1. Introduction
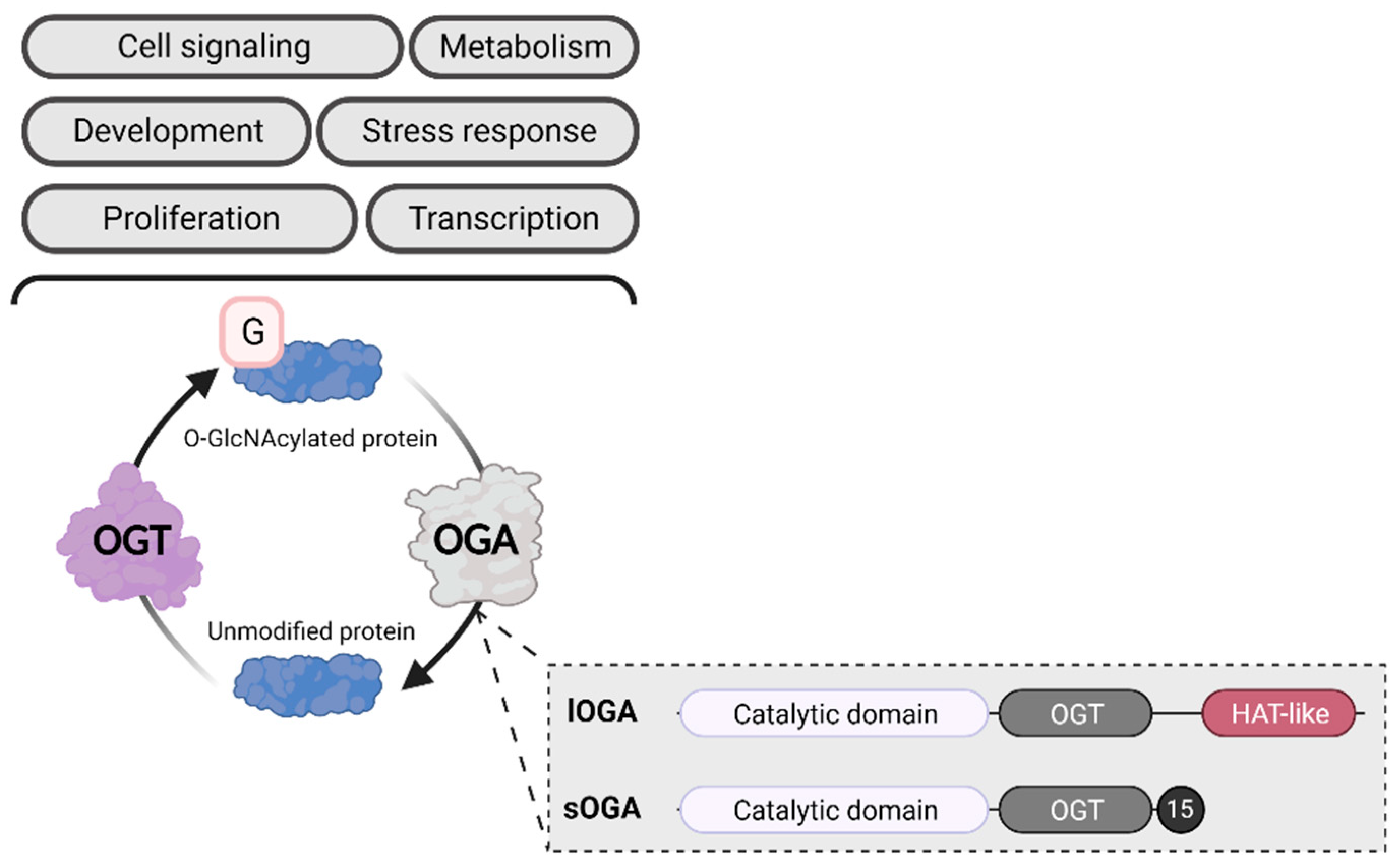
Gene expression is influenced by physiological (e.g., cell differentiation, development and aging, external stressors) and pathological (e.g., cancer, neurodegenerative diseases) factors1. Several cellular processes can also impact gene expression, including transcription, mRNA stability and transport, and translation2. Epigenetic modifications provide an important layer of regulation, altering gene expression without changing the DNA sequence3. The best-described epigenetic mechanism is the addition of biochemical marks directly to the DNA or the histone proteins that organize it. Cytosine methylation to form 5-methylcytosine is the most common chemical DNA base modification, although additional changes (e.g., 5-hydroxymethylcytosine, N6-methyladenine) have been recently discovered4. Covalent changes to histones, known as post-translational modifications (PTMs), include methylation (me), phosphorylation, acetylation (ac), ubiquitylation, SUMOylation, glycosylation, and ADP-ribosylation5. In 2010, Sakabe et al. added a new histone PTM: O-GlcNAcylation (O-GlcNAc)6—the ubiquitous, dynamic, and reversible addition of a sugar motif (β-D-N-acetylglucosamine) to serine and threonine residues. The O-GlcNAcylation cycle is controlled by a single pair of enzymes: O-linked N-acetyl-glucosaminyltransferase (OGT) adds the GlcNAc moiety to proteins, while O-linked N-acetyl β-D-glucosaminidase (OGA) removes it (Figure 1)7.
Figure 1.
O-GlcNAcylation. OGA exists in two isoforms: long OGA (lOGA) has a histone-like acetyltransferase domain (HAT-like), while short OGA (sOGA) does not. Both isoforms contain a catalytic domain and an OGT binding domain (OGT). The C-terminal of sOGA contains a specific sequence of 15 amino acids (15). Created with BioRender.com.
Figure 1.
O-GlcNAcylation. OGA exists in two isoforms: long OGA (lOGA) has a histone-like acetyltransferase domain (HAT-like), while short OGA (sOGA) does not. Both isoforms contain a catalytic domain and an OGT binding domain (OGT). The C-terminal of sOGA contains a specific sequence of 15 amino acids (15). Created with BioRender.com.
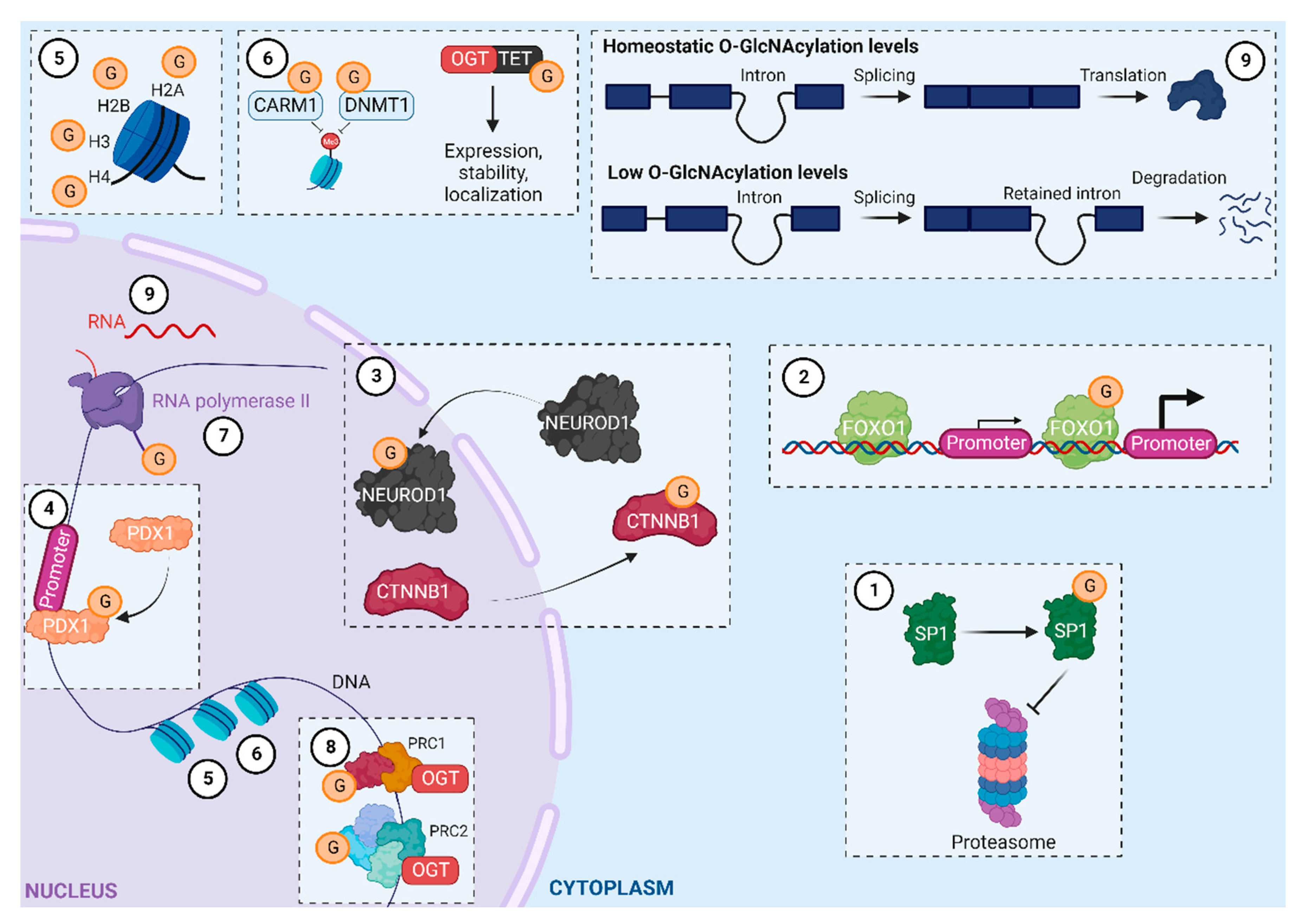
O-GlcNAcylation helps regulate gene expression by 1) changing the properties of transcription factors (localization, stability, DNA binding, and transcriptional activity; Figure 2.1–2.4); 2) directly or indirectly modifying histones (Figure 2.5 and 2.6); and 3) regulating RNA polymerase II transcription at the initiation and elongation stages (Figure 2.7 and 2.8)8,9. Moreover, OGT interacts with and regulates proteins in polycomb repressive complexes (PRCs) 1 and 210, and a recent study reported that O-GlcNAcylation levels contribute to the intron retention process (Figure 2.9 and 2.10)11. Recently developed approaches have enabled considerable progress in i) identifying O-GlcNAcylated proteins and ii) unraveling the role of O-GlcNAcylation in numerous biological processes12. To date, the set of O-GlcNAcylated proteins in humans, known as the O-GlcNAcylome, consists of 8000 proteins and continues to grow (The O-GlcNAc Database, v1.3)13. This review provides an updated look at its role as an epigenetic marker, focusing on histone modifications.
Figure 2.
O-GlcNAcylation in the regulation of gene expression. PRC1, PRC2: Polycomb repressive complex 1, 2; CARM1: coactivator-associated arginine methyltransferase 1; DNMT1: DNA methyltransferase 1; TET: ten-eleven translocation; SP1: transcription factor SP1; PDX1: Pancreas/duodenum homeobox protein 1; CTNNB1: catenin beta-1; NEUROD1: transcription factor NEUROD1; FOXO1: forkhead box protein O1. Adapted from Brimble et al., Tan et al., and Dehennaut et al.8,11,14. Created with BioRender.com.
Figure 2.
O-GlcNAcylation in the regulation of gene expression. PRC1, PRC2: Polycomb repressive complex 1, 2; CARM1: coactivator-associated arginine methyltransferase 1; DNMT1: DNA methyltransferase 1; TET: ten-eleven translocation; SP1: transcription factor SP1; PDX1: Pancreas/duodenum homeobox protein 1; CTNNB1: catenin beta-1; NEUROD1: transcription factor NEUROD1; FOXO1: forkhead box protein O1. Adapted from Brimble et al., Tan et al., and Dehennaut et al.8,11,14. Created with BioRender.com.
2. Histone O-GlcNAcylation
In eukaryotic nuclei, the DNA is wrapped around a histone octamer (containing two copies each of histones H2A, H2B, H3, and H4), to form a nucleosome, and is locked by histone H1. The broad spectrum of histone PTMs constitutes the “histone code”, which not only modulates the recruitment of key enzymes involved in gene expression but also impacts the condensation of chromatin. This results in distinct areas of euchromatin, which is only slightly condensed and transcriptionally active, and highly condensed and transcriptionally silent heterochromatin15,16.
2.1. Initial evidence
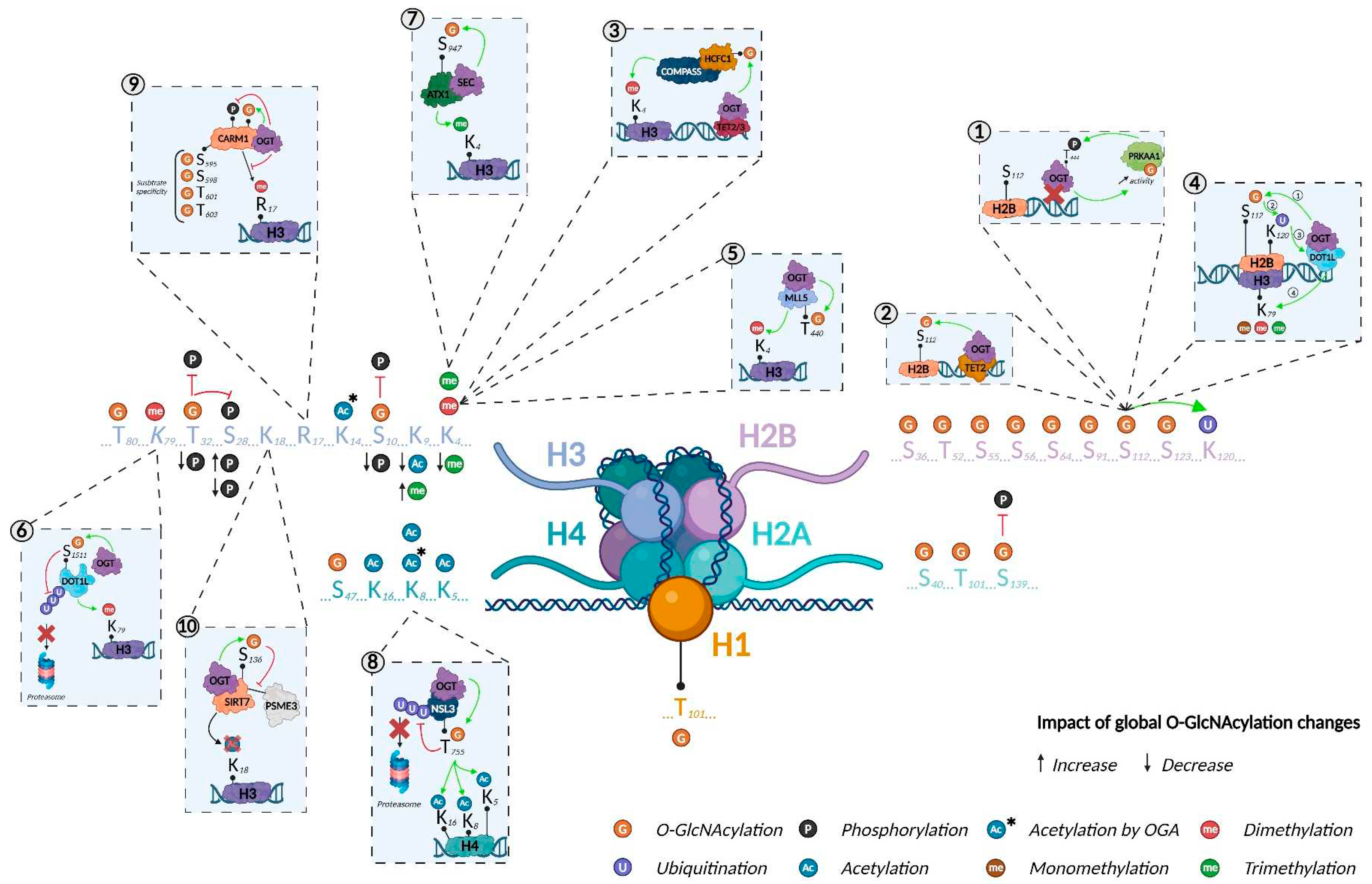
Histone O-GlcNAcylation was first reported in 2010. Using several biochemical and mass spectrometry (MS) approaches, Sakabe et al. revealed that H2A, H2B, H3, and H4 were O-GlcNAcylated (at T101, S36, and S47 in H2A, H2B, and H4, respectively; the modified site on H3 was not identified) in HeLa cells6. They demonstrated that heat stress was associated with increased histone O-GlcNAcylation, concomitant with DNA condensation. This discovery created a novel field of research on O-GlcNAcylation-mediated stress responses and added a new layer of complexity to the histone code. The following year, Hahne et al. mapped additional O-GlcNAcylated sites on H2B (T52, S55, S56, and S64) using a bioinformatics analysis tool called Oscore, which detects and ranks tandem MS spectra by their probability of containing O-GlcNAc peptides (Figure 3, Table 1)17. However, these O-GlcNAc sites have not yet been confirmed by other studies.
2.2. Histone O-GlcNAcylation throughout evolution
O-GlcNAcylated histones have also been reported in plants indicating that the mechanism is conserved across diverse phyla. Schouppe et al. identified three new O-GlcNAcylated sites in cultured cells from Nicotiana tabacum cv. Xanthi, on H1 (T101), H2B (S65), and H3 (T80)18. However, O-GlcNAcylation sites can differ among species. Using MS, Hirosawa et al. mapped O-GlcNAcylation to S40 of H2A. This PTM occurred specifically in viviparous species, which expressed both H2A S40 and H2A A40 isoforms, while more phylogenetically distant species expressed only the A40 isoform19. This study demonstrated that epigenetic processes/machineries are not fully conserved between vertebrates, pinpointing the existence of species-dependent regulatory mechanisms and limiting the use of particular animal models, depending on the scientific hypothesis (e.g., zebrafish are commonly used as a model for epigenetic studies but lack the H2A S40 isoform; Figure 3, Table 1)20
Figure 3.
O-GlcNAcylation as an essential component of the histone code. Created with BioRender.com.
Figure 3.
O-GlcNAcylation as an essential component of the histone code. Created with BioRender.com.
Table 1.
Identified O-GlcNAcylated histone residues.
Table 1.
Identified O-GlcNAcylated histone residues.
| Histone |
Amino acid |
Biological impact |
Identification method |
Location |
Reference |
| H1 |
T101 |
– |
LC-MS/MS; β-elimination |
Nicotiana tabacum L. cv Xanthi cells |
18 |
| H2A |
T101 |
– |
LC-MS/MS; chemoenzymatic labeling (UDP-GalNAz, UDP-[3H]-GlcNAc) |
HeLa cells |
6 |
| S40 |
O-GlcNAc site specific to viviparous species; involved in DNA damage repair (interactions with γH2AX and AcH2AZ, recruitment of PRKDC and RAD51) |
LC-MS/MS; monoclonal antibody 20B2 against O-GlcNAcylated H2A S40 |
HeLa cells; mESCs; Ptk2 and CyEF cells |
19,21 |
| H2AX |
S139 |
Decreases phosphorylation of H2AX (S139); involved in DNA damage repair (restrains γH2AX expansion) |
Recombinant protein (H2AX S139A) |
HeLa cells |
22 |
| H2B |
S36 |
– |
LC-MS/MS; chemoenzymatic labeling (UDP-GalNAz, UDP-[3H]-GlcNAc) |
HeLa cells |
6 |
| T52 |
Oscore (bioinformatics tool based on MS analysis) |
HeLa, hES, iPS, A549, GAMG, HEK293, HepG2, K562, MCF7, RKO, and U2OS cells |
17 |
| S55 |
| S56 |
| S64 |
| S65 |
LC-MS/MS; β-elimination |
Nicotiana tabacum L. cv Xanthi cells |
18 |
| S91 |
ETD-MS/MS; chemoenzymatic labeling (UDP-[3H]-GlcNAc) |
HeLa cells |
24,26,28 |
| S112 |
Involved in DNA damage repair (interaction with NBN and regulation of its foci formation) |
Polyclonal antibody against O-GlcNAcylated H2B S112 |
HEK293T cells |
23 |
| S112 |
Increases monoubiquitination of H2B K120 by recruiting H2B ubiquitin ligase and increases H3K79me (DOT1L); favors TET2-dependent gene transcription |
ETD-MS/MS; chemoenzymatic labeling (UDP-[3H]-GlcNAc) |
HeLa cells |
24,26,28 |
| S123 |
- |
| H3 |
S10 |
Decreases phosphorylation of H3 S10 |
β-elimination and WGA-HRP western blot |
HEK293 cells |
34 |
| T32 |
Decreases phosphorylation of H3 S28 and S28 |
LC-MS/MS |
HeLa cells |
35 |
| H3.3 |
T80 |
– |
LC-MS/MS; β-elimination |
Nicotiana tabacum L. cv Xanthi cells |
18 |
| H4 |
S47 |
– |
LC-MS/MS; chemoenzymatic labeling (UDP-GalNAz, UDP-[3H]-GlcNAc) |
HeLa cells |
6 |
2.3. Histone O-GlcNAcylation and DNA damage repair process
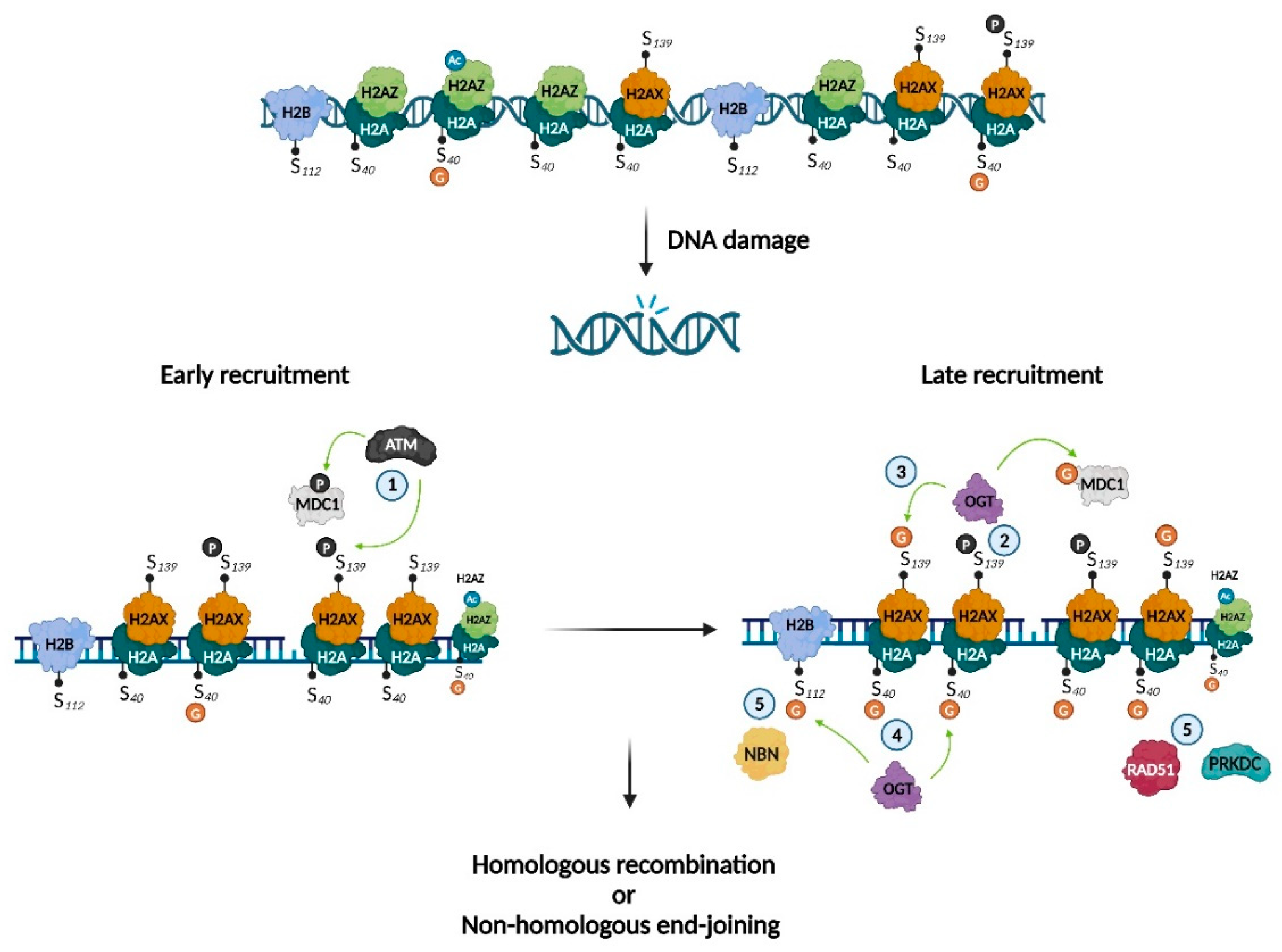
DNA damage, caused by endogenous (e.g., reactive oxygen species, water) or exogenous (e.g., UV radiation or ionizing radiation) sources, can impact health. Accordingly, cells have developed several response mechanisms to maintain the DNA’s integrity. Hayakawa et al. provided evidence that O-GlcNAcylation of H2A S40 is involved in DNA damage repair, by interacting with phosphorylated H2AX (γH2AX) and acetylated H2AZ (AcH2AZ) and recruiting the key DNA repair enzymes protein kinase, DNA-activated, catalytic subunit (PRKDC) and RAD51 recombinase (RAD51)21. As the H2A S40 isoform is species-dependent, this study reinforces the existence of distinct DNA repair mechanisms between species. S139 on H2AX can also undergo O-GlcNAcylation22. Interestingly, Chen et al. showed that OGT was recruited by S139-phosphorylated H2AX, promoting O-GlcNAcylation of H2AX close to sites of damage, thus delimiting the expansion territory of γH2AX. They also determined that mediator of DNA damage checkpoint 1 (MDC1) was O-GlcNAcylated. As phosphorylation of both MDC1 and H2AX prolongs G2/M arrest and can eventually cause apoptosis, the authors suggested that O-GlcNAcylation of MDC1 and H2AX helps cells recover from DNA damage. Finally, Wang et al. demonstrated that DNA damage induction led to local increases in the O-GlcNAcylation of H2B S112. They suggested a mechanism in which H2B S112 O-GlcNAcylation regulates DNA damage repair via interaction with nibrin (NBN), which is involved in DNA double-strand break repair and DNA damage-induced checkpoint activation. They also showed that H2B S112 O-GlcNAcylation promoted NBN accumulation at damaged DNA sites but was not involved in the interaction with γH2AX23. Collectively, these results indicate several mechanisms that could explain the beneficial effects of O-GlcNAcylation in DNA repair (Figure 4, Table 1).
Figure 4.
Histone O-GlcNAcylation as a key mechanism in DNA damage repair. Illustration of the involvement of histone O-GlcNAcylation in DNA-damage repair. ① Kinase ATM phosphorylates H2AX and MDC1; ② γH2AX recruits OGT and favors O-GlcNAcylation of H2AX on S139 and MDC1 ; ③ GlcNAc-H2AX restrains γH2AX expansion around the DNA-damaged site; ④ OGT O-GlcNAcylates H2A on S40 and H2B on S112; ⑤ GlcNAc-H2A favors accumulation of PRKDC and RAD51, while GlcNAc-H2B favors the accumulation of NBN. Adapted from Chen and Yu, Hayakawa et al. and Wang et al.21–23. Created with BioRender.com.
Figure 4.
Histone O-GlcNAcylation as a key mechanism in DNA damage repair. Illustration of the involvement of histone O-GlcNAcylation in DNA-damage repair. ① Kinase ATM phosphorylates H2AX and MDC1; ② γH2AX recruits OGT and favors O-GlcNAcylation of H2AX on S139 and MDC1 ; ③ GlcNAc-H2AX restrains γH2AX expansion around the DNA-damaged site; ④ OGT O-GlcNAcylates H2A on S40 and H2B on S112; ⑤ GlcNAc-H2A favors accumulation of PRKDC and RAD51, while GlcNAc-H2B favors the accumulation of NBN. Adapted from Chen and Yu, Hayakawa et al. and Wang et al.21–23. Created with BioRender.com.
2.4. Impact of histone O-GlcNAcylation on gene expression
Like other PTMs in the histone code, O-GlcNAcylation serves to regulate gene expression. Using electron transfer dissociation (ETD)-MS/MS, Fujijki and colleagues identified several O-GlcNAcylation sites on H2B (S91, S112, and S123) and H2A (T101). Surprisingly, they were unable to detect the previously reported sites on H2B (S36) and H4 (S47). They showed that H2B (S112) O-GlcNAcylation facilitates the recruitment of the H2B ubiquitin ligase, which led to H2B monoubiquitination on lysine 120 (K120). More interestingly, they found that O-GlcNAcylation of H2B S112 (H2BS112G) can co-occur with the active H3K4me2 mark, suggesting that H2BS112G is involved in transcriptional activation24. Consistent with this hypothesis, Xu et al. showed that despite not impacting OGT’s activity, protein kinase AMP-activated catalytic subunit alpha 1 (PRKAA1)-dependent phosphorylation of T444 inhibits OGT’s association with the chromatin, and therefore, H2BS112G deposition and gene expression. They also revealed the existence of a positive feedback loop, in which O-GlcNAcylation of PRKAA1 increases its activity (Figure 3.1)25.
Ten-eleven translocation (TET) proteins are key enzymes implicated in removing DNA methylation marks that impact gene expression. Chen et al. demonstrated that O-GlcNAcylation of H2B S112 occurs after TET2 recruits OGT, and thus, that histone O-GlcNAcylation participates in TET2-dependent gene transcription (Figure 3.2)26. This result was supported by Deplus et al., who reported that TET2/3-mediated OGT recruitment promoted O-GlcNAcylation of host cell factor C1 (HCFC1), an important protein for the formation of COMPASS, a methyltransferase that deposits the active epigenetic mark H3K4me3 (Figure 3.3)27. Recently, Xu et al. reported that DOT1 like histone lysine methyltransferase (DOT1L), which deposits mono-, di-, and tri-methylated marks on H3K79, acts as a scaffold protein that enables OGT’s recruitment to the chromatin. They suggested that OGT recruitment via DOT1L favors H2BS112G deposition, which facilitates the ubiquitination of H2BK120, a modification that stimulates DOT1L activity to increase H3K79me (Figure 3.4)28. Overall, these results highlighted that O-GlcNAcylation plays a pivotal role in gene transcription through histone modification through complex, multi-layered mechanisms (Table 1)
In addition, O-GlcNAcylation actively influences gene accessibility by modulating the open/closed state of chromatin and the recruitment of key enzymes. Lercher et al. demonstrated that O-GlcNAcylation of H2A T101 decreased nucleosome stability, favoring an open state and thus promoting the recruitment of proteins involved in nucleosome remodelling (e.g., mutS homologs 2 and 6)29. Taken together, these studies illustrate that OGT and O-GlcNAcylation are full-fledged players in histone modification.
2.5. Interplay between histone O-GlcNAcylation and phosphorylation
As they target the same amino acids (serine and threonine), phosphorylation and O-GlcNAcylation are closely linked and can compete against each other30. Moreover, O-GlcNAcylation can positively or negatively regulate the phosphorylation of nearby residues, and OGT/OGA can interact with kinases/phosphatases, creating multi-enzyme complexes that can phosphorylate/O-GlcNAcylate proteins31–33. Lowndes’ group revealed that H3 was O-GlcNAcylated, which partially supressed its phosphorylation. They also highlighted that increasing O-GlcNAc levels via glucosamine was associated with decreases in both H3K9ac and H3K4me3—both active marks—and increases in H3 S28 phosphorylation and H3K9me3, which are active and repressive, respectively34. In 2012, Fong et al. determined that H3 T32 was O-GlcNAcylated. As aurora B, the kinase that phosphorylates H3 S10 and S28, is physically associated with OGT/OGA, they evaluated the impact of O-GlcNAc levels on these phosphosites. Mitotic cells that overexpressed OGT or were treated with OGA inhibitors (PUGNAc or thiamet G) displayed reduced H3 S10, S28, and T32 phosphorylation (Figure 3, Table 1)35. Interestingly, no changes in H3S28 phosphorylation were observed in PUGNAc-treated asynchronous cells. This inconsistency with the previous study may stem from the treatments used to increase O-GlcNAcylation levels. Compared with PUGNAc or thiamet G, glucosamine is less specific and has been associated with off-target effects that interfere with proteoglycan and ATP production36. Regardless, considering that H3 S10 and S28 phosphosites are associated with chromatin condensation during mitosis, these two studies provide general evidence that O-GlcNAcylation regulates the cell cycle by competing with phosphorylation of H3 at different sites.
2.6. O-GlcNAcase as a histone acetyltransferase?
The major role of O-GlcNAcylation in histone modification is reinforced by OGA’s C-terminal HAT activity37. Toleman et al. demonstrated that mammalian OGA can acetylate all four core histones in synthetic nucleosomes in vitro. They also identified an O-GlcNAcylated site for H4 (K8) and H3 (K14) (Figure 3). Interestingly, bacterial OGA lacked acetyltransferase activity, except when the enzyme was incubated with mammalian proteins, suggesting the existence of mammalian-specific regulation. Two years later and through a series of biochemical strategies, the researchers involved in the previous study discovered that the OGA contains a zinc finger-like domain that ensures histone binding38. However, Butkinaree et al. extensively demonstrated that OGA lacked histone acetyltransferase activity39. Consistent with this, Rao et al. demonstrated that human OGA lacks the key amino acids for both histone acetyltransferase and acetyl-CoA binding40. Considering these controversial results, the C-terminal extremity is now qualified as a histone acetyltransferase (HAT)-like domain. Interestingly, this domain is only present as the long OGA isoform (in the short isoform, the HAT-like domain is deleted and replaced by a specific 15-residue sequence), suggesting a specific role; however, this role remains unknown (Figure 1)41
3. O-GlcNAcylation indirectly affects histones via chromatin modifying enzymes
Like many proteins, chromatin modifying enzymes are regulated in part by PTMs such as O-GlcNAcylation, impacting their expression, activity, interactomes, and stability (Table 2). Several proteins that add (“writers”) and remove (“erasers”) various histone or DNA marks interact with OGT and/or are O-GlcNAcylated, highlighting the importance of O-GlcNAcylation14,28,42–45.
3.1. O-GlcNAcylation of histone writers
The protein mixed leukemia lineage 5 (MLL5) is a histone lysine methyltransferase (HKMT) involved in regulating cell cycle progression, spermatogenesis, hematopoiesis, and the maintenance of genomic stability46. Fujiki et al. demonstrated that OGT binds and O-GlcNAcylates MLL5 on T440. This increased MLL5’s H3K4 methyltransferase activity to potentiate retinoic acid (RA)-induced granulopoiesis (Figure 3.5). Interestingly, HL60-R2 cells, which are resistant to granulopoiesis, displayed high O-GlcNAcase activity compared with granulopoiesis-sensitive HL60 cells. In addition, the inhibition of OGA with PUGNAc restored responses to RA and thus the methylation of H3K4, suggesting a direct role of O-GlcNAcylation in granulopoiesis47. More recently, Ding et al. showed that MLL5 stability was cooperatively controlled by OGT and ubiquitin-specific protease 7 (USP7). They showed that the three proteins interacted, limiting the ubiquitination and thus the degradation of MLL5. Although the authors identified two O-GlcNAcylated sites on MLL5 (S435 and T440), they did not determine their roles in protein stability. Moreover, as USP7 can be phosphorylated48, it would be relevant to evaluate if it is also O-GlcNAcylated and how that would impact its functions49. Finally, the authors demonstrated that the increased MLL5 levels were similar to increase in both OGT and USP7 observed in cervical adenocarcinomas50,51. Consistently, Nin et al. demonstrated that O-GlcNAcylation of MLL5 on T440 favors the recruitment of MLL5β to the MLL5β-AP-1 complex, which allows the transcription of human papillomavirus E6/E7 oncogenes implicated in the pathogenesis of cervical cancer52. In addition to being implicated in cervical cancer, chromosome translocations involving MLL can cause MLL-fusion leukemia, in which an MLL N terminus is fused to another protein. AF9, AF10, and ENL which all interact with the H3K79 methyltransferase DOT1L, are the most common MLL fusion partners53–55. In 2021, Song et al. demonstrated that DOT1L was O-GlcNAcylated on S1511, which promotes its stability by protecting it from UBE3C-mediated ubiquitination (Figure 3.6).
Table 2.
Summary of O-GlcNAcylated histone writers/erasers.
Table 2.
Summary of O-GlcNAcylated histone writers/erasers.
| Protein class |
Targeted protein (O-GlcNAc site) |
Biological impact |
Biological context |
Reference |
| HKMT |
MLL5 (T440) |
Increases H3K4 methyltransferase activity, potentiates RA-dependent granulopoiesis via co-activation of RARA, and restores sensitivity of HL60-R2 cells |
HL60 cells |
47 |
| MLL5 (S435, T440) |
OGT interacts with USP7 and MLL5, inhibiting ubiquitination and increasing protein stability |
HEK293 and HeLa cells |
49 |
| MLL5β (T440) |
Recruitment and formation of the MLL5β-AP-1 complex, which promotes the transcription of HPV genes involved in cervical cancer pathogenesis |
HPV16/18+ cells |
52 |
| DOT1L (S1511) |
Stabilizes DOT1L by protecting against ubiquitination by UBE3C. Promotes H3K79me2 and the transcription of genes involved in MLL-fusion leukemia |
HEK293T cells |
56 |
| ATX1 (S947) |
Increases ATX1 stability and H3K4me3 activity; promotes the expression of genes that negatively regulate flowering |
Arabidopsis thaliana |
57 |
| PRMT |
CARM1 |
OGT overexpression decreases H3R17me2 and CARM1 phosphorylation, impacting its subcellular localization and causes DNA abnormalities |
HeLa cells |
58 |
| CARM1 (S595, S598,T601, T603) |
Modifies substrate specificity without affecting its function, cellular localization, stability, or dimerization capability |
HEK293T cells |
59 |
| HAT |
NSL3 |
Increases stability and H4K5, 8, and 16 acetyltransferase activity |
HEK293T and HeLa cells |
60 |
| NSL3 (T755) |
Increase stability through blocking ubiquitination by UBE2S; promotes proliferation of A549 cells |
HEK293T cells |
61 |
| HDAC |
SIRT1 (S549) |
Increases deacetylation activity, modifies substrate affinity, and controls cell survival under stress conditions |
NCI H1299 cells |
62 |
| SIRT1 (T160/S161) |
Impacts its localization and degradation in a nutrition-dependent manner |
HeLa and HepG2 cells |
63 |
| SIRT7 (S136) |
Increases its stability by decreasing its interaction with PSME3, thus decreasing H3K18 acetylation; promotes the progression of tumors in cancer cell lines |
HEK293T cells |
64 |
| HDAC1 (T114, S263) |
Inhibits its activity and regulates cell migration, proliferation, and invasion |
HepG2 cells |
65 |
| HDAC4 (S642) |
Exerts a cardioprotective effect by counteracting CAMKII signaling |
HEK2 cells, ventricular cardiomyocytes from neonatal rats |
66 |
| HDAC6 |
Enhances its activity |
hTERT-RPE1 and IMCD3 cells |
67 |
| PRC1 |
RING1B (T250/S251, S278) |
Modifies gene targeting and is involved in the differentiation |
hESCs |
68 |
| BMI1 (S255) |
Increases stability by blocking ubiquitination; promotes tumorigenesis |
Prostate cancer cell lines (C4-2, PC-3 and DU145) |
69 |
| PRC2 |
EZH2 (S76) |
Increases EZH2 stability; the OGT/EZH2 axis limits the expression of tumor suppressor genes |
Breast cancer cell line MCF7 |
70 |
| EZH2 (S73, S76, S84, S87, T313, S729) |
S73, S84, and T313 increase EZH2 stability; S729 stimulates di- and trimethyltransferase activity |
HEK293T cells |
71 |
| EZH2 |
Consolidates fear memories |
CA1 neurons from Sprague Dawley rats |
72 |
| PR-DUB |
BAP1 |
Promotes the pluripotent state |
HEK293 cells |
73 |
| HCFC1 |
Catalyzes proteolysis |
HeLa cells |
74 |
| ASXL1 (S199) |
Promotes ASXL1 stability; potential tumor suppressive role in myeloid malignancies |
HEK293T cells |
75 |
The authors also showed that OGT knockdown was associated with a decrease in H3K79me2 levels and the enrichment of HOXA9/MEIS1 mRNA (genes involved in the initiation and progression of the disease) and H3K79me2/DOT1L on the HOXA9/MEIS1 promoter, illustrating the role of DOT1L O-GlcNAcylation in MLL-fusion leukemia pathogenesis56. Taken together, these studies highlight the important role of O-GlcNAcylation in cancer pathogenesis—regulating ubiquitin-mediated degradation and gene expression.
O-GlcNAcylation also controls HKMTs in plants. The Arabidopsis homolog of trithorax (ATX1) is a H3K4 methyltransferase. Xing et al. demonstrated that secret agent (SEC), the OGT in Arabidopsis, regulated both the stability and activity of ATX1 through O-GlcNAcylation of S947. They also demonstrated that O-GlcNAcylation of ATX1 promotes H3K4me3 deposition on FLOWERING LOCUS C, which encodes key negative regulators of flowering (Figure 3.7)57. Considering O-GlcNAcylation’s reported roles in protein degradation, it would be interesting to identify the mechanism by which it regulates ATX1.
In addition to HKMTs, O-GlcNAcylation can also modify the properties of other histone writers. The histone lysine acetyltransferase 8 (KAT8) contains male-specific lethal and nonspecific lethal (NSL) complexes. Interestingly, OGT1 is a component of the NSL complex76. In 2017, Wu et al. demonstrated that OGT1 interacted with and O-GlcNAcylated the NSL complex subunit NSL3, which was associated with increased stability and activity, thereby facilitating H4 acetylation on K5, K8, and K1660. More recently, the same group used MS and several biochemical methods to identify O-GlcNAcylation of T755 of NSL3, which increased NSL3 stability by blocking UBE2S-dependent ubiquitination. Even more importantly, O-GlcNAcylated T755 was required to maintain the integrity and holoenzyme activity of the NSL complex. In type II epithelium-like lung carcinoma (A549) cells, NSL3 O-GlcNAcylation promoted proliferation, leading them to conclude that O-GlcNAcylation acts as a link between oncogenic signals and the epigenetic changes that occur in cancer (Figure 3.8)61.
Coactivator-associated arginine methyltransferase 1 (CARM1), as also known as protein arginine N-methyltransferase 4 (PRMT4), is an enzyme that asymmetrically dimethylates proteins on arginine residues. It is both a substrate and interactor of OGT77. In 2010, Sakabe and Hart demonstrated that OGT overexpression not only decreased H3R17me2 (a CARM1-specific target) and that CARM1 phosphorylation impacted its subcellular localization, resulting in DNA abnormalities (e.g., errors in chromosomal separation, chromosomal bridges). Interestingly, increasing O-GlcNAcylation levels via thiamet G or N-acetyl-glucosamine-thiazoline did not impact H3R17me2 levels or CARM1 phosphorylation, suggesting the direct involvement of OGT58. Later, Charoensuksai et al. identified four O-GlcNAcylated sites on CARM1: S595, S598, T601, and T603. They showed that these modifications impacted CARM1’s substrate specificity without affecting its function, cellular localization, stability, or dimerization (Figure 3.9)59. As dysregulated CARM1 expression and/or activity of CARM1 has been described in various pathologies78, it would be interesting to determine the impact of O-GlcNAcylation and OGT on CARM1.
3.2. O-GlcNAcylation of histone erasers
Histone deacetylases (HDACs) catalyze the removal of acetyl groups from both histones and non-histone proteins. Humans express 18 HDACs (HDAC1–11, SIRT1–7)79. To date, three (HDAC1, 4, and 6) have been identified as O-GlcNAcylated. Zhu et al. showed that OGT interacted with and O-GlcNAcylated HDAC1 on T114 and S263, which suppresses HDAC1 enzymatic activity. They also showed that HDAC1 O-GlcNAcylation regulates the migration, proliferation, and invasion of HepG2 cells, thus identifying a new potential therapeutic strategy for hepatocellular carcinoma65. More recently, HDAC4 was reported to be O-GlcNAcylated on S642. In the diabetes mellitus mouse model, this O-GlcNAcylation event counteracted pathological CAMKII signaling and thus was deemed cardioprotective66. Considering these important effects, it would be relevant to evaluate if and how HDAC4 O-GlcNAcylation impacts the histone acetylation landscape. HDAC6 plays a pivotal role in cilia assembly and is regulated by phosphorylation80. Considering the widely reported crosstalk between phosphorylation and O-GlcNAcylation81, Tian and Qin examined if HDAC6 was also regulated by O-GlcNAcylation. They discovered that OGT interacted with and O-GlcNAcylated HDAC6 in hTERT-RPE1 cells, resulting in ciliary shortening, and demonstrated that treatment with the OGT inhibitors thiamet G or GlcNAcstatin enhanced HDAC6’s deacetylase activity67. To date, the effects of these O-GlcNAcylated HDACs on histone acetylation have not been examined.
Sirtuins (SIRTs) are nicotine adenine dinucleotide(+)-dependent HDACs that regulate a wide variety of biological processes, such as metabolism, oxidative stress, apoptosis, and inflammation82. SIRT1 is a critical stress sensor that regulates both histones and non-histone proteins (e.g., p53, NFκB, eIF2α). Interestingly, O-GlcNAcylation is also pivotal in the stress response83. Consistent with this overlap, Han et al. demonstrated that SIRT1 interacted directly with OGT. Using chemoenzymatic and metabolic labeling coupled with MS, they showed that SIRT1 was O-GlcNAcylated on S549, which enhanced its deacetylase activity (evaluated on histone H3 and cellular tumor antigen p53 (p53)) and its substrate affinity (evaluated on p53). They found that under stress, SIRT1 O-GlcNAcylation allowed some targets to be deacetylated, including p53 and FOXO3, which regulate the decisions governing cell death and survival62. Han et al. found that SIRT1 O-GlcNAcylation on S549 did not affect its subcellular localization, and a recent study reported complementary results. In fact, Chattopadhyay et al. revealed that the nutrient-dependent SIRT1 O-GlcNAcylation of T160/S161 exerts spatiotemporal control by promoting its localization to the cytosol, where it undergoes ubiquitin-mediated degradation63. Considering these results, evaluating if SIRT1 O-GlcNAcylation impacts the acetylation status of histones would be worth exploring. SIRT7, which catalyzes the selective deacetylation of H3K18, was recently identified as O-GlcNAcylated by OGT at S136, which stabilizes SIRT7 by decreasing its interaction with proteasome activator subunit 3 (PSME3), a core molecule of a new ubiquitin-independent pathway84. By reducing H3K18Ac, SIRT7 O-GlcNAcylation has been associated with repressing tumor suppressor genes to promoting tumor progression in nude mice (Figure 3.10)64.
4. Focus on O-GlcNAcylation of Polycomb repressive complexes
The polycomb group (PcG) proteins form three complexes known as PRC1, PRC2 and PR DeUBiquitinase (PR-DUB). PRCs play a pivotal role in development by repressing homeotic genes. In mice, deleting PcG genes causes embryonic lethality85. PRC1 monoubiquitinates K119 of histone H2, which can be removed by PR-DUB. PRC2 is a HKMT that mono-, di-, and trimethylates H3K27 to form H3K27me1, me2, and me3, respectively. Mutations in the subunits of PRCs are associated with human neurodevelopmental disorders and cancer86,87. O-GlcNAcylation plays an important role in gene regulation in Drosophila. As proof, OGT is encoded by the PcG gene super sex combs (sxc)88, and loss of OGA leads to global perturbation of the epigenetic machinery89. Although PRC1, PRC2, and PR-DUB exist in Drosophila, no major features have been identified recently10.
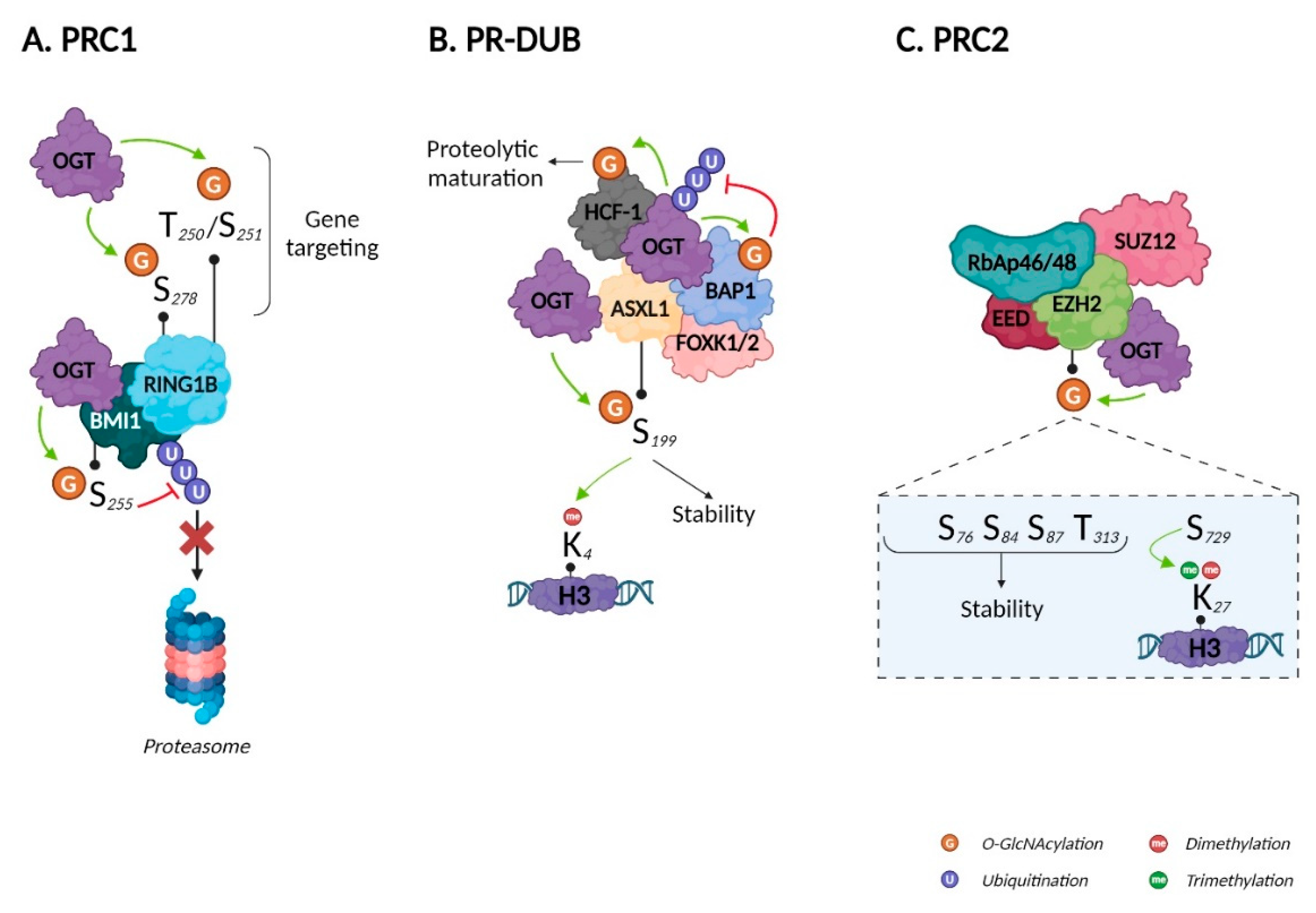
The PRC1 E3 ubiquitin protein ligase can be either RING1A or RING1B, which is associated with one of six PcG ring finger proteins (PCGF1–6). In contrast to RING1A, RING1B can be O-GlcNAcylated. Using MS coupled to beta-elimination and Michael addition with dithiothreitol, Maury et al. identified S278 as a O-GlcNAcylation site. They also found that T250 and S251 were important for O-GlcNAcylation but could not discriminate which residue harbored the GlcNAc moiety. Using chromatin immunoprecipitation coupled with sequencing (ChIP-Seq), the authors revealed that RING1B O-GlcNAcylation impacted its target genes: O-GlcNAcylated RING1B was bound to genes involved in neuronal differentiation, while unmodified RING1B was bound to genes related to cell cycle and metabolism. Accordingly, unmodified RING1B decreased throughout human embryonic stem cell (hESC) differentiation68, reinforcing the described role of O-GlcNAcylation in differentiation90. RING1B, and BMI1 (also known as PGCF4) are all closely related to prostate cancer91. BMI1 phosphorylation protects it from proteasomal degradation92. In 2017, Li et al. demonstrated that OGT interacts with BMI1 and O-GlcNAcylates it on S255. Like phosphorylation, O-GlcNAcylation increased BMI1’s stability by inhibiting its polyubiquitination and proteasomal degradation. They also illustrated the negative role of BMI1 O-GlcNAcylation in prostate cancer tumorigenesis, as it inhibits p53, PTEN, and CDKN1A/CDKN2A signaling, thus favorizing apoptosis, invasion and proliferation respectively (Figure 5A)69. Although both RING1B and BMI1 are O-GlcNAcylated, there is no evidence that this impacts ubiquitin ligase activity; however, BMI1 was recently shown to regulate PRC1 ubiquitin ligase activity, would could be modulated by O-GlcNAcylation93.
Figure 5.
Impact of O-GlcNAcylation on Polycomb repressive complexes. Created with BioRender.com.
Figure 5.
Impact of O-GlcNAcylation on Polycomb repressive complexes. Created with BioRender.com.
PR-DUB is a complex composed of BAP1, HCFC1, FOXK1/2, either ASXL1, 2, or 3, and interestingly, OGT94. In splenocytes, PR-DUB’s catalytic subunit BAP1 regulates HCFC1 and OGT and thus O-GlcNAcylation levels through its deubiquitinase activity95. Since OGT and O-GlcNAcylation are indispensable for the proteolytic maturation of HCFC174, BAP1 i) directly regulates HCFC1 expression and ii) favors its maturation by stabilizing OGT. BAP1 O-GlcNAcylation was demonstrated only recently, with no evidence of impacts on its expression/stability or its deubiquitinase activity73. The close relationships between OGT, HCFC1, and BAP1 are exemplified by their roles modulating peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PRGC1), a master regulator of gluconeogenesis. Ruan et al. demonstrated that HCFC1 recruits OGT to PRGC1 and that O-GlcNAcylation facilitates the binding, thus stabilizing PRGC1 and promoting gluconeogenesis96. ASXL1 is a component of PR-DUB, and also regulates H3K4me397. Recently, Inoue et al. identified as an OGT substrate that displayed increased stability after O-GlcNAcylation of S19975. Disrupting the ASXL1-OGT complex reduced H3K4 methylation, indicating a pivotal tumor suppressive role for this signaling axis in myeloid malignancies (Figure 5B).
PRC2’s catalytic activity is provided by three subunits: enhancer of zeste homolog 2 (EZH2), embryonic ectoderm development (EED), and suppressor of zeste 12 (SUZ12). The first evidence of the impact of EZH2 O-GlcNAcylation in humans was provided by Chu et al. in 2014. By treating two different cell lines with small interfering RNA, they demonstrated that OGT knockdown was associated with a decrease in H3K27me3 only. They showed that OGT i) interacted with the EZH2/PRC2 complex, ii) was essential for EZH2—and therefore, PRC2—stability, and iii) O-GlcNAcylated EZH2 on S76. Finally, the authors demonstrated that the OGT/EZH2 axis downregulated tumor suppressor genes in breast cancer cells, thus identifying a new therapeutic target70. In 2018, using MS coupled to Click-iT® O-GlcNAc enzymatic labeling system, Wong’s group identified five new O-GlcNAcylated sites on EZH2: S73, S84, S87, T313, and S729. After excluding S87, since O-GlcNAcylation at this site was very low, they showed that the N-terminal O-GlcNAcylated sites (S73, S84, and T313) increased the stability of EZH2 by limiting its ubiquitination, while the C-terminal O-GlcNAcylated site (S729) stimulates its di- and trimethyltransferase activity. None of the sites altered the affinity of EZH2 for other PRC2 components71. EZH2 has been shown to be involved in cancer98. In addition, Butler et al. recently demonstrated that OGT exerted control on histone regulation via EZH2-dependent H3K27me3 during the consolidation of fear memories72. To date, EZH2 is the only component of PRC2 identified as O-GlcNAcylated; therefore, it would be relevant to study whether O-GlcNAcylation also impacts SUZ12, EED, and RbAp46/48 (Figure 5C).
5. Perspectives and future directions
While many studies over the past decade have established that H2A, H2B, H3, and H4 are O-GlcNAcylated, evidence for H1 O-GlcNAcylation remains limited. In 2011, an in silico study proposed O-GlcNAcylation of H1’s serine and threonine residues; however, this study relied on YinOYang 1.2 predictive tools, which remain controversial99,100. As H1 plays an important role in chromatin organization and its phosphorylation can destabilize its bond with the DNA101, further exploration of the potential impacts of H1 O-GlcNAcylation in this context is warranted.
The influence of O-GlcNAcylation goes beyond histone modifying enzymes. The different TET isoforms (TET1, 2, and 3), which catalyze the oxidation of 5-methylcytosine to remove DNA methylation102, also interact with OGT27,103. These interactions and/or the O-GlcNAcylation of TET proteins affects their stability, phosphorylation, and DNA binding, and thus ability to remove DNA methylation27,103–106. While TET O-GlcNAcylation is well-documented, the first evidence of O-GlcNAcylation of a DNA writer, DNA methyltransferase (DNMT), only emerged in 2020107. Interestingly, OGT is enriched at the promoter of DNMT3B, which encodes one of two enzymes regulating de novo methylation, suggesting that its expression might be controlled42. Moreover, O-GlcNAcylation of DNMT1, the enzyme that ensures the transmission of DNA methylation patterns during replication, was recently shown to reduce its methyltransferase activity108. Given the central role of DNMTs in physiological (e.g., stem cell fate, cardiac metabolism, and contractility) and pathological (e.g., cancers, Tatton-Brown syndrome) conditions, unravelling how O-GlcNAcylation regulates these proteins could lead to promising research avenues109–112.
Developing tools and approaches to enhance our understanding of O-GlcNAcylation as an epigenetic mark is a major challenge in the field. Currently, only two antibodies against O-GlcNAcylated histones are available (for H2BS112 and H2AS40)19,23. Although they are less expensive and labor-intensive than specific antibodies, pan-O-GlcNAcylation antibodies have different selectivity and poor specificity12. Thus, expanding the antibody collection to detect all possible O-GlcNAcylated histone residues would allow better mapping of the modification using ChIP-Seq- or MS-based approaches. Current strategies to understand the role of O-GlcNAcylation include tissue-specific knockouts, RNA interference, and OGT and OGA inhibitors12. Considering the implications of O-GlcNAcylation in pathophysiological situations (e.g., developmental defects, sepsis), pairing these strategies with advanced epigenetic methods could clarify how O-GlcNAcylation interacts with other histone and DNA marks in various contexts1,113,114. Finally, identifying new O-GlcNAcylated sites on histones and epigenetic enzymes using MS, then preventing their modification via mutation, will enhance our understanding of the broad implications of O-GlcNAcylation12.
6. Conclusion
In this review, we have highlighted the important roles of O-GlcNAcylation on core histones and its cross-talk with the other nucleosomal PTMs. It is very likely that known O-GlcNAcylated proteins represent only a fraction of the broader role it plays in epigenetics. By developing more refined methods, we will identify more proteins influenced by this modification. Notably, some studies have taken a more critical look at histone O-GlcNAcylation. For instance, Gambetta et al. emphasized the need to treat previous results on the role of O-GlcNAcylation in epigenetics with caution115. Moreover, certain recent findings raise questions of abundance and occurrence of O-GlcNAcylation on histones as well as presence of other factors for efficient O-GlcNAcylation116,117. There is still much to uncover to understand the intricacies of O-GlcNAcylation’s roles in epigenetic regulation and determine how these roles can be targeted to improve human health, underscoring the importance of continued research in this area.
Author Contributions
Article conceptualization: T.D.; Literature search and data analysis: T.D.; Writing – original draft preparation: T.D.; Writing – review and editing: T.D., B.L., and S.M. All authors have read and approved the final manuscript.
Funding
This work was supported by the Sauve ton Coeur Association (France) and the Canadian Institutes of Health Research (CIHR). T.D. is a postdoctoral researcher supported by Takeda Canada and CIHR’s Institute of Genetics (Rare Diseases). S.M. is supported by a Scientist Career Award (Fonds de Recherche du Québec – Santé, Fundamental Research Junior 2).
Acknowledgments
The authors thank the Olivier-Van Stichelen Lab for providing the O-GlcNAc Database (v1.3) and High-Fidelity Science Communications for manuscript editing.
Declaration of Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
References
- Dupas, T., Persello, A., Blangy-Letheule, A., Denis, M., Erraud, A., Aillerie, V., Leroux, A.A., Rivière, M., Lebreton, J., Tessier, A., et al. (2022). Beneficial Effects of O-GlcNAc Stimulation in a Young Rat Model of Sepsis: Beyond Modulation of Gene Expression. Int. J. Mol. Sci. 23, 6430. [CrossRef]
- Yilmaz, A., and Grotewold, E. (2010). Components and Mechanisms of Regulation of Gene Expression. In Computational Biology of Transcription Factor Binding Methods in Molecular Biology., I. Ladunga, ed. (Humana Press), pp. 23–32. [CrossRef]
- Berger, S.L., Kouzarides, T., Shiekhattar, R., and Shilatifard, A. (2009). An operational definition of epigenetics. Genes Dev. 23, 781. [CrossRef]
- Breiling, A., and Lyko, F. (2015). Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin 8, 24. [CrossRef]
- Bannister, A.J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. [CrossRef]
- Sakabe, K., Wang, Z., and Hart, G.W. (2010). β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. 107, 19915–19920. [CrossRef]
- Yang, X., and Qian, K. (2017). Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465. [CrossRef]
- Brimble, S., Wollaston-Hayden, E.E., Teo, C.F., Morris, A.C., and Wells, L. (2010). The Role of the O-GlcNAc Modification in Regulating Eukaryotic Gene Expression. Curr. Signal Transduct. Ther. 5, 12–24. [CrossRef]
- Parker, M.P., Peterson, K.R., and Slawson, C. (2021). O-GlcNAcylation and O-GlcNAc Cycling Regulate Gene Transcription: Emerging Roles in Cancer. Cancers 13, 1666. [CrossRef]
- Decourcelle, A., Leprince, D., and Dehennaut, V. (2019). Regulation of Polycomb Repression by O-GlcNAcylation: Linking Nutrition to Epigenetic Reprogramming in Embryonic Development and Cancer. Front. Endocrinol. 10, 117. [CrossRef]
- Tan, Z.-W., Fei, G., Paulo, J.A., Bellaousov, S., Martin, S.E.S., Duveau, D.Y., Thomas, C.J., Gygi, S.P., Boutz, P.L., and Walker, S. (2020). O-GlcNAc regulates gene expression by controlling detained intron splicing. Nucleic Acids Res. 48, 5656–5669. [CrossRef]
- Dupas, T., Betus, C., Blangy-Letheule, A., Pelé, T., Persello, A., Denis, M., and Lauzier, B. (2022). An overview of tools to decipher O-GlcNAcylation from historical approaches to new insights. Int. J. Biochem. Cell Biol. 151, 106289. [CrossRef]
- Wulff-Fuentes, E., Berendt, R.R., Massman, L., Danner, L., Malard, F., Vora, J., Kahsay, R., and Olivier-Van Stichelen, S. (2021). The human O-GlcNAcome database and meta-analysis. Sci. Data 8, 25. [CrossRef]
- Dehennaut, V., Leprince, D., and Lefebvre, T. (2014). O-GlcNAcylation, an Epigenetic Mark. Focus on the Histone Code, TET Family Proteins, and Polycomb Group Proteins. Front. Endocrinol. 5. [CrossRef]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403, 41–45. [CrossRef]
- Morrison, O., and Thakur, J. (2021). Molecular Complexes at Euchromatin, Heterochromatin and Centromeric Chromatin. Int. J. Mol. Sci. 22, 6922. [CrossRef]
- Hahne, H., Gholami, A.M., and Kuster, B. (2012). Discovery of O-GlcNAc-modified Proteins in Published Large-scale Proteome Data*. Mol. Cell. Proteomics 11, 843–850. [CrossRef]
- Schouppe, D., Ghesquière, B., Menschaert, G., De Vos, W.H., Bourque, S., Trooskens, G., Proost, P., Gevaert, K., and Van Damme, E.J.M. (2011). Interaction of the Tobacco Lectin with Histone Proteins. Plant Physiol. 155, 1091–1102. [CrossRef]
- Hirosawa, M., Hayakawa, K., Yoneda, C., Arai, D., Shiota, H., Suzuki, T., Tanaka, S., Dohmae, N., and Shiota, K. (2016). Novel O-GlcNAcylation on Ser40 of canonical H2A isoforms specific to viviparity. Sci. Rep. 6, 31785. [CrossRef]
- Cavalieri, V., and Kathrein, K.L. (2022). Editorial: Zebrafish Epigenetics. Front. Cell Dev. Biol. [CrossRef]
- Hayakawa, K., Hirosawa, M., Tani, R., Yoneda, C., Tanaka, S., and Shiota, K. (2017). H2A O-GlcNAcylation at serine 40 functions genomic protection in association with acetylated H2AZ or γH2AX. Epigenetics Chromatin 10, 51. [CrossRef]
- Chen, Q., and Yu, X. (2016). OGT restrains the expansion of DNA damage signaling. Nucleic Acids Res. 44, 9266–9278. [CrossRef]
- Wang, P., Peng, C., Liu, X., Liu, H., Chen, Y., Zheng, L., Han, B., and Pei, H. (2015). OGT Mediated Histone H2B S112 GlcNAcylation Regulates DNA Damage Response. J. Genet. Genomics 42, 467–475. [CrossRef]
- Fujiki, R., Hashiba, W., Sekine, H., Yokoyama, A., Chikanishi, T., Ito, S., Imai, Y., Kim, J., He, H.H., Igarashi, K., et al. (2011). GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480, 557–560. [CrossRef]
- Xu, Q., Yang, C., Du, Y., Chen, Y., Liu, H., Deng, M., Zhang, H., Zhang, L., Liu, T., Liu, Q., et al. (2014). AMPK regulates histone H2B O-GlcNAcylation. Nucleic Acids Res. 42, 5594–5604. [CrossRef]
- Chen, Q., Chen, Y., Bian, C., Fujiki, R., and Yu, X. (2013). TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564. [CrossRef]
- Deplus, R., Delatte, B., Schwinn, M.K., Defrance, M., Méndez, J., Murphy, N., Dawson, M.A., Volkmar, M., Putmans, P., Calonne, E., et al. (2013). TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655. [CrossRef]
- Xu, B., Zhang, C., Jiang, A., Zhang, X., Liang, F., Wang, X., Li, D., Liu, C., Liu, X., Xia, J., et al. (2022). Histone methyltransferase Dot1L recruits O-GlcNAc transferase to target chromatin sites to regulate histone O-GlcNAcylation. J. Biol. Chem. 298, 102115. [CrossRef]
- Lercher, L., Raj, R., Patel, N.A., Price, J., Mohammed, S., Robinson, C.V., Schofield, C.J., and Davis, B.G. (2015). Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat. Commun. 6, 7978. [CrossRef]
- Musicki, B., Kramer, M.F., Becker, R.E., and Burnett, A.L. (2005). Inactivation of phosphorylated endothelial nitric oxide synthase (Ser-1177) by O -GlcNAc in diabetes-associated erectile dysfunction. Proc. Natl. Acad. Sci. 102, 11870–11875. [CrossRef]
- Dubois-Deruy, E., Belliard, A., Mulder, P., Bouvet, M., Smet-Nocca, C., Janel, S., Lafont, F., Beseme, O., Amouyel, P., Richard, V., et al. (2015). Interplay between troponin T phosphorylation and O-N-acetylglucosaminylation in ischaemic heart failure. Cardiovasc. Res. 107, 56–65. [CrossRef]
- Kakade, P.S., Budnar, S., Kalraiya, R.D., and Vaidya, M.M. (2016). Functional Implications of O-GlcNAcylation-dependent Phosphorylation at a Proximal Site on Keratin 18. J. Biol. Chem. 291, 12003–12013. [CrossRef]
- Cieniewski-Bernard, C., Dupont, E., Richard, E., and Bastide, B. (2014). Phospho-GlcNAc modulation of slow MLC2 during soleus atrophy through a multienzymatic and sarcomeric complex. Pflüg. Arch. - Eur. J. Physiol. 466, 2139–2151. [CrossRef]
- Zhang, S., Roche, K., Nasheuer, H.-P., and Lowndes, N.F. (2011). Modification of histones by sugar β-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J. Biol. Chem. 286, 37483–37495. [CrossRef]
- Fong, J.J., Nguyen, B.L., Bridger, R., Medrano, E.E., Wells, L., Pan, S., and Sifers, R.N. (2012). β-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J. Biol. Chem. 287, 12195–12203. [CrossRef]
- Ferron, M., Denis, M., Persello, A., Rathagirishnan, R., and Lauzier, B. (2019). Protein O-GlcNAcylation in Cardiac Pathologies: Past, Present, Future. Front. Endocrinol. 9, 819. [CrossRef]
- Toleman, C., Paterson, A.J., Whisenhunt, T.R., and Kudlow, J.E. (2004). Characterization of the Histone Acetyltransferase (HAT) Domain of a Bifunctional Protein with Activable O-GlcNAcase and HAT Activities*♦. J. Biol. Chem. 279, 53665–53673. [CrossRef]
- Toleman, C.A., Paterson, A.J., and Kudlow, J.E. (2006). The Histone Acetyltransferase NCOAT Contains a Zinc Finger-like Motif Involved in Substrate Recognition*. J. Biol. Chem. 281, 3918–3925. [CrossRef]
- Butkinaree, C., Cheung, W.D., Park, S., Park, K., Barber, M., and Hart, G.W. (2008). Characterization of β-N-Acetylglucosaminidase Cleavage by Caspase-3 during Apoptosis. J. Biol. Chem. 283, 23557–23566. [CrossRef]
- Rao, F.V., Schüttelkopf, A.W., Dorfmueller, H.C., Ferenbach, A.T., Navratilova, I., and Van Aalten, D.M.F. (2013). Structure of a bacterial putative acetyltransferase defines the fold of the human O -GlcNAcase C-terminal domain. Open Biol. 3, 130021. [CrossRef]
- Pagesy, P., Bouaboud, A., Feng, Z., Hulin, P., and Issad, T. (2022). Short O-GlcNAcase Is Targeted to the Mitochondria and Regulates Mitochondrial Reactive Oxygen Species Level. Cells 11, 1827. [CrossRef]
- Gao, J., Yang, Y., Qiu, R., Zhang, K., Teng, X., Liu, R., and Wang, Y. (2018). Proteomic analysis of the OGT interactome: novel links to epithelial–mesenchymal transition and metastasis of cervical cancer. Carcinogenesis 39, 1222–1234. [CrossRef]
- Martinez, M., Renuse, S., Kreimer, S., O’Meally, R., Natov, P., Madugundu, A.K., Nirujogi, R.S., Tahir, R., Cole, R., Pandey, A., et al. (2021). Quantitative Proteomics Reveals that the OGT Interactome Is Remodeled in Response to Oxidative Stress. Mol. Cell. Proteomics 20, 100069. [CrossRef]
- Ma, J., Hou, C., Li, Y., Chen, S., and Wu, C. (2021). OGT Protein Interaction Network (OGT-PIN): A Curated Database of Experimentally Identified Interaction Proteins of OGT. Int. J. Mol. Sci. 22, 9620. [CrossRef]
- Deng, R.-P., He, X., Guo, S.-J., Liu, W.-F., Tao, Y., and Tao, S.-C. (2014). Global identification of O-GlcNAc transferase (OGT) interactors by a human proteome microarray and the construction of an OGT interactome. PROTEOMICS 14, 1020–1030. [CrossRef]
- Zhang, X., Novera, W., Zhang, Y., and Deng, L.-W. (2017). MLL5 (KMT2E): structure, function, and clinical relevance. Cell. Mol. Life Sci. 74, 2333–2344. [CrossRef]
- Fujiki, R., Chikanishi, T., Hashiba, W., Ito, H., Takada, I., Roeder, R.G., Kitagawa, H., and Kato, S. (2009). GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature 459, 455–459. [CrossRef]
- Fernández-Montalván, A., Bouwmeester, T., Joberty, G., Mader, R., Mahnke, M., Pierrat, B., Schlaeppi, J.-M., Worpenberg, S., and Gerhartz, B. (2007). Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. FEBS J. 274, 4256–4270. [CrossRef]
- Ding, X., Jiang, W., Zhou, P., Liu, L., Wan, X., Yuan, X., Wang, X., Chen, M., Chen, J., Yang, J., et al. (2015). Mixed Lineage Leukemia 5 (MLL5) Protein Stability Is Cooperatively Regulated by O-GlcNac Transferase (OGT) and Ubiquitin Specific Protease 7 (USP7). PloS One 10, e0145023. [CrossRef]
- Kim, M., Kim, Y.S., Kim, H., Kang, M.Y., Park, J., Lee, D.H., Roh, G.S., Kim, H.J., Kang, S.S., Cho, G.J., et al. (2016). O-linked N-acetylglucosamine transferase promotes cervical cancer tumorigenesis through human papillomaviruses E6 and E7 oncogenes. Oncotarget 7, 44596–44607. [CrossRef]
- Su, D., Ma, S., Shan, L., Wang, Y., Wang, Y., Cao, C., Liu, B., Yang, C., Wang, L., Tian, S., et al. (2018). Ubiquitin-specific protease 7 sustains DNA damage response and promotes cervical carcinogenesis. J. Clin. Invest. 128, 4280–4296. [CrossRef]
- Nin, D.S., Huang, W., Ali, M., Yew, C.W., Kutateladze, T.G., and Deng, L.-W. (2015). O-GlcNAcylation of MLL5β is essential for MLL5β–AP-1 transcription complex assembly at the HPV16/18-long control region. J. Mol. Cell Biol. 7, 180–183. [CrossRef]
- Okada, Y., Feng, Q., Lin, Y., Jiang, Q., Li, Y., Coffield, V.M., Su, L., Xu, G., and Zhang, Y. (2005). hDOT1L links histone methylation to leukemogenesis. Cell 121, 167–178. [CrossRef]
- Mueller, D., Bach, C., Zeisig, D., Garcia-Cuellar, M.-P., Monroe, S., Sreekumar, A., Zhou, R., Nesvizhskii, A., Chinnaiyan, A., Hess, J.L., et al. (2007). A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 110, 4445–4454. [CrossRef]
- Nguyen, A.T., Taranova, O., He, J., and Zhang, Y. (2011). DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood 117, 6912–6922. [CrossRef]
- Song, T., Zou, Q., Yan, Y., Lv, S., Li, N., Zhao, X., Ma, X., Liu, H., Tang, B., and Sun, L. (2021). DOT1L O-GlcNAcylation promotes its protein stability and MLL-fusion leukemia cell proliferation. Cell Rep. 36, 109739. [CrossRef]
- Xing, L., Liu, Y., Xu, S., Xiao, J., Wang, B., Deng, H., Lu, Z., Xu, Y., and Chong, K. (2018). Arabidopsis O-GlcNAc transferase SEC activates histone methyltransferase ATX1 to regulate flowering. EMBO J. 37, e98115. [CrossRef]
- Sakabe, K., and Hart, G.W. (2010). O-GlcNAc Transferase Regulates Mitotic Chromatin Dynamics. J. Biol. Chem. 285, 34460–34468. [CrossRef]
- Charoensuksai, P., Kuhn, P., Wang, L., Sherer, N., and Xu, W. (2015). O-GlcNAcylation of co-activator-associated arginine methyltransferase 1 regulates its protein substrate specificity. Biochem. J. 466, 587–599. [CrossRef]
- Wu, D., Zhao, L., Feng, Z., Yu, C., Ding, J., Wang, L., Wang, F., Liu, D., Zhu, H., Xing, F., et al. (2017). O-Linked N-acetylglucosamine transferase 1 regulates global histone H4 acetylation via stabilization of the nonspecific lethal protein NSL3. J. Biol. Chem. 292, 10014–10025. [CrossRef]
- Zhao, L., Li, M., Wei, T., Feng, C., Wu, T., Shah, J.A., Liu, H., Wang, F., Cai, Y., and Jin, J. (2020). O-GlcNAc-Modification of NSL3 at Thr755 Site Maintains the Holoenzyme Activity of MOF/NSL Histone Acetyltransferase Complex. Int. J. Mol. Sci. 21, 173. [CrossRef]
- Han, C., Gu, Y., Shan, H., Mi, W., Sun, J., Shi, M., Zhang, X., Lu, X., Han, F., Gong, Q., et al. (2017). O-GlcNAcylation of SIRT1 enhances its deacetylase activity and promotes cytoprotection under stress. Nat. Commun. 8, 1491. [CrossRef]
- Chattopadhyay, T., Maniyadath, B., Bagul, H.P., Chakraborty, A., Shukla, N., Budnar, S., Rajendran, A., Shukla, A., Kamat, S.S., and Kolthur-Seetharam, U. (2020). Spatiotemporal gating of SIRT1 functions by O-GlcNAcylation is essential for liver metabolic switching and prevents hyperglycemia. Proc. Natl. Acad. Sci. 117, 6890–6900. [CrossRef]
- He, X., Li, Y., Chen, Q., Zheng, L., Lou, J., Lin, C., Gong, J., Zhu, Y., and Wu, Y. (2022). O-GlcNAcylation and stablization of SIRT7 promote pancreatic cancer progression by blocking the SIRT7-REGγ interaction. Cell Death Differ. 29, 1970–1981. [CrossRef]
- Zhu, G., Tao, T., Zhang, D., Liu, X., Qiu, H., Han, L., Xu, Z., Xiao, Y., Cheng, C., and Shen, A. (2016). O-GlcNAcylation of histone deacetylases 1 in hepatocellular carcinoma promotes cancer progression. Glycobiology 26, 820–833. [CrossRef]
- Kronlage, M., Dewenter, M., Grosso, J., Fleming, T., Oehl, U., Lehmann, L.H., Falcão-Pires, I., Leite-Moreira, A.F., Volk, N., Gröne, H.-J., et al. (2019). O-GlcNAcylation of Histone Deacetylase 4 Protects the Diabetic Heart From Failure. Circulation 140, 580–594. [CrossRef]
- Tian, J.L., and Qin, H. (2019). O-GlcNAcylation Regulates Primary Ciliary Length by Promoting Microtubule Disassembly. iScience 12, 379–391. [CrossRef]
- Maury, J.J.P., El Farran, C.A., Ng, D., Loh, Y.-H., Bi, X., Bardor, M., and Choo, A.B.-H. (2015). RING1B O-GlcNAcylation regulates gene targeting of polycomb repressive complex 1 in human embryonic stem cells. Stem Cell Res. 15, 182–189. [CrossRef]
- Li, Y., Wang, L., Liu, J., Zhang, P., An, M., Han, C., Li, Y., Guan, X., and Zhang, K. (2017). O-GlcNAcylation modulates Bmi-1 protein stability and potential oncogenic function in prostate cancer. Oncogene 36, 6293–6305. [CrossRef]
- Chu, C.-S., Lo, P.-W., Yeh, Y.-H., Hsu, P.-H., Peng, S.-H., Teng, Y.-C., Kang, M.-L., Wong, C.-H., and Juan, L.-J. (2014). O-GlcNAcylation regulates EZH2 protein stability and function. Proc. Natl. Acad. Sci. U. S. A. 111, 1355–1360. [CrossRef]
- Lo, P.-W., Shie, J.-J., Chen, C.-H., Wu, C.-Y., Hsu, T.-L., and Wong, C.-H. (2018). O -GlcNAcylation regulates the stability and enzymatic activity of the histone methyltransferase EZH2. Proc. Natl. Acad. Sci. 115, 7302–7307. [CrossRef]
- Butler, A.A., Sanchez, R.G., Jarome, T.J., Webb, W.M., and Lubin, F.D. (2019). O-GlcNAc and EZH2-mediated epigenetic regulation of gene expression during consolidation of fear memories. Learn. Mem. 26, 373–379. [CrossRef]
- Moon, S., Lee, Y.-K., Lee, S.-W., and Um, S.-J. (2017). Suppressive role of OGT-mediated O-GlcNAcylation of BAP1 in retinoic acid signaling. Biochem. Biophys. Res. Commun. 492, 89–95. [CrossRef]
- Capotosti, F., Guernier, S., Lammers, F., Waridel, P., Cai, Y., Jin, J., Conaway, J.W., Conaway, R.C., and Herr, W. (2011). O-GlcNAc Transferase Catalyzes Site-Specific Proteolysis of HCF-1. Cell 144, 376–388. [CrossRef]
- Inoue, D., Fujino, T., Sheridan, P., Zhang, Y., Nagase, R., Horikawa, S., Li, Z., Matsui, H., Kanai, A., Saika, M., et al. (2018). A novel ASXL1–OGT axis plays roles in H3K4 methylation and tumor suppression in myeloid malignancies. Leukemia 32, 1327–1337. [CrossRef]
- Hoe, M., and Nicholas, H.R. (2014). Evidence of a MOF histone acetyltransferase-containing NSL complex in C. elegans. Worm 3, e982967. [CrossRef]
- Cheung, W.D., Sakabe, K., Housley, M.P., Dias, W.B., and Hart, G.W. (2008). O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J. Biol. Chem. 283, 33935–33941. [CrossRef]
- vanLieshout, T.L., Stouth, D.W., Hartel, N.G., Vasam, G., Ng, S.Y., Webb, E.K., Rebalka, I.A., Mikhail, A.I., Graham, N.A., Menzies, K.J., et al. (2022). The CARM1 transcriptome and arginine methylproteome mediate skeletal muscle integrative biology. Mol. Metab. 64, 101555. [CrossRef]
- Seto, E., and Yoshida, M. (2014). Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 6, a018713. [CrossRef]
- Pugacheva, E.N., Jablonski, S.A., Hartman, T.R., Henske, E.P., and Golemis, E.A. (2007). HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129, 1351–1363. [CrossRef]
- Hart, G.W., Slawson, C., Ramirez-Correa, G., and Lagerlof, O. (2011). Cross Talk Between O-GlcNAcylation and Phosphorylation: Roles in Signaling, Transcription, and Chronic Disease. Annu. Rev. Biochem. 80, 825–858. [CrossRef]
- Wu, Q.-J., Zhang, T.-N., Chen, H.-H., Yu, X.-F., Lv, J.-L., Liu, Y.-Y., Liu, Y.-S., Zheng, G., Zhao, J.-Q., Wei, Y.-F., et al. (2022). The sirtuin family in health and disease. Signal Transduct. Target. Ther. 7, 1–74. [CrossRef]
- Fahie, K.M.M., Papanicolaou, K.N., and Zachara, N.E. (2022). Integration of O-GlcNAc into Stress Response Pathways. Cells 11, 3509. [CrossRef]
- Son, S.H., Kim, M.Y., Lim, Y.S., Jin, H.C., Shin, J.H., Yi, J.K., Choi, S., Park, M.A., Chae, J.H., Kang, H.C., et al. (2023). SUMOylation-mediated PSME3-20S proteasomal degradation of transcription factor CP2c is crucial for cell cycle progression. Sci. Adv. 9, eadd4969. [CrossRef]
- Margueron, R., and Reinberg, D. (2011). The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349. [CrossRef]
- Luo, X., Schoch, K., Jangam, S.V., Bhavana, V.H., Graves, H.K., Kansagra, S., Jasien, J.M., Stong, N., Keren, B., Mignot, C., et al. (2021). Rare deleterious de novo missense variants in Rnf2/Ring2 are associated with a neurodevelopmental disorder with unique clinical features. Hum. Mol. Genet. 30, 1283–1292. [CrossRef]
- Parreno, V., Martinez, A.-M., and Cavalli, G. (2022). Mechanisms of Polycomb group protein function in cancer. Cell Res. 32, 231–253. [CrossRef]
- Sinclair, D.A.R., Syrzycka, M., Macauley, M.S., Rastgardani, T., Komljenovic, I., Vocadlo, D.J., Brock, H.W., and Honda, B.M. (2009). Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U. S. A. 106, 13427–13432. [CrossRef]
- Akan, I., Love, D.C., Harwood, K.R., Bond, M.R., and Hanover, J.A. (2016). Drosophila O-GlcNAcase Deletion Globally Perturbs Chromatin O-GlcNAcylation*. J. Biol. Chem. 291, 9906–9919. [CrossRef]
- Sun, C., Shang, J., Yao, Y., Yin, X., Liu, M., Liu, H., and Zhou, Y. (2016). O-GlcNAcylation: a bridge between glucose and cell differentiation. J. Cell. Mol. Med. 20, 769–781. [CrossRef]
- van Leenders, G.J.L.H., Dukers, D., Hessels, D., van den Kieboom, S.W.M., Hulsbergen, C.A., Witjes, J.A., Otte, A.P., Meijer, C.J., and Raaphorst, F.M. (2007). Polycomb-group oncogenes EZH2, BMI1, and RING1 are overexpressed in prostate cancer with adverse pathologic and clinical features. Eur. Urol. 52, 455–463. [CrossRef]
- Voncken, J.W., Niessen, H., Neufeld, B., Rennefahrt, U., Dahlmans, V., Kubben, N., Holzer, B., Ludwig, S., and Rapp, U.R. (2005). MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J. Biol. Chem. 280, 5178–5187. [CrossRef]
- Gray, F., Cho, H.J., Shukla, S., He, S., Harris, A., Boytsov, B., Jaremko, Ł., Jaremko, M., Demeler, B., Lawlor, E.R., et al. (2016). BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization. Nat. Commun. 7, 13343. [CrossRef]
- Kolovos, P., Nishimura, K., Sankar, A., Sidoli, S., Cloos, P.A., Helin, K., and Christensen, J. (2020). PR-DUB maintains the expression of critical genes through FOXK1/2- and ASXL1/2/3-dependent recruitment to chromatin and H2AK119ub1 deubiquitination. Genome Res. 30, 1119–1130. [CrossRef]
- Dey, A., Seshasayee, D., Noubade, R., French, D.M., Liu, J., Chaurushiya, M.S., Kirkpatrick, D.S., Pham, V.C., Lill, J.R., Bakalarski, C.E., et al. (2012). Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546. [CrossRef]
- Ruan, H.-B., Han, X., Li, M.-D., Singh, J.P., Qian, K., Azarhoush, S., Zhao, L., Bennett, A.M., Samuel, V.T., Wu, J., et al. (2012). O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 16, 226–237. [CrossRef]
- Fujino, T., and Kitamura, T. (2020). ASXL1 mutation in clonal hematopoiesis. Exp. Hematol. 83, 74–84. [CrossRef]
- Li, Z., Li, M., Wang, D., Hou, P., Chen, X., Chu, S., Chai, D., Zheng, J., and Bai, J. (2020). Post-translational modifications of EZH2 in cancer. Cell Biosci. 10, 143. [CrossRef]
- Ahmad, W., Shabbiri, K., Nazar, N., Nazar, S., Qaiser, S., and Shabbir Mughal, M.A. (2011). Human linker histones: interplay between phosphorylation and O-β-GlcNAc to mediate chromatin structural modifications. Cell Div. 6, 15. [CrossRef]
- Mauri, T., Menu-Bouaouiche, L., Bardor, M., Lefebvre, T., Lensink, M.F., and Brysbaert, G. (2021). <p><em>O</em>-GlcNAcylation Prediction: An Unattained Objective</p>. Adv. Appl. Bioinforma. Chem. 14, 87–102. [CrossRef]
- Gréen, A., Lönn, A., Peterson, K.H., Öllinger, K., and Rundquist, I. (2010). Translocation of histone H1 subtypes between chromatin and cytoplasm during mitosis in normal human fibroblasts. Cytometry A 77A, 478–484. [CrossRef]
- Rasmussen, K.D., and Helin, K. (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750. [CrossRef]
- Vella, P., Scelfo, A., Jammula, S., Chiacchiera, F., Williams, K., Cuomo, A., Roberto, A., Christensen, J., Bonaldi, T., Helin, K., et al. (2013). Tet Proteins Connect the O-Linked N-acetylglucosamine Transferase Ogt to Chromatin in Embryonic Stem Cells. Mol. Cell 49, 645–656. [CrossRef]
- Shi, F.-T., Kim, H., Lu, W., He, Q., Liu, D., Goodell, M.A., Wan, M., and Songyang, Z. (2013). Ten-Eleven Translocation 1 (Tet1) Is Regulated by O-Linked N-Acetylglucosamine Transferase (Ogt) for Target Gene Repression in Mouse Embryonic Stem Cells *. J. Biol. Chem. 288, 20776–20784. [CrossRef]
- Bauer, C., Göbel, K., Nagaraj, N., Colantuoni, C., Wang, M., Müller, U., Kremmer, E., Rottach, A., and Leonhardt, H. (2015). Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT). J. Biol. Chem. 290, 4801–4812. [CrossRef]
- Wang, X., Rosikiewicz, W., Sedkov, Y., Martinez, T., Hansen, B.S., Schreiner, P., Christensen, J., Xu, B., Pruett-Miller, S.M., Helin, K., et al. (2022). PROSER1 mediates TET2 O-GlcNAcylation to regulate DNA demethylation on UTX-dependent enhancers and CpG islands. Life Sci. Alliance 5. [CrossRef]
- Boulard, M., Rucli, S., Edwards, J.R., and Bestor, T.H. (2020). Methylation-directed glycosylation of chromatin factors represses retrotransposon promoters. Proc. Natl. Acad. Sci. 117, 14292–14298. [CrossRef]
- Shin, H., Leung, A., Costello, K.R., Senapati, P., Kato, H., Moore, R.E., Lee, M., Lin, D., Tang, X., Pirrotte, P., et al. (2023). Inhibition of DNMT1 methyltransferase activity via glucose-regulated O-GlcNAcylation alters the epigenome. eLife 12, e85595. [CrossRef]
- Kinoshita, M., Li, M.A., Barber, M., Mansfield, W., Dietmann, S., and Smith, A. (2021). Disabling de novo DNA methylation in embryonic stem cells allows an illegitimate fate trajectory. Proc. Natl. Acad. Sci. 118, e2109475118. [CrossRef]
- Madsen, A., Höppner, G., Krause, J., Hirt, M.N., Laufer, S.D., Schweizer, M., Tan, W.L.W., Mosqueira, D., Anene-Nzelu, C.G., Lim, I., et al. (2020). An Important Role for DNMT3A-Mediated DNA Methylation in Cardiomyocyte Metabolism and Contractility. Circulation 142, 1562–1578. [CrossRef]
- Tatton-Brown, K., Seal, S., Ruark, E., Harmer, J., Ramsay, E., del Vecchio Duarte, S., Zachariou, A., Hanks, S., O’Brien, E., Aksglaede, L., et al. (2014). Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 46, 385–388. [CrossRef]
- Zhang, W., and Xu, J. (2017). DNA methyltransferases and their roles in tumorigenesis. Biomark. Res. 5, 1. [CrossRef]
- Dupas, T., Denis, M., Dontaine, J., Persello, A., Bultot, L., Erraud, A., Vertommen, D., Bouchard, B., Tessier, A., Rivière, M., et al. (2021). Protein O-GlcNAcylation levels are regulated independently of dietary intake in a tissue and time-specific manner during rat postnatal development. Acta Physiol. Oxf. Engl. 231, e13566. [CrossRef]
- Denis, M., Dupas, T., Persello, A., Dontaine, J., Bultot, L., Betus, C., Pelé, T., Dhot, J., Erraud, A., Maillard, A., et al. (2021). An O-GlcNAcylomic Approach Reveals ACLY as a Potential Target in Sepsis in the Young Rat. Int. J. Mol. Sci. 22, 9236. [CrossRef]
- Gambetta, M.C., and Müller, J. (2015). A critical perspective of the diverse roles of O-GlcNAc transferase in chromatin. Chromosoma 124, 429–442. [CrossRef]
- Gagnon, J., Daou, S., Zamorano, N., Iannantuono, N.V., Hammond-Martel, I., Mashtalir, N., Bonneil, E., Wurtele, H., Thibault, P., and Affar, E.B. (2015). Undetectable histone O-GlcNAcylation in mammalian cells. Epigenetics 10, 677–691. [CrossRef]
- Merx, J., Hintzen, J.C.J., Proietti, G., Elferink, H., Wang, Y., Porzberg, M.R.B., Sondag, D., Bilgin, N., Park, J., Mecinović, J., et al. (2022). Investigation of in vitro histone H3 glycosylation using H3 tail peptides. Sci. Rep. 12, 19251. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).