Submitted:
03 October 2023
Posted:
04 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
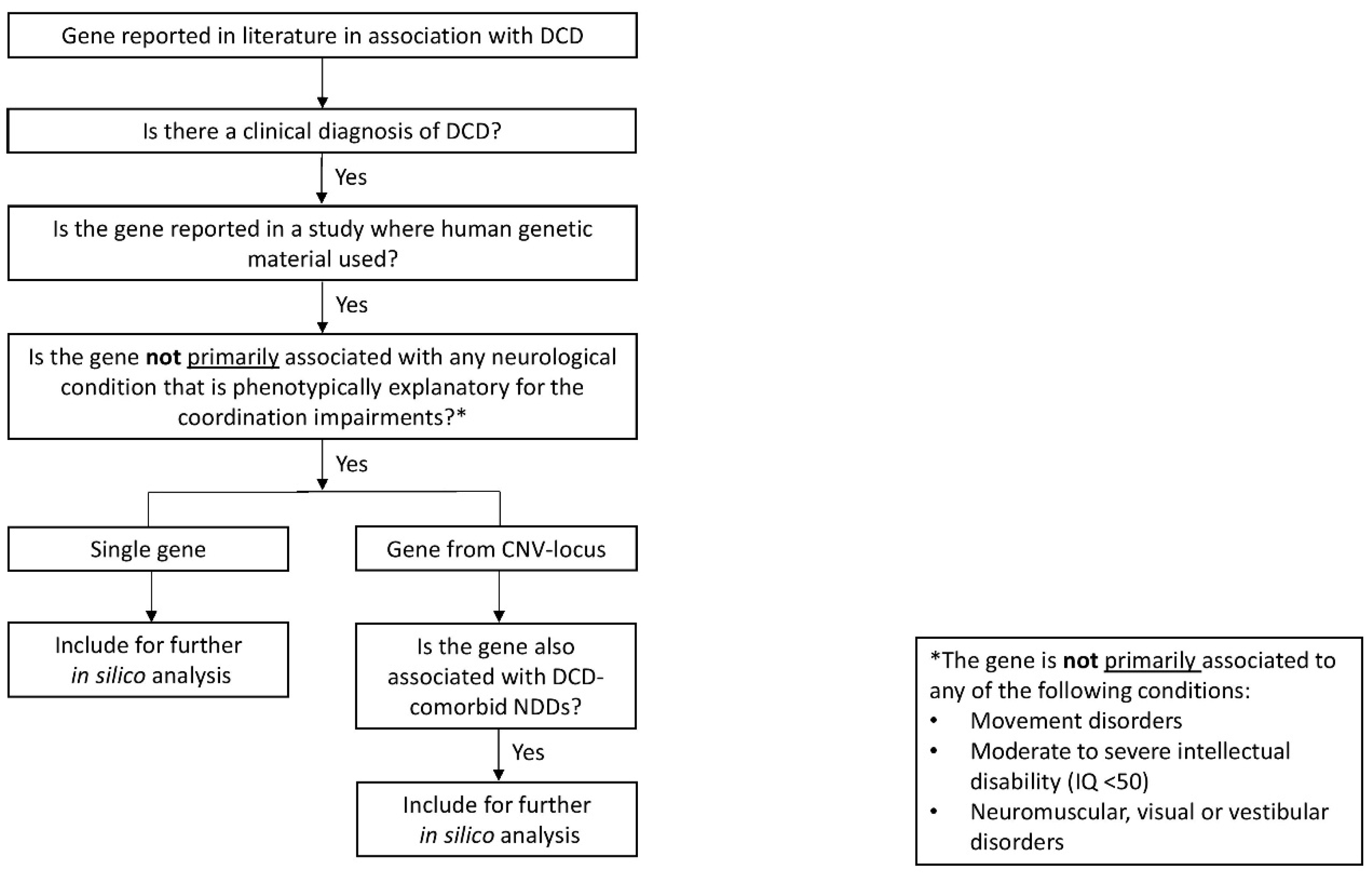
2.1. First aim: Gene identification in literature
2.1.1. Comprehensive literature review and gene inclusion criteria
2.2. Second aim: Analysis of pathogenetic mechanisms underlying DCD
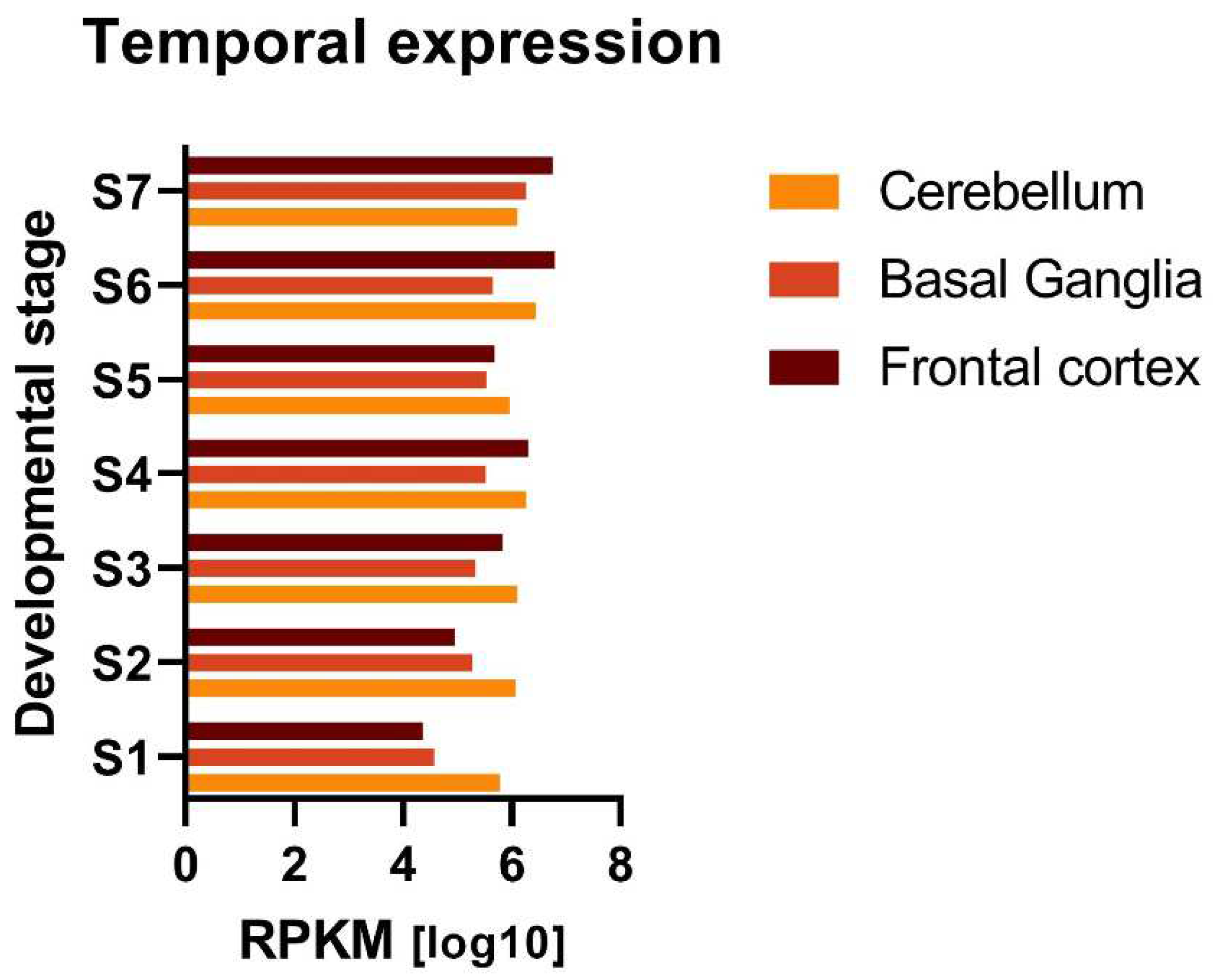
2.2.1. Temporal gene expression analysis
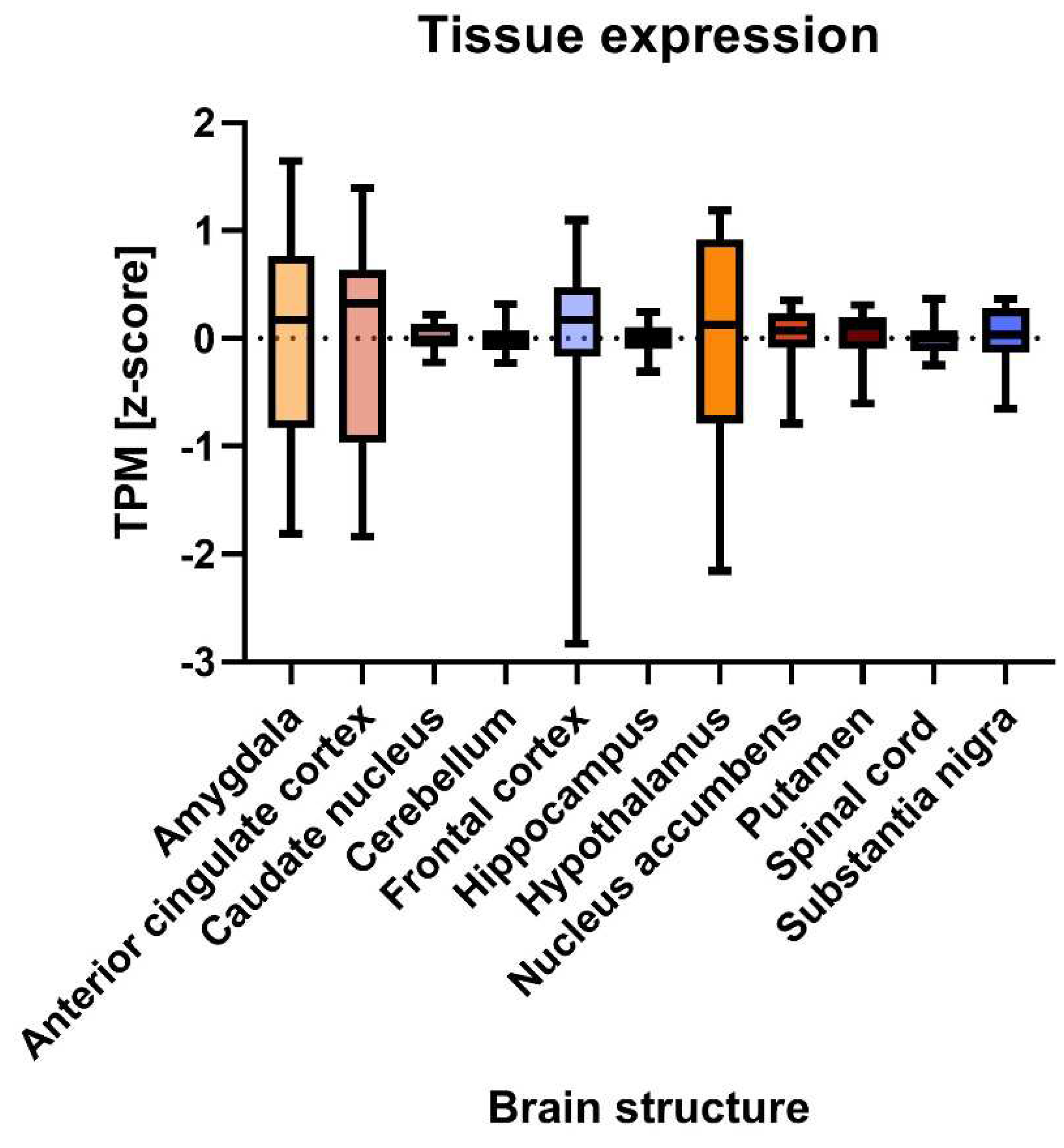
2.2.2. Tissue gene expression analysis
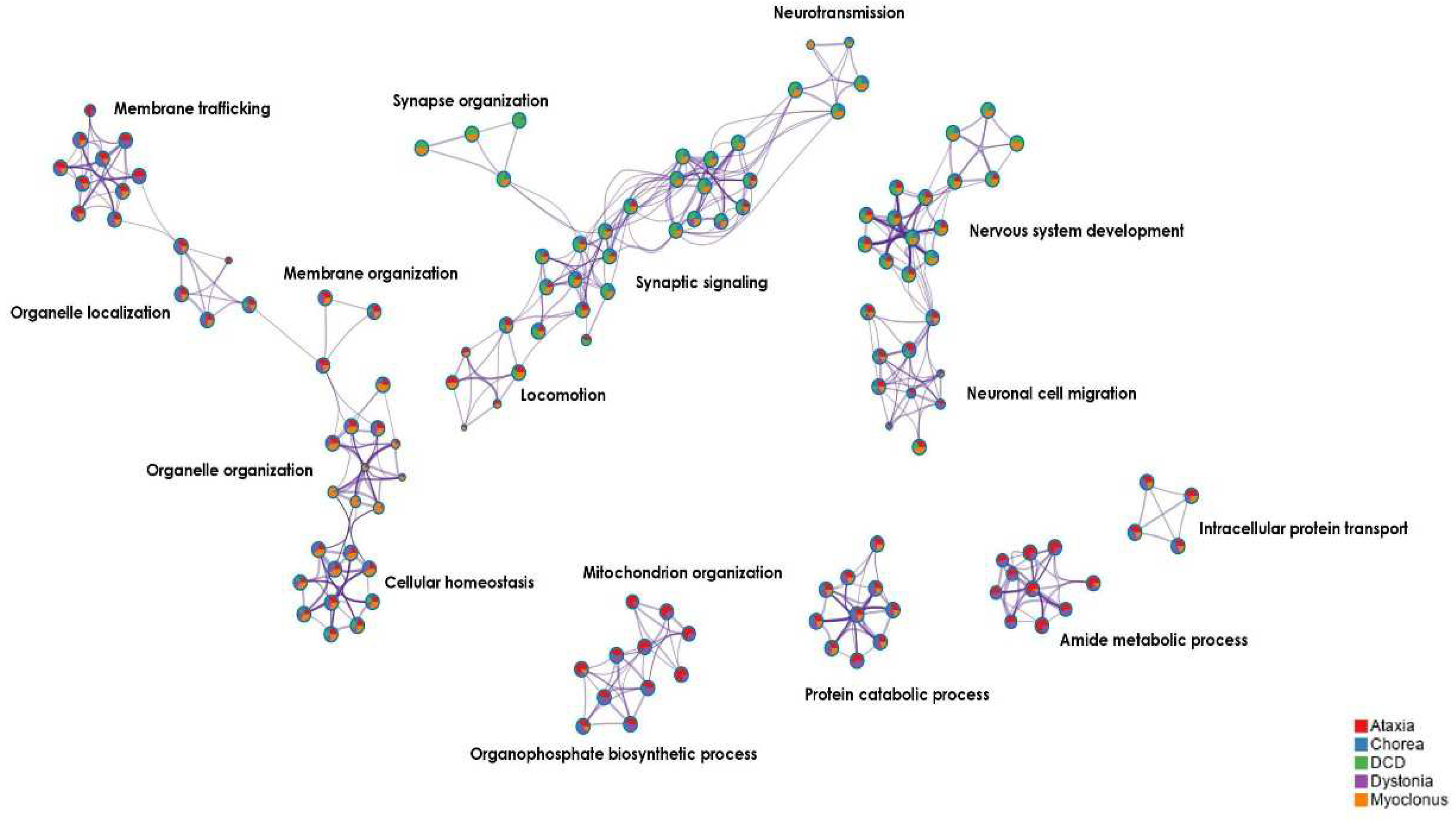
2.2.3. Functional enrichment and biological pathway analysis in the DCD-associated gene co-expression network
2.3. Third aim: Analysis of putative pathogenetic overlap between DCD and ataxia, chorea, dystonia and/or myoclonus
2.3.1. Functional enrichment and biological pathway analysis in the shared DCD-associated/MD gene co-expression network
2.4. Statistical analyses
3. Results
3.1. First aim: Gene identification in literature
3.2. Second aim: Analysis of pathogenetic mechanisms underlying DCD
3.2.1. Ubiquitous expression of DCD-associated genes in brain throughout development
3.2.2. Three main biological themes in the DCD-associated gene co-expression network
3.3. Third aim: Analysis of putative pathogenetic overlap between DCD and ataxia, chorea, dystonia and/or myoclonus
3.3.1. Three main biological themes in the shared DCD/MD gene co-expression network
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Francés, L.; Quintero, J.; Fernández, A.; Ruiz, A.; Caules, J.; Fillon, G.; Hervás, A.; Soler, C.V. Current state of knowledge on the prevalence of neurodevelopmental disorders in childhood according to the DSM-5: a systematic review in accordance with the PRISMA criteria. Child Adolesc. Psychiatry Ment. Health. 2022, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. Neurodevelopmental Disorders. In Diagnostic and Statistical Manual of Mental Disorders, 5th ed; American Psychiatric Association Publishing: Arlington, VA, United States, 2013; pp. 74–77. [Google Scholar]
- Blank, R.; Barnett, A.L.; Cairney, J.; Green, D.; Kirby, A.; Polatajko, H.; Rosenblum, S.; Smits-Engelsman, B.; Sugden, D.; Wilson, P.; et al. International clinical practice recommendations on the definition, diagnosis, assessment, intervention, and psychosocial aspects of developmental coordination disorder. Dev. Med. Child Neurol. 2019, 61, 242–285. [Google Scholar] [CrossRef] [PubMed]
- Tal Saban, M.; Kirby, A. Adulthood in Developmental Coordination Disorder (DCD): a Review of Current Literature Based on ICF Perspective. Curr. Dev. Disord. Reports. 2018, 5, 9–17. [Google Scholar] [CrossRef]
- Ip, A.; Mickelson, E.C.R.; Zwicker, J.G. Assessment, diagnosis, and management of developmental coordination disorder. Paediatr. Child Health. 2021, 26, 375–378. [Google Scholar] [CrossRef]
- Visser, J. Developmental coordination disorder: a review of research on subtypes and comorbidities. Hum. Mov. Sci. 2003, 22, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Visser, L.; Röschinger, J.; Barck, K.; Büttner, G.; Hasselhorn, M. Learning Difficulties in Children with Symptoms of DCD And/or ADHD: Analyses from a Categorical and a Continuous Approach. Int. J. Disabil. Dev. Educ. 2022, 69, 69,1505–1521. [Google Scholar] [CrossRef]
- Hunt, J.; Zwicker, J.; Godecke, E.; Raynor, A. Assessing children to identify developmental coordination disorder: A survey of occupational therapists in Australia. Aust. Occup. Ther. J. 2023, 70, 420–433. [Google Scholar] [CrossRef]
- Dominguez-Vega, Z.T.; Dubber, D.; Elting, J.W.J.; Sival, D.A.; Maurits, N.M. Instrumented classification of patients with early onset ataxia or developmental coordination disorder and healthy control children combining information from three upper limb SARA tests. Eur. J. Paediatr. Neurol. 2021, 34, 74–83. [Google Scholar] [CrossRef]
- Tang, W.; van Ooijen, P.M.A.; Sival, D.A.; Maurits, N.M. 2D Gait Skeleton Data Normalization for Quantitative Assessment of Movement Disorders from Freehand Single Camera Video Recordings. Sensors. 2022, 22, 4245. [Google Scholar] [CrossRef]
- Lawerman, T.F.; Brandsma, R.; Maurits, N.M.; Martinez-Manzanera, O.; Verschuuren-Bemelmans, C.C.; Lunsing, R.J.; Brouwer, O.F.; Kremer, H.P.H.; Sival, D.A. Paediatric motor phenotypes in early-onset ataxia, developmental coordination disorder, and central hypotonia. Dev. Med. Child. Neurol. 2020, 62, 75–82. [Google Scholar] [CrossRef]
- Kuiper, M.J.; Brandsma, R.; Lunsing, R.J.; Eggink, H.; Ter Horst, H.J.; Bos, A.F.; Sival, D.A. The neurological phenotype of developmental motor patterns during early childhood. Brain Behav. 2019, 9, e01153. [Google Scholar] [CrossRef]
- van Hoorn, J.F.; Schoemaker, M.M.; Stuive, I.; Dijkstra, P.U.; Rodrigues Trigo Pereira, F.; Van der Sluis, C.K.; Hadders-Algra, M. Risk factors in early life for developmental coordination disorder:a scoping review. Dev. Med. Child. Neurol. 2021, 63, 511–519. [Google Scholar] [CrossRef]
- Hadders-Algra, M. Developmental Coordination Disorder: Is Clumsy Motor Behavior Caused By a Lesion of the Brain At Early Age? Neural Plast. 2003, 10, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.; Sirigu, A. Developmental coordination disorder: core sensori-motor deficits, neurobiology and etiology. Neuropsychologia. 2015, 79, 272–287. [Google Scholar] [CrossRef] [PubMed]
- Gaines, R.; Collins, D.; Boycott, K.; Missiuna, C.; DeLaat, D.; Soucie, H. Clinical expression of developmental coordination disorder in a large Canadian family. Paediatr. Child Health. 2008, 13, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Anckarsäter, H.; Lundström, S.; Kollber, L.; Kerekes, N.; Palm, C.; Carlström, E.; Langström, N.; Magnusson, P.K.E.; Halldner, L.; Bölte, S.; et al. The child and adolescent twin study in Sweden (CATSS). Twin Res. Hum. Genet. 2011, 14, 495–508. [Google Scholar] [CrossRef]
- Lichtenstein, P.; Carlström, E.; Råstam, M.; Gillberg, C.; Anckarsäter, H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am. J. Psychiatry. 2010, 167, 1357–1363. [Google Scholar] [CrossRef]
- Martin, N.C.; Piek, J.P.; Hay, D. DCD and ADHD: A genetic study of their shared aetiology. Hum. Mov. Sci. 2006, 25, 110–124. [Google Scholar] [CrossRef]
- Ketcheson, L.R.; Pitchford, E.A.; Wentz, C.F. The Relationship Between Developmental Coordination Disorder and Concurrent Deficits in Social Communication and Repetitive Behaviors Among Children with Autism Spectrum Disorder. Autism Res. 2021, 14, 804–816. [Google Scholar] [CrossRef]
- Kopp, S.; Beckung, E.; Gillberg, C. Developmental coordination disorder and other motor control problems in girls with autism spectrum disorder and/or attention-deficit/hyperactivity disorder. Res. Dev. Disabil. 2010, 31, 350–361. [Google Scholar] [CrossRef]
- Mosca, S.J.; Langevin, L.M.; Dewey, D.; Innes, A.M.; Lionel, A.C.; Marshall, C.C.; Scherer, S.W.; Parboosingh, J.S.; Bernier, F.P. Copy-number variations are enriched for neurodevelopmental genes in children with developmental coordination disorder. J. Med. Genet. 2016, 53, 812–819. [Google Scholar] [CrossRef]
- Dewey, D. What Is Comorbidity and Why Does It Matter in Neurodevelopmental Disorders? Curr. Dev. Disord. Reports. 2018, 5, 235–242. [Google Scholar] [CrossRef]
- Mountford, H.S.; Hill, A.; Barnett, A.L.; Newbury, D.F. Genome-Wide Association Study of Motor Coordination. Front. Hum. Neurosci. 2021, 286, 669902. [Google Scholar] [CrossRef]
- Fliers, E.A.; Vasquez, A.A.; Poelmans, G.; Rommelse, N.; Altink, M.; Buschgens, C.; Asherson, P.; Banaschewski, T.; Ebstein, R.; Gill, M.; et al. Genome-wide association study of motor coordination problems in ADHD identifies genes for brain and muscle function. World J. Biol. Psychiatry. 2012, 13, 211–222. [Google Scholar] [CrossRef]
- Gill, K.; Rajan, J.R.S.; Chow, E.; Ashbrook, D.; Williams, R.W.; Zwicker, J.G.; Goldowitz, D. Investigating mouse motor coordination using quantitative trait locus analysis to model the genetic underpinnings of developmental coordination disorder. bioRxiv. 2022, 495138. [Google Scholar]
- Pérez-Dueñas, B.; Gorman, K.; Marcé-Grau, A.; Ortigoza-Escobar, J.D.; Macaya, A.; Danti, F. R.; Barwick, K.; Papandreou, A.; Ng, J.; Meyer, E.; et al. The Genetic Landscape of Complex Childhood-Onset Hyperkinetic Movement Disorders. Mov. Dis. 2022, 37, 2197–2209. [Google Scholar] [CrossRef] [PubMed]
- Brandsma, R.; van Egmond, M.E.; Tijssen, M.A.; the Groningen Movement Disorder Expertise Centre. Diagnostic approach to paediatric movement disorders: a clinical practice guide. Dev. Med. Child. Neurol. 2021, 63, 63–252. [Google Scholar] [CrossRef] [PubMed]
- ICD-10 Version:2019. Available online: https://icd.who.int/browse10/2019/en (accessed on 22/05/2023).
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Gao, Z.; Davis, C.; Thomas, A.M.; Economo, M.N.; Abrego, A.M.; Svoboda, K.; De Zeeuw, C.I.; Li, N. A cortico-cerebellar loop for motor planning. Nature. 2018, 563, 113–116. [Google Scholar] [CrossRef]
- Leisman, G.; Braun-Benjamin, O.; Melillo, R. Cognitive-motor interactions of the basal ganglia in development. Front. Syst. Neurosci. 2014, 8, 16. [Google Scholar] [CrossRef]
- Wierenga, L.; Langen, M.; Ambrosino, S.; van Dijk, S.; Oranje, B.; Durston, S. Typical development of basal ganglia, hippocampus, amygdala and cerebellum from age 7 to 24. Neuroimage. 2014, 96, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Eidhof, I, van de Warrenburg, B.P.; Schenck, A. Integrative network and brain expression analysis reveals mechanistic modules in ataxia. J. Med. Genet. 2019, 56, 283–292. [CrossRef] [PubMed]
- Garofalo, M.; Vansenne, F.; Verbeek, D.S.; Sival, D.A. The Pathogenetic Basis for a Disease Continuum in Early- and Late-onset Ataxia-Dystonia supports a Unified Genetic Diagnostic Approach. Eur. J. Paediatr. Neurol. 2023, 43, 44–51. [Google Scholar] [CrossRef]
- Zwicker, J.G.; Missiuna, C.; Harris, S.R.; Boyd, L.A. Developmental coordination disorder: a review and update. Eur. J. Paediatr. Neurol. 2012, 16, 573–581. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020, 369, 1318–133. [Google Scholar] [CrossRef] [PubMed]
- de Klein, N.; Tsai, E.A.; Vochteloo, M.; Baird, D.; Huang, Y.; Chen, C.Y.; van Dam, S.; Oelen, R.; Deelen, P.; Bakker, O.B.; et al. Brain expression quantitative trait locus and network analysis reveals downstream effects and putative drivers for brain-related diseases. bioRxiv. 2023, 55, 377–388. [Google Scholar]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulous, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.A.; Kutmon, M.; Bohler, A.; Waagmeester, A.; Evelo, C.T.; Willighagen, E.L. Understanding signaling and metabolic paths using semantified and harmonized information about biological interactions. PLoS One. 2022, 17, e0263057. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N. S.; Wang, J. T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Lange, L.M.; Gonzalez-Latapi, P.; Rajalingam, R.; Tijssen, M.A.J.; Ebrahimi-Fakhari, D.; Gabbert, C.; Ganos, C.; Ghosh, R.; Kumar, K.R.; Lang, A.E.; et al. Nomenclature of Genetic Movement Disorders: Recommendations of the International Parkinson and Movement Disorder Society Task Force–An Update. Mov. Disord. 2022, 37, 905–935. [Google Scholar] [CrossRef]
- Genoomdiagnostiek UMCG. LAB-F0701 Genenlijst panels. Available online: https://www.umcg.nl/-/afdeling/genetica/aanvragen-genoomdiagnostiek (accessed on 13/03/2023).
- Busiah, K.; Drunat, S.; Vaivre-Douret, L.; Bonnefond, A.; Simon, A.; Flechtner, I.; Gérard, B.; Pouvreau, N.; Elie, C.; Nimri, R.; et al. Neuropsychological dysfunction and developmental defects associated with genetic changes in infants with neonatal diabetes mellitus: a prospective cohort study. Lancet Diabetes Endocrinol. 2013, 1, 199–207. [Google Scholar] [CrossRef]
- Bernier, R.; Steinman, K.J.; Reilly, B.; Stevens Wallace, A.; Sherr, E.H.; Pojman, N.; Mefford, H.C.; Gerdts, J.; Earl, R.; Hanson, R.; et al. Clinical phenotype of the recurrent 1q21.1 copy-number variant. Genet. Med. 2016, 18, 341–349. [Google Scholar] [CrossRef]
- Bernier, R.; Mudac, C.M.; Chen, Q.; Zeng, C.; Stevens Wallace, A.; Gerdts, A.; Earl, R. , Peterson, J.; Wolken, A.; Peters, A.; et al. Developmental trajectories for young children with 16p11.2 copy number variation. Am. J. Med. Genet. Part B. 2017, 174, 367–380. [Google Scholar] [CrossRef]
- De Cinque, M.; Palumbo, O.; Mazzucco, E.; Simone, A.; Palumbo, P.; Ciavatta, R.; Maria, G.; Ferese, R.; Gambardella, S.; Angiolillo, A.; et al. Developmental Coordination Disorder in a Patient with Mental Disability and a Mild Phenotype Carrying Terminal 6q26-qter Deletion. Front. Genet. 2017, 8, 206. [Google Scholar] [CrossRef]
- Coton, J.; Labalme, A.; Till, M.; Bussy, G.; Krifi Papoz, S.; Lesca, G.; Heron, D.; Sanlaville, D.; Edery, P.; des Portes, V.; et al. Characterization of two familial cases presenting with a syndromic specific learning disorder and carrying (17q;21q) unbalanced translocations. Clin. case reports. 2018, 6, 827. [Google Scholar] [CrossRef]
- Morris, C.A.; Mervis, C.B.; Paciorkowski, A.P.; Abdul-Rahman, O.; Dugan, S.L.; Rope, A.F.; Bader, P.; Hendon, L.G.; Velleman, S.L.; Klein-Tasman, B.P.; et al. 7q11.23 Duplication syndrome: Physical characteristics and natural history. Am. J. Med. Genet. Part A. 2015, 167, 2916–2935. [Google Scholar] [CrossRef]
- Hanson, E.; Bernier, R.; Porche, K.; Jackson, F.I.; Goin-Kochel, R.P.; Green Snyder, L.; Snow, A.V.; Stevens Wallace, A.; Campe, K.L.; Zhang, Y.; et al. The Cognitive and Behavioral Phenotype of the 16p11.2 Deletion in a Clinically Ascertained Population. Biol. Psychiatry. 2015, 77, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Sival, D.A.; Garofalo, M.; Brandsma, R.; Bokkers, T.A.; van den Berg, M.; de Koning, T.J.; Tijssen, M.A.J.; Verbeek, D.S. Early onset ataxia with comorbid dystonia: Clinical, anatomical and biological pathway analysis expose shared pathophysiology. Diagnostics. 2020, 10, 997. [Google Scholar] [CrossRef] [PubMed]
- van Noort, S.A.M.; van der Veen, S.; de Koning, T.J.; de Koning-Tijssen, M.A.J.; Verbeek, D.S.; Sival, D.A. Early onset ataxia with comorbid myoclonus and epilepsy: A disease spectrum with shared molecular pathways and cortico-thalamo-cerebellar network involvement. Eur. J. Paediatr. Neurol. 2023, 45, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Mariën, P.; Wackenier, P.; De Surgeloose, D.; De Deyn, P.P.; Verhoeven, J. Developmental Coordination Disorder: Disruption of the Cerebello-Cerebral Network evidenced by SPECT. Cerebellum. 2010, 9, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Hyde, C.; Fuelscher, I.; Williams, J. Neurophysiological Approaches to Understanding Motor Control in DCD: Current Trends and Future Directions. Curr. Dev. Disord. Reports. 2019, 6, 78–86. [Google Scholar] [CrossRef]
- Grohs, M.N.; Lebel, C.; Carlson, H.L.; Craig, B.T.; Dewey, D. Subcortical brain structure in children with developmental coordination disorder: A T1-weighted volumetric study. Brain Imaging Behav. 2021, 15, 2756–2765. [Google Scholar] [CrossRef]
- Biotteau, M.; Chaix, Y.; Blais, M.; Tallet, J.; Péran, P.; Alabaret, J.M. Neural Signature of DCD: A Critical Review of MRI Neuroimaging Studies. Front. Neurol. 2016, 7, 227. [Google Scholar] [CrossRef] [PubMed]
- Dewey, D.; Thompson, D.K.; Kelly, C.E.; Spittle, A.J.; Cheong, J.L.; Doyle, L.W.; Anderson, P. Very preterm children at risk for developmental coordination disorder have brain alterations in motor areas. Acta Paediatr. 2019, 108, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Sival, D.A.; van Noort, S.A.M.; Tijssen, M.A.J.; de Koning, T.J.; Verbeek, D.S. Developmental neurobiology of cerebellar and Basal Ganglia connections. Eur. J. Paediatr. Neurol. 2022, 36, 123–129. [Google Scholar] [CrossRef]
- Neychev, V.K.; Gross, R.E.; Lehéricy, S.; Hess, E.J.; Jinnah, H.A. The functional neuroanatomy of dystonia. Neurobiol. Dis. 2011, 42, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Nibbeling, E.A.R.; Delnooz, C.C.S.; de Koning, T.J.; Sinke, R.J.; Jinnah, H.A.; Tijssen, M.A.J.; Verbeek, D.S. Using the shared genetics of dystonia and ataxia to unravel their pathogenesis. Neurosci. Biobehav. Rev. 2017, 75, 22–39. [Google Scholar] [CrossRef] [PubMed]
- Marapin, R.S.; van der Horn, H.J.; van der Stouwe, A.M.M.; Dalenberg, J.R.; de Jong, B.M.; Tijssen, M.A.J. Altered brain connectivity in hyperkinetic movement disorders: A review of resting-state fMRI. Neuroimage Clin. 2023, 37, 103302. [Google Scholar] [CrossRef]
- Oguro-Ando, A.; Zuko, A.; Kleijer, K.T.E.; Burbach, J.P.H. A current view on contactin-4, -5, and -6: Implications in neurodevelopmental disorders. Mol. Cell. Neurosci. 2017, 81, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yuan, M.; Lau, B.W.M.; Li, Y. SHANK family on stem cell fate and development. Cell Death Dis. 2022, 13, 880. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.R.; Penzes, P. Ankyrins: roles in synaptic biology and pathology. Mol. Cell. Neurosci. 2018, 91, 131–139. [Google Scholar] [CrossRef]
- Mercati, O.; Huguet, G.; Danckaert, A.; André-Leroux, G.; Maruani, A.; Bellinzoni, M.; Rolland, T.; Gouder, L.; Mathieu, A.; Buratti, J.; et al. CNTN6 mutations are risk factors for abnormal auditory sensory perception in autism spectrum disorders. Mol. Psychiatry. 2017, 22, 625–633. [Google Scholar] [CrossRef]
- Zuko, A.; Kleijer, K.T.E.; Oguro-Ando, A.; Kas, M.J.H.; van Daalen, E.; van der Zwaag, B.; Burbach, J.P.H. Contactins in the neurobiology of autism. Eur. J. Pharmacol. 2013, 719, 63–74. [Google Scholar] [CrossRef]
- Hortsch, M.; Nagaraj, K.; Godenschwege, T. The interaction between L1-type proteins and ankyrins-a master switch for L1-type CAM function. Cell. Mol. Biol. Lett. 2009, 14, 57–69. [Google Scholar] [CrossRef]
- Stevens, S.R.; Rasband, M.N. Anyrinks and neurological disease. Curr. Opin. Neurobiol. 2021, 69, 51–57. [Google Scholar] [CrossRef]
- Woike, D.; Wang, E.; Tibbe, D.; Hassani Nia, F.; Failla, A.V.; Kibæk, M.; Overgård, T.M.; Larsen, M.J.; Fagerberg, C.R.; Barsukov, I.; Kreienkamp, H.J. Mutations affecting the N-terminal domains of SHANK3 point to different pathomechanisms in neurodevelopmental disorders. Sci. Rep. 2022, 12, 902. [Google Scholar] [CrossRef] [PubMed]
- Phelan, K; McDermid, H.E. The 22q13.3 Deletion Syndrome (Phelan-McDermid Syndrome). Mol. Syndromol. 2012, 2, 186–201. [Google Scholar]
- Matas, E.; Maisterrena, A.; Thabault, M.; Balado, E.; Francheteau, M.; Balbous, A.; Galvan, L.; Jaber, M. Major motor and gait deficits with sexual dimorphism in a Shank3 mutant mouse model. Mol. Autism. 2021, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; McCoy, P.A.; Rodriguiz, R.M.; Pan, Y.; Je, H.S.; Roberts, A.C.; Kim, C.J.; Berrios, J.; Colvin, J.S.; Bousquet-Moore, D.; et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum. Mol. Genet. 2011, 20, 3093–3108. [Google Scholar] [CrossRef]
- Steinman, K. J.; Spence, S.J.; Ramocki, M.B.; Proud, M.B.; Kessler, S.K.; Marco, E.J.; Green Snyder, L.; D'Angelo, D.; Chen, Q.; Chung, W.K.; et al. 16p11. 2 deletion and duplication: characterizing neurologic phenotypes in a large clinically ascertained cohort. Am. J. Med. Genet. Part A. 2016, 170, 2943–2955. [Google Scholar] [CrossRef]
- Bertrand, R.E.; Wang, J.; Xiong, K.H.; Thangavel, C.; Qian, X.; Ba-Abbad, R.; Liang, Q.; Simões, R.T.; Sampaio, S.A.M.; Carss, K.J.; et al. Ceramide synthase TLCD3B is a novel gene associated with human recessive retinal dystrophy. Genet. Med. 2021, 23, 488–497. [Google Scholar] [CrossRef] [PubMed]
| Gene | Genetic variant information | Clinical information |
|---|---|---|
| ABCC8 | See Supplementary Tables S3 and S7 of Busiah et al. (2013) [49] | 11 patients with nDM and DCD |
| CNTN4 | CNV (deletion) in locus 3p26.3, unknown inheritance [22] | 1 patient with isolated DCD |
| CTNNA3 | CNV (deletion) in locus 10q21.3, paternally inherited [22] | 1 patient with isolated DCD *1 1 patient with DCD and ADHD |
| FHIT | CNVs (deletion) in locus 3p14.2, unknown inheritance [22] | 2 patients with isolated DCD |
| GAP43 | CNV (deletion) in locus 3q13.31, de novo [22] | 1 patient with DCD and ADHD *2 |
| KCNJ11 | See Supplementary Tables S2 and S8 of Busiah et al. (2013) [49] | 11 patients with nDM and DCD |
| KLF7 | Not available [11] | 1 patient with DCD |
| LSAMP | CNV (deletion) in locus 3q13.31, de novo [22] | 1 patient with DCD and ADHD *2 |
| PTPRN2 | CNV (duplication) in locus 7q36.3, maternally inherited [22] | 1 patient with DCD, ADHD, and RD |
| RBFOX1 | CNV (deletion) in locus 16p13.3, maternally inherited [22] | 1 patient with isolated DCD *1 |
| SHANK3 | CNV (duplication) in locus 22q13.33, maternally inherited [22] | 1 patient with isolated DCD |
| VIPR2 | CNV (deletion) in locus 7q36.3, maternally inherited [22] | 1 patient with DCD and ADHD |
| Biological cluster | Parental biological term | p-value (Log10) |
|---|---|---|
| Modulation of chemical synaptic transmission | Synaptic signaling | -26.82 |
| Synaptic signaling | Synaptic signaling | -21.91 |
| Cell junction organization | Cellular process – Cellular component organization | -15.87 |
| Neuronal system | Synaptic signaling – Chemical synaptic transmission | -14.69 |
| Behavior | Multicellular organismal process | -13.99 |
| Regulation of cell projection organization | Cellular process – Cellular component organization | -13.61 |
| Neuron projection development | Nervous system development | -12.77 |
| Regulation of ion transport | Cellular process – Transport | -9.48 |
| Protein localization to synapse | Cell process – Cellular localization | -8.94 |
| L1CAM interactions | Nervous system development – Axon guidance | -8.89 |
| Metal ion transport | Cellular process – Transport | -7.57 |
| Actin filament-based process | Cellular process | -7.00 |
| Cell-cell adhesion | Cellular process – Cell adhesion | -6.35 |
| Action potential | Biological regulation –Regulation of biological quality | -6.30 |
| Neuromuscular process | Nervous system process | -5.94 |
| Brain development | Nervous system development | -5.42 |
| Protein localization to membrane | Cellular process- Cellular localization | -5.32 |
| Calcium-ion regulated exocytosis | Cellular process – Export from cell | -5.32 |
| Regulation of glutamatergic synaptic transmission | Synaptic signaling | -4.91 |
| Locomotory behavior | Multicellular organismal process -Behavior | -4.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





