Introduction
Chemokines are cytokines that appeared with the emergence of chordates. There are 41 ligands and 21 receptor genes in the human genome[
1]. The chemotactic action of chemokines induces the effector cells to move against the gradient through the concentration gradient formed by the local tissue expression and secretion. This system plays an important role in embryonic development and immune cell homing. In addition to initial chemotaxis, chemokines are also involved in metabolic regulation[
2], angiogenesis[
3], and tissue development [
4].
CXCL14 is one of the earliest members appeared in the chemokine family [
5]. Compared with other members, it evolves extremely slowly and nonsense SNPs are rare, indicating its significant function. In healthy people, CXCL14 is usually highly expressed in tissues with a rapid renewal rate, such as epithelial cells of small intestine, colorectal, gastric mucosa, endometrium, mammary gland, skin, and other tissues. and it is low in resting state cells, such as the liver, muscle, connective tissue, brain, and other parenchymal cells. CXCL14 has shown abnormal expression levels in some immune diseases and a large number of tumor samples [6-8]. CXCL14 gene knockout has effects on the survival rate and metabolism of sugar and lipids in mouse embryos [
9].
Contrary to its important function, the receptor for CXCL14 has not yet been discovered. Ariadni Kouzeli et al [
10] systematically studied the interaction between CXCL14 and all genes of the chemokine receptor family and found that CXCL14 could not activate anyone. However, it is interesting that CXCL14 can synergistically promote the physiological activity of several other chemokines. Philipp von Hundelshausen et al. [
11] systematically studied the pattern of heterodimers formed by chemokines and found that a large number of heterodimers existed in the chemokine family. It is suggested that CXCL14 functions by promoting or inhibiting the activity of other chemokines. This may explain why the effects of CXCL14 may be opposite in different tissues and tumor types [
12], because the chemokines that partner with CXCL14 in different tissues can vary greatly in both type and concentration. CXCL14 appears as a tumor suppressor in colon cancer.
Tumor development is the result of activation of oncogenes and/or inactivation of tumor suppressor genes. It was previously thought that these genetic variations were all due to genetic mutations [
13]. However, it is now known that epigenetic variation is another pathway that leads to the silencing of tumor suppressor genes, and abnormal expression of genes caused by abnormal DNA methylation is more frequent than changes caused by gene copy number [
14]. We investigated the methylation level variation and gene mutation frequency of colon cancer tumors and found that the CXCL14 promoter was particularly hypermethylated in colon cancer samples.
The relationship between CXCL14 hypermethylation and gene silencing was also found in some tumor types. Tessema et al. found that CXCL14 was epigenetically silenced in lung cancer and considered this gene to be an important epigenetic therapeutic site [
7]. Cao et al. observed that the epigenetic silencing of CXCL14 in colon cancer is a mechanism leading to tumor metastasis and invasion [
15]. Hu et al. also found that abnormal methylation of promoters in gastric tumors resulted in inhibition of CXCL14 expression [
16].
To further reveal the changes of CXCL14 in colon cancer, this paper studied the changes of CXCL14 expression from normal tissue to colitis, to the primary tumor, and eventually to metastatic cancer by mining multiple datasets. We explored the changes of CXCL14 methylation in colon cancer and its correlation with CXCL14 gene silencing, prognosis, and important clinical markers, and revealed the silencing mechanism of CXCL14 and its important significance in the development of colon cancer.
Methods
mRNA Expression Data
The four groups of human colon cancer expression profiles analyzed in this study are as follows. (1) Memorial Sloan-Kettering Cancer Center, a total of 390 microarray data (E-GEOD-41258), Includes primary colon adenocarcinomas, adenomas, metastasis, and corresponding normal mucosae[
17]. (2) Different stage colorectal carcinomas (CRC) and inflammatory bowel diseases (IBD), (E-GEOD-4183), including biopsies of 15 patients with CRC, 15 with adenoma, 15 with IBD, and 8 healthy normal controls[
18]. (3) 36 CRC tissues and 24 non-cancerous colorectal tissue. (E-GEOD-23878). (4)Pairs of primary tumors, hepatic metastases, and normal controls [
19].
Two mouse primary colorectal cancer microarray databet was analyzed. They are about AOM/DSS induced colitis-associated cancer (CAC) model[
20,
21] and ApcMin/+/J transgenic mice-based spontaneous tumor model[
21].
DNA Methylation Data
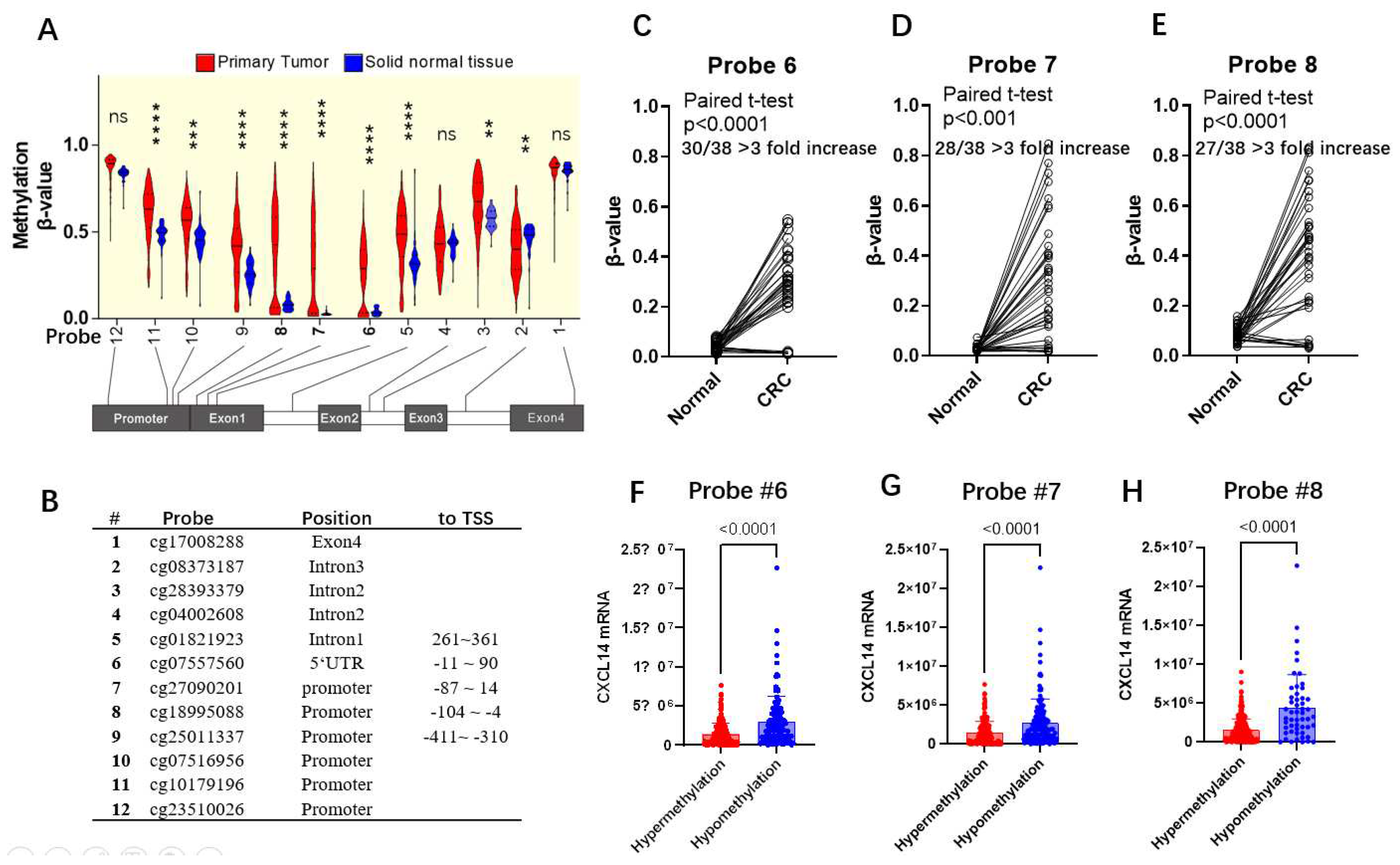
We used the mexpress database (
http://mexpress.be) to study the methylation status of the CXCL14 gene in the TCGA-COAD dataset [
22]. The methylation of sites of probe,6, 7, and 8, that was significantly hypermethylated in clinical colorectal carcinoma, were used to analyze associations with clinical indexes in
Table 2.
Survival Analysis
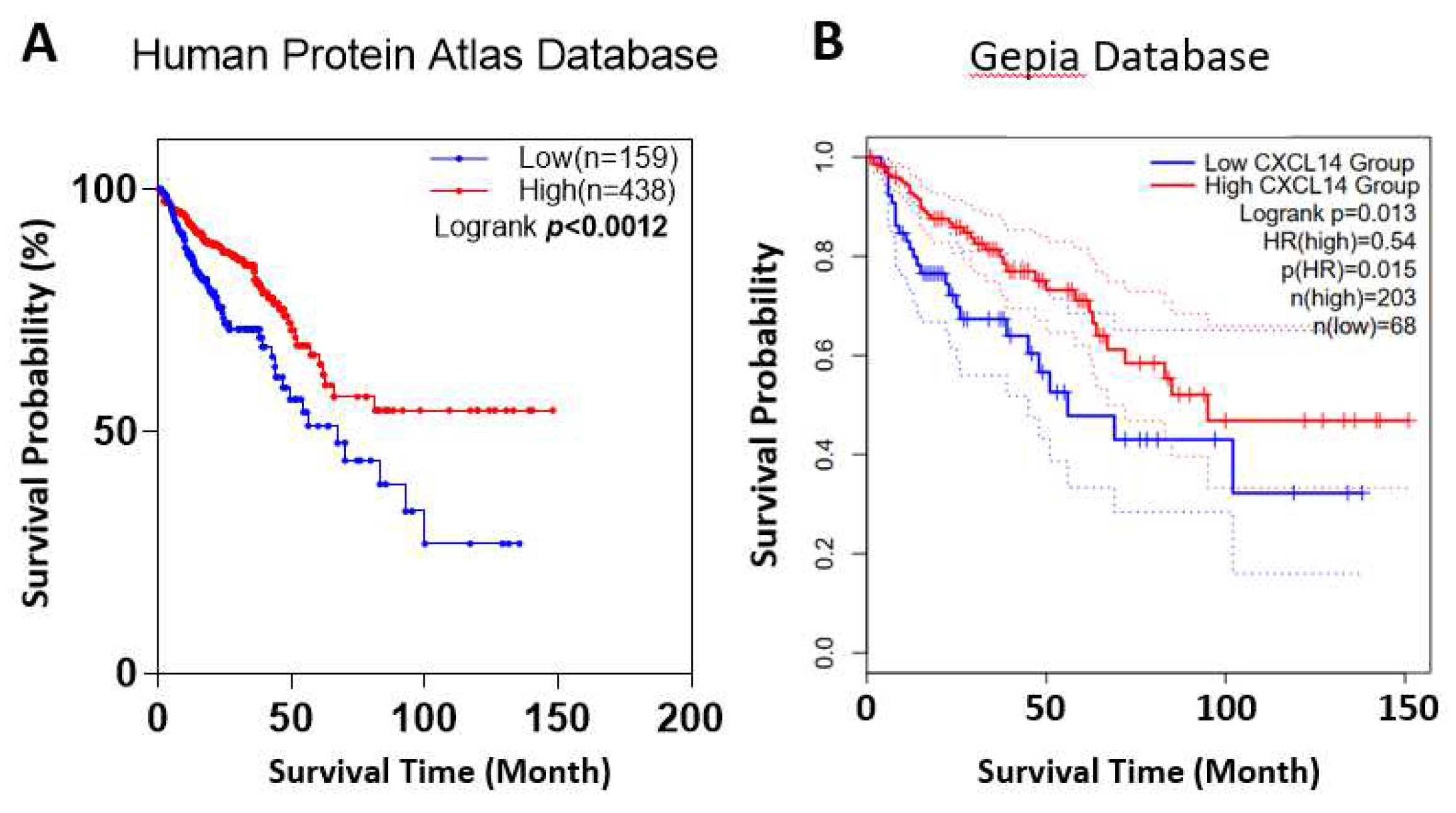
Cases were separated into groups of higher or lower CXCL14 mRNA expression by the best expression cut-off of the default setting of the Human Protein Atlas (
https://www.proteinatlas.org/) and Gepia2 (
http://gepia2.cancer-pku.cn) platform. For survival analysis of CXCL14 methylation, the cases were split into groups of hypermethylation and hypomethylation by the maxstat algorithm. Overall survival curves were achieved using the Kaplan-Meier assessment and the log-rank test.
Cell Culture
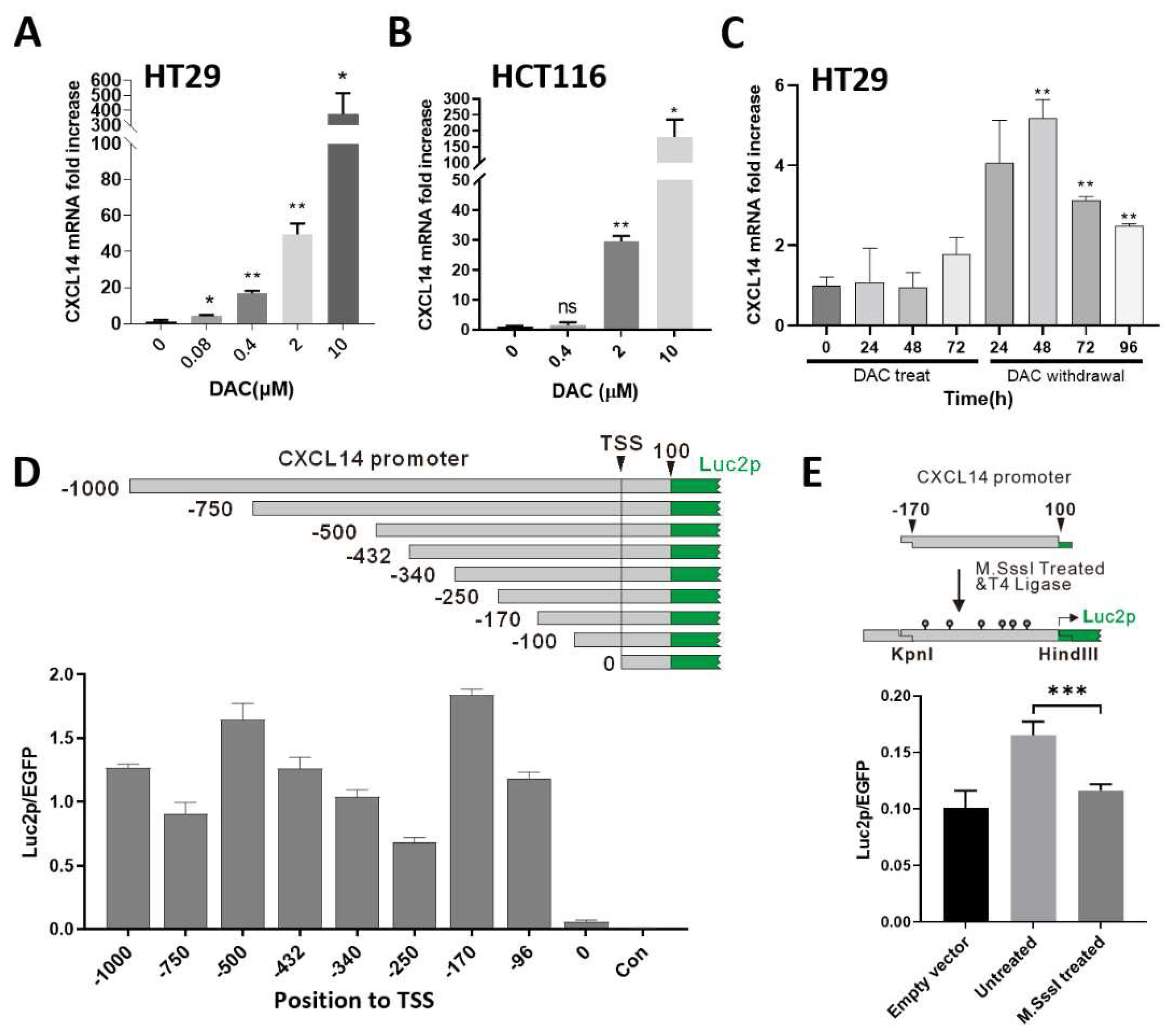
293T, HCT15 and HT29 cells were cultured in RPMI1640 medium containing 10% FBS, and HCT116 cells were cultured in McCoy’s 5A containing 10% FBS. The cell culture conditions were 37℃, 5% carbon dioxide, and 70% humidity. Decitabine (DAC) was purchased from MCE (HY-A0004, shanghai, China) and formulated with DMSO for 10mM storage. When DAC is treated with cells, DAC is diluted to the desired concentration in the medium and added to the cell culture dish.
Real-Time Quantitative PCR
Cell total RNA was extracted using the TaKaRa MiniBEST Universal RNA Extraction Kit (Cat 9767, TAKARA, Japan). Reverse transcription was performed using HiScript II 1st Strand cDNA Synthesis Kit (Cat R212-01, Vazyme, China). Quantitative PCR was performed using the TAKARA SybrGreen kit (RR820Q, TAKARA, Japan). The quantitative PCR primer for human CXCL14 was 5’-AAGCCAAAGTACCCGCACTG (Forward), 5’-GACCTCGGTACCTGGACACG (Reverse), the reference gene β-actin primers are: 5’-GTGAAGGTGACAGCAGTCGGTT(Forward), 5’-GAAGTGGGGTGGCTTTTAGGAT(Reverse). The relative expression of target genes was calculated by the ΔΔCt method.
Statistical Analysis
SPSS software was used for statistical analysis in this study. The student T-test is used to assess the significance between two sets of data. Paired T-tests are used to test the significance of differences between paired data from the same individual. Chi-square tests were used to examine the association between CXCL14 methylation and different clinical indexes. Spearman’s correlation approach was examined in methylation β-value and patient ages. *: p<0.05, **: p<0.01, ***: p<0.001.
Result
CXCL14 Silences as the Colorectal Cancer Progresses
We mined four groups of human colon cancer expression profile data sets and found that CXCL14 expression levels had obvious changes in different stages of colon cancer development.
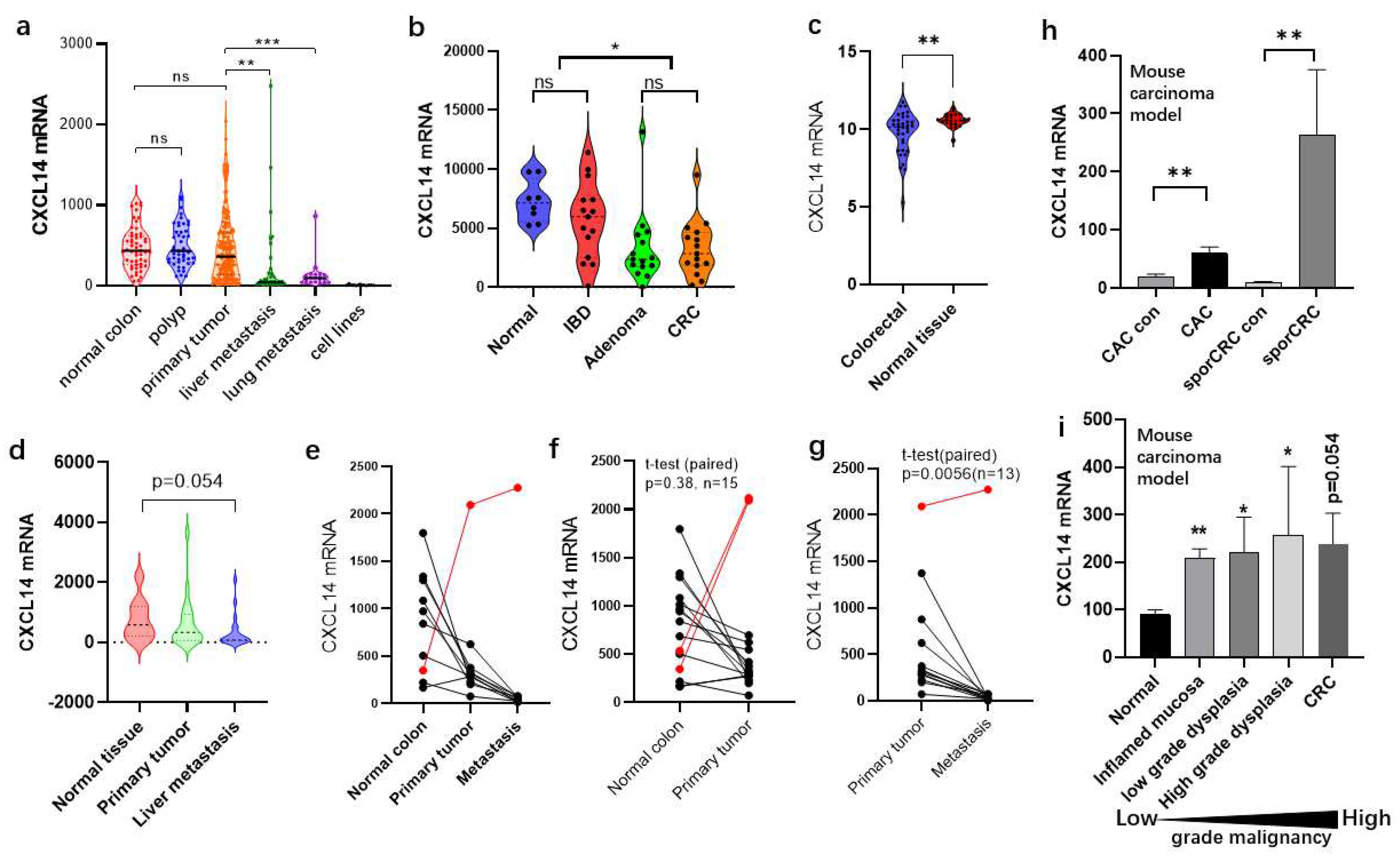
Figure 1A is RNA microarray data from 259 colon cancer samples and 12 human cell lines collected at Memorial Sloan-Kettering Cancer Center between 1992 and 2004[
18]. From the data, we can clearly see the expression changes of CXCL14 in different cancer stages. Firstly, CXCL14 expression levels were normally distributed in healthy tissues, but after the occurrence of intestinal polyps, the expression level dispersion increased, indicating a slight dysregulation in the expression level. After further development into primary tumors, the dispersion of expression levels increased further and the median decreased, indicating that the dysregulation of the expression level was intensified, and the selection of cancer cells in vivo was progressing in the direction of CXCL14 silencing. The phenomenon of gene silencing became more significant after liver metastasis or lung metastasis, indicating that CXCL14 silencing has certain advantages for tumor metastasis or tumor formation after metastasis. CXCL14 silencing occurred completely in 12 tumor cell lines.
In the dataset, E-GEOD-4183 (
Figure 1B), Orsolya Galamb, et al. collected 15 patients with CRC, 15 with adenoma, 15 with IBD, and 8 healthy normal controls[
18]. It was also found that in inflammatory bowel disease, although the mean expression of CXCL14 did not change significantly, the dispersion increased, and the CV% increased from 24% in healthy tissues to 83% in colitis tissues, indicating dysregulation of CXCL14 expression. CXCL14 levels decreased significantly with adenoma and colorectal cancer (CRC) (p<0.001).
In another set of expression profile data of colon cancer and adjacent tissues (
Figure 1C), it was also found that the expression of CXCL14 in colon cancer tissues decreased and was accompanied by an increase in dispersion.
In the last set of data, which was about a clinical follow-up investigation, Pierre Martineau et al. tracked and collected primary cancer tissue, metastatic tissue and healthy colon tissues from 19 tumor patients, and performed RNA microarray analysis[
17]. We analyzed the expression of CXCL14 and found that CXCL14 expression continued to decline during tumor progression (
Figure 1D-G).
From the above four sets of expression profile data, it can be seen that CXCL14 silencing becomes more and more significant with the increase of colon cancer malignancy.
After that, we investigated the expression profiles of two independent mouse colon cancer models. In the first experiment (
Figure 1H), two modeling methods were used to establish the primary model of colon cancer in mice [
20]. In the first, the CAC model was induced by AOM/DSS, and the second was spontaneous colon cancer modeling by C57BL/6-ApcMinC/Nju transgenic mice (SporCRC). The expression profiles of cancer tissues and adjacent tissues in the CAC group and SporCRC group were detected respectively, and it was found that the CXCL14 expression level in tumor tissues was higher than that in corresponding healthy control tissues. In the second model (
Figure 1I) [
21], the authors used a single AOM/DSS model, and it was observed that the expression level of CXCL14 increased.
Therefore, the expression changes of CXCL14 in human colon cancer and in mouse colon cancer development seem to be opposite, whether they are contradictory, we will discuss in the following.
Clinical data show that patients with higher CXCL14 expression have a better prognosis. Two colon cancer datasets from two databases, Protein Atlas and Gepia, respectively, showed that higher CXCL14 expression was associated with longer survival.
CXCL14 Methylation and Senescence
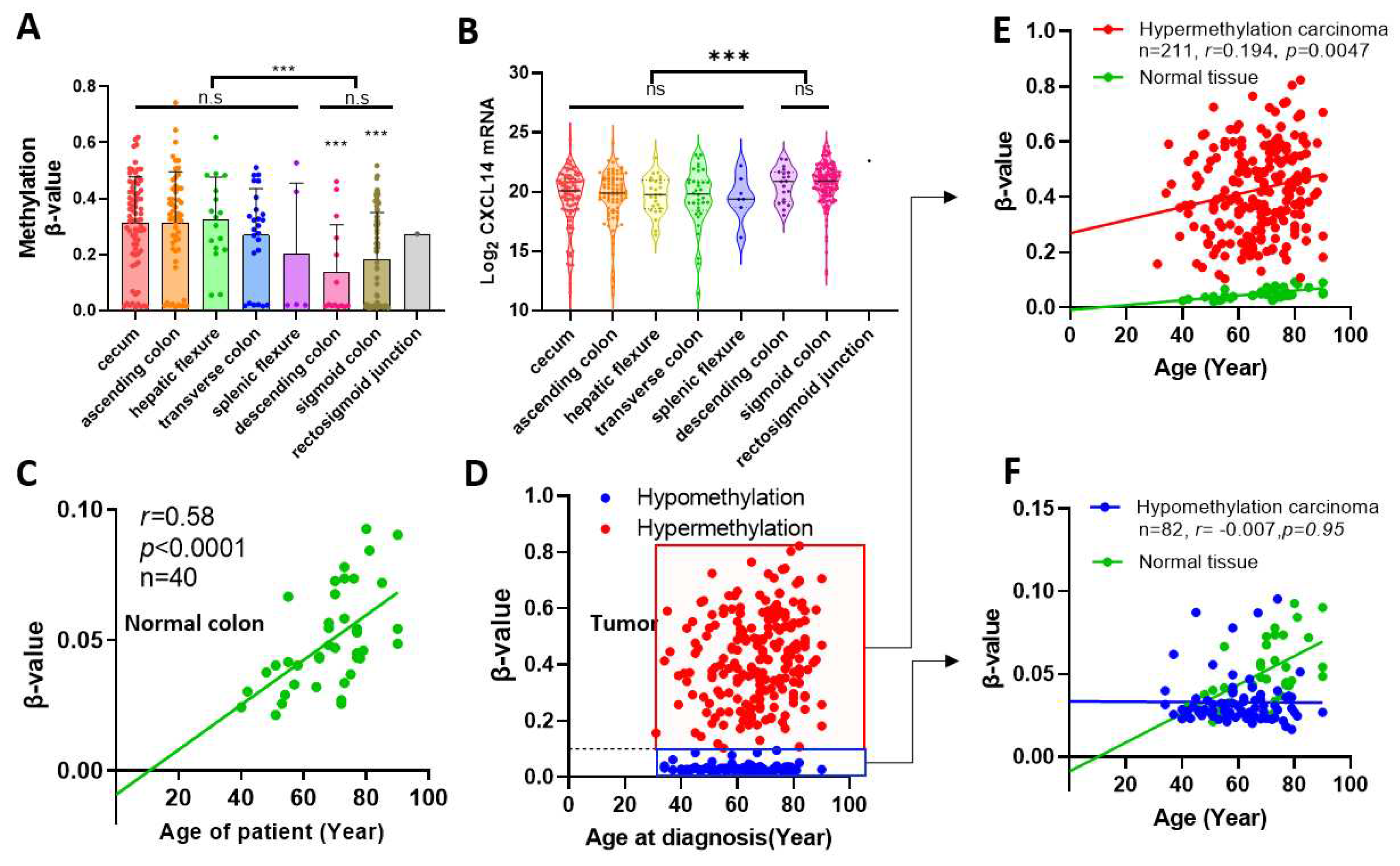
By correlation analysis, it was found that CXCL14 methylation increased slowly with age in normal colon tissue, and the correlation was very strong (r=0.58, p<0.0001). However, the methylation trend of CXCL14 in tumor samples showed a distinct differentiation. We used the upper limit of CXCL14 methylation level in normal colon samples, β-value=0.1, as the boundary to divide tumor samples into hypermethylated and hypomethylated groups. The hypermethylated group accounted for 72% of the total samples. Although elevated methylation was still linearly correlated with age in this sample, the degree of correlation was significantly reduced (r=0.194, p=0.0047). On the contrary, in the hypomethylated group (28% of total tumor samples), the methylation accumulation process tended to stall and did not increase with age, which was a different mode from both normal tissue and hypermethylated tumor samples.
CXCL14 Methylation was Associated with Survival
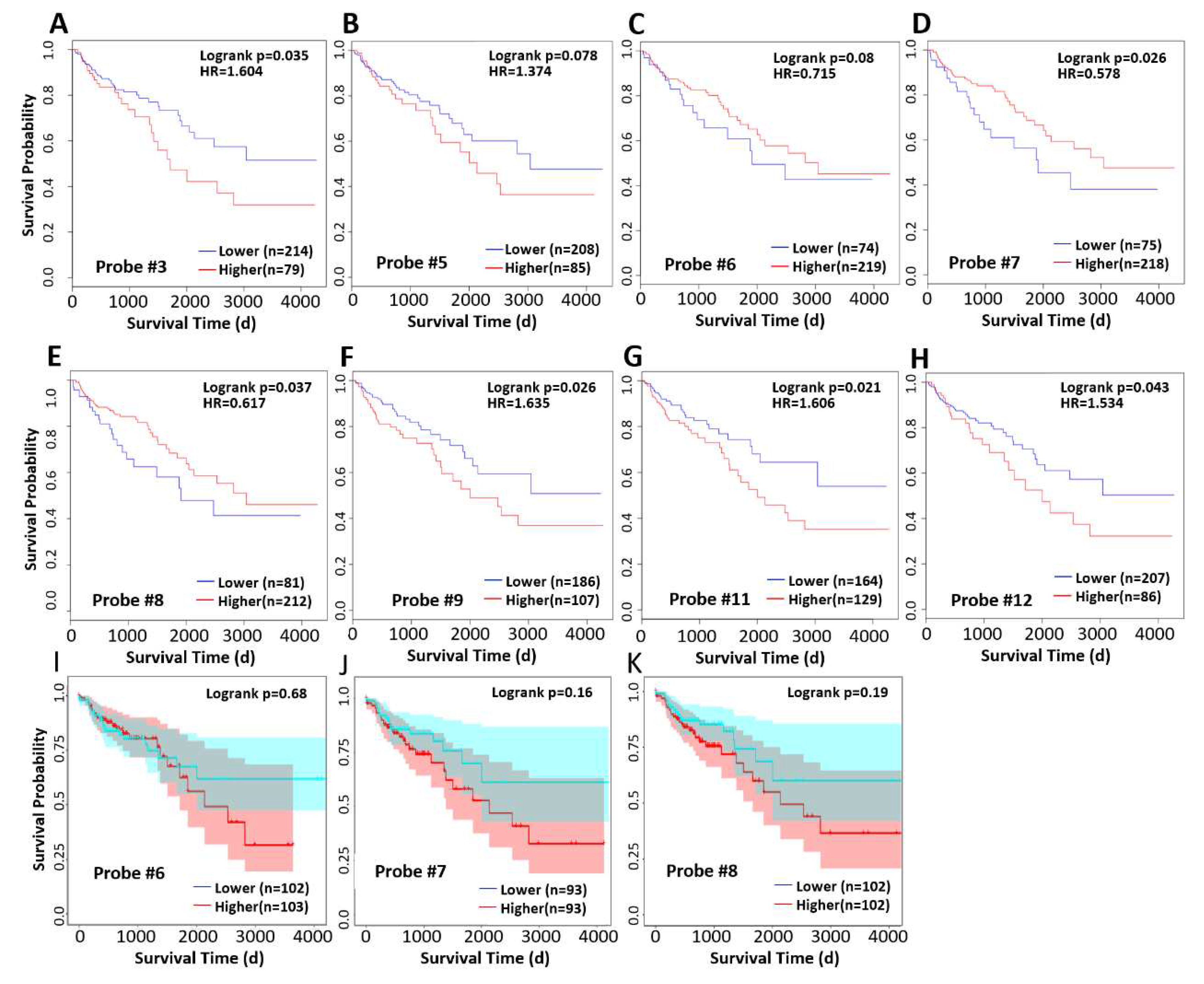
We analyzed the correlation between methylation levels and survival at different probe sites and found that higher methylation level at probe 3, 9, 11, and 12 was associated with worse prognosis, while high methylation levels at probe 6,7, and 8, in contrast, associated with longer survival (
Figure 5D-E). Since we had observed abnormally low methylation in some cases in
Figure 4D, we excluded these cases and reanalyzed the survival time, and found that high methylation level was associated with poor prognosis (
Figure 5I~J), although the correlation was not significant.
Discussion
In this study, we found significant differences in the regulation of CXCL14 expression between human colon cancer tumors and mouse colon cancer models. CXCL14 is generally silenced in clinical tumor tissues, while it is upregulated in primary mouse tumors. Whether this contradiction is based on the inherent differences between humans and mice or some other reason deserves further discussion. First, we know that human tumor tissues progress for years to decades before being discovered, so the clinical tumor samples we observe are the result of many mutations and clonal differentiation, and at the same time, many years of selection in vivo. While, the mouse tumor samples were all primary tumors that developed within a few months, with limited time for mutation and selection. So, there’s a difference in the stage between the human and mouse tumors. Secondly, it should be noted that although the expression of CXCL14 decreased with the increase of cancer cell malignancy in a general trend, the expression of CXCL14 increased in some tumors (
Figure 1A~D).
Taking the above two points together, we propose that the up-regulation of CXCL14 in the primary tumor of mice is not inconsistent with its silencing in clinical samples, but with its expression at different stages of tumor development. The mouse tumor model revealed the gene regulation pattern of CXCL14 in the early stage of tumor formation. Because of some currently unknown mechanisms, CXCL14 is activated during tumor formation, possibly by recruiting immune cells to participate in the tumor immune response. The clinical tumor samples reflect the CXCL14 status in the middle and late stages of tumor development. It is the result of genetic and/or epigenetic variation and has undergone long-term evolution and subclonal selection. However, this hypothesis is difficult to confirm in clinical samples, because early latent tumors are difficult to obtain clinically, so it is difficult to prove the hypothesis that CXCL14 is activated and then silenced during tumorigenesis. This hypothesis may be proved by long-term systematic animal tumor research.
The silencing of CXCL14 in colon cancer is mainly due to promoter hypermethylation. Since the database shows that the mutation rate of CXCL14 gene is very low in various tumor samples (not shown), the gene mutation is not the cause of CXCL14 silencing. Why in the tumor genome, some genes are more prone to change their expression levels through gene mutations, such as the well-known p53, while others, such as CXCL14, are more prone to epigenetic silencing? We believe that compared with simple deletion of genes, gene missense mutation can produce new phenotypes, one of the important aspects of which is to promote tumor metastasis. For example, some p53 mutants not only lose the cancer-suppressing function of the wild-type gene but also interfere with other signaling pathways, thus obtains survival advantages through the gain-of-function mechanism [
23]. With chemokines like CXCL14, tumor cells could gain a survival advantage by simply reducing immune surveillance through gene silencing. Because epigenetic modification is less stable than genetic information, in the process of tumor progression, epigenetic silencing becomes the fate of the genes that are more sensitive to epigenetic regulation in tumor cells.
CXCL14 methylation levels were correlated with tumor anatomical location. Firstly, we report that the level of CXCL14 promoter methylation in colon cancer is correlated with the anatomical site of the primary tumor. The data showed that tumors from the anterior segment of the colon had a greater methylation increase. Although the methylation level from the posterior segment of colon cancer is still elevated compared with normal paracancer tissue, the extent of the methylation level increase is significantly decreased compared with the tumor from the anterior segment. This result is interesting, but we can’t yet explain why this phenomenon occurs.
We also found a relationship between the degree of CXCL14 methylation and age. In healthy colon tissue, CXCL14 methylation was significantly positively correlated with age (r=0.58), the regression line almost passes through the origin of coordinates. This fits well with the epigenetic clock model [
24]. For tumor tissue, the samples can be divided into two subgroups, one group showed hypermethylation, while the other one showed hypomethylation. Hypermethylated groups exhibit faster methylation accumulation than healthy tissues, which is consistent with what has been reported in the literature [
25]. However in the hypomethylated group, the epigenetic clock stopped completely, and methylation levels did not accumulate with age. Whether this group of tumors shows a reversal of epigenetic aging, and which kind of tumors have better prognosis compared with those that accelerate epigenetic aging, is a question worthy of attention.
We investigated the correlation between methylation level and survival, and found that methylation level was significantly negatively correlated with overall survival at probe positions 3-5, 9-12, but was opposite at probe positions 6, 7, and 8 (
Figure 5C-E). To explain this contradiction, we specifically excluded the abnormal hypomethylated samples at 6, 7, and 8 probe locations and re-performed correlation analysis. We found that after such treatment, methylation levels at these three probe locations were negatively correlated with survival, which was consistent with the trend in the other locations. Therefore, we believe that the hypomethylated cases at the 6,7,8 probe location show different characteristics from the hypermethylated cases, and they have a worse prognosis than the CXCL14 promoter hypermethylated cases, and the mechanism needs to be further studied.
In summary, this paper describes the dynamic profile of CXCL14 expression with tumor development from the time axis through bioinformatics mining. The results revealed a dynamic process in which CXCL14 expression is up-regulated in early cancerous tissues and goes to silencing in metastatic tumors, suggesting that colon cancer cells gain proliferation and metastasis advantages through CXCL14 silencing that caused by its promoter methylation. Survival analysis confirmed that the gene silencing of CXCL14 in clinical colon cancer samples was associated with poor prognosis and revealed its anti-colon cancer function. In vitro experiments have shown that CXCL14 promoter methylation is an important cause of gene silencing. However, the mechanism of CXCL14 silencing promoting the progression of colon cancer is still unclear, which may be related to the immune surveillance of CXCL14[3,26-28] or/and antiangiogenic function [
29].
Funding& Acknowledgements
B. Ma thanks support from Natural Science Foundation of China (Grant No. 32171246) and Shanghai Municipal Government Science Innovation grant 21JC1403700. Y. Wang thanks support from the grants from the National Natural Science Foundation of China (No.32200531), the Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2022QN114 and YG2022QN082), and Startup Fund for Young Faculty at SJTU (SFYF at SJTU).
References
- Shields, D.C. Molecular evolution of CXC chemokines and receptors. Trends Immunol 2003, 24, 355. [Google Scholar] [CrossRef] [PubMed]
- Nara, N.; Nakayama, Y.; Okamoto, S.; Tamura, H.; Kiyono, M.; Muraoka, M.; Tanaka, K.; Taya, C.; Shitara, H.; Ishii, R.; et al. Disruption of CXC motif chemokine ligand-14 in mice ameliorates obesity-induced insulin resistance. J Biol Chem 2007, 282, 30794–30803. [Google Scholar] [CrossRef]
- Shellenberger, T.D.; Wang, M.; Gujrati, M.; Jayakumar, A.; Strieter, R.M.; Burdick, M.D.; Ioannides, C.G.; Efferson, C.L.; El-Naggar, A.K.; Roberts, D.; et al. BRAK/CXCL14 is a potent inhibitor of angiogenesis and a chemotactic factor for immature dendritic cells. Cancer Res 2004, 64, 8262–8270. [Google Scholar] [CrossRef]
- Kuang, H.; Chen, Q.; Zhang, Y.; Zhang, L.; Peng, H.; Ning, L.; Cao, Y.; Duan, E. The cytokine gene CXCL14 restricts human trophoblast cell invasion by suppressing gelatinase activity. Endocrinology 2009, 150, 5596–5605. [Google Scholar] [CrossRef] [PubMed]
- Huising, M.O.; Stet, R.J.; Kruiswijk, C.P.; Savelkoul, H.F.; Lidy Verburg-van Kemenade, B.M. Molecular evolution of CXC chemokines: extant CXC chemokines originate from the CNS. Trends Immunol 2003, 24, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Guo, L.; Tian, J.; He, H.; Marinova, E.; Zhang, P.; Zheng, B.; Han, S. Overexpression of CXC chemokine ligand 14 exacerbates collagen-induced arthritis. J Immunol 2010, 184, 4455–4459. [Google Scholar] [CrossRef]
- Tessema, M.; Klinge, D.M.; Yingling, C.M.; Do, K.; Van Neste, L.; Belinsky, S.A. Re-expression of CXCL14, a common target for epigenetic silencing in lung cancer, induces tumor necrosis. Oncogene 2010, 29, 5159–5170. [Google Scholar] [CrossRef]
- Song, E.Y.; Shurin, M.R.; Tourkova, I.L.; Gutkin, D.W.; Shurin, G.V. Epigenetic mechanisms of promigratory chemokine CXCL14 regulation in human prostate cancer cells. Cancer Res 2010, 70, 4394–4401. [Google Scholar] [CrossRef]
- Hara, T.; Nakayama, Y. CXCL14 and insulin action. Vitam Horm 2009, 80, 107–123. [Google Scholar] [CrossRef] [PubMed]
- Kouzeli, A.; Collins, P.J.; Metzemaekers, M.; Meyrath, M.; Szpakowska, M.; Artinger, M.; Struyf, S.; Proost, P.; Chevigne, A.; Legler, D.F.; et al. CXCL14 Preferentially Synergizes With Homeostatic Chemokine Receptor Systems. Frontiers in immunology 2020, 11, 561404. [Google Scholar] [CrossRef]
- von Hundelshausen, P.; Agten, S.M.; Eckardt, V.; Blanchet, X.; Schmitt, M.M.; Ippel, H.; Neideck, C.; Bidzhekov, K.; Leberzammer, J.; Wichapong, K.; et al. Chemokine interactome mapping enables tailored intervention in acute and chronic inflammation. Sci Transl Med 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Tanegashima, K. Pleiotropic functions of the CXC-type chemokine CXCL14 in mammals. J Biochem 2012, 151, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat Med 2004, 10, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Zardo, G.; Tiirikainen, M.I.; Hong, C.; Misra, A.; Feuerstein, B.G.; Volik, S.; Collins, C.C.; Lamborn, K.R.; Bollen, A.; Pinkel, D.; et al. Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nat Genet 2002, 32, 453–458. [Google Scholar] [CrossRef]
- Cao, B.; Yang, Y.; Pan, Y.; Jia, Y.; Brock, M.V.; Herman, J.G.; Guo, M. Epigenetic silencing of CXCL14 induced colorectal cancer migration and invasion. Discov Med 2013, 16, 137–147. [Google Scholar]
- Hu, C.; Lin, F.; Zhu, G.; Xue, X.; Ding, Y.; Zhao, Z.; Zhang, L.; Shen, X. Abnormal hypermethylation of promoter region downregulates chemokine CXC ligand 14 expression in gastric cancer. Int J Oncol 2013, 43, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Sheffer, M.; Bacolod, M.D.; Zuk, O.; Giardina, S.F.; Pincas, H.; Barany, F.; Paty, P.B.; Gerald, W.L.; Notterman, D.A.; Domany, E. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A 2009, 106, 7131–7136. [Google Scholar] [CrossRef]
- Galamb, O.; Gyorffy, B.; Sipos, F.; Spisak, S.; Nemeth, A.M.; Miheller, P.; Tulassay, Z.; Dinya, E.; Molnar, B. Inflammation, adenoma and cancer: objective classification of colon biopsy specimens with gene expression signature. Dis Markers 2008, 25, 1–16. [Google Scholar] [CrossRef]
- Del Rio, M.; Mollevi, C.; Vezzio-Vie, N.; Bibeau, F.; Ychou, M.; Martineau, P. Specific extracellular matrix remodeling signature of colon hepatic metastases. PLoS One 2013, 8, e74599. [Google Scholar] [CrossRef]
- Neufert, C.; Becker, C.; Tureci, O.; Waldner, M.J.; Backert, I.; Floh, K.; Atreya, I.; Leppkes, M.; Jefremow, A.; Vieth, M.; et al. Tumor fibroblast-derived epiregulin promotes growth of colitis-associated neoplasms through ERK. J Clin Invest 2013, 123, 1428–1443. [Google Scholar] [CrossRef]
- Tang, A.; Li, N.; Li, X.; Yang, H.; Wang, W.; Zhang, L.; Li, G.; Xiong, W.; Ma, J.; Shen, S. Dynamic activation of the key pathways: linking colitis to colorectal cancer in a mouse model. Carcinogenesis 2012, 33, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Kon, N.; Yi, J.; Zhao, H.; Zhang, W.; Tang, Q.; Li, H.; Kobayashi, H.; Li, Z.; Duan, S.; et al. Specific regulation of BACH1 by the hotspot mutant p53(R175H) reveals a distinct gain-of-function mechanism. Nature cancer 2023, 4, 564–581. [Google Scholar] [CrossRef]
- Seale, K.; Horvath, S.; Teschendorff, A.; Eynon, N.; Voisin, S. Making sense of the ageing methylome. Nat Rev Genet 2022, 23, 585–605. [Google Scholar] [CrossRef] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013, 49, 359–367. [Google Scholar] [CrossRef]
- Starnes, T.; Rasila, K.K.; Robertson, M.J.; Brahmi, Z.; Dahl, R.; Christopherson, K.; Hromas, R. The chemokine CXCL14 (BRAK) stimulates activated NK cell migration: implications for the downregulation of CXCL14 in malignancy. Exp Hematol 2006, 34, 1101–1105. [Google Scholar] [CrossRef]
- Ozawa, S.; Kato, Y.; Kubota, E.; Hata, R. BRAK/CXCL14 expression in oral carcinoma cells completely suppresses tumor cell xenografts in SCID mouse. Biomed Res 2009, 30, 315–318. [Google Scholar] [CrossRef]
- Shurin, G.V.; Ferris, R.L.; Tourkova, I.L.; Perez, L.; Lokshin, A.; Balkir, L.; Collins, B.; Chatta, G.S.; Shurin, M.R. Loss of new chemokine CXCL14 in tumor tissue is associated with low infiltration by dendritic cells (DC), while restoration of human CXCL14 expression in tumor cells causes attraction of DC both in vitro and in vivo. J Immunol 2005, 174, 5490–5498. [Google Scholar] [CrossRef]
- Liu, Y.; Chang, Q.; Wu, X.; Yu, Y.; Zhang, H. Effect of chemokine CXCL14 on in vitro angiogenesis of human hepatocellular carcinoma cells. Arch Physiol Biochem 2022, 128, 1316–1322. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).