Submitted:
26 September 2023
Posted:
28 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Clinical information and mRNA expression dataset of patients
2.2. Gene Set Enrichment Analysis
2.3. Statistical analysis
3. Results
3.1. Initial screening of the genes using GSEA
3.2. Identification of glycolysis-related genes associated with survival in CRC patients
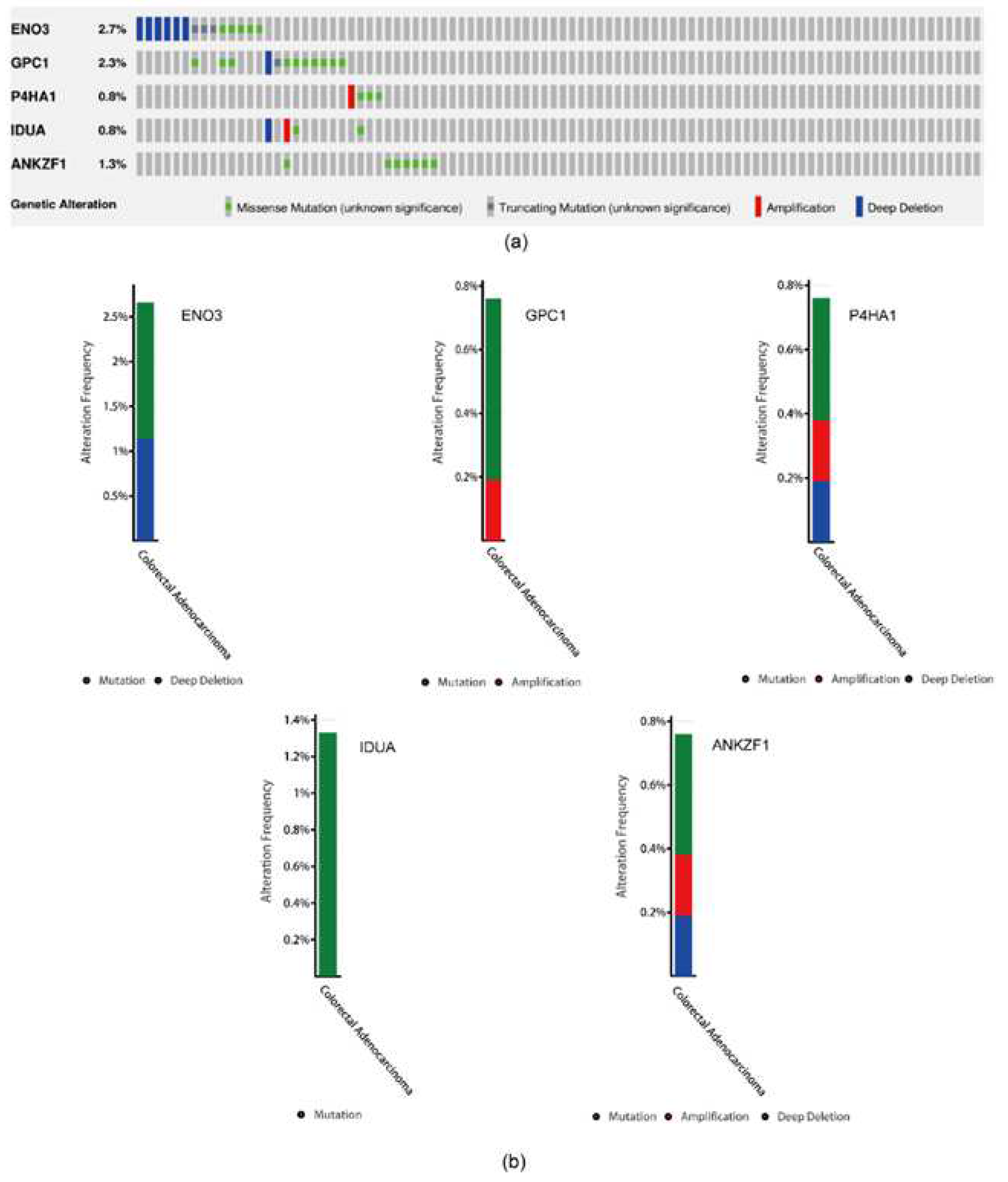
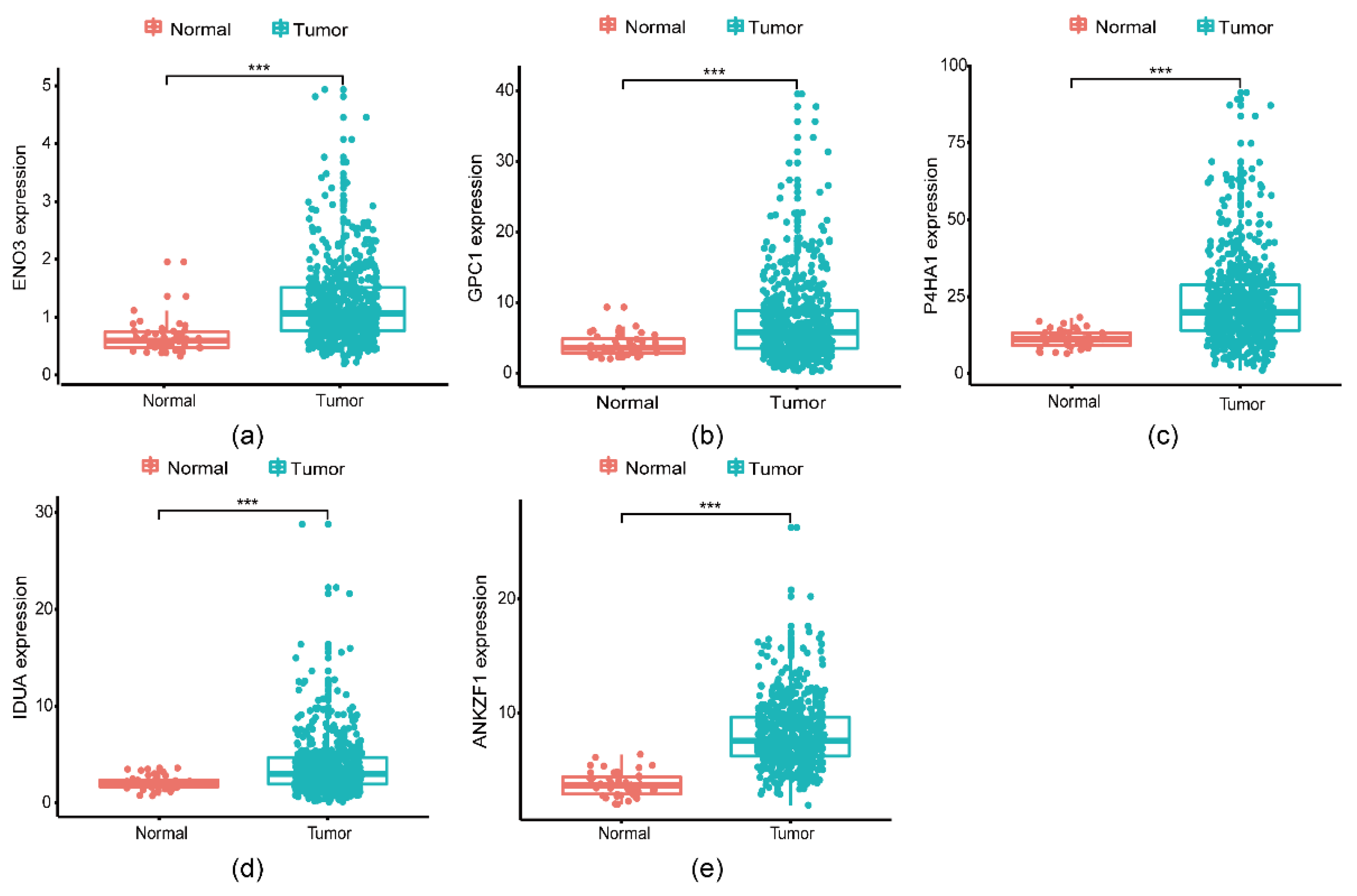
3.3. A five genes signature for predicting patients’ outcome
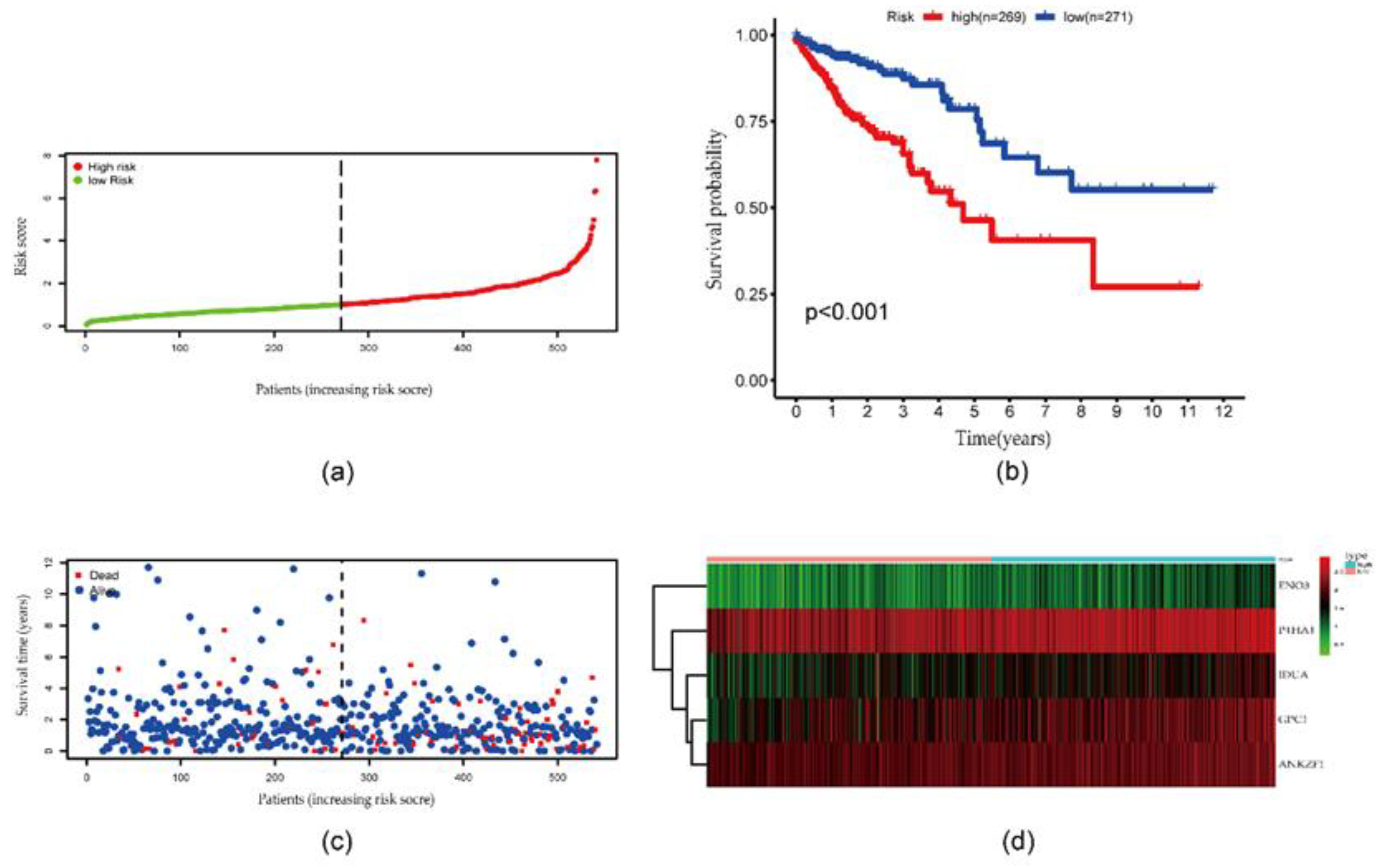
3.4. Risk score derived from the signature of five genes
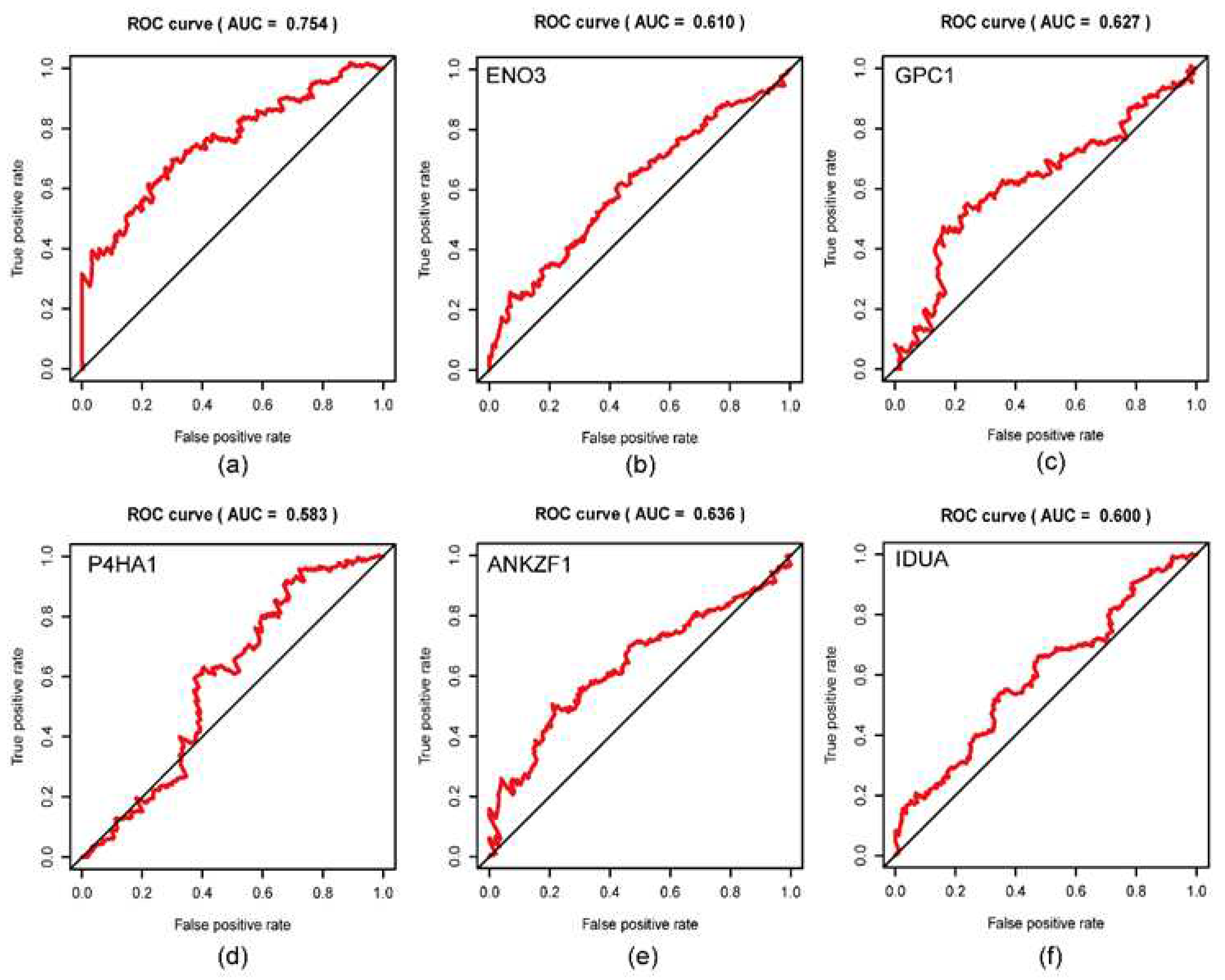
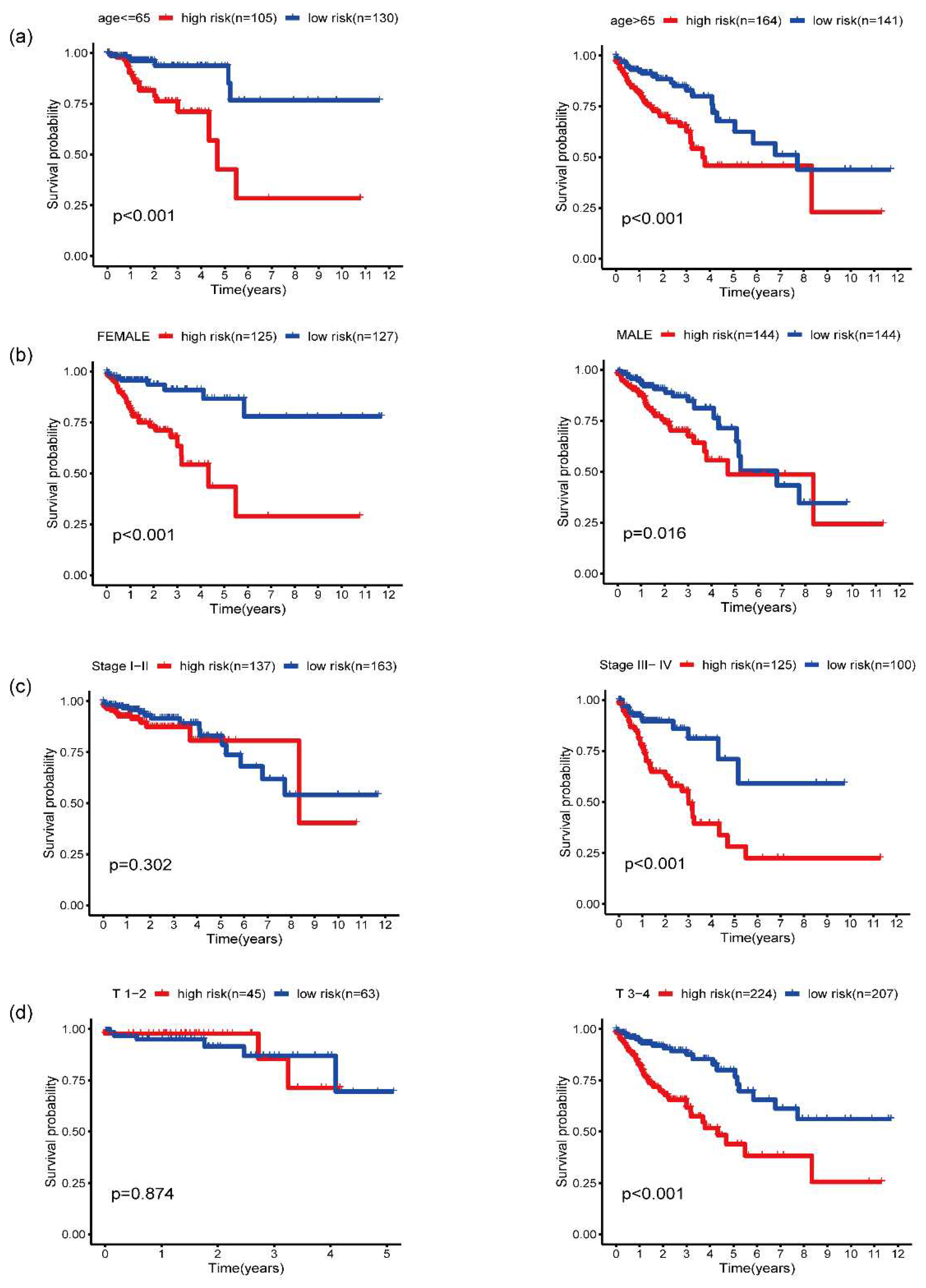
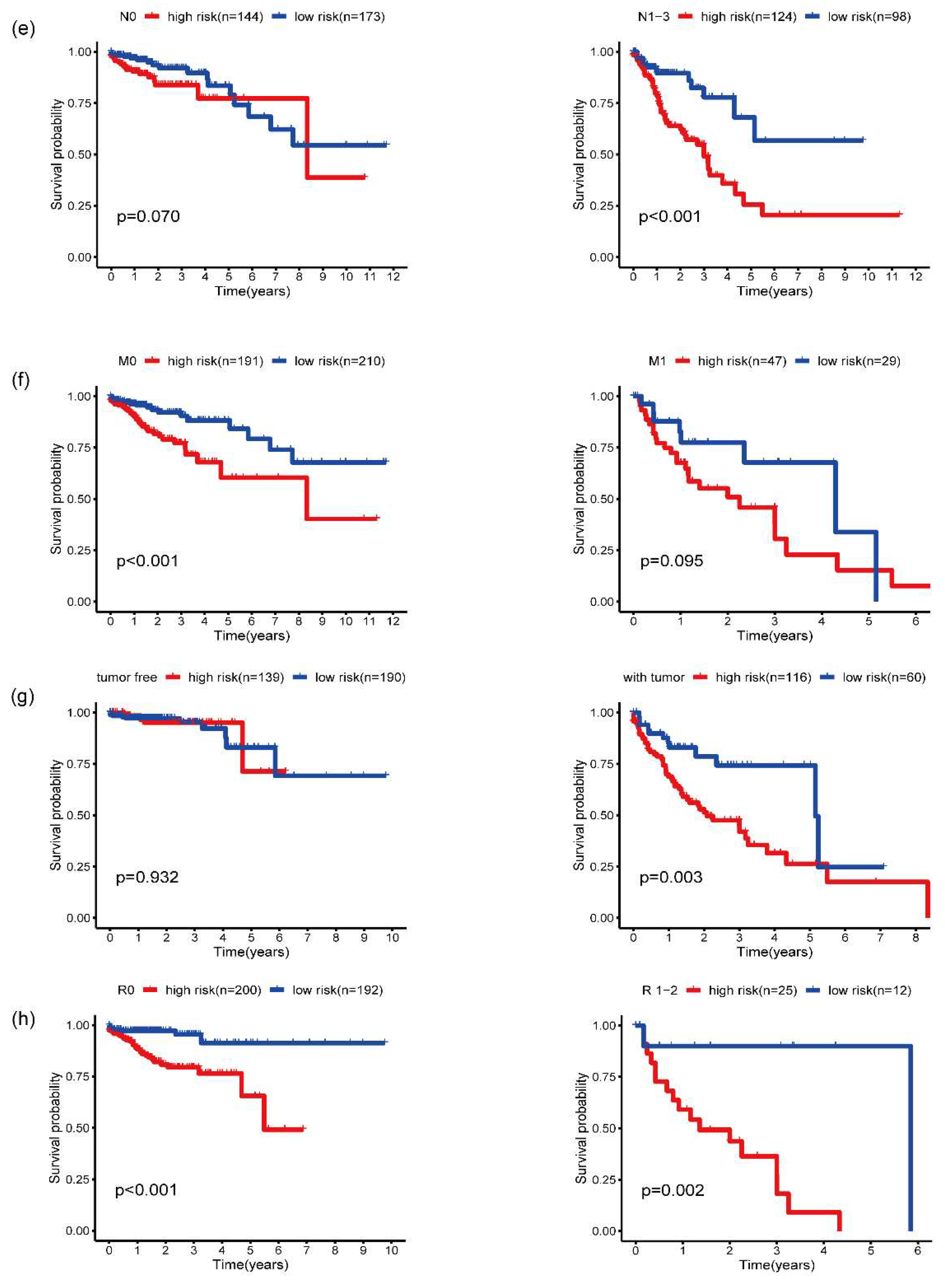
3.5. Validation of five-gene signature for survival prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science (80-. ). 2009, 324, 1029–1033. [CrossRef]
- Vander Heiden, M.G. Targeting Cancer Metabolism: A Therapeutic Window Opens. Nat. Rev. Drug Discov. 2011, 10, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.E.; Short, S.P.; Williams, C.S. Colorectal Cancer and Metabolism. Curr. Colorectal Cancer Rep. 2018, 14, 226–241. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Shen, Z.Y.; Zhan, Y.Z.; Feng, X.C.; Chen, K.L.; Li, Y.S.; Deng, H.J.; Pan, S.M.; Wu, D.H.; Ding, Y. CD36 Inhibits β-Catenin/c-Myc-Mediated Glycolysis through Ubiquitination of GPC4 to Repress Colorectal Tumorigenesis. Nat. Commun. 2019, 10, 3981. [Google Scholar] [CrossRef] [PubMed]
- Naseri, M.; Saeednejad Zanjani, L.; Vafaei, S.; Gheytanchi, E.; Abolhasani, M.; Bozorgmehr, M.; Ranaei Pirmardan, E.; Ghods, R.; Madjd, Z. Increased Cytoplasmic Expression of DLL4 Is Associated with Favorable Prognosis in Colorectal Cancer. Futur. Oncol. 2021, 17, 3231–3242. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, X.; Li, M.; Gong, G.; Liu, W.; Li, T.; Zuo, H.; Li, W.; Gao, F.; Liu, H. Cdh1-Mediated Skp2 Degradation by Dioscin Reprogrammes Aerobic Glycolysis and Inhibits Colorectal Cancer Cells Growth. EBioMedicine 2020, 51. [Google Scholar] [CrossRef]
- Fitzmaurice, C.; Abate, D.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdel-Rahman, O.; Abdelalim, A.; Abdoli, A.; Abdollahpour, I.; Abdulle, A.S.M.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA. Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Frampton, M.; Houlston, R.S. Modeling the Prevention of Colorectal Cancer from the Combined Impact of Host and Behavioral Risk Factors. Genet. Med. 2017, 19, 314–321. [Google Scholar] [CrossRef]
- Saeednejad Zanjani, L.; Vafaei, S.; Abolhasani, M.; Fattahi, F.; Madjd, Z. Prognostic Value of Talin-1 in Renal Cell Carcinoma and Its Association with B7-H3. Cancer Biomarkers 2022, 35, 269–292. [Google Scholar] [CrossRef]
- Bretthauer, M. Colorectal Cancer Screening. J. Intern. Med. 2011, 270, 87–98. [Google Scholar] [CrossRef]
- Roseweir, A.K.; Clark, J.; McSorley, S.T.; vanWyk, H.C.; Quinn, J.A.; Horgan, P.G.; McMillan, D.C.; Park, J.H.; Edwards, J. The Association between Markers of Tumour Cell Metabolism, the Tumour Microenvironment and Outcomes in Patients with Colorectal Cancer. Int. J. Cancer 2019, 144, 2320–2329. [Google Scholar] [CrossRef]
- Thomas, M.A.; Yang, L.; Carter, B.J.; Klaper, R.D. Gene Set Enrichment Analysis of Microarray Data from Pimephales Promelas (Rafinesque), a Non-Mammalian Model Organism. BMC Genomics 2011, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jary, M.; Hasanova, R.; Vienot, A.; Asgarov, K.; Loyon, R.; Tirole, C.; Bouard, A.; Orillard, E.; Klajer, E.; Kim, S.; et al. Molecular Description of ANGPT2 Associated Colorectal Carcinoma. Int. J. Cancer 2020, 147, 2007–2018. [Google Scholar] [CrossRef] [PubMed]
- Sharifzad, F.; Yasavoli-Sharahi, H.; Mardpour, S.; Fakharian, E.; Nikuinejad, H.; Heydari, Y.; Mardpour, S.; Taghikhani, A.; khellat, R.; Vafaei, S.; et al. Neuropathological and Genomic Characterization of Glioblastoma-Induced Rat Model: How Similar Is It to Humans for Targeted Therapy? J. Cell. Physiol. 2019, 234, 22493–22504. [Google Scholar] [CrossRef] [PubMed]
- Koncina, E.; Haan, S.; Rauh, S.; Letellier, E. Prognostic and Predictive Molecular Biomarkers for Colorectal Cancer: Updates and Challenges. Cancers (Basel). 2020, 12, 319. [CrossRef]
- Kubota, N.; Taniguchi, F.; Nyuya, A.; Umeda, Y.; Mori, Y.; Fujiwara, T.; Tanioka, H.; Tsuruta, A.; Yamaguchi, Y.; Nagasaka, T. Upregulation of MicroRNA-31 Is Associated with Poor Prognosis in Patients with Advanced Colorectal Cancer. Oncol. Lett. 2020, 19, 2685–2694. [Google Scholar] [CrossRef]
- Wang, X.; Feng, Q.; Yu, H.; Zhou, X.; Shan, C.; Zhang, Q.; Liu, S. HBXIP: A Potential Prognosis Biomarker of Colorectal Cancer Which Promotes Invasion and Migration via Epithelial-Mesenchymal Transition. Life Sci. 2020, 245, 117354. [Google Scholar] [CrossRef]
- Beaulieu, J.F. Integrin A6β4 in Colorectal Cancer: Expression, Regulation, Functional Alterations and Use as a Biomarker. Cancers (Basel). 2020, 12, 41. [CrossRef]
- Fattahi, F.; Kiani, J.; Khosravi, M.; Vafaei, S.; Mohammadi, A.; Madjd, Z.; Najafi, M. Enrichment of Up-Regulated and Down-Regulated Gene Clusters Using Gene Ontology, MiRNAs and LncRNAs in Colorectal Cancer. Comb. Chem. High Throughput Screen. 2019, 22, 534–545. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Q.; Zhao, L.; Jiang, L.; Qi, A.; Wei, Q.; Song, X.; Wang, L.; Zhang, L.; Zhao, Y.; et al. A Combined Four-MRNA Signature Associated with Lymphatic Metastasis for Prognosis of Colorectal Cancer. J. Cancer 2020, 11, 2139–2149. [Google Scholar] [CrossRef]
- Dai, W.; Li, Y.; Mo, S.; Feng, Y.; Zhang, L.; Xu, Y.; Li, Q.; Cai, G. A Robust Gene Signature for the Prediction of Early Relapse in Stage I–III Colon Cancer. Mol. Oncol. 2018, 12, 463–475. [Google Scholar] [CrossRef]
- Tarnopolsky, M.A. Myopathies Related to Glycogen Metabolism Disorders. Neurotherapeutics 2018, 15, 915–927. [Google Scholar] [CrossRef]
- Park, C.; Lee, Y.; Je, S.; Chang, S.; Kim, N.; Jeong, E.; Yoon, S. Overexpression and Selective Anticancer Efficacy of ENO3 in STK11 Mutant Lung Cancers. Mol. Cells 2019, 42, 804–809. [Google Scholar] [CrossRef] [PubMed]
- Filmus, J.; Capurro, M.; Rast, J. Glypicans. Genome Biol. 2008, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Lei, Y.; Lv, G.; Liu, Y.; Zhao, W.; Yang, Q.; Su, X.; Song, Z.; Lu, L.; Shi, Y. Identification of a Novel Autophagy Signature for Predicting Survival in Patients with Lung Adenocarcinoma. PeerJ 2021, 9, 1–13. [Google Scholar] [CrossRef]
- Jiang, F.; Wu, C.; Wang, M.; Wei, K.; Wang, J. Identification of Novel Cell Glycolysis Related Gene Signature Predicting Survival in Patients with Breast Cancer. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Li, J.; Li, B.; Ren, C.; Chen, Y.; Guo, X.; Zhou, L.; Peng, Z.; Tang, Y.; Chen, Y.; Liu, W.; et al. The Clinical Significance of Circulating GPC1 Positive Exosomes and Its Regulative MiRNAs in Colon Cancer Patients. Oncotarget 2017, 8, 101189–101202. [Google Scholar] [CrossRef]
- Gilkes, D.M.; Chaturvedi, P.; Bajpai, S.; Wong, C.C.; Wei, H.; Pitcairn, S.; Hubbi, M.E.; Wirtz, D.; Semenza, G.L. Collagen Prolyl Hydroxylases Are Essential for Breast Cancer Metastasis. Cancer Res. 2013, 73, 3285–3296. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gao, Y.; Vafaei, S.; Yu, Q.; Zhang, J.; Wang, L. A Chemoresistance LncRNA Signature for Recurrence Risk Stratification of Colon Cancer Patients with Chemotherapy. Mol. Ther. - Nucleic Acids 2022, 27, 427–438. [Google Scholar] [CrossRef]
- Agarwal, S.; Behring, M.; Kim, H.G.; Bajpai, P.; Chakravarthi, B.V.S.K.; Gupta, N.; Elkholy, A.; Al Diffalha, S.; Varambally, S.; Manne, U. Targeting P4HA1 with a Small Molecule Inhibitor in a Colorectal Cancer PDX Model. Transl. Oncol. 2020, 13, 100754. [Google Scholar] [CrossRef]
- Maita, N.; Tsukimura, T.; Taniguchi, T.; Saito, S.; Ohno, K.; Taniguchi, H.; Sakuraba, H. Human α-L-Iduronidase Uses Its Own N-Glycan as a Substrate-Binding and Catalytic Module. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 14628–14633. [Google Scholar] [CrossRef]
- Bie, H.; Yin, J.; He, X.; Kermode, A.R.; Goddard-Borger, E.D.; Withers, S.G.; James, M.N.G. Insights into Mucopolysaccharidosis I from the Structure and Action of α-L-Iduronidase. Nat. Chem. Biol. 2013, 9, 739–745. [Google Scholar] [CrossRef]
- Van Haaften-Visser, D.Y.; Harakalova, M.; Mocholi, E.; Van Montfrans, J.M.; Elkadri, A.; Rieter, E.; Fiedler, K.; Van Hasselt, P.M.; Triffaux, E.M.M.; Van Haelst, M.M.; et al. Ankyrin Repeat and Zinc-Finger Domain-Containing 1 Mutations Are Associated with Infantile-Onset Inflammatory Bowel Disease. J. Biol. Chem. 2017, 292, 7904–7920. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Shang, Y.N.; Lu, R.; Fan, C.W.; Mo, X.M. High ANKZF1 Expression Is Associated with Poor Overall Survival and Recurrence-Free Survival in Colon Cancer. Futur. Oncol. 2019, 15, 2093–2106. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



