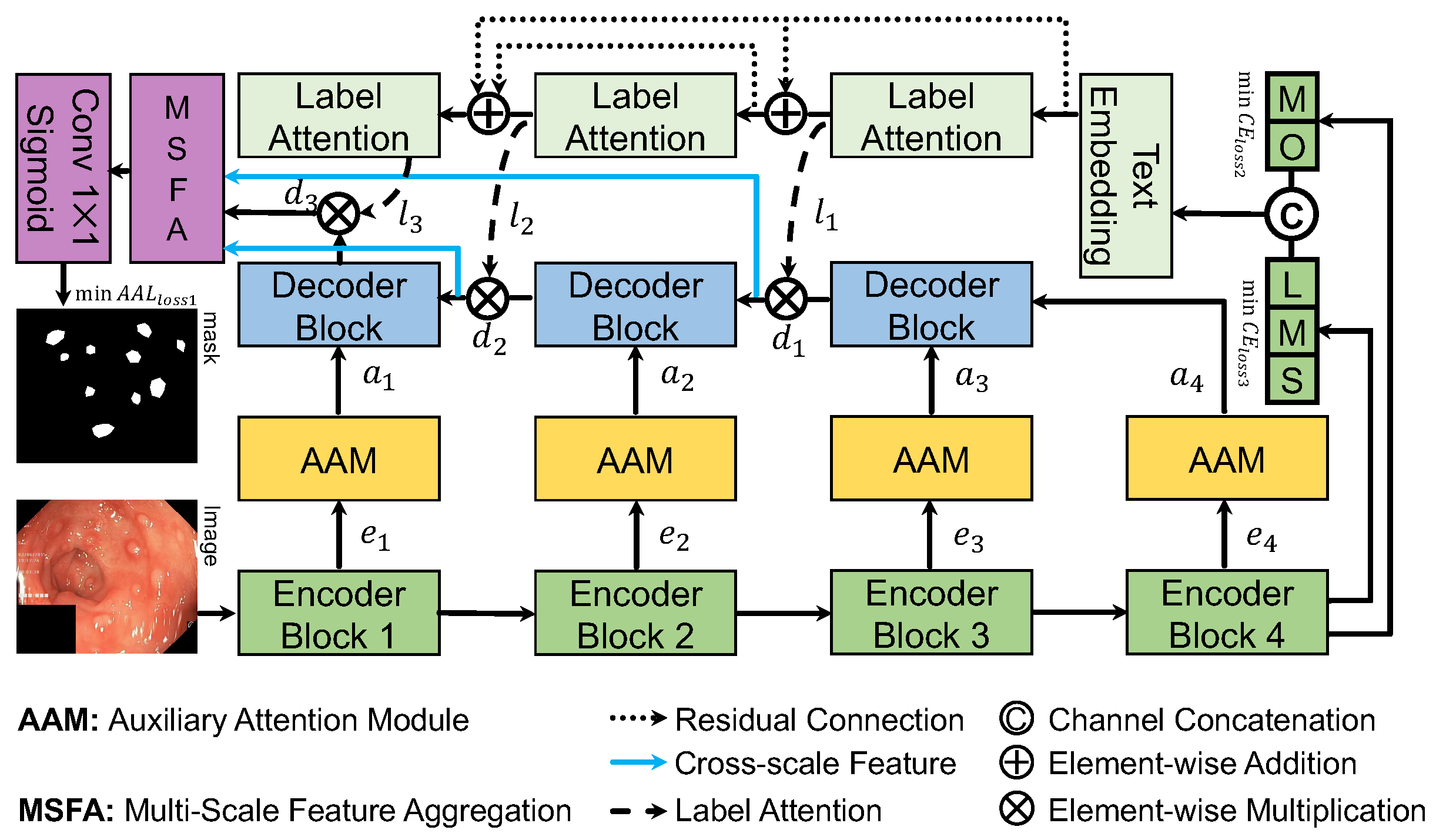
Figure 1 illustrates the architecture of our proposed DoubleAANet. The ResNet50 is applied as the sub-networks for the encoders, which consists of four different encoder blocks. For the fourth encoder block, we use the text-based attention of TGANet to focus more on number of polyps and their size. The decoder in our proposed DoubleAANet upsamples the input features to achieve multi-scale information and generates the output ones using a number of convolutional layers.
The AAM module are placed between encoder blocks and decoder blocks. Specifically, the AAM module suppresses irrelevant region and accordingly aggregates the feature from multiple branches. The whole network adopts AAL loss to train in an encoder-to-decoder manner, which make model mine multiple diminutive polyps attention.
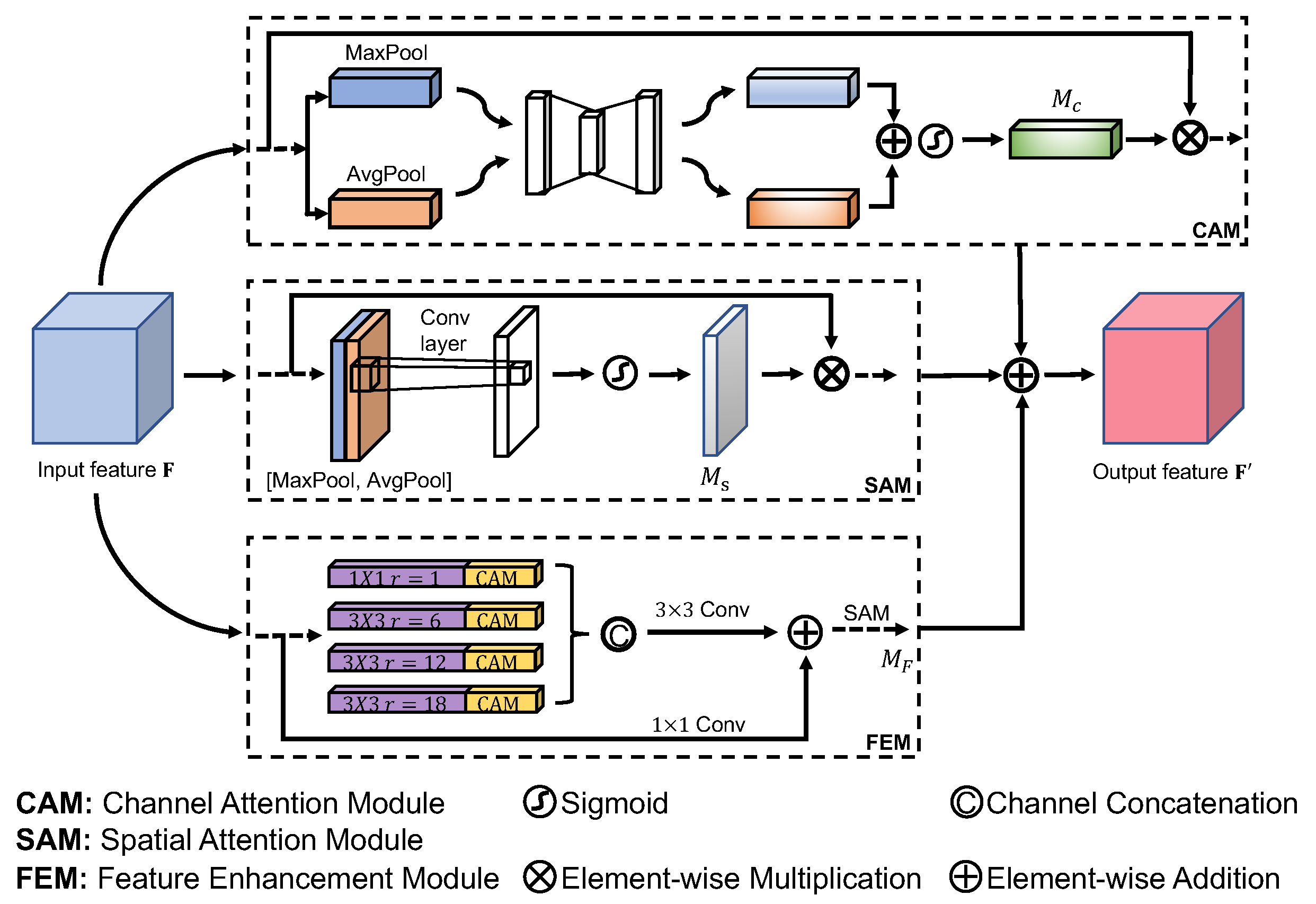
3.2. Auxiliary Attention Module
The
Auxiliary Attention Module is developed to supply regional and global information to the input feature of the decoding block in our DoubleAANet. We utilize four auxiliary attention modules,
, to the input of three different decoder blocks that allows the relevant features to attain higher weights and eliminates the irrelevant features. Each module includes channel attention module(CAM), spatial attention module(SAM) and feature enhancement module(FEM) which we refer as AAM shown in
Figure 2. We place the three modules in a parallel manner and merge the output features using element-wise addition.
Feature enhancement module utilizes four dilated convolutions in a parallel with a dilation rate of , which is the same as the setup of TGANet. A batch normalization layer and rectified linear unit follow each dilated convlution. The dilated convolution is able to achieve larger receptive field and learn higher semantic information, which make model easily ignore localized and detailed feature. Therefore, we must solve the issue by introducing channel attention module and spatial attention module learning extensive localized and detailed information.
Channel attention module generates a map through the utilization of the inter-channel feature relationship. Channel attention module concentrates on what are detected by feature detector consisted of each channel. We compute the channel attention by squeezing the spatial dimension of the input feature map. To aggregate spatial information and gather finer object features, We adopt average-pooling and max-pooling to achieve feature in a parallel manner. We gather two different finer object featutes
and
, which represent features of average-pooling and max-pooling separately. To attain the channel attention map
, we put the two object features through multilayer perceptron(MLP) and add up the output features. We can compute the channel attention as follow:
where
represents the sigmoid function and MLP is a shared network including a hidden layer.
Spatial attention module generates a map through the utilization of the inter-spatial feature relationship. spatial attention module focuses on where objects are detected, which is in addition to the channel attention module. we utilize max-pooling and average-pooling operation along the channel axis to compute the spatial attention and attain two effective object features. Similarly, we gather two different finer object featutes
and
, which represent features of average-pooling and max-pooling across the channel. To attain the spatial attention map
, we concatenate two object features and put the output through a convolution layer with
filter. We can compute the spatial attention as follow:
where
refers to the sigmoid function which normalizes the result between 0 and 1,
represents a convolution operation using fixed filter size.
With feature enhancement module, the model can achieve higher semantic feature in a large receptive filed. Meanwhile,Two attention modules compute complementary attention focusing on spatial and channel respectively, which capture more localized and detailed feature. Considering this, we design to stack the three modules in a parallel manner, which mines more feature of multiple and diminutive polyps without losing the big ones. The AAM module is formulated as follow:
Note that the FEM includes a number of convolution operations and reuses modules from CAM and SAM.
3.3. Area Adaptive Loss
The
Area Adaptive Loss is designed to address polyp segmentation scenario in which there are many difficult samples for small polyp and more than one polyp. We introduce the area adaptive loss starting from the dice loss and the binary cross entropy(BCE) loss:
In the above
and
represent the number of pixels in ground true and predict mask respectively.
represents the ground-truth class and
is the predicted probability for ground-truth class. The dice loss mainly measures the pixel similarity. A common method for improving performance is to combine dice loss with BCE loss:
While the loss function does not differentiate between small/large examples and one/many examples. Therefore, we redesign the loss function and establish the relationship between weight, size and quantity of polyps, which decreases weight of individual large polyp and focus training on multiple diminutive polyps.
More formally, we consider introducing an area adaptive weight factor
to the DiceBCE loss. We define the area adaptive loss as :
where
are normalized into the range from 0 to 1.
represents the degree of attention paid to the DiceBCE loss. Whether the multiple diminutive polyps achieve more attention depends on the obtained value of DiceBCE loss.
In order to realize that
is related to size and quantity of polyps, we design a fusion function combining number of polyps with pixel area of polyps. We define the fusion function as follow:
where
is a scaling factor that converts the number of polyps into area information.
C is a constant avoiding too small value for
.
Q refers to the number of polyps embedded in the corresponding text message.
denotes the pixel area corresponding to all predicted polyp number
i.
In fact, we adjust the range of value of
by using the sofmax function that normalizes the result. Meanwhile, we introduce temperature coefficient
T to make the classification smoother. The area adaptive weight
for the
k-th example is computed as follow:
Obviously, the multiple diminutive polyps will obtain higher value of , which makes the model focus more on hard examples. We use the area adaptive loss in our experiments as it yields greatly improved performance over the DiceBCE loss.
4. Experiments
4.1. Datasets
To evaluate the outcomes of our DoubleAANet, we performed both qualitative and quantitative experiments on three benchmark colonoscopy datasets:
Kvasir-Sessile [
16] : This dataset contains 196 pairs of colonoscopy images including diminutive polyps, sessile polyps and flat polyps.
Kvasir-SEG [
17]: The dataset consisted of 1000 pairs of colonoscopy images, which has various resolutions. Meanwhile, the dataset presents the ground truth segmentation mask of polyp.
CVC-ClinicDB [
18]: This dataset includes 612 pairs of colonoscopy images. It is collected from real-time colonoscopy videos with 288 × 384 resolution.
4.2. Evaluation Metrics
We adopted the five most commonly used metrics for polyp segmentation to evaluate the performance of DoubleAANet and other methods. We used the mean intersection over union (mIoU), mean Srensen-dice coefficient (mDSC), recall, precision and F2-score.
4.3. Implementation Details
We implement our method by Pytorch. The model is optimized by Adam with batch size of 16 and learning rate of . Additionally, we adopt an early stopping mechanism to prevent model from overfitting and attain the best model. All datasets other than Kvasir-SEG are split to training, validation, and testing 80:10:10 with 256 × 256 resolution. In the case of the Kvasir-SEG, we used the original split for training and testing. Meanwhile, some data augmentation strategies, such as random rotation, vertical flipping and horizontal flipping, etc., are adopted to improve generalization of model. All experiments are conducted using an NVIDIA GeForce RTX 3090 GPU.
4.4. Result
We perform a series of experiments in comparisiom with the proposed DoubleAANet using six SOTA methods (i.e., U-Net, HarDNet-MSEG [
35], ColonSegNet [
36], DeepLabV3+[
28], PraNet and TGANet). These algorithms are developed for polyp segmentation or medical image segmentation. The results of extensive quantitative and qualitative experiments are presented below.
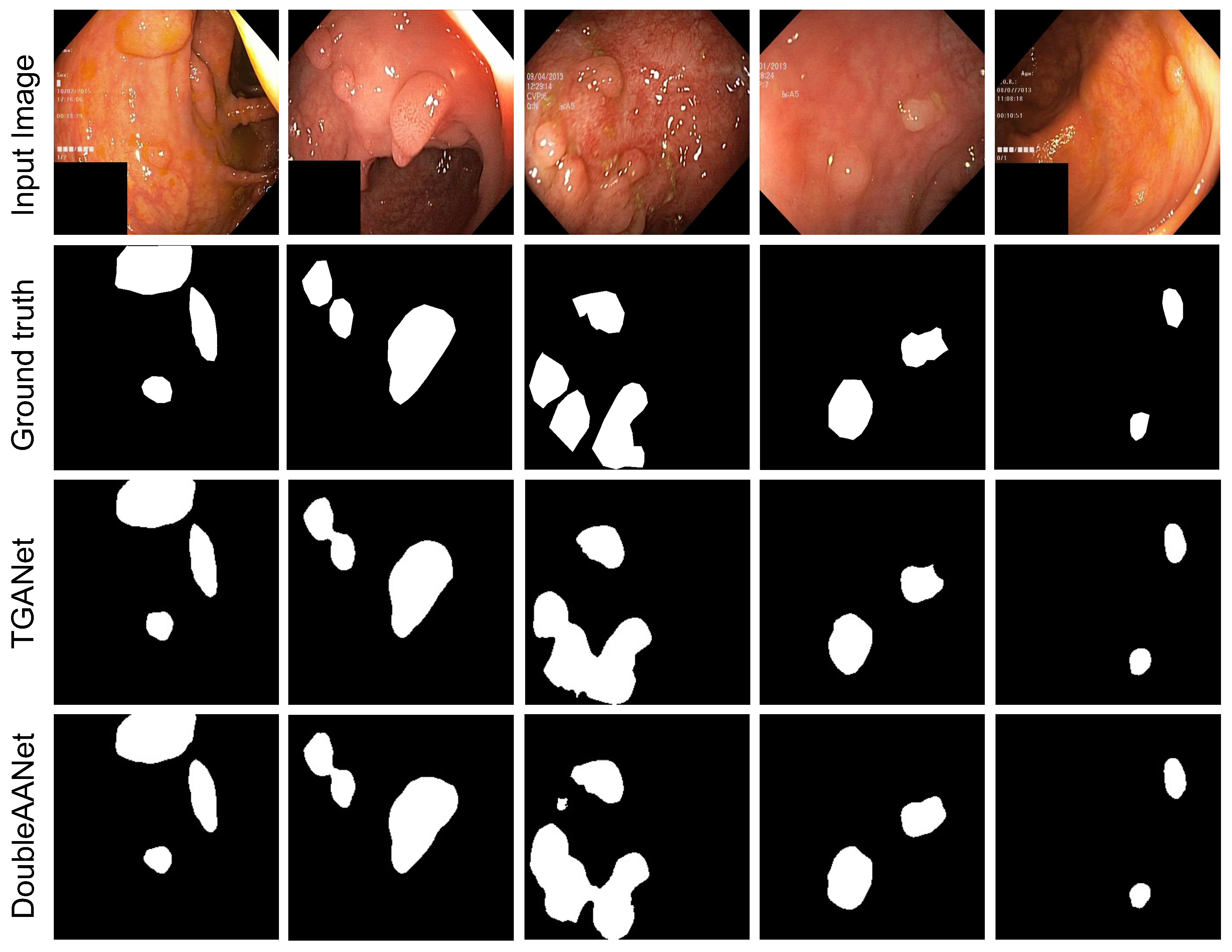
Experiments on Kvasir-SEG Dataset. To evaluate the performance of our proposed model, we conducted extensive experiments using five evaluation metrics and comparing it with six state-of-the-art methods on the Kvasir-SEG dataset. As shown in
Table 1, it can be noted that all compared models are inferior to our proposed method. The PraNet obtain relatively superior segmentation performance comparing the other models. In addition, the TGANet introduces label attention which guides network according to size and quantity of polyps, but it still produces missed polyps. The
Figure 3 shows that our proposed method can partly suppress the issue and attain the best performance. This is because our proposed method can mine more localized and detailed information to address the diminutive polyps, and the area adaptive loss function provides higher weights for multiple polyps. The proposed DoubleAANet achieves the highest mIoU of 86.65% and mDSC of 92.17%, which overcomes most competitive TGANet by 3.35% in mIoU and 2.35% in mDSC.
Experiments on CVC-ClinicDB Dataset. Similarly, we conducted the same experiments comparing it with six state-of-the-art methods on the CVC-ClinicDB dataset in order to verify the effectiveness of our proposed method. Compared to the other segmentation models, our proposed method achieves distinct advantage as shown in
Table 2. In terms of precision, the TGANet achieves the best performance compared to the other models. On the other hand, the PraNet and the DeepLabV3+ obtain slightly higher precision than our proposed method. In terms of the other four metrics, Our proposed method surpasses the other models overall. From the results, it can indicate the proposed DoubleAANet achieves the highest mIoU of 90.00% and mDSC of 94.58%, which outperforms the compared SOTA methods.
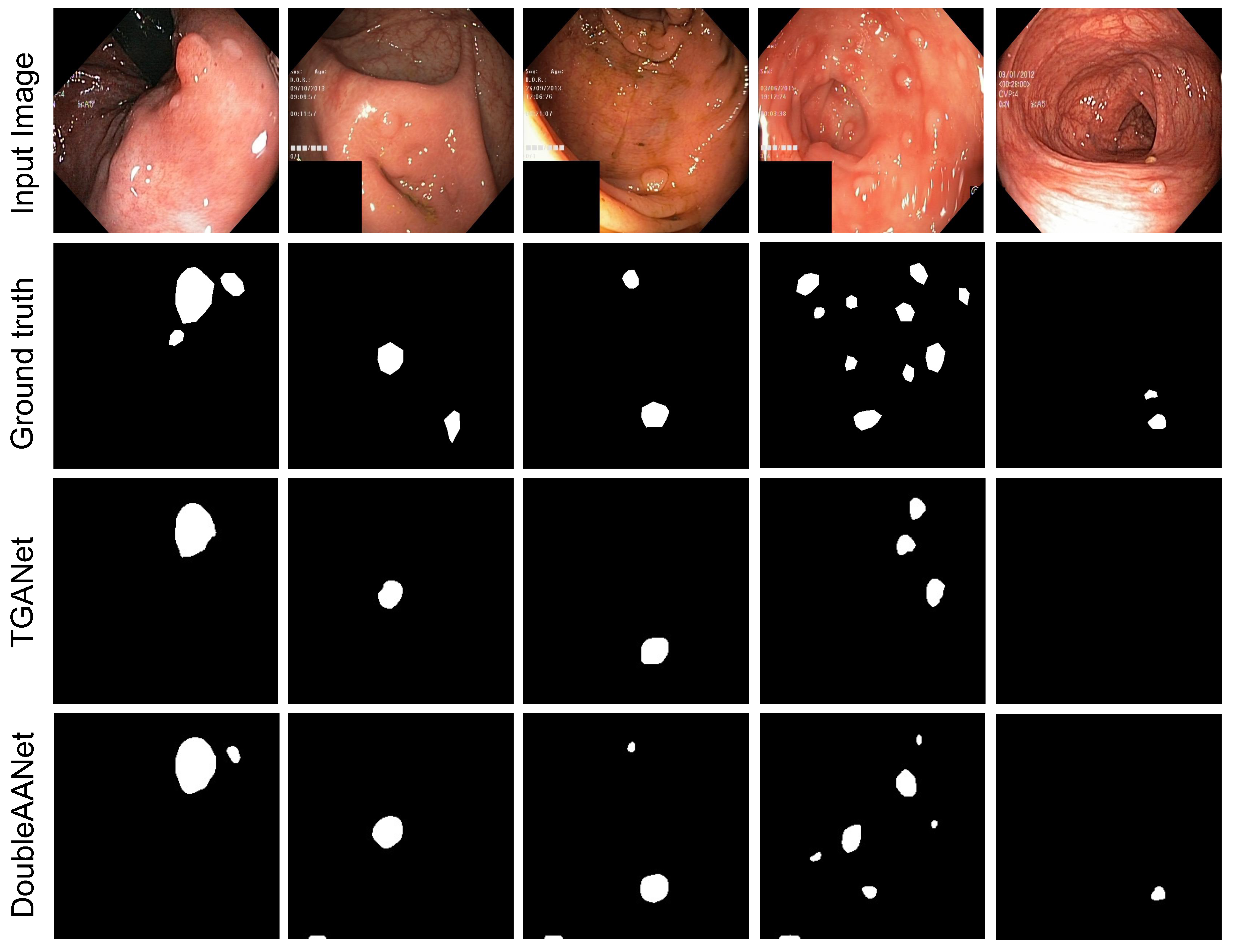
Experiments on Kvasir-Sessile Dataset. To further verify the efficient of our proposed model in multiple and diminutive polyps, we designed extensive experiments on the Kvasir-Sessile dataset including small, flat and diminutive polyps. Kvasir-Sessile dataset mainly consists of small and flat polyps, which greatly increases the difficulty of segmentation. As shown in
Table 3, the PraNet and TGANet obtain relatively superior segmentation performance comparing the other models. However, our proposed method achieve significant improvement in segmentation of multiple and diminutive polyps compared to the TGANet approaches, as shown in
Figure 4. These results witness our proposed method achieves effective performance in diminutive and flat polyps and surpasses all SOTA methods with an increase of 3.12% in mIoU and 2.59% in mDSC.
Experiments on Cross Dataset. For cross dataset, the DoubleAANet achieves the highest mIoU of 76.90% and mDSC of 84.77%, surpassing all compared SOTA methods and demonstrating remarkable generalization ability. This is because that our proposed DoubleAANet focuses more on localized and detailed information that is vital feature for polyps in various size dataset. As shown in
Table 4, our proposed method improve 2.46% of mIoU and 2.81% of mDSC in comparison to the TGANet.
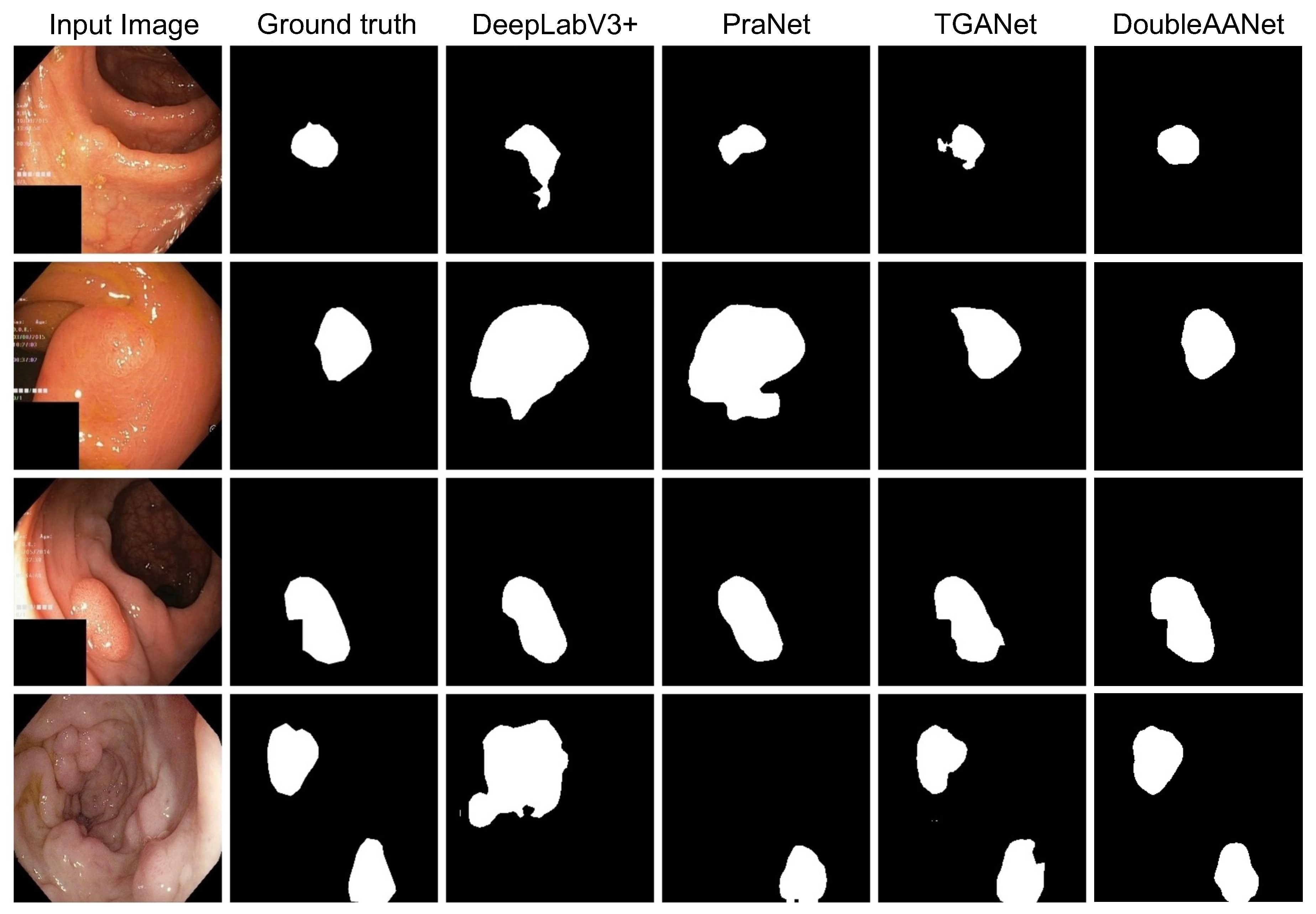
To better visualize the superiority of our proposed DoubleAANet, we compare segmentation masks of four different methods. As shown in
Figure 5, both the DeepLabV3+ and the PraNet exhibit missed polyps and generate incorrect morphology in camparision with the ground truth. For the TGANet, it can better segment polyps and generate the improvement result closing to the ground truth. However, the segmentation masks of the TGANet appear different degrees of marginal loss. In contrast, our proposed method not only produces accurate segmentation but also attains excellent edge matching the ground truth. In short, the DoubleAANet achieves more accurate segmentation in terms of morphology and quantity, which dues to introduce of the area adaptive loss and the auxiliary attention module.
4.5. Robust Estimation
To validate the robustness of model, we conduct robust experiments by introducing loss function hyperparameters. We propose to introduce two modulating factors
to the loss function. The loss function with modulating factors can be formulated as follow:
To estimate the robust of our proposed method, we intuitively use a variety of values to train and test on the Kvasir-SEG dataset.
As shown in
Table 5, our proposed DoubleAANet still overcomes all SOTA methods by using different modulating factors. In other word, our method has highly robustness and low parameter sensitivity. Meanwhile, it demonstrates that module and loss introduced our proposed method is beneficial for improving performance. This is not a random result.
4.6. Ablation Study
To further verify the efficiency and necessity of the area adaptive loss and the auxiliary attention module, we design ablation experiments on the Kvasir-SEG. We conduct the relationship between weights, size and quantity by designing an area adaptive loss function that controls the network to concentrate on multiple and diminutive polyps. However, the loss function only changes the level of attention aimed at learned features and can not capture unlearned features, resulting in a slight improvement in the performance of our proposed model. To address the issue mentioned above, we design the auxiliary attention module to learn more localized and detailed features at the same time, which allows the model to obtain extensive useful information and strongly boosts the performance of our method. As can be seen in
Table 6, the results further show that we significantly improves the performance of the whole network by implementing the auxiliary attention module. In addition, our algorithm has performance degradation without either AAL or AAM, decreasing the mIoU by 1.52% and 3.48% respectively.
5. Discussion
With the novel DoubleAANet, the mIoU and mDSC were significantly improved by 3.12% and 2.59% respectively compared to the TGANet on the Kvasir-Sessile dataset, which reveals the effectiveness of the algorithm for multiple and diminutive polyps. For the other datasets, our method still achieve noticeable improvement. This indicates that the strategy introduced SAM, CAM and FEM in a parallel manner and trained with AAL is a reasonable idea to optimize our proposed model.
Colonoscopy is a necessary procedure that is optimized to detect and remove polyps. The effectiveness of colonoscopy depends heavily on the experience and technology of the endoscopist. Therefore, there must be missed polyps in colonoscopy, especially for multiple and diminutive polyps, which causes great potential danger to patients. In order to focus more on localized and detailed feature, the DoubleAANet model was proposed. In this study, the proposed DoubleAANet aims to overcome these challenges in real medical scene. It has outperformed existing medical image segmentation algorithms in terms of polyp segmentation accuracy.
However, this model exhibited poor performance in real-time segmentation for all polyp datasets, which requires extensive computational capability to train and infer a model. We expect to lighten the weight of the model and optimize the architecture of the algorithm in our future work, which allow our method to be applied to different devices.
6. Conclusions
Our proposed DoubleAANet improve the performance of segmentation for multiple diminutive polyps. To reduce lost of features for multiple and small polyps, we propose an auxiliary attention module that focuses on localized and detailed information. The design of the area adaptive loss is aimed to rationalize the assignment of weights and enable multiple small polyps to attain higher loss. Finally, our experimental results demonstrated the proposed DoubleAAet achieved effective performance for various polyp sizes, especially for multiple and diminutive polyps.
Author Contributions
Conceptualization, Zhongliang Wang and F.D.; methodology, F.D.; software, F.D. and X.Y.; validation, F.D., and Y.L.; formal analysis, F.D.; investigation, W.W and L.X.; data curation, N.Q. and Zhenxing Wang; writing—original draft preparation, F.D.; writing—review and editing, Zhongliang Wang; visualization, F.D.; supervision, Zhongliang Wang and C.H.; project administration, Zhongliang Wang and S.Z.; All authors have read and agreed to the published version of the manuscript.
Acknowledgments
This work was partially supported by Natural Science Research Project of Tongling University(2022tlxy41&2022tlxy43) and College Students’ Innovative Entrepreneurial Training Plan Program(2023103830016&2023103830004). Furthermore,this publication has also been partially supported by the University Synergy Innovation Program of Anhui Province(GXXT-2022-022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Holme, .; Bretthauer, M.; Fretheim, A.; Odgaard-Jensen, J.; Hoff, G. Flexible sigmoidoscopy versus faecal occult blood testing for colorectal cancer screening in asymptomatic individuals. Cochrane Database of Systematic Reviews 2013.
- Kaminski, M.F.; Regula, J.; Kraszewska, E.; Polkowski, M.; Wojciechowska, U.; Didkowska, J.; Zwierko, M.; Rupinski, M.; Nowacki, M.P.; Butruk, E. Quality indicators for colonoscopy and the risk of interval cancer. New England journal of medicine 2010, 362, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Mamonov, A.V.; Figueiredo, I.N.; Figueiredo, P.N.; Tsai, Y.H.R. Automated polyp detection in colon capsule endoscopy. IEEE transactions on medical imaging 2014, 33, 1488–1502. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.H.; Yoon, K.J. Polyp detection via imbalanced learning and discriminative feature learning. IEEE transactions on medical imaging 2015, 34, 2379–2393. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, N.; Gurudu, S.R.; Liang, J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE transactions on medical imaging 2015, 35, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp.; pp. 3431–3440.
- Brandao, P.; Mazomenos, E.; Ciuti, G.; Caliò, R.; Bianchi, F.; Menciassi, A.; Dario, P.; Koulaouzidis, A.; Arezzo, A.; Stoyanov, D. Fully convolutional neural networks for polyp segmentation in colonoscopy. In Proceedings of the Medical Imaging 2017: Computer-Aided Diagnosis. Spie, Vol. 10134; 2017; pp. 101–107. [Google Scholar]
- Akbari, M.; Mohrekesh, M.; Nasr-Esfahani, E.; Soroushmehr, S.R.; Karimi, N.; Samavi, S.; Najarian, K. Polyp segmentation in colonoscopy images using fully convolutional network. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE; 2018; pp. 69–72. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 2015, Proceedings, Part III 18. Springer, 2015, October 5-9; pp. 234–241.
- Mohammed, A.; Yildirim, S.; Farup, I.; Pedersen, M.; Hovde, . Y-net: A deep convolutional neural network for polyp detection. arXiv preprint arXiv:1806.01907 2018. arXiv:1806.01907 2018.
- Fan, D.P.; Ji, G.P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Pranet: Parallel reverse attention network for polyp segmentation. In Proceedings of the International conference on medical image computing and computer-assisted intervention. Springer; 2020; pp. 263–273. [Google Scholar]
- Shen, Y.; Jia, X.; Meng, M.Q.H. Hrenet: A hard region enhancement network for polyp segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 2021, Proceedings, Part I 24. Springer, 2021, September 27–October 1; pp. 559–568.
- Zhong, J.; Wang, W.; Wu, H.; Wen, Z.; Qin, J. PolypSeg: An efficient context-aware network for polyp segmentation from colonoscopy videos. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 2020, Proceedings, Part VI 23. Springer, 2020, October 4–8; pp. 285–294.
- Tomar, N.K.; Jha, D.; Bagci, U.; Ali, S. TGANet: Text-guided attention for improved polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2022; pp. 151–160. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Johansen, D.; de Lange, T.; Johansen, H.D.; Halvorsen, P.; Riegler, M.A. A comprehensive study on colorectal polyp segmentation with ResUNet++, conditional random field and test-time augmentation. IEEE journal of biomedical and health informatics 2021, 25, 2029–2040. [Google Scholar] [CrossRef] [PubMed]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; de Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented polyp dataset. In Proceedings of the MultiMedia Modeling: 26th International Conference, MMM 2020, Daejeon, South Korea, 2020, Proceedings, Part II 26. Springer, 2020, January 5–8; pp. 451–462.
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Computerized medical imaging and graphics 2015, 43, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE transactions on medical imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Zhou, F.; Braden, B.; Bailey, A.; Yang, S.; Cheng, G.; Zhang, P.; Li, X.; Kayser, M.; Soberanis-Mukul, R.D.; et al. An objective comparison of detection and segmentation algorithms for artefacts in clinical endoscopy. Scientific reports 2020, 10, 2748. [Google Scholar] [CrossRef] [PubMed]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE transactions on pattern analysis and machine intelligence 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Wang, D.; Shelhamer, E.; Darrell, T. Deep layer aggregation. In Proceedings of the Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp.; pp. 2403–2412.
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 2018, Proceedings 4. Springer, 2018, September 20; pp. 3–11.
- Guo, Y.B.; Matuszewski, B. Giana polyp segmentation with fully convolutional dilation neural networks. In Proceedings of the Proceedings of the 14th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications.
- Yang, X.; Li, X.; Ye, Y.; Lau, R.Y.; Zhang, X.; Huang, X. Road detection and centerline extraction via deep recurrent convolutional neural network U-Net. IEEE Transactions on Geoscience and Remote Sensing 2019, 57, 7209–7220. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; De Lange, T.; Halvorsen, P.; Johansen, H.D. Resunet++: An advanced architecture for medical image segmentation. In Proceedings of the 2019 IEEE international symposium on multimedia (ISM). IEEE; 2019; pp. 225–2255. [Google Scholar]
- Jha, D.; Riegler, M.A.; Johansen, D.; Halvorsen, P.; Johansen, H.D. Doubleu-net: A deep convolutional neural network for medical image segmentation. In Proceedings of the 2020 IEEE 33rd International symposium on computer-based medical systems (CBMS). IEEE; 2020; pp. 558–564. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the Proceedings of the European conference on computer vision (ECCV), 2018, pp.; pp. 801–818.
- Liu, S.; Huang, D.; et al. Receptive field block net for accurate and fast object detection. In Proceedings of the Proceedings of the European conference on computer vision (ECCV), 2018, pp.; pp. 385–400.
- Fang, Y.; Zhu, D.; Yao, J.; Yuan, Y.; Tong, K.Y. Abc-net: Area-boundary constraint network with dynamical feature selection for colorectal polyp segmentation. IEEE Sensors Journal 2020, 21, 11799–11809. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp.; pp. 7132–7141.
- Chen, L.; Zhang, H.; Xiao, J.; Nie, L.; Shao, J.; Liu, W.; Chua, T.S. Sca-cnn: Spatial and channel-wise attention in convolutional networks for image captioning. In Proceedings of the Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp.; pp. 5659–5667.
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the Proceedings of the European conference on computer vision (ECCV), 2018, pp.; pp. 3–19.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp.; pp. 770–778.
- Huang, C.H.; Wu, H.Y.; Lin, Y.L. Hardnet-mseg: A simple encoder-decoder polyp segmentation neural network that achieves over 0.9 mean dice and 86 fps. arXiv preprint arXiv:2101.07172 2021. arXiv:2101.07172 2021.
- Jha, D.; Ali, S.; Tomar, N.K.; Johansen, H.D.; Johansen, D.; Rittscher, J.; Riegler, M.A.; Halvorsen, P. Real-time polyp detection, localization and segmentation in colonoscopy using deep learning. Ieee Access 2021, 9, 40496–40510. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).