1. Introduction
Desmoglein 3 (DSG3) is a cell adhesion molecule that plays a crucial role in desmosome assembly. Desmosomes are specialized intercellular junctions found in epithelial tissues that anchor keratin intermediate filament and provide strong adhesion between cells, and thus are the key to the maintenance of tissue structure and integrity. The assembly of desmosomes is a complex and coordinated process involving the synthesis, trafficking, and interaction of various proteins, with DSG3 playing a central role in initiating desmosome formation through its homophilic and heterophilic interactions with itself and other desmosomal isoforms located on the neighbouring cell as well as with those on the same cells. Although DSG3 is known primarily to serve as a cell adhesion protein in desmosomes, mounting evidence suggests that it also participates in cell signalling processes beyond adhesion. For instance, DSG3 is involved in regulating indirectly the Wnt/β-catenin signalling pathway via its interaction with plakoglobin [
10]. Increased DSG3 levels were shown to be correlated with reduced nuclear plakoglobin accompanied with elevated expression of the LEF/TCF transcriptional targets, cyclin D1, c-Myc and MMP7 in the tissues of head and neck cancer patients and oral squamous cell carcinoma lines [
7,
11,
37]. DSG3 is also reported in participating in regulating kinases and phosphatases and one example is its interaction with the Src kinase, which is involved in multiple signalling pathways that have an impact on cell proliferation, migration, and survival [
29,
35,
36]. Additionally, DSG3 has been shown to interact with various scaffolding proteins and signalling adaptors, such as plakoglobin and p0071, within desmosomes. These interactions can potentially mediate crosstalk between cell adhesion and signalling pathways, allowing DSG3 to influence signal transduction events. In particular, relevant to this study is that DSG3 can regulate epidermal growth factor receptor (EGFR), a receptor involved in cell growth, proliferation, and survival, and regulate its signalling [
3,
5,
20,
23,
25,
28] and thus, affecting downstream pathways such as MAPK/ERK, PI3K/Akt and YAP [
2,
3,
30]. It is important to note that the understanding of DSG3's role in cell signalling is still evolving, and further research is needed to elucidate the precise mechanisms and functional implications of its involvement in signalling pathways.
Due to the important roles in cell biology, DSG3 has been implicated in various diseases and pathological conditions. The most well-known is Pemphigus Vulgaris, an autoimmune blistering disorder characterized by the production of autoantibodies against DSG3 and DSG1, another desmoglein isoform. The autoantibodies disrupt the interaction between desmogleins, leading to weakened cell-cell adhesion in the epidermis and mucous membranes, leading to the formation of skin blisters and mucosal erosions. DSG3 is also one of the autoantibody targets in Paraneoplastic pemphigus, a rare autoimmune condition associated with malignancies and lymphoproliferative disorders often affecting multiple organ systems [
4]. Another example is the Hailey-Hailey disease, known as familial benign pemphigus and caused by mutations in the ATP2C1 gene that encodes for a calcium pump responsible for maintaining calcium homeostasis in keratinocytes [
14,
16]. Dysfunctional ATP2C1 leads to impaired desmosome function and reduced levels of DSG3, resulting in compromised cell-cell adhesion and the formation of persistent blisters and erosions.
Alterations in DSG3 expression have also been observed in squamous cell carcinoma (SCC) in multiple organ systems. DSG3 is often upregulated in SCCs, and its overexpression has been associated with increased tumour invasiveness and metastasis [
2,
7,
26,
39]. However, the role of DSG3 in cancer remains not fully understood and contradictory findings have been reported in the literature [
3,
12,
15,
34]. For instance, a recent investigation discovered that contrary to expectations, DSG3 inhibits the collective cell migration in oral SCC cell lines [
3]. Mechanistically, it was found that DSG3 limits the activity of the EGFR signaling pathway that in turn triggers the phosphorylation and subsequent export of YAP (Yes-associated protein) from the cell nucleus, leading to its inactivation and inhibition of cell migration. Despite these findings, a comprehensive understanding of how DSG3 contributes to the regulation of gene expression in epithelial cells is still lacking. Using RNA-Seq, this study explored in depth the role of DSG3 in the regulation of the transcriptome network in oral SCC cells. The study reveals that DSG3 is involved in various cellular processes, including extracellular matrix (ECM) organization, focal adhesion, intercellular junction assembly, and keratinocyte functions. Interestingly, the results indicate that DSG3 has a dual role. While it promotes intercellular junction assembly, it also somehow inhibits the association of desmosomes with keratin intermediate filaments, suggesting that DSG3 may exert a function in finely tuning cellular adhesion and keratinocyte maturation.
2. Materials and Methods
2.1. Cell Lines
Myc-tagged DSG3 was ectopically and stably expressed in human oral carcinoma H413 cell line (D), which was used in a previous study [
3]. The matched control cell line was H413 cells with transduction of empty vector (V). Cells were thawed from liquid nitrogen and recovered in growth medium, that is Dulbecco’s Modified Eagle Medium (DMEM, Lonza, Basel, Switzerland)/Ham’s F-12 (Thermo Fisher Scientific) 1:1 supplemented with 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific) and 0.5 μg/mL hydrocortisone (Sigma, Dorset, UK) for at least a couple of weeks before being used for experiments. Cells were incubated at 37 °C in a humidified atmosphere of 5% CO2/95% air, changing medium every 1–2 days.
2.2. Sample Preparation
For RNA-Seq analysis, four samples of two conditions, i.e. empty vector control (V) and DSG3 transduced cell line (D) were prepared and each having two different passage numbers. Cell stocks from liquid nitrogen were thawed and recovered in culture with growth medium for at least a couple of weeks before cell counting and plating in a 12-well plate at confluent density (0.5x106 cells/well). Cells reached fresh confluence the next day and manually scrapped off from the wells, spun down and stored at -80°C. Similar procedures were used for the extraction of cell lysates for RT-qPCR analysis with lysis buffer as described previously [
3].
For transmission electron microscopy (TEM), H413-V and D cell lines were seeded at confluent density in a 6-well plate in duplicate and after two days of culture, the epithelial monolayers were established. Cells were washed briefly with PBS and treated with dispase (2.4U/ml) for 20 minutes to detach the epithelial sheets from the substrate. They were washed briefly with phosphate buffer before fixation with freshly prepared 4% glutaraldehyde in phosphate buffer overnight. After washing with phosphate buffer, samples were stored in a fridge before being proceeded for TEM sample preparation and analysis by a specially trained technician.
2.3. Extraction, RNA Library Preparation and NovaSeq Sequencing
Total RNA was extracted from frozen cell pellets using a Qiagen RNeasy Mini kit following the manufacturer’s instructions (Qiagen, Hilden, Germany).
RNA samples were quantified using Qubit 4.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and RNA integrity was checked with RNA Kit on Agilent 5300 Fragment Analyzer (Agilent Technologies, Palo Alto, CA, USA). RNA sequencing libraries were prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina following the manufacturer’s instructions (NEB, Ipswich, MA, USA). Briefly, mRNAs were first enriched with Oligo(dT) beads. Enriched mRNAs were fragmented according to the manufacturer’s instructions. First-strand and second-strand cDNAs were subsequently synthesized. cDNA fragments were end-repaired and adenylated at 3’ends, and universal adapters were ligated to cDNA fragments, followed by index addition and library enrichment by limited-cycle PCR. Sequencing libraries were validated using NGS Kit on the Agilent 5300 Fragment Analyzer (Agilent Technologies, Palo Alto, CA, USA), and quantified by using Qubit 4.0 Fluorometer (Invitrogen, Carlsbad, CA). The sequencing libraries were multiplexed and loaded on the flow cell on the Illumina NovaSeq 6000 instrument according to the manufacturer’s instructions. The samples were sequenced using a 2x150 Pair-End (PE) configuration v1.5. Image analysis and base calling were conducted by the NovaSeq Control Software v1.7 on the NovaSeq instrument. Raw sequence data (.bcl files) generated from Illumina NovaSeq was converted into fastq files and de-multiplexed using Illumina bcl2fastq program version 2.20. One mismatch was allowed for index sequence identification. Heatmap generation was achieved using the R package pHeatmap. The scale is relative gene expression showed as rLog-transformed normalized counts.
2.4. GO and Pathway Enrichment Analysis
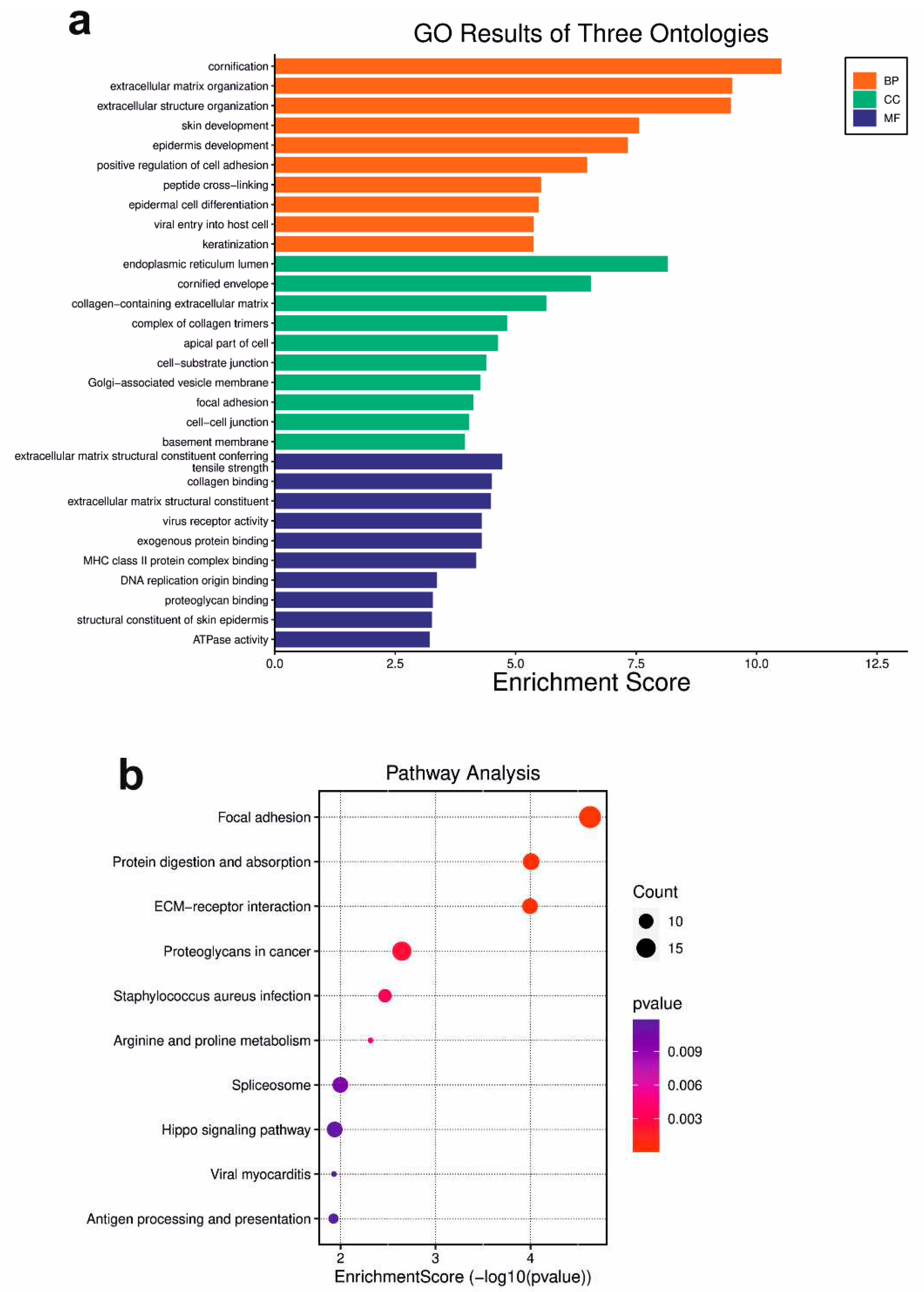
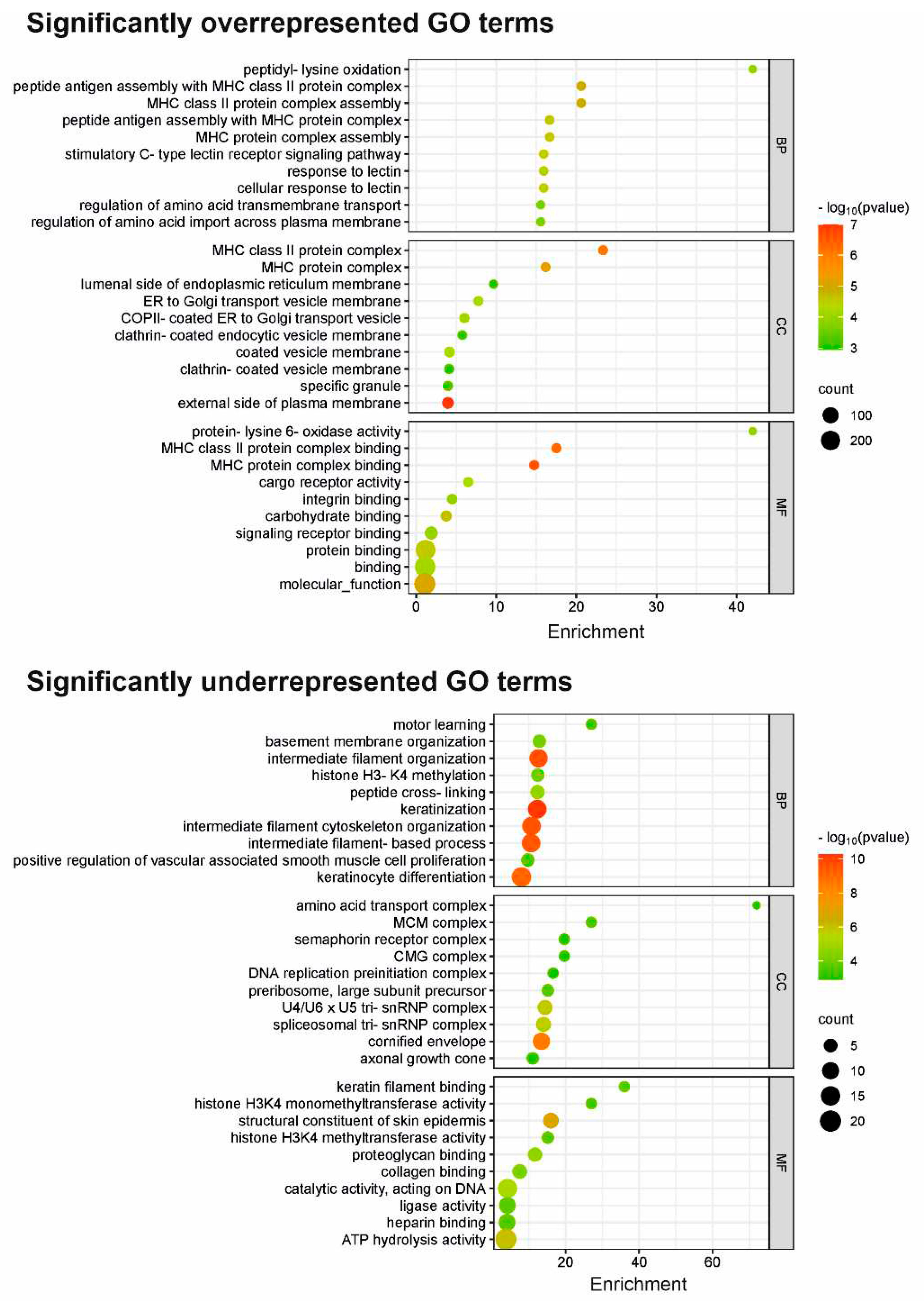
The gene ontology (GO) analysis was performed on the statistically significant set (padj < 0.05) of genes by implementing the software GeneSCF v.1.1-p2. The goa_human GO list was used to cluster the set of genes based on their biological processes and determine their statistical significance. Significantly DEGs were clustered by their GO and the enrichment of GO terms was tested using Fisher exact test (GeneSCF v1.1-p2). Up to 40 GO terms are significantly enriched with an adjusted p-value less than 0.05 in the DEG sets. Additionally, the Gene Ontology (GO) database (
http://geneontology.org/) was employed in a separate analysis approach to characterize gene products based on their associated biological processes (BP), cellular compartments (CC), and molecular functions (MF) for the 599 differentially expressed genes (DEGs) with statistical significance (non-adj p-value less than 0.05) in the DEGs dataset (
Table S1). The same approach was also done for the subsets of underrepresented and overrepresented DEGs, respectively. The top 10 GO terms from each category (BP, CC, and MF) were extracted and incorporated in various enrichment plots (SRplot - Free online GO:
http://bioinformatics.com.cn/en).
2.5. Reverse Transcription-Quantitative PCR (RT-qPCR)
RT-qPCR assays were performed as described previously [
3,
33] with minor modifications. Briefly, mRNA was purified directly from cells using the Dynabeads™ mRNA DIRECT™ Purification Kit (61012; ThermoFisher Scientific, UK) and then used directly in RT-qPCR reaction containing qPCRBIO SyGrene 1-Step Go Lo-ROX (PB25.31-12; PCRBiosystems, UK) and gene-specific primers for one-step reverse transcription and qPCR to quantify gene expression in the LightCycler 480 qPCR system (Roche, UK). Thermocycling begins at 45ºC for 10 mins (for reverse transcription) followed by 95ºC for 30s prior to 45 cycles of amplification at 95ºC for 1s, 60ºC for 1s, 72ºC for 1s, 78ºC for 1s (data acquisition). A ‘touch-down’ annealing temperature intervention (66ºC starting temperature with a stepwise reduction of 0.6ºC/cycle; 8 cycles) was introduced prior to the amplification step to maximise primer specificity. Melting analysis (95ºC for 30s, 75ºC for 30s, 75-99ºC at a ramp rate of 0.57ºC/s) was performed at the end of qPCR amplification to validate single product amplification in each well. Relative quantification of mRNA transcripts was calculated based on the second derivative maximum algorithm (Roche). Primer sequences are provided in
Supplementary Table S2. All target genes were normalised to two stable reference genes.
2.6. Immunofluorescence
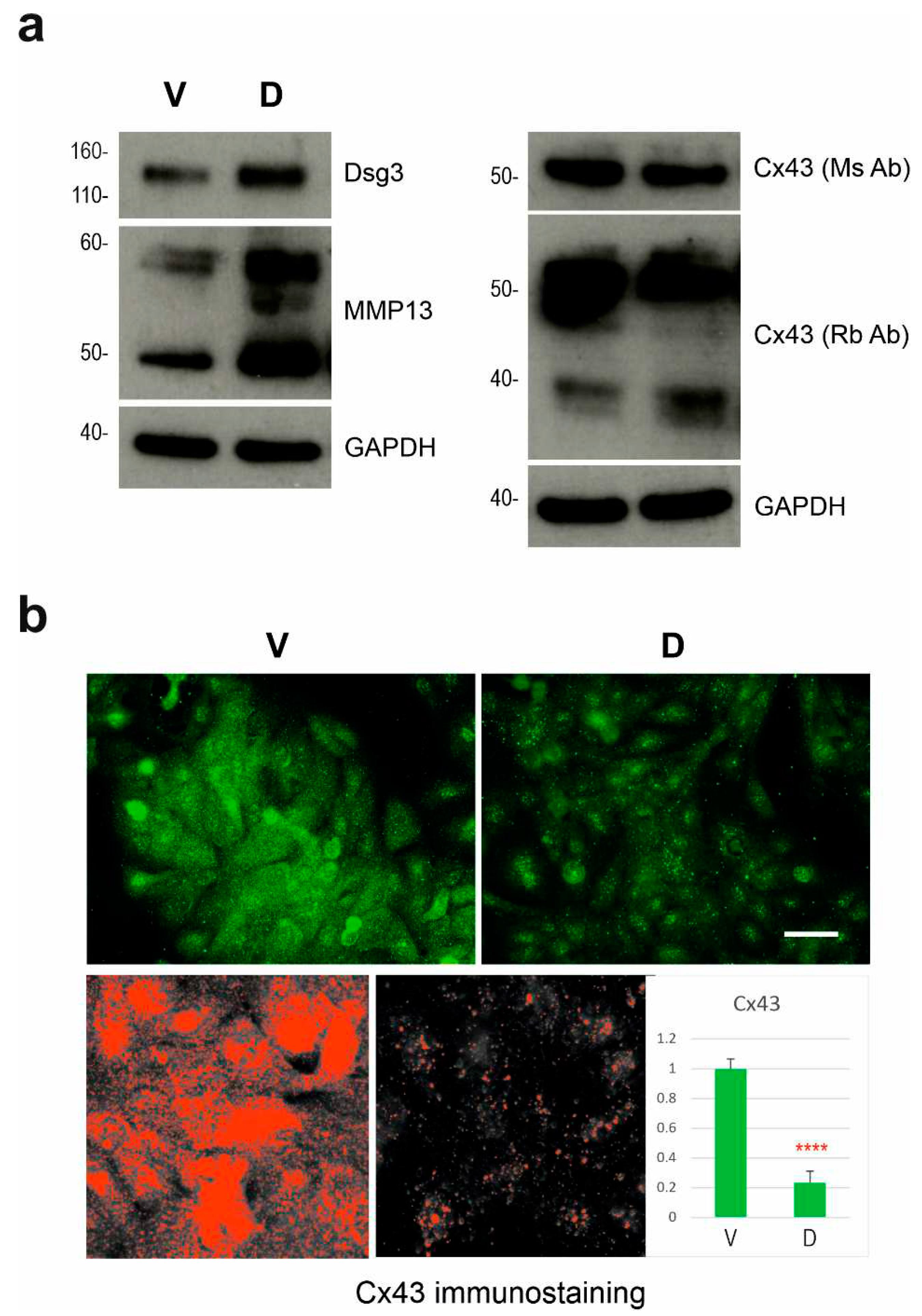
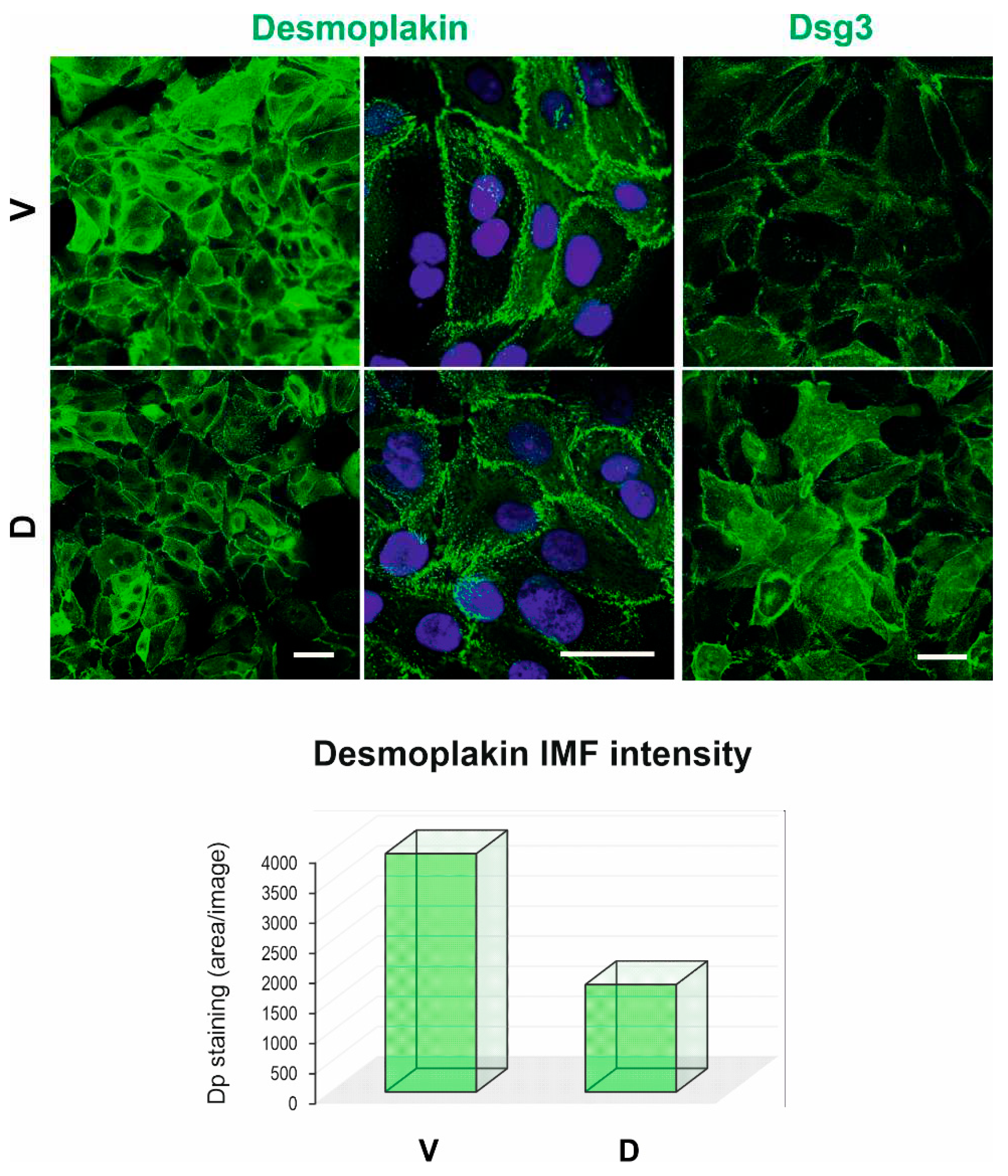
Cells were seeded and grown on coverslips overnight before fixation in either 3.6% formaldehyde (for Cx43 staining, Sigma: C6219, rabbit anti-Cx43, gift from Diana Blaydon) in PBS or ice-cold methanol (for desmoplakin staining, mouse monoclonal 115F, gift from David Garrod) for 10 min. The samples fixed with formaldehyde were subsequently permeabilized with 0.1% Triton X-100 for 5 min. After blocking with 10% goat serum (Sigma) for 20 min, coverslips were incubated with the primary and secondary antibodies, respectively, each lasting for 1 hour and washed 3 times at room temperature. Finally, coverslips were counterstained with DAPI for 8–10 min and were mounted on slides before image acquisition with Leica DM5000 Epi-Fluorescence Microscope or Zeiss 710 Laser Scanning Confocal Microscope (Blizard Institute Core Facility). Images were analyzed with FIJI Software (version 1.53). Student’s t-test was used to determine the p-values. p<0.05 was considered statistically significant between two group comparisons.
2.7. Western Blotting Analysis
This method was also described in our previous study [
3]. Briefly, cells grown to fresh confluence in a 6-well plate were extracted with sodium dodecyl sulfate (SDS) Laemmli sample buffer (0.5 M Tris-Cl pH 6.8, 4% SDS, 20% Glycerol; 10% (v/v) 2-mercaptoethanol was added after protein assay). Protein concentration for each sample was determined by the DC Protein Assay (Bio-Rad, Hercules, CA, USA), and equal amounts of protein samples were loaded and resolved by SDS/PAGE, and then transferred to a nitrocellulose membrane. After blocking the nonspecific binding sites on the membrane for 30 min in blocking buffer (5% w/v nonfat dry milk in TTBS containing 0.1% Tween 20), the membrane was incubated with primary antibody against specific proteins (Abcam: ab3208, mouse anti-MMP13 antibody (gift from Michael Allen); Sigma: C6219, rabbit anti-Cx43 and mouse anti-Cx43, clone 4E6.2, gifts from Diana Blaydon) at appropriate dilutions overnight at 4°C. After three washes in TTBS, the membrane was incubated with the secondary antibody conjugated with HRP (rabbit-SAB3700846, mouse-A0168; Sigma) at appropriate dilutions in blocking buffer for 1 hour. After three washes, the membrane was subjected to ECL Western Blotting Substrate (Thermo Fisher Scientific). The membrane was then exposed to Amersham Hyperfilm ECL (VWR, Leicestershire, UK) and developed in an AGFA Curix 60 Developer (Blizard Institute Core Facility) in a dark room to detect target proteins.
2.8. Transmission Electron Microscopy (TEM)
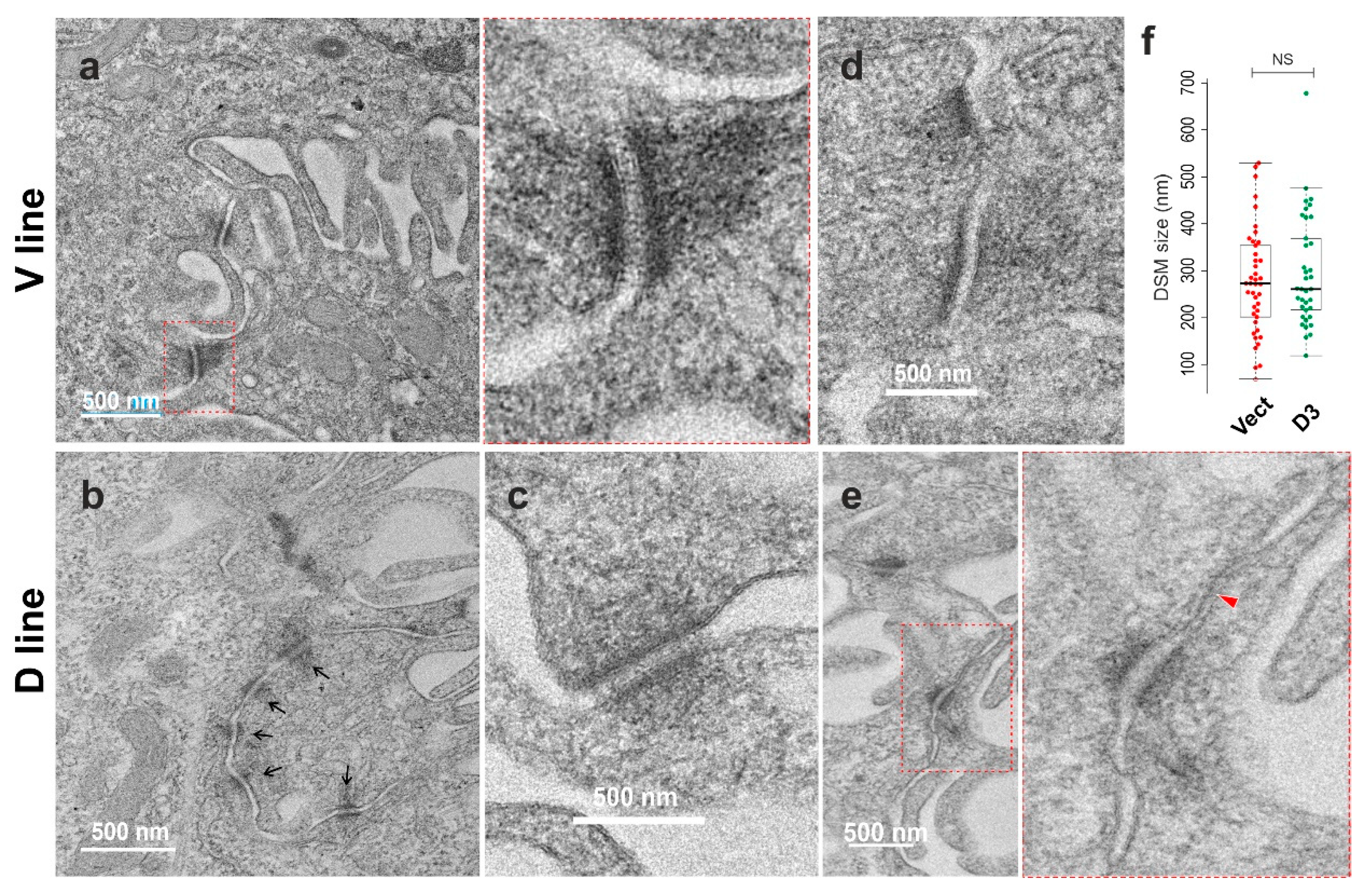
The sample was embedded in Araldite resin (Agar Scientific) and ultrathin sectioned for TEM analysis in JEM1400F (JEOL) at 120 kV. Images taken with Rio 16 CMOS camera (Gatan).
4. Discussion
DSG3 exhibits increased expression in SCC across various organ systems, yet a complete understanding of its intricate biological effects remains unexplored. Our prior in vitro study found that heightened DSG3 expression in oral SCC H413 cells restrains synchronized cell migration by inducing YAP phosphorylation and subsequently attenuating its transcriptional activity in the nucleus and this is achieved via inhibiting EGFR signalling [
3]. RNA-Seq, a comprehensive technique capturing the entire transcriptome, offers insights into gene expression, functional diversity, and disease mechanisms. The study aimed to uncover differentially expressed genes (DEGs) between DSG3-overexpressing and vector control-expressing oral SCC cells (H413) [
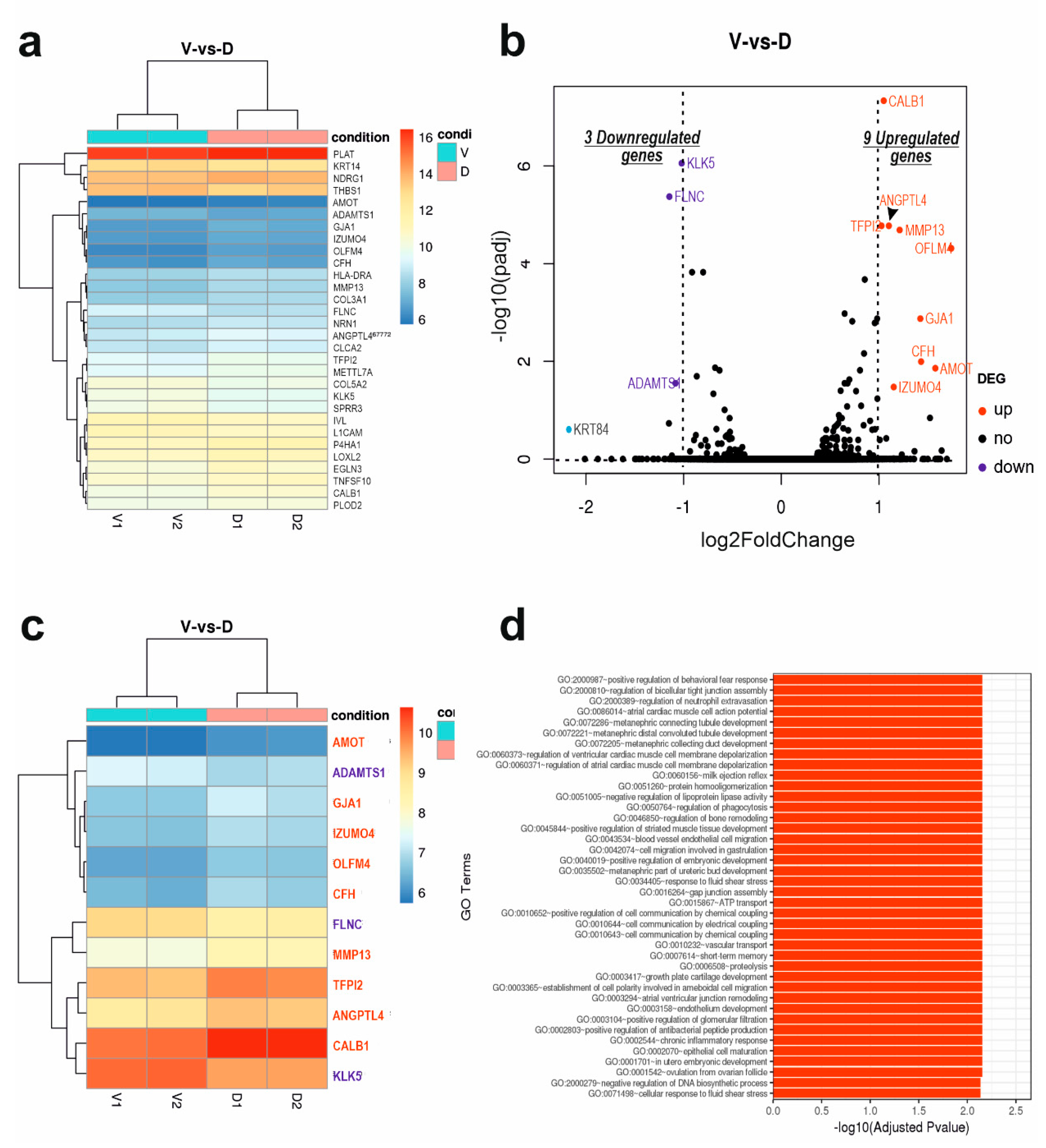
1]. The analysis identified 599 DEGs, with 286 genes down-regulated and 313 up-regulated, which are associated with DSG3 expression. These DEGs are involved in various biological processes, including collagen formation, cell adhesion, immune response, amino acid transport, keratinocyte function, and more.
The study unveils DSG3's pivotal role in critical cellular processes like cell adhesion, collagen formation, and ECM organization, primarily through the integrin-mediated focal adhesion pathway. We confirm DSG3's active promotion of MMP-13 expression, an enzyme that degrades ECM components, including key collagens found in skin, tendons, and cartilage, crucial for tissue integrity. MMPs transcend normal physiology, influencing cancer progression. Heightened MMP-13 expression, especially in cancer, facilitates tumour invasion into nearby tissues. MMP-13's enzymatic activity reshapes the tumour microenvironment, favouring tumour advancement. Research indicates elevated MMP-13 in various cancers is linked to increased invasiveness, metastasis, and poor prognosis [
29]. Thus, targeting MMP-13 holds promise for anti-cancer therapies. Interestingly, DSG3-overexpressing cells display enriched peptidyl lysine oxidation (GO:0018057), vital for collagen synthesis and tissue strength. This process hydroxylates lysine in procollagen, forming cross-links that bolster collagen-rich tissues. Moreover, the study highlights integrins, essential signalling receptors, in the DSG3 context. Integrins drive intracellular cascades upon ligand binding, supporting adhesion, migration, and other vital cellular processes. This bears relevance for normal functions and diseases like cancer metastasis. Unravelling DSG3's impact on focal adhesion and integrin complex assembly awaits further exploration in future research.
GJA1, a member of the highly conserved connexin family, encodes the protein connexin-43 (Cx43). Connexins form hexameric complexes called connexons on the cell membrane, acting as hemichannels for direct communication and ion/molecule exchange between neighbouring cells. This gap junction function is crucial for various physiological processes, including skin development, cornified envelope formation related to skin barrier function, and overall skin homeostasis. Disruptions in gap junctions are implicated in diverse diseases, including cancers [
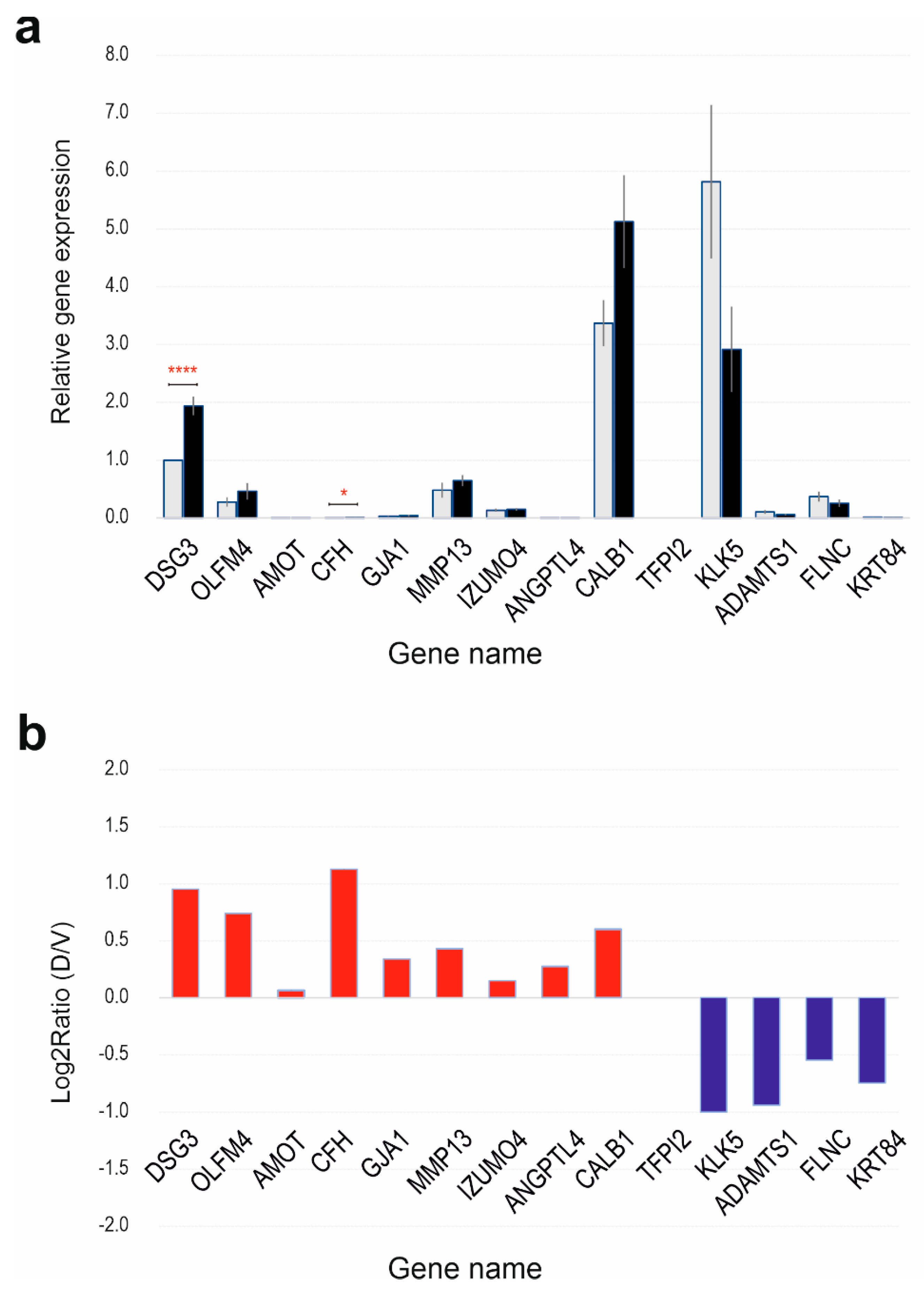
1]. Cx43 notably facilitates gap junction-mediated intercellular communication in the skin, contributing to processes like epidermal maturation (GO:0002070~epithelial cell maturation). Applying rigorous criteria (padj < 0.05 and log2 fold change > 1), we identified, for the first time, GJA1 as one of the 9 upregulated DEGs in the context of DSG3 influence (GO:0016264~gap junction assembly). Enhanced GJA1 expression was confirmed using RT-qPCR. However, Western blot analysis revealed a modest decrease in Cx43 protein levels in DSG3-overexpressing cells compared to control and this finding was further corroborated by immunostaining and quantification of CX43 expression. This finding is in line with the notion that connexins function as cancer suppressor [
1]. Intriguingly, Cx43 distribution in H413 cells appeared concentrated in the cytoplasm and nucleus, rather than the cell periphery, suggesting a potential irregular gap junction function or the specific gap junction-independent mechanism operating in this oral cancer cell line that awaits further illucidations [
1].
While DSG3 is known to enhance junction assembly as expected, analysis of biological processes and functions reveals that DSG3 appears to inhibit intermediate filament organization (GO:0045109; GO:0045104; GO:0045103), keratinocyte differentiation (GO:0030216; GO:0009913), cornified envelope formation (GO:0001533), skin development (GO:0031424; GO:0043588), and hair cycle (GO:0042633). Such inhibitions are associated with the downregulation of KLK5 (kallikrein-related peptidase 5) and other differentiation markers (such as IVL) linked to DSG3 expression. KLK5 plays a role in processing profilaggrin, contributing to keratinocyte differentiation and the functional skin barrier. All these concepts listed here are interconnected, collectively influencing keratinocyte differentiation and the protective skin barrier. Skin development is intricate, involving multiple layers, cell types, and structures. The cornified envelope, formed during late keratinocyte differentiation, serves as a vital shield against water loss and pathogens, contributing to overall skin barrier function. Intermediate filaments are part of the cytoskeleton, providing structural support for keratinocytes and maintaining epidermal integrity. Notably, our previous research indicated DSG3 overexpression does not evoke significant enhancement in cell-cell adhesion strength in H413 cells compared to controls [
3]. Additionally, in this study, we conducted transmission electron microscopy to investigate the morphology of intercellular junctions, in particular gap junction and desmosome. While the characteristic gap junctions were scarce in both samples, scattered desmosomes were detectable along plasma membrane projections. We noticed no apparent increase in desmosome frequency in D cells, though occasional clusters were observed, with lower electron density than control samples. Nevertheless, although the desmosomes appeared slightly smaller in D cells, there was no indication of statistical significance for desmosome size between the two conditions. Consistent with the downregulation of KRT14 (Keratin 14), immunostaining for Dp, which anchors keratin intermediate filaments to desmosomes, showed reduced Dp expression in D compared to V control cells, suggesting less mature desmosomes in DSG3-overexpressing cells than V control. Collectively, integrating RNA-Seq, bioinformatics, and laboratory experimental work prompted us to propose that while DSG3 facilitates junction assembly, it may not be necessarily essential for junction maturation.
The study identifies changes in several genes with their roles in cancer being controversial. It showed an association between DSG3 expression and AMOT, a component of the Hippo network. While AMOT family members typically promote cancer cell proliferation and invasion in most cancers, they have opposing effects in lung cancer [
22]. The regulation of YAP by AMOT remains unclear, with literature controversies [
22]. Other DSG3-associated cancer-related genes include upregulated OLFM4 (anti-apoptotic factor), downregulated ADAMTS1 (anti-angiogenic activity), downregulated KRT84 (cancer suppressor in oral SCC) [
21] and FLNC [
32]. OLFM4, olfactomedin-4, is reported to be significantly increased in head and neck squamous cell carinoma (75% tested), suggesting a potential biomarker in this type of cancer [
24]. ADAMTS1 can have either pro-tumourigenic or anti-tumour progressive role depending on the cell context [
32]. KRT84 has been implicated as a tumour supporessor and good prognostic indicator for oral SCC. Low KRT84 expression level is shown to have inferior overall survival independently of multiple factors [
21]. Filamin (FLNC), an actin-binding protein of the filamin family and anchoring several transmembrane proteins to the actin cytoskeleton, serves diverse structural and signalling roles [
38]. It plays contrasting roles in contractile myocytes and non-contractile cancer cells. In contractile myocytes, FLNC is linked with Hoppo, regulating migration and differentiation. Conversely, in cancer, FLNC promotes cell proliferation and migration [
18]. In general, these findings suggest a potential oncogenic role of DSG3 in oral SCC, as proposed previously [
7,
8,
9]. Another interesting finding from this study is that DSG3 plays a significant role in MHC complex assembly and binding (GO:0002399, GO:0002501, GO:0002396, GO:0042613, GO:0042611), pointing to its involvement in immune regulation, as demonstrated in pemphigus autoimmune blistering disease [
13,
27,
31].