Submitted:
25 September 2023
Posted:
26 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Host Plant Material
2.2. Samples of the Pathogen Population
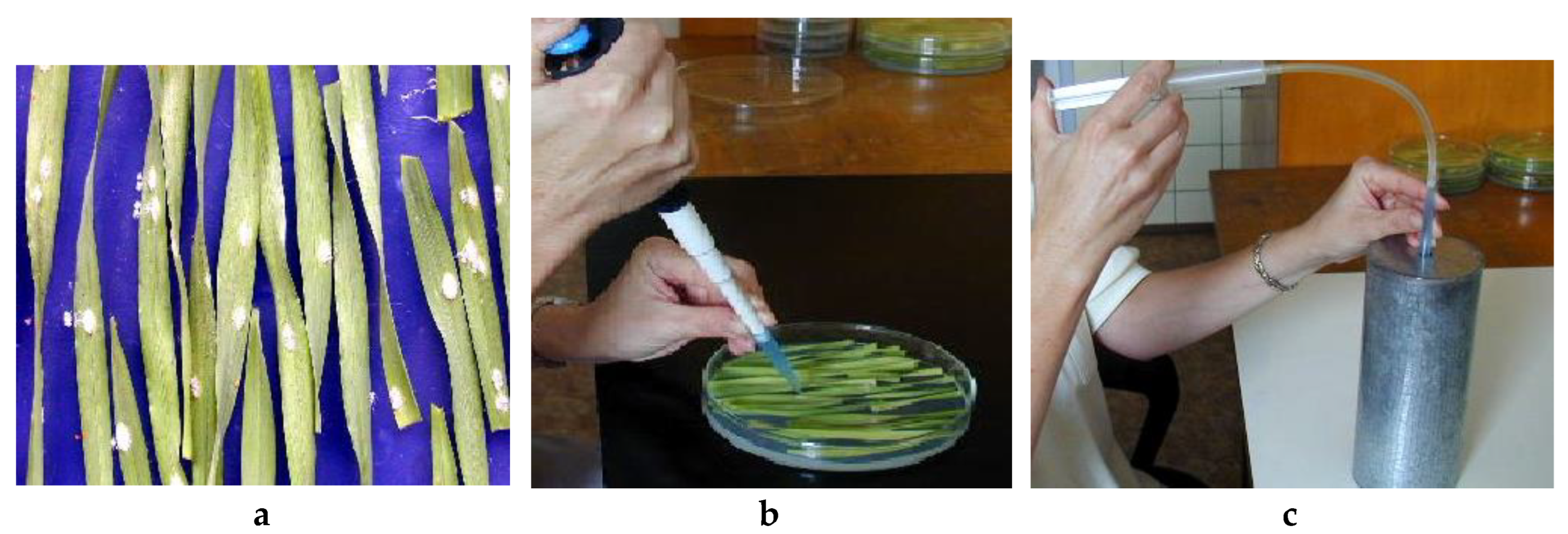
2.3. Testing Procedure
2.4. Evaluation
2.5. Pathotype Classification
3. Results
3.1. Virulence of Isolates and Virulence Frequency of the Population
3.2. Pathotype Diversity
3.3. Complexity of Virulences
3.4. Selection of Isolates for Resistance Gene Postulation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, M.; Braun, U.; Takamatsu, S.; Hambleton, S.; Shoukouhi, P.; Bisson, K.R.; Hubbard, K. Taxonomic revision of Blumeria based on multi-gene DNA sequences, host preferences and morphology. Mycoscience 2021, 62, 143–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Zhang, G.X.; Wang, F.T.; Lang, X.W.; Zhao, X.Q.; Zhu, J.H.; Hu, C.Y.; et al. Virulence variability and genetic diversity in Blumeria graminis f. sp. hordei in Southeastern and Southwestern China. Plant Dis. 2023, 107, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Zhuoma, Q.; Xu, Z.; Peng, Y.L.; Wang, M. Virulence and genetic types of Blumeria graminis f. sp. hordei in Tibet and surrounding areas. J. Fungi 2023, 9, 363. [Google Scholar] [CrossRef]
- Murray, G.M.; Brennan, J.P. Estimating disease losses to the Australian barley industry. Aust. Plant Pathol. 2010, 39, 85–96. [Google Scholar] [CrossRef]
- Marzani, Q.A.; Amin, M.M.; Fateh, S.A. Evaluation the effects of powdery mildew caused by Blumeria graminis f. sp. hordei and cultivar on the barley lodging. Eur. J. Plant Pathol. 2023, 165, 233–240. [Google Scholar] [CrossRef]
- Jensen, H.P.; Christensen, E.; Jørgensen, J.H. Powdery mildew resistance genes in 127 northwest European spring barley varieties. Plant Breed. 1992, 108, 210–228. [Google Scholar] [CrossRef]
- Dreiseitl, A. Diferences in powdery mildew epidemics in spring and winter barley based on 30-year variety trials. Ann. Appl. Biol. 2011, 159, 49–57. [Google Scholar] [CrossRef]
- Jørgensen, J.H.; Jensen, H.P. Powdery mildew resistance in barley landrace material 1. Screening for resistance. Euphytica 1997, 97, 227–233. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, H.J. Powdery mildew resistance in selections from Moroccan barley landraces. Phytoparasitica 2000, 28, 65–78. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, H.J. Selections from barley landrace collected in Libya as new sources of efective resistance to powdery mildew (Blumeria graminis f.sp. hordei). Rostl. Vyrob. 2002, 48, 217–223. [Google Scholar] [CrossRef]
- Fischbeck, G.; Schwarzbach, E.; Sobel, Z.; Wahl, I. Mildew resistance in Israeli populations of 2-rowed wild barley (Hordeum spontaneum). Z. Pflanz. 1976, 76, 163–166. [Google Scholar]
- Dreiseitl, A.; Dinoor, A. Phenotypic diversity of barley powdery mildew resistance sources. Genet. Resour. Crop Evol. 2004, 51, 251–258. [Google Scholar] [CrossRef]
- Dreiseitl, A. Heterogeneity of powdery mildew resistance revealed in accessions of the ICARDA wild barley collection. Front. Plant Sci. 2017, 8, 202. [Google Scholar] [CrossRef]
- Brown, J.K.M.; Jørgensen, J.H. A catalogue of mildew resistance genes in European barley varieties. In Integrated Control of Cereal Mildews: Virulence and Their Change, Proceedings of the Second European Workshop on Integrated Control of Cereal Mildews, Risø National Laboratory, Roskilde, Denmark, 23–25 January 1990; Jørgensen, J.H., Ed.; Risø National Laboratory: Roskilde, Denmark, 1991; pp. 263–286. [Google Scholar]
- Jørgensen, J.H. Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 1994, 13, 97–119. [Google Scholar] [CrossRef]
- Dreiseitl, A. Specific resistance of barley to powdery mildew, its use and beyond. A concise critical review. Genes 2020, 11, 971. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef]
- Praz, C.R.; Menardo, F.; Robinson, M.D.; Mueller, M.C.; Wicker, T.; Bourras, S.; Keller, B. Non-parent of origin expression of numerous effector genes indicates a role of gene regulation in host adaption of the hybrid triticale powdery mildew pathogen. Front. Plant Sci. 2018, 9, 49. [Google Scholar] [CrossRef]
- Muller, M.C.; Kunz, L.; Graf, J.; Schudel, S.; Keller, B. Host adaptation through hybridization: Genome analysis of triticale powdery mildew reveals unique combination of lineage-specific effectors. Molec. Plant-Microbe Interact. 2021, 34, 1350–1357. [Google Scholar] [CrossRef]
- Kusch, S.; Qian, J.; Loos, A.; Kuemmel, F.; Spanu, P.D.; Panstruga, R. Long-term and rapid evolution in powdery mildew fungi. Molec. Ecology Early Access. 2023. [Google Scholar] [CrossRef]
- Dreiseitl, A. Great pathotype diversity and reduced virulence complexity in a Central European population of Blumeria graminis f. sp. hordei in 2015–2017. Eur. J. Plant Pathol. 2019, 53, 801–811. [Google Scholar] [CrossRef]
- Dreiseitl, A. Virulence frequency to powdery mildew resistances in winter barley cultivars. Czech J. Genet. Plant Breed. 2008, 44, 160–166. [Google Scholar] [CrossRef]
- Czembor, H.J.; Domeradzka, O.; Czembor, J.H.; Mankowski, D.R. Virulence structure of the powdery mildew (Blumeria graminis) population occurring on triticale (x triticosecale) in Poland. J. Phytopathology 2014, 162, 499–512. [Google Scholar] [CrossRef]
- Lalosevic, M.; Jevtic, R.; Zupunski, V.; Masirevic, S.; Orbovic, B. Virulence structure of the wheat powdery mildew population in Serbia. Agronomy 2022, 12, 45. [Google Scholar] [CrossRef]
- Cieplak, M; Nucia, A.; Ociepa, T; Okon, S. Virulence structure and genetic diversity of Blumeria graminis f. sp. avenae from different regions of Europe. Plants 2022, 11, 1358. [CrossRef]
- ovmøller, M.S.; Caffier, V.; Jalli, M.; Anderson, O.; Besenhofer, G.; Czembor, J.H.; Dreiseitl, A.; Felsenstein, F.; Fleck, A.; Heinrics, F. et al. The European barley powdery mildew virulence survey and disease nursery 1993–1999. Agronomie, 2000, 20, 729−743. [CrossRef]
- Dreiseitl, A. Pathogenic divergence of Central European and Australian populations of Blumeria graminis f. sp. hordei. Ann. Appl. Biol. 2014, 165, 364–372. [Google Scholar] [CrossRef]
- Komínková, E.; Dreiseitl, A.; Malečková, E.; Doležel, J.; Valárik, M. Genetic diversity of Blumeria graminis f. sp. hordei in Central Europe and its comparison with Australian population. PLoS ONE 2016, 11, e0167099. [Google Scholar] [CrossRef]
- Dreiseitl, A. Rare virulences of barley powdery mildew found in aerial populations in the Czech Republic from 2009 to 2014. Czech J. Genet. Plant Breed. 2015, 51, 1–8. [Google Scholar] [CrossRef]
- FAOSTAT [on-line]. Fao.org/faostat [cit. 2023-08-09].
- Dreiseitl, A. Postulation of specific powdery mildew resistance genes in cereals: A widely used method and its detailed description. Pathogens 2022, 11, 284. [Google Scholar] [CrossRef]
- Dreiseitl, A.; Platz, G. Powdery mildew resistance genes in barley varieties grown in Australia. Crop Pasture Sci. 2012, 63, 997–1006. [Google Scholar] [CrossRef]
- Kølster, P.; Munk, L.; Stølen, O.; Løhde, J. Near-isogenic barley lines with genes for resistance to powdery mildew. Crop Sci. 1986, 26, 903–907. [Google Scholar] [CrossRef]
- Dreiseitl, A.; Nesvadba, Z. Powdery mildew resistance genes in single-plant progenies derived from accessions of a winter barley core collection. Plants 2021, 10, 1998. [Google Scholar] [CrossRef]
- Schwarzbach, E. A high throughput jet trap for collecting mildew spores on living leaves. Phytopathol. Z. 1979, 94, 165–171. [Google Scholar] [CrossRef]
- Torp, J.; Jensen, H.P.; Jørgensen, J.H. Powdery Mildew Resistance Genes in 106 Northwest European Spring Barley Cultivars. Year-book, 1978; Royal Veterinary and Agricultural University: Copenhagen, Denmark, 1978; pp. 75–102. [Google Scholar]
- Kosman, E.; Chen, X.; Dreiseitl, A.; McCallum, B.; Lebeda, A.; Ben-Yehuda, P.; Gultyaeva, E.; Manisterski, J. Functional variation of plant-pathogen interactions: New concept and methods for virulence data analyses. Phytopathology 2019, 109, 1324–1330. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- McVey, D.V.; Roelfs, A.P. Postulation of genes for stem rust resistance in the entries of the Fourth international winter wheat performance nursery. Crop Sci. 1975, 15, 335–337. [Google Scholar] [CrossRef]
- Gilmour, J. Octal notation for designating physiologic races of plant pathogens. Nature 1973, 242, 620. [Google Scholar] [CrossRef]
- Limpert, E.; Müller, K. Designation of pathotypes of plant pathogens. J. Phytopathol. 1994, 140, 346–358. [Google Scholar] [CrossRef]
- Herrmann, A.; Löwer, C.F.; Schachtel, G.A. A new tool for entry and analysis of virulence data for plant pathogens. Plant Pathol. 1999, 48, 154–158. [Google Scholar] [CrossRef]
- Brückner, F. Powdery mildew (Erysiphe graminis DC.) on barley. III. Investigation of physiological races of Erysiphe graminis DC. Detected in Czechoslovakia in 1960-61. Rostl. Vyr. 1963, 9, 1–8. [Google Scholar]
- Brückner, F. The finding of powdery mildew (Erysiphe graminis DC. var. hordei Marchal) race on barley: a race virulent to resistance genes Mla9 and Mla14. Ochrana Rostl. 1982, 18, 101–105. [Google Scholar]
- Dreiseitl, A. Resistance of ‘Roxana’ to powdery mildew and its presence in some European spring barley cultivars. Plant Breed. 2011, 130, 419–422. [Google Scholar] [CrossRef]
- Brückner, F. Powdery mildew (Erysiphe graminis DC.) on barley. V. The resistance of barley varieties to physiological races of Erysiphe graminis DC. detected in Czechoslovakia and the possibility to use it in breeding for resistance. Rostl. Vyr. 1964, 10, 395–408. [Google Scholar]
- Dreiseitl, A. Genes for resistance to powdery mildew in European barley cultivars registered in the Czech Republic from 2011 to 2015. Plant Breed. 2017, 136, 351–356. [Google Scholar] [CrossRef]
- Dreiseitl, A. Emerging Blumeria graminis f. sp. hordei pathotypes reveal ‘Psaknon’ resistance in European barley varieties. J. Agric. Sci. 2016, 154, 1082–1089. [Google Scholar] [CrossRef]
- Dreiseitl, A. Resistance of barley variety ‘Venezia’ and its reflection in Blumeria graminis f. sp. hordei population. Euphytica 2018, 214, UNSP 40. [Google Scholar] [CrossRef]
- Dreiseitl, A. Powdery mildew resistance genes in European barley cultivars registered in the Czech Republic from 2016 to 2020. Genes 2022, 13, 1274. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Cereals 2023. List of recommended varieties. Central Institure for Supervising and Testing in Agriculture, Brno 2023.
- Brown, J.K.M. Recombination and selection in populations of plant pathogens. Plant Pathol. 1995, 44, 279–293. [Google Scholar] [CrossRef]
- Huang, R.; Kranz, J; Welz, H.G. Virulence gene-frequency change in Erysiphe graminis f. sp. hordei due to selection by non-corresponding barley mildew resistance gene and hitchhiking. J. Phytopathology 1995, 143, 287–294. [CrossRef]
- Jahoor, A.; Stephan, U.; Fischbeck, G. Study of powdery mildew resistance gene from ´Engledow India´. Barley Genet. Newslett. 1990, 20, 41–42. [Google Scholar]
- Giese, H.; Jensen, H.P.; Jørgensen, J.H. Allelism of genes in the Ml-a locus. Barley Genet. Newslett. 1980, 10, 22–24. [Google Scholar]
- Dreiseitl, A. , Yang, J. Powdery mildew resistance in a collection of Chinese barley varieties. Genet. Resour. Crop Evol. 2007, 54, 259–266. [Google Scholar] [CrossRef]
- Dreiseitl, A.; Fowler, R.A.; Platz, G.J. Pathogenicity of Blumeria graminis f. sp. hordei in Australia in 2010 and 2011. Australas. Plant Pathol. 2013, 42, 713–721. [Google Scholar] [CrossRef]
- Rsaliyev, A.; Pahratdinova, Z.; Rsaliyev, S. Characterizing the pathotype structure of barley powdery mildew and effectiveness of resistance genes to this pathogen in Kazakhstan. BMC Plant Biol. 2017, 17, 178. [Google Scholar] [CrossRef] [PubMed]
- Zeybek, A.; Khan, M.K.; Pandey, A.; Gunel, A.; Erdogan, O.; Akkaya, M.S. Genetic structure of powdery mildew disease pathogen Blumeria graminis f. sp. hordei in the barley fields of Cukurova in Turkey. Fresenius Environ. Bull. 2017, 26, 906–912. [Google Scholar]
- Tucker, M.A.; Jayasena, K.; Ellwood, S.R.; Oliver, R.P. Pathotype variation of barley powdery mildew in Western Australia. Australas. Plant Pathol. 2013, 42, 617–623. [Google Scholar] [CrossRef]
- Hiura, U.; Heta, H. Studies on the disease resistance in barley. III. Further studies on the physiologic races of Erysiphe graminis hordei in Japan. Berichte des Ohara Instituts für landwirtschaftliche Biologie 1955, 10, 135–156. [Google Scholar]
- Bettgenhaeuser, J.; Hernández-Pinzón, I., Dawson, A.M.; Gardiner, M.; Green, P.; Taylor, J. et al. The barley immune receptor Mla recognizes multiple pathogens and contributes to host range dynamics. Nature Commun. 12, 2021, 6915.
- Roelfs, A.; McVey, D.V. Wheat stem rust races in Yaqui valley of Mexico during 1972. Plant Dis. Report. 1972, 56, 1038–1039. [Google Scholar]
- Limpert. E.; Clifford, B.; Dreiseitl, A.; Johnson, R.; Müller, K.; Roelfs, A.; Wellings, C. Systems of designation of pathotypes of plant pathogens. J. Phytopathol. 1994, 140, 359–362. [CrossRef]
- Okon, S; Cieplak, M; Kuzdralinski, A; Ociepa, T. New pathotype nomenclature for better characterisation the virulence and diversity of Blumeria graminis f. sp. avenae populations. Agronomy 2021, 11, 1852. [CrossRef]
- Kusch, S; Panstruga, R. mlo-based resistance: an apparently universal “weapon” to defeat powdery mildew disease. Molec. Plant-Microbe Interact. 2017, 30, 179–189. [CrossRef]
- Panstruga, R.; Moscou, M. What is the molecular basis of nonhost resistance? Molec. Plant-Microbe Interact. 2020, 33, 1253–1264. [CrossRef]
- Dreiseitl, A. Adaptation of Blumeria graminis f.sp. hordei to barley resistance genes in the Czech Republic in 1971-2000. Plant Soil Environ. 2003, 49, 241–248. [CrossRef]

| Section of sampling route | Distance | Number of isolates | ||||
|---|---|---|---|---|---|---|
| km | 2019 | 2021 | 2023 | Sum | ||
| Brno - Břeclav | C | 54 | 9 | 13 | 2 | 24 |
| Brno - Kroměříž | A | 68 | 9 | 9 | 17 | 35 |
| Brno - Pardubice | S | 70 | 17 | 39 | 56 | |
| Brno - Praha direction, motorway D1, 95. km | X | 95 | 12 | 9 | 12 | 33 |
| Brno - Znojmo | B | 65 | 4 | 2 | 5 | 11 |
| Kroměříž - Olomouc - Velký Újezd | R | 92 | 37 | 7 | 5 | 49 |
| Kroměříž - Otrokovice - Přerov | G | 82 | 10 | 1 | 11 | |
| Olomouc - Šumperk | F | 51 | 1 | 1 | 2 | 4 |
| Ostrava - Velký Újezd | E | 62 | 4 | 3 | 1 | 8 |
| Praha - Brno direction, motorway D1, 95. km | I | 90 | 1 | 2 | 3 | 6 |
| Praha - Chomutov | U | 75 | 12 | 12 | ||
| Praha - Karlovy Vary | L | 65 | 6 | 0 | 5 | 11 |
| Praha - Pardubice | O | 61 | 2 | 5 | 6 | 13 |
| Praha - Plzeň | K | 76 | 4 | 0 | 4 | |
| Praha - Turnov | N | 68 | 1 | 2 | 2 | 5 |
| Praha - Ústí nad Labem | M | 70 | 5 | 2 | 10 | 17 |
| Sum | 1 144 | 105 | 72 | 122 | 299 | |
| No. | Differential variety | Main | Virulence frequency | |||
|---|---|---|---|---|---|---|
| Ml gene(s) | 2019 | 2021 | 2023 | Total | ||
| 1 | Ab 1128 | ab | 0 | 0 | 0 | 0 |
| 2 | Black Russian | a2 | 0 | 0 | 0 | 0 |
| 3 | Bonita | u | 0 | 0 | 0 | 0 |
| 4 | Gopal | a5 | 0 | 0 | 0 | 0 |
| 5 | Hb-81882/83 | hb1 | 0 | 0 | 0 | 0 |
| 6 | Hb-BC1-D5 | hb2 | 0 | 0 | 0 | 0 |
| 7 | LP 1506.1.96 | a3, aTu2 | 0 | 0 | 0 | 0 |
| 8 | Nigrate | a30 | 0 | 0 | 0 | 0 |
| 9 | NORD 07017/69 | u | 0 | 0 | 0 | 0 |
| 10 | NORD 18/2622 | u | 0 | 0 | 0 | 0 |
| 11 | P13 | a23 | 0 | 0 | 0 | 0 |
| 12 | Sara | a3, aTu2 | 0 | 0 | 0 | 0 |
| 13 | SK-4770-7 | g, u | 0 | 0 | 0 | 0 |
| 14 | Spilka | u | 0 | 0 | 0 | 0 |
| 15 | Zeppelin selection | SI-1 | 0 | 1.4 | 0 | 0.3 |
| 16 | Burštyn selection | g, u | 1.0 | 1.4 | 0 | 0.7 |
| 17 | GK Metal | Ln, g, h | 1.0 | 1.4 | 0 | 0.7 |
| 18 | HMK-8 selection | g, u | 1.9 | 0 | 0.8 | 1.0 |
| 19 | Klarinette | SI-1 | 1.9 | 1.4 | 0 | 1.0 |
| 20 | KM-12/2010 | u | 0 | 1.4 | 1.6 | 1.0 |
| 21 | SZD 3894 | u | 1.0 | 2.8 | 0 | 1.0 |
| 22 | Florian | Ln | 1.0 | 4.2 | 0 | 1.3 |
| 23 | Pop | SI-1 | 0 | 2.8 | 1.6 | 1.3 |
| 24 | Dubai | u | 2.9 | 2.8 | 1.6 | 2.3 |
| 25 | SBCC097 | Sb | 3.8 | 4.2 | 0 | 2.3 |
| 26 | NORD 12101/116 | u | 1.9 | .4.2 | 2.5 | 2.7 |
| 27 | Remark | SI-1 | 0 | 2.8 | 4.9 | 2.7 |
| 28 | SI-1 | SI-1 | 0 | 4.2 | 5.7 | 3.3 |
| 29 | Bente | SI-1 | 1.9 | 4.2 | 6.6 | 4.3 |
| 30 | SU Celly | u | 1.0 | 6.9 | 5.7 | 4.3 |
| 31 | Camilla selection | SI-1 | 1.9 | 4.2 | 7.4 | 4.7 |
| 32 | SU Laubella | u | 1.0 | 6.9 | 7.4 | 5.0 |
| 33 | P08B | a9 | 11.4 | 6.9 | 7.4 | 8.7 |
| 34 | P20 | at | 17.4 | 11.1 | 5.7 | 11.0 |
| 35 | KM-1867 | u | 6.7 | 16.7 | 15.6 | 12.7 |
| 36 | Venezia selection | Ve | 2.9 | 16.7 | 26.2 | 15.7 |
| 37 | Laverda | aLv | 13.3 | 15.3 | 21.3 | 17.1 |
| 38 | P02 | a3 | 17.1 | 19.4 | 16.4 | 17.4 |
| 39 | Signal | aN81 | 23.8 | 19.4 | 22.1 | 22.1 |
| 40 | P11 | a13 | 29.5 | 25.0 | 27.9 | 27.8 |
| 41 | P23 | La | 21.0 | 34.7 | 30.3 | 28.1 |
| 42 | P17 | k1 | 41.9 | 44.4 | 44.3 | 43.5 |
| 43 | P09 | a10 | 40.0 | 54.2 | 54.1 | 47.5 |
| 44 | P19 | p1 | 5.7 | 58.3 | 77.0 | 47.5 |
| 45 | P12 | a22 | 47.6 | 45.8 | 50.0 | 48.2 |
| 46 | Alinghi | IM9 | 41.0 | 47.2 | 55.7 | 48.5 |
| 47 | Annabell | St | 49.5 | 63.9 | 40.2 | 49.2 |
| 48 | Kangoo | Ro | 69.5 | 61.1 | 43.4 | 56.9 |
| 49 | P01 | a1 | 47.6 | 56.9 | 65.6 | 57.2 |
| 50 | P21 | g | 81.9 | 86.1 | 71.3 | 78.6 |
| 51 | P04B | a7 | 80.0 | 83.3 | 88.5 | 84.3 |
| 52 | P15 | Ru2 | 73.3 | 97.2 | 91.8 | 86.6 |
| 53 | P10 | a12 | 92.4 | 90.3 | 91.8 | 91.6 |
| 54 | P03 | a6 | 96.2 | 100.0 | 100.0 | 98.7 |
| 55 | SJ123063 | SI-1 | 0 | 1.4 | 0 | 0.3 |
| 56 | SY412-329 | SI-1 | 0 | 1.4 | 0 | 0.3 |
| 57 | Landi | Ln, h | 1.0 | 4.2 | 0 | 1.3 |
| 58 | NORD 14/1116 | u | 0 | 2.8 | 1.6 | 1.3 |
| 59 | CH-666 | La | 21.0 | 34.7 | 30.3 | 27.8 |
| 60 | Diabas | a7 | 80.0 | 83.3 | 88.5 | 84.3 |
| 61 | Kompolti 4 | Ru2 | 73.3 | 97.2 | 91.8 | 86.6 |
| 62 | B-141/99 | a17 | 0 | 0 | 0 | |
| 63 | C-213/01 | a26 | 0 | 0 | 0 | |
| 64 | Hs HSY-78 x Aramir | j | 0 | 0 | 0 | |
| 65 | Hs RS 110-4 x Sonja | a29 | 0 | 0 | 0 | |
| 66 | Hs RS 137-28 x Elgina | f1 | 0 | 0 | 0 | |
| 67 | Hs RS 142-29 x Dura | a32 | 0 | 0 | 0 | |
| 68 | Hs RS 145-39 x Kiebitz B | a20 | 0 | 0 | 0 | |
| 69 | Hs RS 170-10 x Piccolo A | a25 | 0 | 0 | 0 | |
| 70 | Hs RS 42-8 x Oriol A | t | 0 | 0 | 0 | |
| 71 | Hs Diamant x 1B-86B | a19 | 1.0 | 0 | 0.6 | |
| 72 | Hs RS 170-47 x Kiebitz B | a17 | 0 | 1.4 | 0.6 | |
| 73 | E-388/01 | u | 0 | 2.8 | 1.1 | |
| 74 | Prosa | u | 10.5 | 23.6 | 15.8 | |
| 75 | KM-1998 | u | 7.6 | 31.9 | 17.5 | |
| 76 | A222 | a11 | 7.6 | 4.9 | 6.2 | |
| 77 | Meltan selection | a13, Hu2 | 16.2 | 10.7 | 13.2 | |
| 78 | Pribina | a13, Hu2 | 15.2 | 11.5 | 13.2 | |
| 79 | Souleyka | aLv | 19.0 | 34.4 | 27.2 | |
| 80 | STRG 576/15 | aLv | 21.0 | 33.6 | 27.8 | |
| 81 | Traminer | St, IM9 | 37.1 | 38.5 | 37.9 | |
| 82 | Psaknon | p1 | 4.8 | 68.8 | 39.2 | |
| 83 | Klimek | p1 | 5.7 | 72.1 | 41.4 | |
| 84 | Amazone | St | 45.7 | 38.5 | 41.8 | |
| 85 | Pionier | Ro | 72.4 | 38.5 | 54.2 | |
| 86 | Adam | mlo | 0 | 0 | 0 | |
| 87 | HOR2573 | La-H | 0 | 0 | 0 | |
| 88 | Kairyobozu-mugi | kb | 0 | 0 | 0 | |
| 89 | LG Nabuco | mlo | 0 | 0 | 0 | |
| 90 | SZD 5014A | u | 0 | 0 | 0 | |
| 91 | Focus | SI-1 | 2.8 | 0 | 1.0 | |
| 92 | NOS 111.336-62 | u | 2.8 | 0 | 1.0 | |
| 93 | SG-S717-18 | u | 4.2 | 2.5 | 3.1 | |
| 94 | Padura | u | 6.9 | 5.7 | 6.2 | |
| 95 | Torpedo | u | 6.9 | 5.7 | 6.2 | |
| 96 | Maridol | aN81, La | 5.6 | 7.4 | 6.7 | |
| 97 | Hulda | a7, k1 | 13.9 | 10.7 | 11.9 | |
| 98 | KM-2161 | u | 20.8 | 11.5 | 15.0 | |
| 99 | AC 07/624/34 | a3, aTu2 | 0 | |||
| 100 | D-535/98 | a17 | 0 | |||
| 101 | Hb-BC1-D27 | hb2 | 0 | |||
| 102 | Hs Diamant x 1B-20 | a26 | 0 | |||
| 103 | KM-1244 | a3, aTu2 | 0 | |||
| 104 | KM-14/2010 | u | 0 | |||
| 105 | HE 1051 | u | 8.6 | |||
| 106 | Oowajao | u | 11.4 | |||
| 107 | Black Heart | u | 23.8 | |||
| 108 | Ricus | u | 59.0 | |||
| 109 | Leenke | mlo | 0 | |||
| 110 | Newton | u | 4.2 | |||
| 111 | SZD 5111 | u | 8.3 | |||
| 112 | HM-407 selection | u | 33.3 | |||
| 113 | SU Lauvira | u | 0.8 | |||
| 114 | Nakaizumi-zairai | k2 | 4.1 | |||
| 115 | SC 21529 PH | u | 5.7 | |||
| 116 | Chinerme | p1 | 14.8 | |||
| 117 | Engledow India | a24 | 27.9 | |||
| 118 | KM-2168 | u | 35.2 | |||
| 119 | Gilberta | u | 62.3 | |||
| 120 | Mirko | u | 79.5 | |||
| 121 | Tadmor | aLo | 96.7 | |||
| No. Differential Varieties | 95 | 92 | 93 | |||
| Virulence | No. | Sum of | Virulence | No. | Sum of |
|---|---|---|---|---|---|
| Complexity | Isolates | Isolate | Complexity | Isolates | Isolate |
| of Isolates | Virulences | of Isolates | Virulences | ||
| 4 | 2 | 8 | 13 | 26 | 338 |
| 6 | 8 | 48 | 14 | 15 | 210 |
| 7 | 28 | 196 | 15 | 5 | 75 |
| 8 | 34 | 272 | 16 | 6 | 96 |
| 9 | 44 | 396 | 17 | 2 | 34 |
| 10 | 44 | 440 | 18 | 1 | 18 |
| 11 | 47 | 517 | 19 | 2 | 38 |
| 12 | 35 | 420 | Sum | 299 | 3 106 |
| Differential | Main | Central | Australia2 | Kazakh- | China | China | Turkey | Turkey |
|---|---|---|---|---|---|---|---|---|
| variety | Ml gene | Europe1 | stan3 | South4 | Tibet5 | Adana6 | Hatay6 | |
| P01 | a1 | 57.2 | 0 | 0 | 0 | 0 | 0 | 0 |
| P03 | a6 | 98.7 | 0 | 0 | 10.6 | 0 | 36.7 | 27.9 |
| P04B | a7 | 84.3 | 0 | 0 | 3.7 | 0 | 12.7 | 4.4 |
| P10 | a12 | 91.6 | 0 | 1.9 | 34.6 | 0.7 | 31.0 | 33.8 |
| P11 | a13 | 27.8 | 0 | 0 | 0.5 | 23.2 | 2.8 | 4.4 |
| P21 | g | 78.6 | 79.5 | 0.9 | 21.8 | 20.8 | 19.7 | 8.8 |
| P15 | Ru2 | 86.6 | 95.7 | 76.9 | 25.3 | 16.2 | ||
| P22 | mlo | 0 | 0 | 0 | 4.8 | 8.6 | 4.2 | 14.7 |
| Pallas | a8 | 100 | 100 | 100 | 100 | 100 | 94.3 | 91.1 |
| Variety | Year of | Ml resistance | Average resistance in field trials | |||
|---|---|---|---|---|---|---|
| Registration | gene(s) | Highest | Lowest | |||
| Ametyst | 1972 | a6 | 1971 | 7.20 | 1977 | 4.33 |
| Trumpf | 1976 | a7, aTr3, Ab | 1975 | 8.86 | 1979 | 5.44 |
| Spartan | 1977 | a9 | 1976 | 8.60 | 1983 | 3.38 |
| Zefir | 1981 | a12 | 1978 | 7.00 | 1986 | 2.50 |
| Koral | 1978 | a13, g | 1982 | 9.00 | 1986 | 5.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





