Submitted:
26 September 2023
Posted:
28 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Variance components estimation
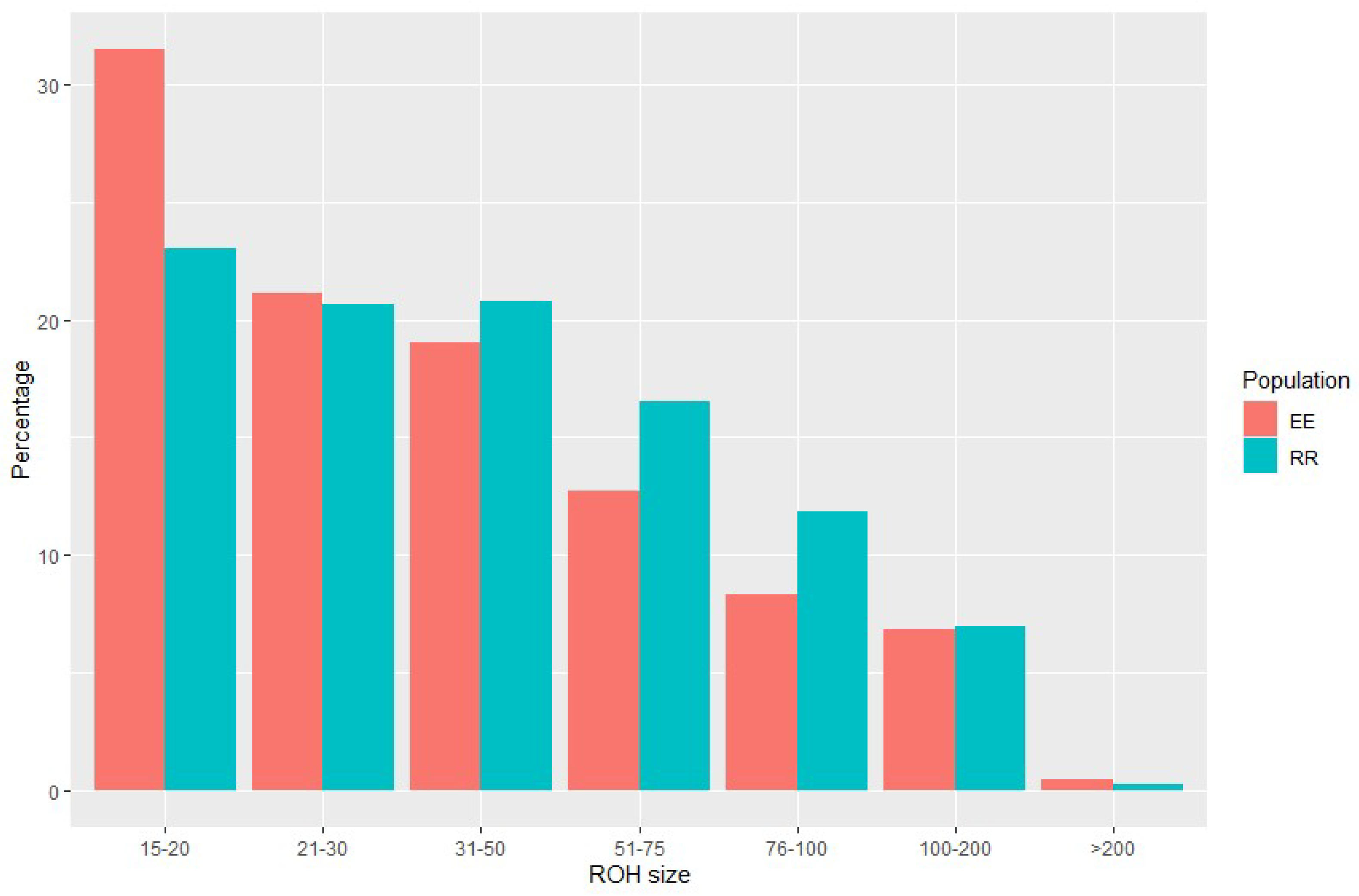
3.2. Runs of Homozygosity (ROH) identification
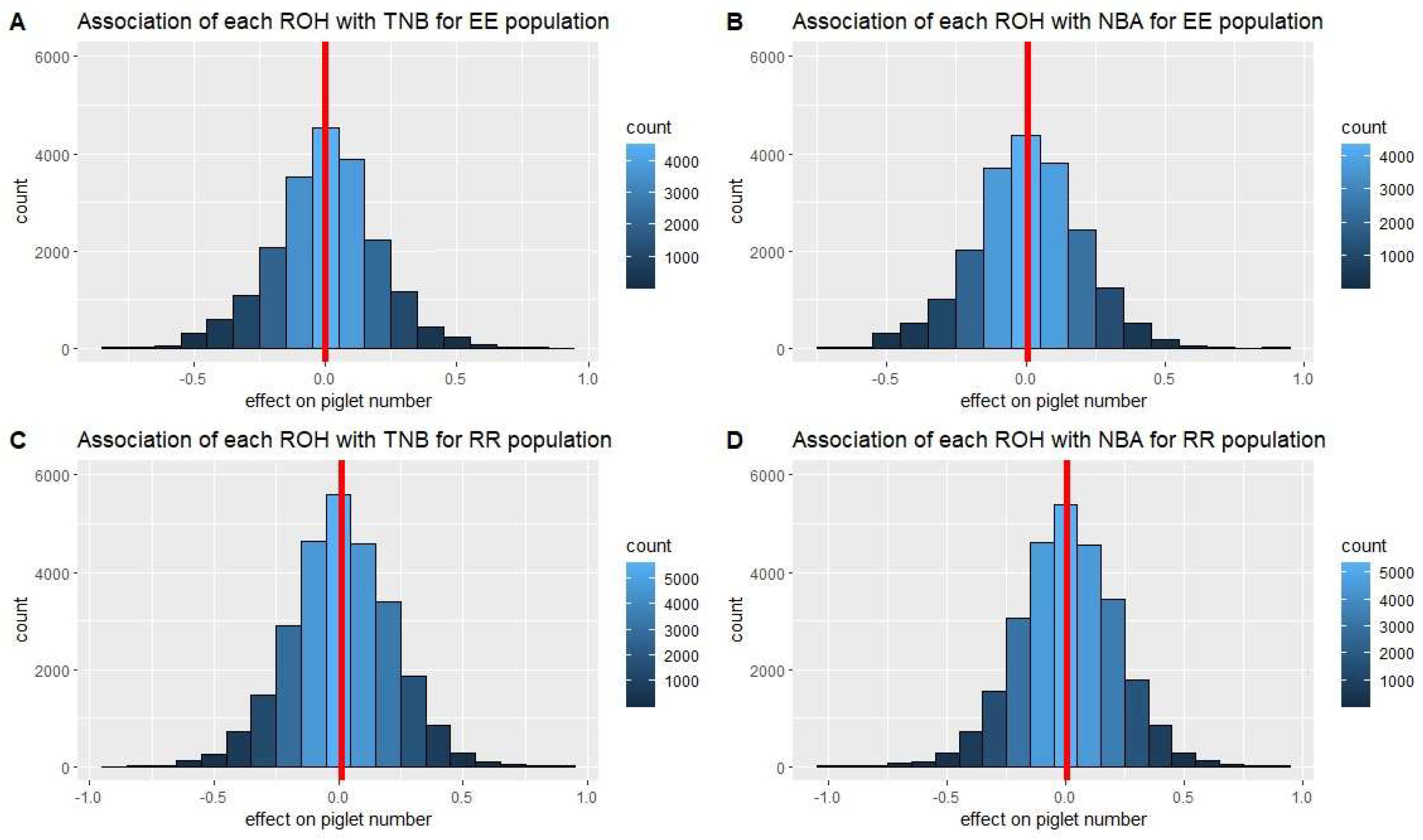
3.3. ROH segments and inbreeding depression
4. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leroy, G. Inbreeding depression in livestock species: review and meta-analysis. Anim. Genet. 2014, 45, 618–628. [Google Scholar] [CrossRef]
- Charlesworth, B.; Charlesworth, D. The genetic basis of inbreeding depression. Genet. Res. 1999, 74, 329–340. [Google Scholar] [CrossRef]
- Farkas, J.; Curik, I.; Csató, L.; Csörnyei, Z.; Baumung, R.; Nagy, I. Bayesian inference of inbreeding effects on litter size and gestation length in Hungarian Landrace and Hungarian Large White pigs. Livest. Sci. 2007, 112, 109–114. [Google Scholar] [CrossRef]
- Silió, L.; Barragán, C.; Fernández, A.I.; García-Casco, J.; Rodríguez, M.C. Assessing effective population size, coancestry and inbreeding effects on litter size using the pedigree and SNP data in closed lines of the Iberian pig breed. J. Anim. Breed. Genet. 2016, 133, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics; Longman Group, Harlow, UK, 1996.
- Martikainen, K.; Sironen, A.; Uimari, P. Estimation of intrachromosomal inbreeding depression on female fertility using runs of homozygosity in Finnish Ayrshire cattle. J. Dairy Sci. 2018, 101, 11097–11107. [Google Scholar] [CrossRef] [PubMed]
- Saura, M.; Fernández, A.; Varona, L.; Fernández, A.I.; De Cara, M.Á.R.; Barragán, C.; Villanueva, B. Detecting inbreeding depression for reproductive traits in Iberian pigs using genome-wide data. Genet. Sel. Evol. 2015, 47, 1. [Google Scholar] [CrossRef]
- Pryce, J.E.; Haile-Mariam, M.; Goddard, M.E.; Hayes, B.J. Identification of genomic regions associated with inbreeding depression in Holstein and Jersey dairy cattle. Genet. Sel. Evol. 2014, 46, 71. [Google Scholar] [CrossRef]
- Curik, I.; Ferenčaković, M.; Sölkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Howard, J.T.; Tiezzi, F.; Huang, Y.; Gray, K.A.; Maltecca, C. A heuristic method to identify runs of homozygosity associated with reduced performance in livestock. J. Anim. Sci. 2017, 95, 4318–4332. [Google Scholar] [CrossRef]
- Henderson, C.R. Applications of Linear Models in Animal Breeding.; University of Guelph: Ontario, Canada, 1984. [Google Scholar]
- Fabuel, E.; Barragán, C.; Silió, L.; Rodríguez, M.C.; Toro, M.A. Analysis of genetic diversity and conservation priorities in Iberian pigs based on microsatellite markers. Heredity (Edinb). 2004, 93, 104–113. [Google Scholar] [CrossRef]
- Alonso, I.; Ibáñez-Escriche, N.; Noguera, J.L.; Casellas, J.; Martín de Hijas-Villalba, M.; Gracia-Santana, M.J.; Varona, L. Genomic differentiation among varieties of Iberian pig. Spanish J. Agric. Res. 2020, 18, e0401. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Sargolzaei, M.; Chesnais, J.P.; Schenkel, F.S. A new approach for efficient genotype imputation using information from relatives. BMC Genomics 2014, 15, 478. [Google Scholar] [CrossRef]
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef]
- Gilmour, A.R. Average information residual maximum likelihood in practice. J. Anim. Breed. Genet. 2019, 136, 262–272. [Google Scholar] [CrossRef]
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPF90 family of programs; University of Georgia: Athens, GA (USA), 2018. [Google Scholar]
- Srihi, H.; Noguera, J.L.; Topayan, V.; de Hijas, M.M.; Ibañez-Escriche, N.; Casellas, J.; Vázquez-Gómez, M.; Martínez-Castillero, M.; Rosas, J.P.; Varona, L. Additive and dominance genomic analysis for litter size in purebred and crossbred iberian pigs. Genes (Basel). 2022, 13, 12. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Z.; Wang, Z.; Zhang, Z.; Wang, Q.; Pan, Y. Heterozygosity and homozygosity regions affect reproductive success and the loss of reproduction: A case study with litter traits in pigs. Comput. Struct. Biotechnol. J. 2022, 20, 4060–4071. [Google Scholar] [CrossRef]
- He, L.C.; Li, P.H.; Ma, X.; Sui, S.P.; Gao, S.; Kim, S.W.; Gu, Y.Q.; Huang, Y.; Ding, N.S.; Huang, R.H. Identification of new single nucleotide polymorphisms affecting total number born and candidate genes related to ovulation rate in Chinese Erhualian pigs. Anim. Genet. 2017, 48, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.S.; Lin, F.; Wang, Z.W.; Hu, M.W.; Huang, L.; Meng, T.G.; Zhe Jiang, Z.; Schatten, H.; Wang, Z.B.; Sun, Q.Y. Geminin deletion in mouse oocytes results in impaired embryo development and reduced fertility. Mol. Biol. Cell 2016, 27, 768–775. [Google Scholar] [CrossRef]
- Chen, L.J.; Hu, B.; Han, Z.Q.; Zhu, J.H.; Fan, X.; Chen, X.X.; Li, Z.P.; Zhou, H. BAG2-Mediated Inhibition of CHIP Expression and Overexpression of MDM2 Contribute to the Initiation of Endometriosis by Modulating Estrogen Receptor Status. Front. Cell Dev. Biol. 2021, 8, 554190. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.; Lu, Y.; Zhu, S.; Feng, L.; Qi, W.; Chen, X.; Xie, B.; Chen, B.; Lan, G.; Liang, J. Genome-Wide Association Studies, Runs of Homozygosity Analysis, and Copy Number Variation Detection to Identify Reproduction-Related Genes in Bama Xiang Pigs. Front. Vet. Sci. 2022, 9, 892815. [Google Scholar] [CrossRef]
- Xin, W.S.; Zhang, F.; Yan, G.R.; Xu, W.W.; Xiao, S.J.; Zhang, Z.Y.; Huang, L.S. A whole genome sequence association study for puberty in a large Duroc × Erhualian F2 population. Anim. Genet. 2018, 49, 29–35. [Google Scholar] [CrossRef]
- Tyc, K.M.; Yakoubi, W. El; Bag, A.; Landis, J.; Zhan, Y.; Treff, N.R.; Scott, R.T.; Tao, X.; Schindler, K.; Xing, J. Exome sequencing links CEP120 mutation to maternally derived aneuploid conception risk. Hum. Reprod. 2020, 35, 2134–2148. [Google Scholar] [CrossRef] [PubMed]
- O’Lone, R.; Frith, M.C.; Karlsson, E.K.; Hansen, U. Genomic targets of nuclear estrogen receptors. Mol. Endocrinol. 2004, 18, 1859–1875. [Google Scholar] [CrossRef]
- Oberska, P.; Michałek, K. Aquaporins: New markers for male (in)fertility in livestock and poultry? Anim. Reprod. Sci. 2021, 231, 106807. [Google Scholar] [CrossRef] [PubMed]
- Skowronski, M.T.; Kwon, T.H.; Nielsen, S. Immunolocalization of aquaporin 1, 5, and 9 in the female pig reproductive system. J. Histochem. Cytochem. 2009, 57, 61–67. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, M.; Gu, W.; Chen, A.; Liu, J.; Li, L.; Zhang, S.; Liu, G. Downregulation of Aquaporins (AQP1 and AQP5) and Na,K-ATPase in Porcine Reproductive and Respiratory Syndrome Virus-Infected Pig Lungs. Inflammation 2018, 41, 1104–1114. [Google Scholar] [CrossRef]
- Grewal, S.; Carver, J.G.; Ridley, A.J.; Mardon, H.J. Implantation of the human embryo requires Rac1-dependent endometrial stromal cell migration. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 16189–16194. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, P.H.; Zhu, M.X.; He, L.C.; Sui, S.P.; Gao, S.; Su, G.S.; Ding, N.S.; Huang, Y.; Lu, Z.Q.; et al. Genome-wide association analysis reveals genomic regions on Chromosome 13 affecting litter size and candidate genes for uterine horn length in Erhualian pigs. Animal 2018, 12, 2453–2461. [Google Scholar] [CrossRef]
- Hernández-Montiel, W.; Martínez-Núñez, M.A.; Ramón-Ugalde, J.P.; Román-Ponce, S.I.; Calderón-Chagoya, R.; Zamora-Bustillos, R. Genome-wide association study reveals candidate genes for litter size traits in pelibuey sheep. Animals 2020, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Krsmanović, L.Z.; Stojilković, S.S.; Merelli, F.; Dufour, S.M.; Virmani, M.A.; Catt, K.J. Calcium signaling and episodic secretion of gonadotropin-releasing hormone in hypothalamic neurons. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 8462–8466. [Google Scholar] [CrossRef] [PubMed]
- Spergel, D.J.; Krsmanovic, L.Z.; Stojilkovic, S.S.; Cali, K.J. L-type Ca2+ channels mediate joint modulation by gamma-amino-butyric acid and glutamate of [Ca2+]i and neuropeptide secretion in immortalized gonadodropin-releasing hormone neurons. Neuroendocrinology 1995, 61, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Fukuda, A.; Nabekura, J. The role of GABA in the regulation of GnRH neurons. Front. Neurosci. 2014, 8, 387. [Google Scholar] [CrossRef]
- Kaitu’U-Lino, T.J.; Ye, L.; Tuohey, L.; Dimitriadis, E.; Bulmer, J.; Rogers, P.; Menkhorst, E.; Van Sinderen, M.; Girling, J.E.; Hannan, N.; et al. Corin, an enzyme with a putative role in spiral artery remodeling, is up-regulated in late secretory endometrium and first trimester decidua. Hum. Reprod. 2013, 28, 1172–1180. [Google Scholar] [CrossRef]
- Purfield, D.C.; Bradley, D.G.; Evans, R.D.; Kearney, F.J.; Berry, D.P. Genome-wide association study for calving performance using high-density genotypes in dairy and beef cattle. Genet. Sel. Evol. 2015, 47, 47. [Google Scholar] [CrossRef]
- Lan, Q.; Deng, Q.; Qi, S.; Zhang, Y.; Li, Z.; Yin, S.; Li, Y.; Tan, H.; Wu, M.; Yin, Y.; et al. Genome-Wide Association Analysis Identified Variants Associated with Body Measurement and Reproduction Traits in Shaziling Pigs. Genes (Basel). 2023, 14, 522. [Google Scholar] [CrossRef]
- Huang, M.; Wang, H.; Li, J.; Zhou, Z.; Du, Y.; Lin, M.; Sha, J. Involvement of ALF in human spermatogenesis and male infertility. Int. J. Mol. Med. 2006, 17, 599–604. [Google Scholar] [CrossRef]
- Yang, W.; Lee, Y.H.; Jones, A.E.; Woolnough, J.L.; Zhou, D.; Dai, Q.; Wu, Q.; Giles, K.E.; Townes, T.M.; Wang, H. The histone H2A deubiquitinase Usp16 regulates embryonic stem cell gene expression and lineage commitment. Nat. Commun. 2014, 5, 3818. [Google Scholar] [CrossRef]
- Tinoco, E.M.; Gigante, A.R.; Ferreira, E.; Sanches, I.; Pereira, R.; Sá, R.; Monteiro, R.; Sousa, M.; Pascoal, I. Primary Ciliary Dyskinesia in a Portuguese Bronchiectasis Outpatient Clinic. Genes (Basel). 2023, 14. [Google Scholar] [CrossRef]
| Population | TNB | NBA |
|---|---|---|
| Entrepelado | 7.96 (1.93) | 7.70 (1.88) |
| Retinto | 8.27 (2.18) | 7.99 (2.17) |
| SSC | Nm | bp |
|---|---|---|
| 1 | 5,442 | 274,315,671 |
| 2 | 3,713 | 151,610,480 |
| 3 | 3,308 | 132,657,669 |
| 4 | 3,557 | 130,773,976 |
| 5 | 2,716 | 104,477,606 |
| 6 | 4,368 | 170,802,600 |
| 7 | 3,563 | 121,758,423 |
| 8 | 3,324 | 138,930,735 |
| 9 | 3,513 | 139,386,589 |
| 10 | 2,477 | 69,319,537 |
| 11 | 2,178 | 79,072,521 |
| 12 | 2,295 | 60,834,034 |
| 13 | 4,146 | 208,240,759 |
| 14 | 3,812 | 141,719,266 |
| 15 | 3,281 | 140,404,164 |
| 16 | 2,205 | 79,282,526 |
| 17 | 1,968 | 63,391,207 |
| 18 | 1,607 | 55,752,892 |
| Entrepelado | Retinto | |||
|---|---|---|---|---|
| TNB | NBA | TNB | NBA | |
| 0.145 (0.085) | 0.098 (0.071) | 0.165 (0.092) | 0.225 (0.110) | |
| 0.366 (0.097) | 0.341 (0.089) | 0.283 (0.11) | 0.291(0.116) | |
| 0.170 (0.545) | 0.129 (0.071) | 0.317 (0,085) | 0.172 (0,061) | |
| 2.901 (0.101) | 2.853 (0.099) | 3.908 (0.138) | 3.796 (0.134) | |
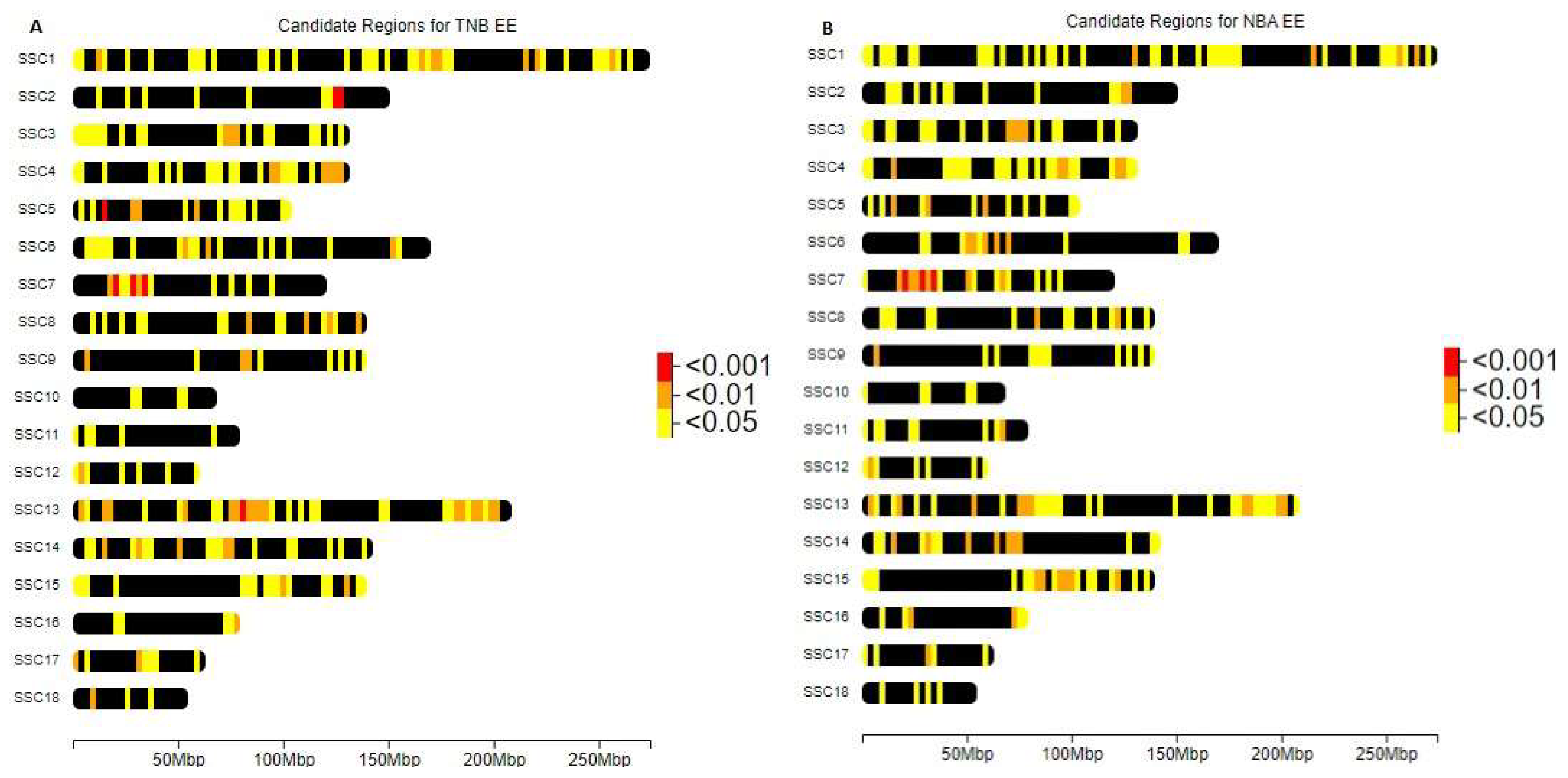
| SSC | bp(c) | Piglets (NBA) | Piglets (TNB) | p-value (NBA) | p-value (TNB) | Genes |
|---|---|---|---|---|---|---|
| SSC2 | 126,506,354 – 126,841,331 | -0.5428 | -0.6264 | 2.23E-03 | 8,83E-04 | CEP120 |
| 126,516,022 – 126,857,203 | -0.5358 | -0.6239 | 1.28E-03 | 4,08E-04 | ||
| 126,562,142 – 126,971,814 | -0.4838 | -0.5840 | 3.46E-03 | 9,82E-04 | ||
| 126,700,457 – 127,000,238 | -0.4935 | -0.5904 | 2.69E-03 | 7,87E-04 | ||
| 126,777,258 – 127,080,032 | -0.4838 | -0.5840 | 3.46E-03 | 9,82E-04 | ||
| 127,096,117 – 127,764,704 | -0.6088 | -0.7179 | 2.75E-03 | 9,18E-04 | ||
| 127,492,532 – 127,890,446 | -0.6305 | -0.7393 | 1.70E-03 | 5,36E-04 | ||
| SSC5 | 15,871,592 – 16,9148,74 | -0.6141 | -0,7758 | 5,00E-03 | 9,12E-04 | ATF1 AQP5 AQP6 RACGAP1 |
| 16,134,195 – 16,852,941 | -0.5377 | -0,7575 | 1,10E-02 | 9,73E-04 | ||
| SSC7 | 20,074,761 – 20,586,603 | -0,5995 | -0,5869 | 7,23E-05 | 2,03E-04 | GMNN |
| 28,252,780 – 28,721,664 | -0,6691 | -0,6795 | 3,54E-05 | 6,34E-05 | ||
| 35,594,623 – 36,062,243 | -0,6175 | -0,6308 | 6,97E-04 | 9,86E-04 | BAG2 RAB23 |
|
| 20,074,761 – 20,586,603 | -0,5995 | -0,5869 | 7,23E-05 | 2,03E-04 | ||
| SSC13 | 80,594,858 – 81,214,645 | -0,4839 | -0,5679 | 2,28E-03 | 7,63E-04 | CLSTN2 |
| 80,682,896 – 81,329,857 | -0,4929 | -0,5693 | 1,72E-03 | 6,53E-04 |
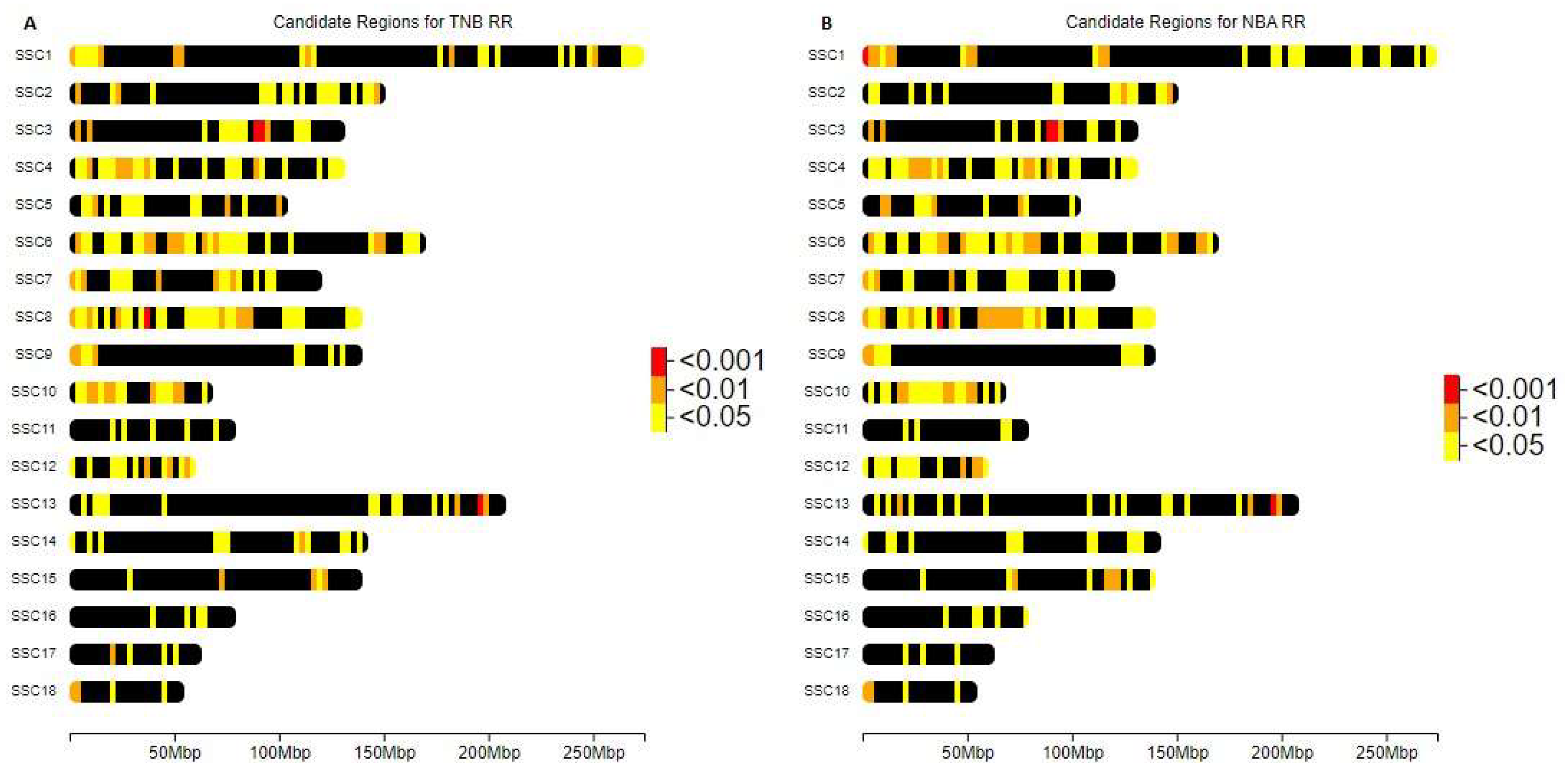
| SSC | bp(c) | Piglets (NBA) | Piglets (TNB) | p-value (NBA) | p-value (TNB) | Genes |
|---|---|---|---|---|---|---|
| SSC1 | 632,758 – 18,69,413 | -0,9609 | -0,7750 | 1,35E-04 | 1,41E-03 | |
| 734,657 – 1,869,413 | -0,9609 | -0,6383 | 1,35E-04 | 4,82E-03 | ||
| 989,159 – 13,58,337 | -0,9609 | -0,7750 | 1,35E-04 | 1,41E-03 | ||
| 1,189,180 – 1,471,069 | -0,8987 | -0,7750 | 2,31E-04 | 1,41E-03 | ||
| 1,189,180 – 2,160,998 | -0,8174 | -0,7244 | 5,59E-04 | 2,06E-03 | ||
| SSC3 | 88,968,396 – 91,465,748 | -0,5929 | -0,5525 | 7,53E-04 | 1,26E-03 | FSHR LHCGR GTF2A1L |
| 90,580,151 – 91,903,672 | -0,6761 | -0,6322 | 1,69E-04 | 3,08E-04 | ||
| 91,082,368 – 92,692,939 | -0,6931 | -0,6421 | 1,60E-04 | 3,29E-04 | ||
| 91,240,698 – 91,903,672 | -0,6388 | -0,6003 | 2,88E-04 | 4,70E-04 | ||
| 91,265,546 – 92,395,629 | -0,6761 | -0,6322 | 1,69E-04 | 3,08E-04 | ||
| 91,381,281 – 92,692,939 | -0,6608 | -0,6010 | 2,64E-04 | 6,37E-04 | ||
| 91,381,281 – 92,437,314 | -0,6458 | -0,5931 | 2,75E-04 | 5,88E-04 | ||
| 91,381,281 – 91,966,239 | -0,6102 | -0,5632 | 4,53E-04 | 8,66E-04 | ||
| SSC8 | 37,024,885 – 37,966,306 | -1,0159 | -0,8876 | 6,77E-05 | 3,58E-04 | GABRB1 CORIN |
| 37,513,284 – 38,036,453 | -0,8992 | -0,7349 | 2,59E-04 | 2,00E-03 | ||
| SSC13 | 196,187,718 - 196,460,966 | -0,6757 | -0,6677 | 9,06E-04 | 8,51E-04 | USP16 CFAP298 |
| 196,216,549 – 196,471,450 | -0,7089 | -0,6826 | 3,89E-04 | 4,98E-04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





