1. Introduction
Hearing loss is a common sensory disorder affecting more than 1.5 billion people worldwide to some degree, including 430 million with moderate or severe hearing loss [
1,
2]. Hereditary factors or predispositions account for approximately 50% of hearing loss [
3,
4]. Genetic hearing loss includes syndromic and non-syndromic types, with marked genetic heterogeneity [
5,
6]. Of the genetic cases, about 70% are non-syndromic deafness, of which around 80% are inherited as an autosomal recessive trait [
4]. To date, 124 genes have been identified for non-syndromic hearing loss and 77 for autosomal recessive non-syndromic hearing loss [
7]. The most common cause of non-syndromic hearing loss is the mutations in the gap junction protein beta 2 (
GJB2) gene (OMIM: 121011), which encodes the gap junction protein connexin 26 (Cx26) on chromosome 13q12.11 [
8,
9,
10]. Based on our systematic research, we suggest that developmental disorders of the organ of Corti in the cochlea due to Cx26 loss are a significant contributor to GJB2-associated hearing loss [
11,
12,
13,
14].
Since hearing loss related to
GJB2 was first described in 1997 [
15,
16,
17], more than 300 pathogenic variants of
GJB2 have been reported [
10,
16]. In different regions of the world, the spectrums and frequencies of mutations in
GJB2 vary widely [
16]. The mutant allele of c.35delG is prevalent in populations of Europe and the Middle East, c.235delC is common in East Asia, and p.W24X is mainly found in India [
16]. In addition, c.167delT and p.R143W are found in Ashkenazim and Ghanaians, respectively [
16]. The p.V37I, a specific variant of
GJB2, is also common among deaf patients in East Asia [
16]. However, the classification of this missense variant has been controversial [
18]. The p.V37I variant was initially reported as a benign polymorphism because of its high prevalence among people with normal hearing [
19]. Later, p.V37I was identified as homozygous or in trans with the known pathogenic
GJB2 mutations in affected individuals [
10,
20,
21]. Currently, p.V37I is considered a pathogenic mutation with incomplete penetrance [
18], and individuals with the homozygous mutation is associated with various hearing phenotypes [
10,
22]. Similarly, the other
GJB2 variants show diverse phenotypes, ranging from mild to profound [
8,
23], which largely depend on the genotype [
8,
10,
24]. Patients with biallelic "truncating" mutations that completely block protein expression generally have severe to profound hearing loss, whereas patients with at least one "non-truncating" mutant allele have a milder hearing threshold due to impaired but not inactivated protein function [
8,
10,
24]. However, even among patients with the same genotype of the
GJB2 mutation, auditory phenotypes show great variability [
8,
10,
25]. Therefore, further investigation of genotype-phenotype correlations is needed to guide clinical evaluation and genetic counseling of patients with hearing loss associated with the
GJB2 mutations.
Given the dominant role of GJB2 mutations in hearing loss and their genetic heterogeneity and phenotypic diversity, it is important to analyze the frequency distribution and genotype-phenotype correlation of GJB2 mutations in hearing loss patients for genetic diagnosis and counseling. This study aimed to: (1) analyze the frequencies of genotypes and alleles of GJB2 mutations in the Chinese population from the Dongfeng Tongji (DFTJ) cohort, and (2) investigate the audiogram shapes in patients with hearing loss under different genotypes of p.V37I and c.235delC mutations in the GJB2 gene.
2. Materials and Methods
2.1. Study Population
The DFTJ cohort is a prospective study based on retired employees of Dongfeng Motor Corporation, launched in September 2008 in Shiyan City, Hubei Province, China [
26]. The baseline survey was conducted between September 2008 and June 2010, with subsequent follow-up every five years [
26]. A total of 27,009 participants completed baseline questionnaires, physical examinations, and blood sample collections. Between April and October 2013, 25,978 participants completed the first follow-up. Pure tone audiometry (PTA) was first added in 2013, with 11,513 participants in the baseline population undergoing the test [
27]. The approval of the DFTJ cohort protocol has been obtained from the Medical Ethics Committee of the School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, and Dongfeng General Hospital, Dongfeng Motor Corporation [
26]. Written informed consent was obtained from each participant.
For the purpose of genetic analysis related to hearing loss, nine common GJB2 mutations in the Chinese population were selected. This study included 9912 participants with complete data on nine GJB2 mutations and audiometry. After removing those with missing data for the age variable (n = 2), 9910 participants were used for the final analysis.
2.2. Measurement of Hearing
In a special soundproof room at Dongfeng General Hospital, participants underwent pure-tone tests by professional audiologists using a calibrated pure-tone audiometer (Micro-DSP ZD21) after an otological examination. Air conduction thresholds at 0.5, 1, 2, 4, and 8 kHz were recorded for each ear and coded as a maximum when the maximum was unresponsive [
28]. Hearing loss was defined as an average threshold at 0.5, 1, 2, and 4 kHz (PTA
0.5–4 kHz) greater than 25 decibels of hearing level (dB HL) in the better ear [
29]. The severity of hearing loss was categorized as mild (25 dB HL < PTA
0.5–4 kHz ≤ 40 dB HL), moderate (40 dB HL < PTA
0.5–4 kHz ≤ 60 dB HL), and severe-profound (PTA
0.5–4 kHz > 60 dB HL).
Based on Liu's [
25] classification criteria, the audiogram shapes of 5742 participants with hearing loss were classified as follows: sloping, where the difference between the mean thresholds at 4 kHz and 8 kHz and the mean thresholds at 0.5 kHz and 1 kHz was greater than 15 dB HL; flat, where the difference between the thresholds at 0. 5 to 8 kHz was 15 less than dB HL; mid-frequency U-shaped, where the difference between the worst hearing thresholds at mid-frequency and those at lower and higher frequencies was greater than 15 dB HL; ascending, where the difference between the low-frequency thresholds and the high-frequency thresholds was greater than 15 dB HL; and residual, where only residual hearing was present at lower frequencies. Those not belonging to any of the above audiogram shapes were classified as "other". Due to the relatively small sample size of patients with mid-frequency U-shaped (0.8%) and ascending (3.7%) hearing loss, the two types were combined into the "special" type.
2.3. Genotyping
Nine
GJB2 variants or single nucleotide polymorphisms were selected as follows: p.G4D (rs111033222), p.V27I (rs2274084), c.*84T>C (rs3751385), p.V37I (rs72474224), c.235delC (rs80338943), p.E114G (rs2274083), p.T123N (rs111033188), p.F191L (rs397516878), and p.I203T (rs76838169). These variants were genotyped in a genome-wide association study scan of the DFTJ cohort using Affymetrix Genome-Wide Human SNP Array 6.0 chips and Illumina Infinium Omni Zhong Hua-8 Chips [
30]. Detailed descriptions of the genotyping and quality control processes are available elsewhere [
31,
32].
2.4. Statistical Analysis
The categorical variables were expressed as numbers with percentages, and the continuous variables as means with standard deviations. Comparisons of differences in genotype and allele frequencies between groups were conducted using the chi-squared test or Fisher's exact test. The threshold for significance was set at a two-sided P-value of less than 0.05. All statistical analyses were done using R software (version 4.2.3, R Foundation for Statistical Computing).
3. Results
3.1. Basic Characteristics of the Study Population
Table 1 shows the characteristics of the total sample. A total of 9910 participants were included in the study, including 52.4% (n = 5197) female and 47.6% (n = 4713) male, respectively. The mean age was 67.2 years, ranging from 38 to 92 years. In total, 5742 participants had hearing loss, including 66.3% (n = 3806) mild, 27.5% (n = 1581) moderate, and 6.2% (n = 355) severe to profound.
3.2. Frequencies of the Genotypes and Alleles of the Mutations in the GJB2 Gene
Table 2 shows the genotype and allele frequencies of the nine mutations of the
GJB2 gene in the cohort and between the hearing-impaired and normal groups. Based on the variant annotation in the ClinVar database (
https://www.ncbi.nlm.nih.gov/clinvar/), two are reported as pathogenic variants p.V37I and c.235delC); five are benign or likely benign variants (p.V27I, c.*84T>C, p.E114G, p.T123N, and p.I203T); one is uncertain (p.F191L); and the last is a conflicting interpretation of pathogenicity (p.G4D). We found a significant difference (
P-value < 0.001) in the genotype frequency distribution of p.V37I between the case-control groups (
Table 2). Compared to the normal hearing group, the frequency of homozygous genotype was higher in the hearing loss group (0.1% in the normal hearing group and 0.5% in the hearing loss group). The homozygous mutation in p.V37I affected a wide range of frequencies, and the detailed characteristics and pure-tone thresholds of the 31 participants with homozygous genotypes in the cohort were shown in
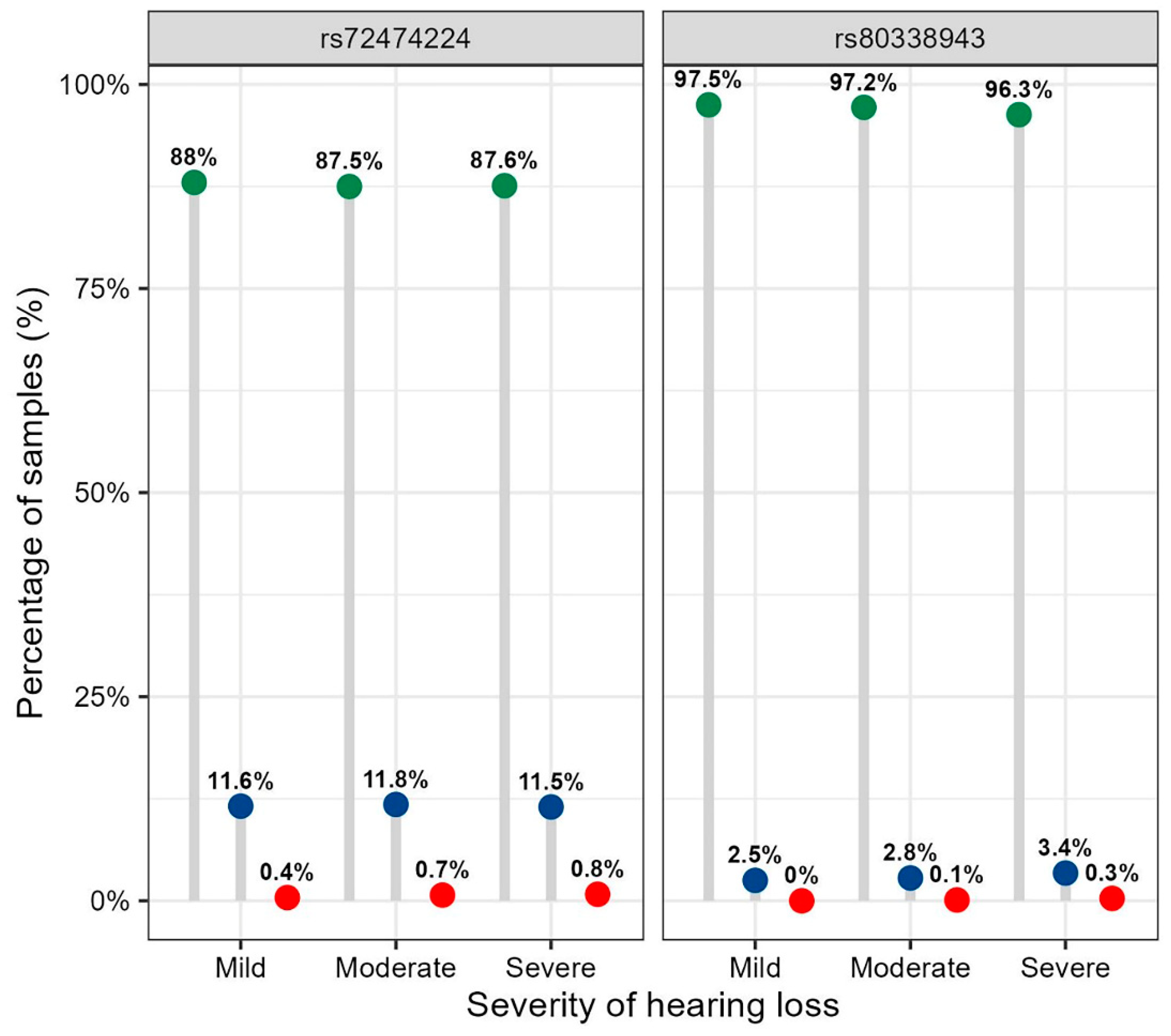
Table 3. The frequency distribution of p.V37 I alleles was nearly the same between the case-control groups (6.0% in controls and 6.3% in the hearing loss group), but not significantly different. Furthermore, the frequency of the p.V37I homozygous genotype increased with hearing threshold in 5742 participants with hearing loss (0.4% in mild, 0.7% in moderate, and 0.8% in severe-profound) (
Figure 1), but no significant difference was observed between groups (
P = 0.369). Likewise, no statistical difference was found for the frequency distribution of p.V37I alleles across hearing loss severity groups (
P = 0.274) (
Figure 2).
However, regarding c.235delC, the most common mutation reported in the Chinese population with hearing loss, the frequency distributions of its genotype and allele were not found to differ significantly between controls and cases (
Table 2). The carrier frequency of c.235delC was 1.1% in the general population controls (
Table 2). Among the 5742 patients with hearing loss, the carrier frequency was 1.4%, and only two patients were identified with homozygous mutations (
Table 2), with proportions of 0.1% and 0.3% in moderate and severe-profound hearing loss, respectively (
Figure 1). Additionally, there was no significant difference in the allele frequency of c.235delC between the different hearing loss severity groups (
P = 0.661) (
Figure 2).
Moreover, among the five benign variants, we found statistically significant differences in genotype and allele frequencies of c.*84T>C between case-control groups (
Table 2). The allele frequency of c.*84T>C was comparable between the case-control groups (53.9% in the control group and 52.5% in the hearing loss group), and a similar distribution was observed for the other four benign variants (p.V27I, p.E114G, p.T123N, p.I203T) (
Table 2). There were no statistical differences in genotype and allele frequencies for the remaining two variants (p.G4D and p.F191L) (
Table 2).
3.3. Distribution of Audiogram Shapes for the Different Genotypes of rs72474224 (p.V37I) and rs80338943 (c.235delC)
Figure 3a shows the distribution of audiogram shapes in the overall hearing loss group: 66.6% were sloped, 11.6% were flat, 4.5% were special, 6.7% were residual, and 10.6% were other. We analyzed the audiogram shape distributions of the two pathogenic variants (p.V37I, and c.235delC) under different genotypes. As shown in
Figure 3b, the p.V37I and c.235delC variants presented various audiogram shapes, mainly showing sloping and flat types. The proportions of the sloping type in the p.V37I heterozygous and homozygous genotypes were 70% and 82.1%, and the proportions of the flat type were 10% and 7.1%. For the c.235delC variant, the proportions of the sloping phenotype were 66% in the heterozygous genotypes and 100% in the homozygous genotypes; the flat phenotype was 14.4% in the heterozygous genotype.
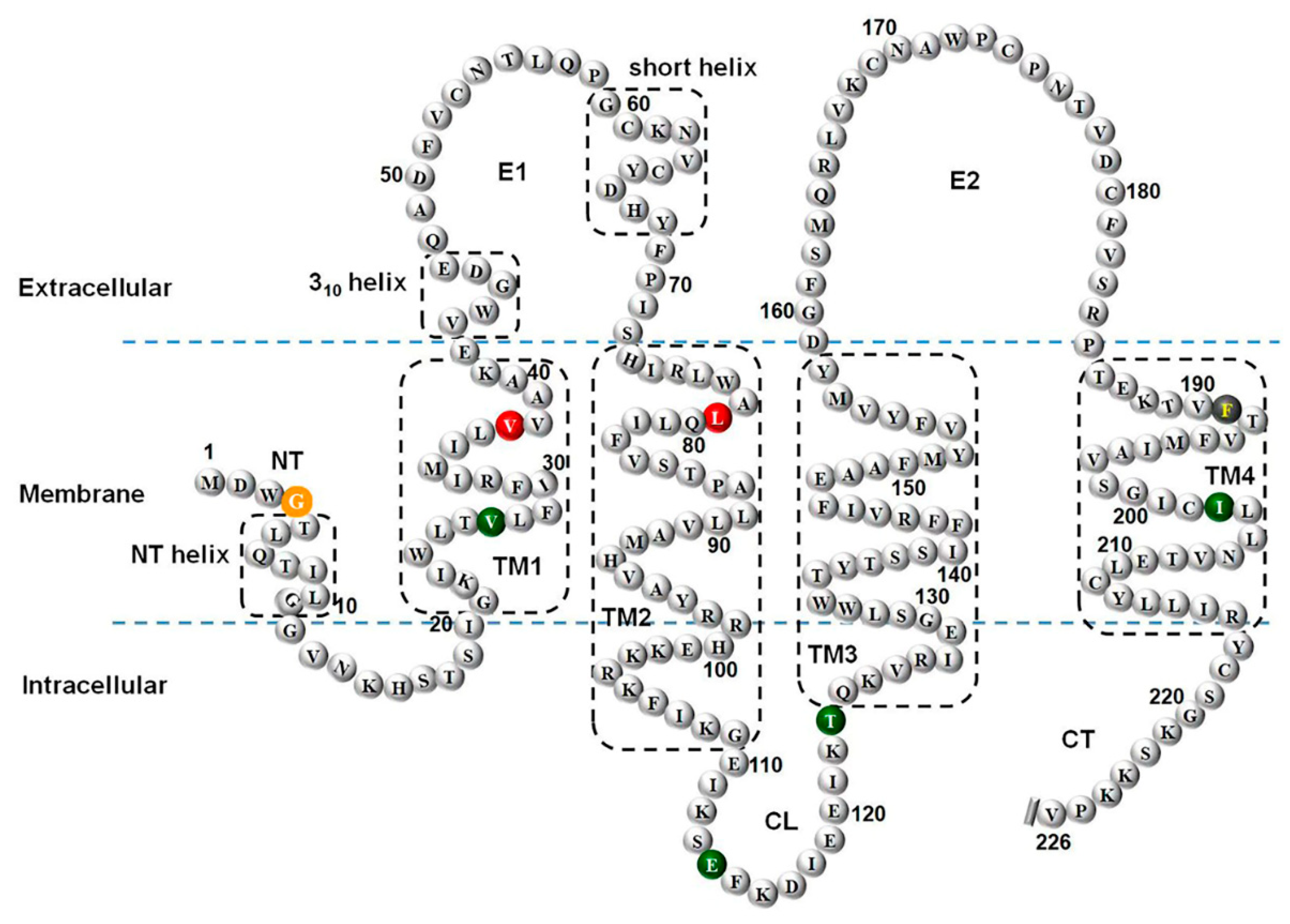
3.4. Positions of the Variants in the Cx26 Protomer
Figure 4 shows the positions of eight variants in the Cx26 protomer. The c.*84T>C, located in the 3' UTR region of the GJB2 gene, was not shown in the figure.
4. Discussion
In this study of 9910 participants from the DFTJ cohort, we described an overview of the genotype and allele frequency distributions of nine variants in the GJB2 gene, and analyzed the audiogram phenotypic profiles of two pathogenic variants for the hearing loss group. A significant difference in genotype frequencies between cases and controls was observed for the variants c.*84T>C and p.V37I, while the other seven variants (including c.235delC) were not statistically different. Homozygous mutations in p.V37I and c.235delC exhibited different degrees of hearing loss and various shapes of audiogram, mainly sloping and flat. This study was a large-scale population analysis of GJB2-related mutations (including 4168 participants with normal hearing and 5742 patients with hearing loss), and contained objective pure-tone audiometric measurements that could effectively characterize the hearing phenotype.
Consistent with our expectations, we found that the genotype frequencies of the p.V37I variant in
GJB2 were statistically significantly distributed between cases and controls. The frequency of homozygous genotypes in the hearing impaired group was 0.5%, lower than that reported in other Chinese populations in a recent study [
22], where p.V37I homozygous genotypes accounted for 2.6% of patients with hearing loss (there were eleven p.V37I homozygous genotypes in the eighty-eight patients with mild to moderate hearing loss and fourteen in the eight hundred and fifty-seven patients with severe to profound hearing loss). A possible explanation for this difference could be the different populations included. The age of the subjects in our study was generally much older, which might not exclude the contribution of aging to the hearing loss phenotype. In addition, we observed that the allele frequencies of p.V37I were approximately equal between the normal and impaired hearing groups, with no significant difference. This might be because most of the alleles were contributed by unaffected heterozygote carriers, as observed in our study where the proportion of p.V37I heterozygotes approximated between control and case groups (
Table 2). Accordingly, it is more appropriate to compare genotype frequencies rather than allele frequencies in case-control analyses to interpret the variants related to autosomal recessive inheritance patterns in the
GJB2 gene [
18]. In agreement with several previous studies [
10,
22,
33], we observed that patients with the p.V37I homozygous genotype displayed different levels of hearing loss. Notably, Chai et al. further investigated potentially pathogenic variants in fourteen patients with severe to profound hearing loss of the p.V37I homozygous genotype, and they identified another pathogenic variant of
CDH23 coexisting with the p.V37I homozygous mutation in one patient [
22]. Therefore, when p.V37I homozygous mutations are identified in patients with severe to profound hearing loss, there may be concurrent mutations in other genes or other alternative causes, such as infection or aging [
18,
22,
33]. However, because of the lack of such data, alternative etiologies of p.V37I homozygous mutations in patients with severe to profound hearing loss were not investigated in our study, and further studies are needed to elucidate them.
The mutation c.235delC has been reported to be the most prevalent
GJB2-associated variant in Chinese patients with hearing loss [
4,
16,
34,
35]. In addition, Dai et al. [
34] reported that the frequency distributions of 235delC homozygous and heterozygous genotypes in patients with hearing loss varied significantly in different regions of China, ranging from 0% to 14.7% for the homozygous genotype and 1.7% to 16.1% for the heterozygous genotype. In general, the 235delC mutation is more commonly found in northern China [
16]. In our study, we observed two hearing-impaired patients with c.235delC homozygous genotypes, showing moderate and severe to profound hearing loss, respectively. Participants in the DFTJ cohort were retired employees of Dongfeng Motor Corporation who had normal hearing before entering work. However, it should be noted that individuals with severe hearing loss would not be hired. This restriction on hearing performance might explain the inconsistent frequency of c.235delC homozygous mutations observed in our study compared to other studies. The 235delC variant in the
GJB2 gene causes a frameshift mutation that prematurely terminates translation and produces a non-functional gap junction protein [
36,
37]. Patients with the c.235delC homozygous genotype usually present with severe to profound prelingual hearing loss [
25,
38,
39]. However, other studies have shown that patients with the c.235delC homozygous mutation also show mild to moderate hearing phenotypes [
35,
39]. Consistent with these findings, one of the patients in our study with the c.235delC homozygous genotype had moderate hearing loss, suggesting that the hearing phenotype of patients with the c.235delC homozygous mutation was diverse.
We found that the genotype and allele frequencies of c.*84T>C were statistically different between the normal and impaired hearing groups. The c.*84T>C is located in the 3'UTR region of the
GJB2 gene and has been identified as a benign variant. Technological advances in sequencing and many other assays have facilitated the elucidation of the noncoding genome [
40]. Many noncoding elements have been found to be linked to hearing loss, and the identification of noncoding variants in hearing loss is expected to improve diagnostic rates [
40]. The genotype and allele frequency of the c.*84T>C variant reported in our study may provide a molecular epidemiologic basis for future studies on non-coding region variants. In addition, two previous studies reported a significant association of the c.*84T>C with noise-induced hearing loss [
41]. We observed that the allele frequencies of four benign variants (p.V27I, p.E114G, p.T123N, p.I203T) were similar between cases and controls, whereas another study reported that their allele frequencies were greater in controls than in patients [
5].
In agreement with a previous study [
10], the audiogram shapes of the p.V37I and c.235delC variants presented various audiometric configurations in this study, with a dominance of sloping and flat types. Similarly, Liu et al. found that audiogram shapes in most patients with
GJB2-related hearing loss were generally residual, sloping, or flat, but similar to those observed in hearing impaired patients without the
GJB2 mutation, suggesting a possible poor correlation between the type of
GJB2 mutation and audiogram shape [
25].
5. Conclusions
To sum up, we observed a statistically significant distribution of genotype frequencies of the GJB2 variants c.*84T>C and p.V37I between cases and controls in a Chinese population in the large-scale DFTJ cohort, but not for the c.235delC variant. Patients with p.V37I and c.235delC homozygous mutations exhibited diverse hearing loss severities and audiogram shapes. These findings could provide evidence for genetic diagnosis and counseling for hearing loss.
Author Contributions
Y.S., W.K., L.Y., S.C., X.W. and X.L. designed the study. L.Y. analyzed the data and prepared the original draft. Y.S., M.H., W.K., L.Y., S.C,. X.W. and X.L. revised and edited the paper. Y.S., M.H., and W.K. supervised the work. Y.S. acquired funding for the work. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 82071508), and the National Key Research and Development Program of China (No. 2021YFF0702303).
Informed Consent Statement
All procedures conducted in this study involving human participants were in accordance with the ethical standards of the National Research Committee and the 1964 Declaration of Helsinki. The approval of the DFTJ cohort protocol has been obtained from the Medical Ethics Committee of the School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, and Dongfeng General Hospital, Dongfeng Motor Corporation. Written informed consent was obtained from each participant.
Data Availability Statement
Requests may be addressed to the corresponding authors.
Acknowledgments
The contributions of all the participants, the staff of the Dongfeng Central Hospital, and all the members of the study team are greatly appreciated.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
Cx26: Connexin 26; DFTJ, Dongfeng Tongji; PTA, Pure tone audiometry; SD, Standard deviation; SNPs, Single nucleotide polymorphisms.
References
- WHO. World report on hearing; World Health Organization: Geneva, 2021. [Google Scholar]
- Collaborators, G.B.D.H.L. Hearing loss prevalence and years lived with disability, 1990-2019: findings from the Global Burden of Disease Study 2019. Lancet 2021, 397, 996–1009. [Google Scholar] [CrossRef]
- Morton, N.E. Genetic epidemiology of hearing impairment. Annals of the New York Academy of Sciences 1991, 630, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.B.; Chen, Z.B.; Wei, Q.J.; Lu, Y.J.; Xing, G.Q.; Cao, X. Single nucleotide polymorphisms and haplotypes analysis of DFNB1 locus in Chinese sporadic hearing impairment population. Chinese medical journal 2009, 122, 1549–1553. [Google Scholar]
- Dai, P.; Yu, F.; Han, B.; Liu, X.; Wang, G.; Li, Q.; Yuan, Y.; Liu, X.; Huang, D.; Kang, D.; et al. GJB2 mutation spectrum in 2,063 Chinese patients with nonsyndromic hearing impairment. J Transl Med 2009, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Grillo, A.P.; de Oliveira, F.M.; de Carvalho, G.Q.; Medrano, R.F.; da Silva-Costa, S.M.; Sartorato, E.L.; de Oliveira, C.A. Single nucleotide polymorphisms of the GJB2 and GJB6 genes are associated with autosomal recessive nonsyndromic hearing loss. Biomed Res Int 2015, 2015, 318727. [Google Scholar] [CrossRef] [PubMed]
- G, V.C.; RJH, S. Hereditary Hearing Loss Homepage. Available online: https://hereditaryhearingloss.org (accessed in September, 2023).
- Snoeckx, R.L.; Huygen, P.L.; Feldmann, D.; Marlin, S.; Denoyelle, F.; Waligora, J.; Mueller-Malesinska, M.; Pollak, A.; Ploski, R.; Murgia, A.; et al. GJB2 mutations and degree of hearing loss: a multicenter study. American journal of human genetics 2005, 77, 945–957. [Google Scholar] [CrossRef]
- Morton, C.C.; Nance, W.E. Newborn hearing screening--a silent revolution. N Engl J Med 2006, 354, 2151–2164. [Google Scholar] [CrossRef]
- Chen, P.Y.; Lin, Y.H.; Liu, T.C.; Lin, Y.H.; Tseng, L.H.; Yang, T.H.; Chen, P.L.; Wu, C.C.; Hsu, C.J. Prediction Model for Audiological Outcomes in Patients With GJB2 Mutations. Ear and hearing 2020, 41, 143–149. [Google Scholar] [CrossRef]
- Chen, S.; Xie, L.; Xu, K.; Cao, H.Y.; Wu, X.; Xu, X.X.; Sun, Y.; Kong, W.J. Developmental abnormalities in supporting cell phalangeal processes and cytoskeleton in the Gjb2 knockdown mouse model. Disease models & mechanisms 2018, 11. [Google Scholar] [CrossRef]
- Chen, S.; Xu, K.; Xie, L.; Cao, H.Y.; Wu, X.; Du, A.N.; He, Z.H.; Lin, X.; Sun, Y.; Kong, W.J. The spatial distribution pattern of Connexin26 expression in supporting cells and its role in outer hair cell survival. Cell death & disease 2018, 9, 1180. [Google Scholar] [CrossRef]
- Liu, X.Z.; Jin, Y.; Chen, S.; Xu, K.; Xie, L.; Qiu, Y.; Wang, X.H.; Sun, Y.; Kong, W.J. F-Actin Dysplasia Involved in Organ of Corti Deformity in Gjb2 Knockdown Mouse Model. Frontiers in molecular neuroscience 2021, 14, 808553. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Xu, K.; Xie, L.; Chen, S.; Sun, Y. The Reduction in Microtubule Arrays Caused by the Dysplasia of the Non-Centrosomal Microtubule-Organizing Center Leads to a Malformed Organ of Corti in the Cx26-Null Mouse. Biomedicines 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Kelsell, D.P.; Dunlop, J.; Stevens, H.P.; Lench, N.J.; Liang, J.N.; Parry, G.; Mueller, R.F.; Leigh, I.M. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 1997, 387, 80–83. [Google Scholar] [CrossRef]
- Chan, D.K.; Chang, K.W. GJB2-associated hearing loss: systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope 2014, 124, E34–53. [Google Scholar] [CrossRef]
- Kecskeméti, N.; Szönyi, M.; Gáborján, A.; Küstel, M.; Milley, G.M.; Süveges, A.; Illés, A.; Kékesi, A.; Tamás, L.; Molnár, M.J.; et al. Analysis of GJB2 mutations and the clinical manifestation in a large Hungarian cohort. Eur Arch Otorhinolaryngol 2018, 275, 2441–2448. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Oza, A.M.; Del Castillo, I.; Duzkale, H.; Matsunaga, T.; Pandya, A.; Kang, H.P.; Mar-Heyming, R.; Guha, S.; Moyer, K.; et al. Consensus interpretation of the p.Met34Thr and p.Val37Ile variants in GJB2 by the ClinGen Hearing Loss Expert Panel. Genetics in medicine : official journal of the American College of Medical Genetics 2019, 21, 2442–2452. [Google Scholar] [CrossRef]
- Kelley, P.M.; Harris, D.J.; Comer, B.C.; Askew, J.W.; Fowler, T.; Smith, S.D.; Kimberling, W.J. Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. American journal of human genetics 1998, 62, 792–799. [Google Scholar] [CrossRef]
- Abe, S.; Usami, S.; Shinkawa, H.; Kelley, P.M.; Kimberling, W.J. Prevalent connexin 26 gene (GJB2) mutations in Japanese. Journal of medical genetics 2000, 37, 41–43. [Google Scholar] [CrossRef]
- Wilcox, S.A.; Saunders, K.; Osborn, A.H.; Arnold, A.; Wunderlich, J.; Kelly, T.; Collins, V.; Wilcox, L.J.; McKinlay Gardner, R.J.; Kamarinos, M.; et al. High frequency hearing loss correlated with mutations in the GJB2 gene. Human genetics 2000, 106, 399–405. [Google Scholar] [CrossRef]
- Chai, Y.; Chen, D.; Sun, L.; Li, L.; Chen, Y.; Pang, X.; Zhang, L.; Wu, H.; Yang, T. The homozygous p.V37I variant of GJB2 is associated with diverse hearing phenotypes. Clinical genetics 2015, 87, 350–355. [Google Scholar] [CrossRef]
- Xiang, J.; Sun, X.; Song, N.; Ramaswamy, S.; Abou Tayoun, A.N.; Peng, Z. Comprehensive interpretation of single-nucleotide substitutions in GJB2 reveals the genetic and phenotypic landscape of GJB2-related hearing loss. Human genetics 2023, 142, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.K.; Schrijver, I.; Chang, K.W. Connexin-26-associated deafness: phenotypic variability and progression of hearing loss. Genetics in medicine : official journal of the American College of Medical Genetics 2010, 12, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Z.; Pandya, A.; Angeli, S.; Telischi, F.F.; Arnos, K.S.; Nance, W.E.; Balkany, T. Audiological features of GJB2 (connexin 26) deafness. Ear and hearing 2005, 26, 361–369. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, J.; Yao, P.; Li, X.; He, M.; Liu, Y.; Yuan, J.; Chen, W.; Zhou, L.; Min, X.; et al. Cohort Profile: the Dongfeng-Tongji cohort study of retired workers. International journal of epidemiology 2013, 42, 731–740. [Google Scholar] [CrossRef]
- Wang, D.; Zhou, Y.; Ma, J.; Xiao, L.; Cao, L.; Zhou, M.; Kong, W.; Wang, Z.; Li, W.; He, M.; et al. Association between shift work and hearing loss: The Dongfeng-Tongji cohort study. Hearing research 2019, 384, 107827. [Google Scholar] [CrossRef]
- Wang, D.; Xiao, Y.; Feng, X.; Wang, B.; Li, W.; He, M.; Zhang, X.; Yuan, J.; Yi, G.; Chen, Z.; et al. Association of occupational noise exposure, bilateral hearing loss with atherosclerotic cardiovascular disease risk in Chinese adults. International journal of hygiene and environmental health 2021, 235, 113776. [Google Scholar] [CrossRef]
- Report of the Informal Working Group on Prevention of Deafness and Hearing Impairment Programme Planning, Geneva, 18-21 June 1991. Available online: https://apps.who.int/iris/handle/10665/58839.
- Jiang, J.; He, S.; Liu, K.; Yu, K.; Long, P.; Xiao, Y.; Liu, Y.; Yu, Y.; Wang, H.; Zhou, L.; et al. Multiple plasma metals, genetic risk and serum complement C3, C4: A gene-metal interaction study. Chemosphere 2022, 291, 132801. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Wu, C.; Xu, J.; Guo, H.; Yang, H.; Zhang, X.; Sun, J.; Yu, D.; Zhou, L.; Peng, T.; et al. A genome wide association study of genetic loci that influence tumour biomarkers cancer antigen 19-9, carcinoembryonic antigen and α fetoprotein and their associations with cancer risk. Gut 2014, 63, 143–151. [Google Scholar] [CrossRef]
- Long, P.; Liu, X.; Li, J.; He, S.; Chen, H.; Yuan, Y.; Qiu, G.; Yu, K.; Liu, K.; Jiang, J.; et al. Circulating folate concentrations and risk of coronary artery disease: a prospective cohort study in Chinese adults and a Mendelian randomization analysis. Am J Clin Nutr 2020, 111, 635–643. [Google Scholar] [CrossRef]
- Huang, S.; Huang, B.; Wang, G.; Yuan, Y.; Dai, P. The Relationship between the p.V37I Mutation in GJB2 and Hearing Phenotypes in Chinese Individuals. PLoS One 2015, 10, e0129662. [Google Scholar] [CrossRef]
- Dai, P.; Yu, F.; Han, B.; Yuan, Y.; Li, Q.; Wang, G.; Liu, X.; He, J.; Huang, D.; Kang, D.; et al. The prevalence of the 235delC GJB2 mutation in a Chinese deaf population. Genetics in medicine : official journal of the American College of Medical Genetics 2007, 9, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, Y.; Xu, J.; Che, T.; Li, L.; Yang, T.; Wu, H. Molecular epidemiology of Chinese Han deaf patients with bi-allelic and mono-allelic GJB2 mutations. Orphanet journal of rare diseases 2020, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Z.; Xia, X.J.; Ke, X.M.; Ouyang, X.M.; Du, L.L.; Liu, Y.H.; Angeli, S.; Telischi, F.F.; Nance, W.E.; Balkany, T.; et al. The prevalence of connexin 26 ( GJB2) mutations in the Chinese population. Human genetics 2002, 111, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.F.; Lin, H.C.; Tsai, C.L.; Hsu, Y.C. GJB2 mutation spectrum in the Taiwanese population and genotype-phenotype comparisons in patients with hearing loss carrying GJB2 c.109G>A and c.235delC mutations. Hearing research 2022, 413, 108135. [Google Scholar] [CrossRef]
- Zhao, F.F.; Ji, Y.B.; Wang, D.Y.; Lan, L.; Han, M.K.; Li, Q.; Zhao, Y.; Rao, S.; Han, D.; Wang, Q.J. Phenotype-genotype correlation in 295 Chinese deaf subjects with biallelic causative mutations in the GJB2 gene. Genetic testing and molecular biomarkers 2011, 15, 619–625. [Google Scholar] [CrossRef]
- Wang, H.; Gao, Y.; Guan, J.; Lan, L.; Yang, J.; Xiong, W.; Zhao, C.; Xie, L.; Yu, L.; Wang, D.; et al. Phenotypic Heterogeneity of Post-lingual and/or Milder Hearing Loss for the Patients With the GJB2 c.235delC Homozygous Mutation. Frontiers in cell and developmental biology 2021, 9, 647240. [Google Scholar] [CrossRef]
- Avraham, K.B.; Khalaily, L.; Noy, Y.; Kamal, L.; Koffler-Brill, T.; Taiber, S. The noncoding genome and hearing loss. Human genetics 2022, 141, 323–333. [Google Scholar] [CrossRef]
- Miao, L.; Ji, J.; Wan, L.; Zhang, J.; Yin, L.; Pu, Y. An overview of research trends and genetic polymorphisms for noise-induced hearing loss from 2009 to 2018. Environmental science and pollution research international 2019, 26, 34754–34774. [Google Scholar] [CrossRef]
Figure 1.
Frequency distribution of the genotypes for rs72474224 (p.V37I) and rs80338943 (c.235delC) in the GJB2 gene across hearing loss severity groups. Note: The genotypes of wild, heterozygous and homozygous for rs72474224, and rs80338943 are: C/C, T/C and T/T; and AG/AG, A/AG, and A/A, respectively. The distribution of the genotypes of rs72474224 and rs80338943 among different hearing loss severity groups was tested using Fisher's exact test, with P = 0.369 and P = 0.091, respectively.
Figure 1.
Frequency distribution of the genotypes for rs72474224 (p.V37I) and rs80338943 (c.235delC) in the GJB2 gene across hearing loss severity groups. Note: The genotypes of wild, heterozygous and homozygous for rs72474224, and rs80338943 are: C/C, T/C and T/T; and AG/AG, A/AG, and A/A, respectively. The distribution of the genotypes of rs72474224 and rs80338943 among different hearing loss severity groups was tested using Fisher's exact test, with P = 0.369 and P = 0.091, respectively.
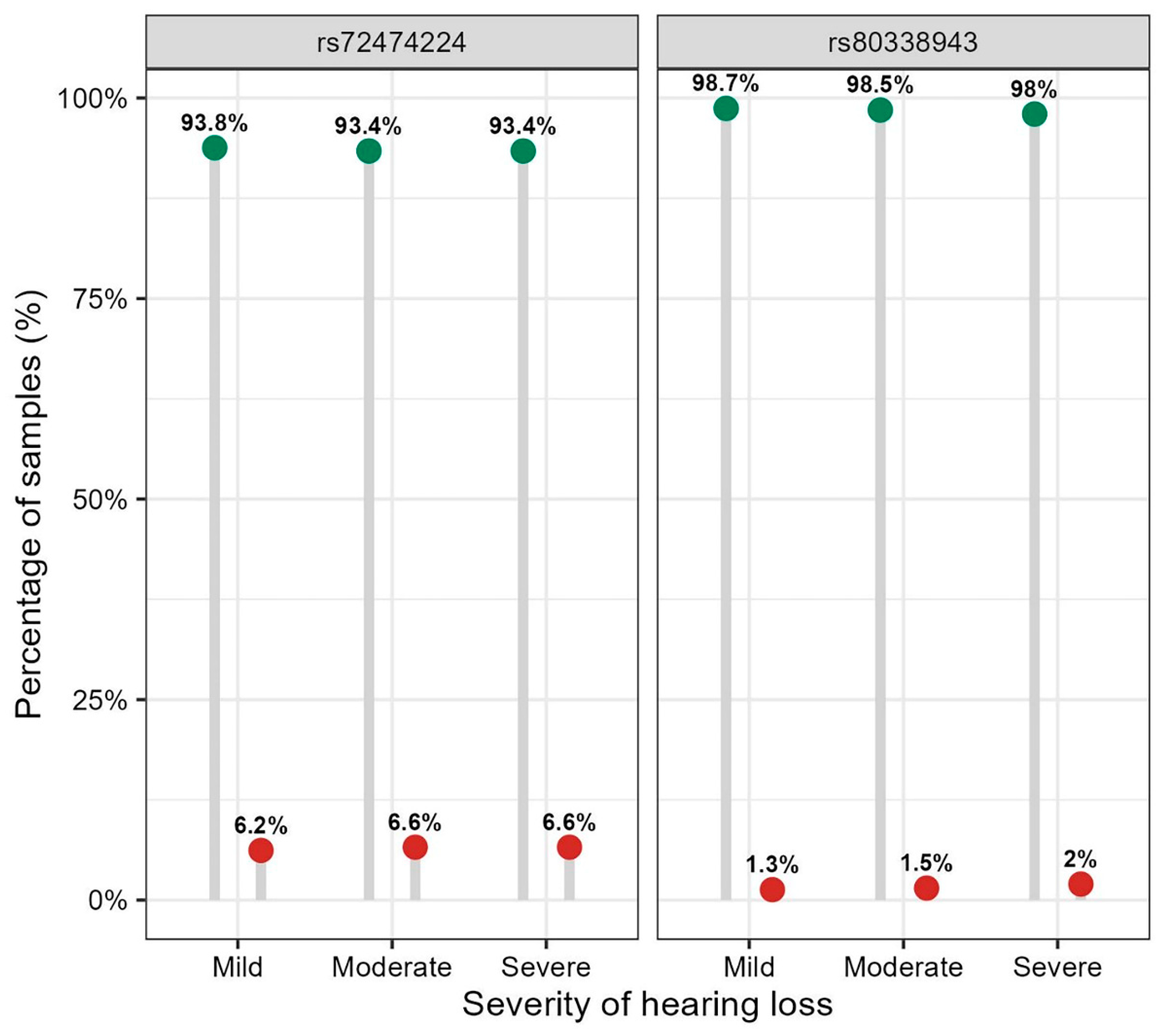
Figure 2.
Frequency distribution of the alleles for rs72474224 (p.V37I) and rs80338943 (c.235delC) in the GJB2 gene across hearing loss severity groups. Note: The wild-type and mutant alleles for rs72474224 and rs80338943 are: C and T; and AG as well as A. The distribution of the alleles of rs72474224 and rs80338943 in different hearing loss severity groups was tested using Chi-squared test, and P values were: P = 0.274; and P = 0.661, respectively.
Figure 2.
Frequency distribution of the alleles for rs72474224 (p.V37I) and rs80338943 (c.235delC) in the GJB2 gene across hearing loss severity groups. Note: The wild-type and mutant alleles for rs72474224 and rs80338943 are: C and T; and AG as well as A. The distribution of the alleles of rs72474224 and rs80338943 in different hearing loss severity groups was tested using Chi-squared test, and P values were: P = 0.274; and P = 0.661, respectively.
Figure 3.
The audiogram shapes (a) in 5742 participants with hearing loss and (b) in the different genotypes of rs72474224 (p.V37I) and rs80338943 (c.235delC). Note: The genotypes of wild, heterozygous and homozygous for rs72474224, and rs80338943 are: C/C, T/C and T/T; and AG/AG, A/AG, and A/A, respectively. The distribution of the types of audiogram in different genotypes of rs72474224 and rs80338943 was tested using Fisher's exact test, and P values were: P = 0.324, and P = 0.576, respectively.
Figure 3.
The audiogram shapes (a) in 5742 participants with hearing loss and (b) in the different genotypes of rs72474224 (p.V37I) and rs80338943 (c.235delC). Note: The genotypes of wild, heterozygous and homozygous for rs72474224, and rs80338943 are: C/C, T/C and T/T; and AG/AG, A/AG, and A/A, respectively. The distribution of the types of audiogram in different genotypes of rs72474224 and rs80338943 was tested using Fisher's exact test, and P values were: P = 0.324, and P = 0.576, respectively.
Figure 4.
Positions of eight variants in the Cx26 protomer. Note: The eight variants are p.G4D, p.V27I, p.V37I, c.235delC, p.E114G, p.T123N, p.F191L, and p.I203T. The c.*84T>C is located in the 3' UTR region of the GJB2 gene and is therefore not included. The orange color represents conflicting variants; the green color represents benign variants; the red color represents pathogenic variants; and the grey-black color represents uncertainty. TM1 to TM4, four transmembrane domains; E1 and E2, two extracellular loops; NT, N-terminal tail; CL cytoplasmic loop; CT carboxy-tail.
Figure 4.
Positions of eight variants in the Cx26 protomer. Note: The eight variants are p.G4D, p.V27I, p.V37I, c.235delC, p.E114G, p.T123N, p.F191L, and p.I203T. The c.*84T>C is located in the 3' UTR region of the GJB2 gene and is therefore not included. The orange color represents conflicting variants; the green color represents benign variants; the red color represents pathogenic variants; and the grey-black color represents uncertainty. TM1 to TM4, four transmembrane domains; E1 and E2, two extracellular loops; NT, N-terminal tail; CL cytoplasmic loop; CT carboxy-tail.
Table 1.
Characteristics of overall participants.
Table 1.
Characteristics of overall participants.
| Characteristic |
Total (n=9910) |
| Age, mean (SD), y |
67.2 (7.6) |
| Age group (%) |
|
| <60 |
1548 (15.6) |
| 60-69 |
4765 (48.1) |
| 70-79 |
3054 (30.8) |
| ≥80 |
543 (5.5) |
| Gender (%) |
|
| Female |
5197 (52.4) |
| Male |
4713 (47.6) |
| Severity of hearing loss (%) |
|
| Normal |
4168 (42.1) |
| Mild |
3806 (38.4) |
| Moderate |
1581 (16.0) |
| Severe |
355 (3.6) |
Table 2.
Frequency distribution of genotypes and alleles of single nucleotide polymorphisms in the GJB2 gene.
Table 2.
Frequency distribution of genotypes and alleles of single nucleotide polymorphisms in the GJB2 gene.
| SNPs |
Nucleotide change |
Protein change |
Clinical significance |
Overall
(n=9910) |
Hearing loss |
P value |
Control group
(n=4168) |
Patients
group
(n= 5742) |
| rs111033222 |
c.11G>A |
p.G4D |
Conflicting |
|
|
|
|
| Genotype |
C/C |
|
|
9854 (99.4) |
4143 (99.4) |
5711 (99.5) |
0.797 a
|
| T/C |
|
|
56 (0.6) |
25 (0.6) |
31 (0.5) |
| Allele |
C |
|
|
19764 (99.7) |
8311 (99.7) |
11453 (99.7) |
0.797 a
|
| T |
|
|
56 (0.3) |
25 (0.3) |
31 (0.3) |
| rs2274084 |
c.79G>A |
p.V27I |
Benign |
|
|
|
|
| Genotype |
C/C |
|
|
4391 (44.3) |
1854 (44.5) |
2537 (44.2) |
0.680 a
|
| T/C |
|
|
4518 (45.6) |
1906 (45.7) |
2612 (45.5) |
| T/T |
|
|
1001 (10.1) |
408 (9.8) |
593 (10.3) |
| Allele |
C |
|
|
13300 (67.1) |
5614 (67.3) |
7686 (66.9) |
0.546 a
|
| T |
|
|
6520 (32.9) |
2723 (32.7) |
3798 (33.1) |
| rs3751385 |
c.*84T>C |
- |
Benign |
|
|
|
|
| Genotype |
A/A |
|
|
2157 (21.8) |
850 (20.4) |
1307 (22.8) |
0.017 a
|
| A/G |
|
|
4985 (50.3) |
2142 (51.4) |
2843 (49.5) |
| G/G |
|
|
2768 (27.9) |
1176 (28.2) |
1592 (27.7) |
| Allele |
A |
|
|
9299 (46.9) |
3842 (46.1) |
5457 (47.5) |
0.048 a
|
| G |
|
|
10521 (53.1) |
4494 (53.9) |
6027 (52.5) |
| rs72474224 |
c.109G>A |
p.V37I |
Pathogenic |
|
|
|
|
| Genotype |
C/C |
|
|
8718 (88.0) |
3674 (88.1) |
5044 (87.8) |
< 0.001 b
|
| T/C |
|
|
1161 (11.7) |
491 (11.8) |
670 (11.7) |
| T/T |
|
|
31 (0.3) |
3 (0.1) |
28 (0.5) |
| Allele |
C |
|
|
18597 (93.8) |
7839 (94.0) |
10758 (93.7) |
0.313 a
|
| T |
|
|
1223 (6.2) |
497 (6.0) |
726 (6.3) |
| rs80338943 |
c.235delC |
p.Leu79fs |
Pathogenic |
|
|
|
|
| Genotype |
AG/AG |
|
|
9665 (97.5) |
4078 (97.8) |
5587 (97.3) |
0.118 b
|
| A/AG |
|
|
243 (2.5) |
90 (2.2) |
153 (2.7) |
| A/A |
|
|
2 (0.02) |
0 (0.0) |
2 (0.03) |
| Allele |
AG |
|
|
19573 (98.8) |
8246 (98.9) |
11327 (98.6) |
0.083 a
|
| A |
|
|
247 (1.2) |
90 (1.1) |
157 (1.4) |
| rs2274083 |
c.341A>G |
p.E114G |
Benign/Likely benign |
|
|
|
|
| Genotype |
T/T |
|
|
5284 (53.3) |
2249 (54.0) |
3035 (52.9) |
0.423 a
|
| C/T |
|
|
4025 (40.6) |
1678 (40.3) |
2347 (40.9) |
| C/C |
|
|
601 (6.1) |
241 (5.8) |
360 (6.3) |
| Allele |
T |
|
|
14593 (73.6) |
6176 (74.1) |
8417 (73.3) |
0.216 a
|
| C |
|
|
5227 (26.3) |
2160 (25.9) |
3067 (26.7) |
| rs111033188 |
c.368C>A |
p.T123N |
Benign/Likely benign |
|
|
|
|
| Genotype |
G/G |
|
|
9828 (99.2) |
4134 (99.2) |
5694 (99.2) |
0.965 b
|
| T/G |
|
|
80 (0.8) |
33 (0.8) |
47 (0.8) |
| T/T |
|
|
2 (0.02) |
1 (0.02) |
1 (0.02) |
| Allele |
G |
|
|
19736 (99.6) |
8301 (99.6) |
11435 (9.6) |
0.942 a
|
| T |
|
|
84 (0.4) |
35 (0.4) |
49 (0.4) |
| rs397516878 |
c.571T>C |
p.F191L |
Uncertain |
|
|
|
|
| Genotype |
A/A |
|
|
9899 (99.9) |
4163 (99.9) |
5736 (99.9) |
0.871 b
|
| A/G |
|
|
11 (0.1) |
5 (0.1) |
6 (0.1) |
| Allele |
A |
|
|
19809 (99.9) |
8331 (99.9) |
11478 (99.95) |
0.871 b
|
| G |
|
|
11 (0.1) |
5 (0.1) |
6 (0.05) |
| rs76838169 |
c.608T>C |
p.I203T |
Benign |
|
|
|
|
| Genotype |
A/A |
|
|
9298 (93.8) |
3929 (94.3) |
5369 (93.5) |
0.289 a
|
| A/G |
|
|
598 (6.0) |
234 (5.6) |
364 (6.3) |
| G/G |
|
|
14 (0.1) |
5 (0.1) |
9 (0.2) |
| Allele |
A |
|
|
19194 (96.8) |
8092 (97.1) |
11102 (96.7) |
0.122 a
|
| G |
|
|
626 (3.2) |
244 (2.9) |
382 (3.3) |
Table 3.
Characteristics and pure tone threshold of participants with the homozygote genotype of rs72474224 (p.V37I) (n=31).
Table 3.
Characteristics and pure tone threshold of participants with the homozygote genotype of rs72474224 (p.V37I) (n=31).
| Hearing loss |
Medical ID |
Age (year) |
Gender |
Better ear (dB) |
| 0.5 kHz |
1 kHz |
2 kHz |
4 kHz |
8 kHz |
PTA
0.5-4 kHz
|
| Normal |
106846 |
58 |
Female |
15 |
15 |
5 |
10 |
10 |
11.25 |
| Normal |
105802 |
59 |
Female |
15 |
5 |
25 |
40 |
60 |
21.25 |
| Normal |
90678 |
68 |
Male |
25 |
15 |
20 |
25 |
30 |
21.25 |
| Mild |
78353 |
60 |
Male |
20 |
25 |
25 |
40 |
65 |
27.5 |
| Mild |
70091 |
61 |
Female |
25 |
25 |
20 |
50 |
45 |
30 |
| Mild |
70740 |
61 |
Female |
30 |
30 |
30 |
40 |
50 |
32.5 |
| Mild |
11011 |
63 |
Female |
15 |
15 |
50 |
50 |
70 |
32.5 |
| Mild |
4888 |
64 |
Female |
25 |
30 |
40 |
45 |
75 |
35 |
| Mild |
60195 |
64 |
Female |
25 |
20 |
30 |
40 |
70 |
28.75 |
| Mild |
107326 |
64 |
Female |
25 |
30 |
35 |
55 |
65 |
36.25 |
| Mild |
54382 |
66 |
Male |
20 |
30 |
40 |
45 |
95 |
33.75 |
| Mild |
101181 |
66 |
Male |
30 |
35 |
35 |
40 |
65 |
35 |
| Mild |
11321 |
67 |
Male |
20 |
30 |
35 |
60 |
85 |
36.25 |
| Mild |
67746 |
69 |
Female |
25 |
25 |
25 |
45 |
90 |
30 |
| Mild |
39311 |
71 |
Male |
15 |
15 |
30 |
60 |
25 |
30 |
| Mild |
59571 |
74 |
Female |
15 |
15 |
40 |
60 |
70 |
32.5 |
| Mild |
53680 |
76 |
Male |
20 |
20 |
30 |
35 |
90 |
26.25 |
| Moderate |
54632 |
56 |
Female |
35 |
35 |
35 |
60 |
90 |
41.25 |
| Moderate |
79049 |
61 |
Female |
35 |
40 |
50 |
45 |
55 |
42.5 |
| Moderate |
16099 |
67 |
Male |
20 |
30 |
60 |
70 |
85 |
45 |
| Moderate |
28532 |
67 |
Male |
35 |
35 |
60 |
70 |
80 |
50 |
| Moderate |
90158 |
68 |
Male |
40 |
50 |
50 |
65 |
55 |
51.25 |
| Moderate |
94371 |
68 |
Male |
30 |
45 |
65 |
75 |
100 |
53.75 |
| Moderate |
42832 |
77 |
Male |
15 |
30 |
60 |
70 |
90 |
43.75 |
| Moderate |
100741 |
77 |
Female |
20 |
40 |
55 |
50 |
80 |
41.25 |
| Moderate |
116341 |
78 |
Male |
40 |
40 |
50 |
60 |
85 |
47.5 |
| Moderate |
107144 |
79 |
Male |
20 |
25 |
55 |
70 |
90 |
42.5 |
| Moderate |
5826 |
80 |
Male |
40 |
50 |
65 |
85 |
90 |
60 |
| Severe |
56456 |
64 |
Male |
50 |
55 |
65 |
90 |
110 |
65 |
| Severe |
115724 |
73 |
Male |
60 |
70 |
70 |
75 |
110 |
68.75 |
| Severe |
93245 |
74 |
Male |
60 |
60 |
70 |
70 |
100 |
65 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).