1. Introduction
The Liliaceae family is one of the most economically and culturally important plants worldwide [
1]. According to Chase et al, [
2] there are approximately 70 genera and over 3,000 species in this family, among them many are grown as ornamental plants, while others have medicinal, culinary, and industrial uses. Lily (
Lilium spp.) is the most popular species of this family and they are commercially cultivated in The Netherlands; France, Chile, USA, Japan, New Zealand and China for cut flowers [
1,
3,
4]. Germplasm collection, characterization and maintenance is the key step prior to the cultivar development of any plant species. Lily germplasm is collected and maintained by many organizations around the globe among them KEW, USDA, The Kunming Institute of Botany in China, The National Institute of Floricultural Science in Korea and The Instituto de Botánica Darwinion in Argentina are notable [
5]. Genomic and genetic resources are also important for crop development. For the Lilium species complete chloroplast genome sequences [
6,
7,
8,
9], RNA sequences [
10,
11] and molecular markers [
1,
3,
12] are available as genetics and genomic resources. To date, there are no whole genome sequences of Lilium species available.
Freely accessible genomic resource databases have been a driving force in advancing plant research over the past decade. There are many publicly available genomic resource databases for various plant species, for example, TAIR (
https://www.arabidopsis.org/); Banana Genome Hub [
13]; Sol Genomics Network [
14]; Ensete knowledge base (
http://www.genomicsres.org/enknbase/); BRAD [
15]; cottonFGD (
https://cottonfgd.net/); GDR [
16] and citrus genome database [
17] are notable. These databases offer comprehensive and diverse data sets, such as whole genome sequences, transcriptomes, and metabolomes, that can be accessed and utilized by researchers to address biological questions. Such databases lacking for the Lilium species.
The development of genomic resource databases provides a centralized and comprehensive collection of genetic information for plant species that typically contains DNA sequences, RNA sequences, gene structures, functional annotations, molecular markers, phenotypic data, germplasm information etc. These data are valuable for the research community and they accelerate the research activity in several ways. Such as it is freely accessible and ready to use so it cut the data production cost, time and labour. It offers a diverse set and large amounts of data that can be used to test new methods for analyzing and interpreting genomic information, which can ultimately lead to new insights into the genetic basis of complex traits and diseases. Freely accessible RNA-seq databases are powerful for comparative genomics, gene identification, gene expression study, alternate splicing event discovery, molecular marker development, and phylogenomic and GWA studies.
Molecular markers have played a potential role in lily breeding compared to traditional methods. Various DNA markers, such as RAPD, ISSR, AFLP, DArT, SSR, and SNP, have been developed for genetic diversity analysis, germplasm characterization, hybrid identification, mutant detection, and genetic mapping in lilies [
18,
19,
20,
21]. However, RAPD and ISSR markers have limited applicability due to their low reproducibility and dominance issues. RAPD markers were utilized as linked markers for Fusarium resistance in 150 Asiatic hybrid individuals, but only three RAPD markers were polymorphic and explained 24% of the resistance, revealing limitations in lily breeding. Varshney et al. [
22] failed to detect variation in micro-propagated Lilium species using RAPD markers. On the other hand, ISSR markers were suggested by Xi et al. [
23] as potentially useful for identifying L.
longiflorum mutants instead of RAPD. However, Yin et al. [
24] were unable to identify mutants in the oriental hybrid ‘Siberia’ using ISSR markers and found very low-frequency polymorphism using AFLP markers among regenerated ‘Siberia.’ Therefore, to enhance Molecular Marker-Assisted Breeding (MMAB), user-friendly, cost-effective, and transferable molecular markers are needed for lily species.
This study was conducted to develop genomic resources for the Lilium sp. which is a comprehensive genomic resource for the Liliaceae family, providing access to a variety of molecular markers, transcription factor genes, gene expression data, and phenotypic data. These resources are expected to play a crucial role in advancing our understanding of the genomics of Lilium species and the improvement of this species.
2. Materials and Methods
2.1. Data Sources and Data Processing
In this study seven sets of RNA sequence data were used among them 4 sets were obtained from the NCBI and the remaining three sets were generated from the RNA-sequencing libraries prepared from three genotypes of Lilium species subjected to different treatments (as listed in
Table S1). Leaf samples from greenhouse-grown plants were collected for each treatment and immediately transferred to liquid nitrogen, where they were stored at −80 °C until RNA extractions. RNA was extracted using the RNeasy mini kit (Qiagen, Hilden, Germany), following the manufacturer’s guidelines. The RNA library was prepared according to Illumina’s guidelines (San Diego, CA), and the library was sequenced using the Illumina Hiseq 2000 platform. The quality of the raw reads was assessed using the FastQC tool [
25], and denovo assembly was performed using Trinity [
26] and RSEM tools [
27]. Afterward, all transcriptomes were pooled based on the treatment and species name (sample) and assembled into non-redundant lily unigenes using CAP3 [
28]. Furthermore, to access publicly available nucleotide sequences of Lilium species, we also downloaded data from the NCBI database (date on 03/09/2023). All the sequences were then categorized into four groups based on the respective Lilium species: Longiflorum (L), Asiatic (A), Oriental (O), and hybrid (H) species sequences and the list are presented in the
Table 1. Phenotypic data, including morphological characters, taxonomic information, and flower images, were collected from 141 lily cultivars at six different research institutes in South Korea.
2.2. Marker Development
In this study, we conducted microsatellite (SSR) mining and primer design using the LSAT pipeline. We identified microsatellite repeat motifs in the Lily transcriptome sequences, with a minimum of 10 repeat units for mononucleotides, 5 for dinucleotides, and 4 for other repeat types. After identifying the SSRs, we extracted flanking regions around them using custom Perl scripts. Primer design was accomplished using Primer3 executables with default parameters: a melting temperature range of 55-56°C, GC content between 40-60%, primer size ranging from 18-25 nucleotides, and a desired product length of 150-280 base pairs. All SSR markers were then
insilico characterized and experimentally validated. Further details can be found in our previous study published by Biswas et al [
29].
SNP discovery and characterization were conducted using seven lily datasets, as detailed in
Table 1. Initially, all data underwent clustering with the cd-hit tool employing a 95% identity threshold and a 5% length difference parameter. Subsequently, clusters containing a minimum of four representative sequences were selected, and the longest representative sequences from these chosen clusters were extracted using a combination of Bash and Perl scripts. These extracted representative sequences served as the reference sequences for SNP mining. To facilitate SNP discovery, the reference sequences were indexed using Bowtie2 with default parameters. Subsequently, all seven datasets were aligned to these reference sequences using Bowtie2 with its default settings. SNP calling was carried out using samtools, following established protocols. The identified SNPs were subjected to comprehensive characterization, considering various attributes such as the base mutation type, allele types, and their distribution across lily species. Additionally, the heterozygosity and homozygosity of these SNPs were estimated as part of the characterization process.
To identify COS loci, the transcriptome data sets of Lilium formolongi cv. “Sinnapal,” Lilium longiflorum cv. “Napal,” and Lilium longiflorum “Easter lily” were BLASTed against each other in a “round-robin” fashion, and reciprocal best BLAST hits were retained. The following blast parameters were used: e-10, Hit-3. Potential COS loci were identified where all comparisons successfully produced a reciprocal best BLAST hit with identity at 90% and query coverage at 85%. The COS sequences were then used to develop COS markers. Primer pairs for COS sequences were designed using primer3 software with default parameters.
2.3. TF Gene Analysis
All the data sets (sequences) were searched against the Plant Transcription Factor Database version 5.0 using an E-value cut-off of e-10. A cut-off value of 50% for query coverage and 80% for identity was set to filter TF gene-encoded transcripts. After that putative TF-encoded lily RNA sequences were analyzed using the online tool iTAK (Plant Transcription Factor & Protein Kinase Identifier and Classifier,
http://itak.feilab.net/cgi-bin/itak/online_itak.cgi).
2.4. Expression Analysis
To ensure quality control, raw reads underwent trimming to remove adapters and eliminate low-quality reads with a Q value of less than 20, utilizing the command-line tools ‘trimmomatic’ [
30] and fastQC [
25]. Subsequently, the high-quality reads (transcriptome) were assembled using the de novo approach with the Trinity program [
26], and their transcript abundances were quantified using RSEM [
27]. Since Lily lacks a reference genome, RSEM was employed for quantification purposes. The transcript expression levels were quantified in terms of FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values, ranging from greater than 0 to over 104, along with the estimation of fold change (FC). Transcripts meeting the criteria of FC ≥ 2 and FC ≤ –2 were considered significantly upregulated and downregulated DEGs (Differentially Expressed Genes), respectively, in at least one of the total comparisons. This rigorous analysis allowed for the identification of key transcripts that showed substantial changes in expression, shedding light on the crucial regulatory mechanisms involved in the studied lily species.
2.5. Database Architecture and Web Interface Design
The Three-Level Schema Architecture has been employed to develop the lily database. This architecture is designed to divide the database into three distinct tiers: the front-end, middle, and back-end. The front-end tier is responsible for the client-side interface of the application and is mostly developed using web technologies such as HTML and JavaScript. Its main purpose is to provide users with a visually attractive and user-friendly interface for interacting with the database. The middle tier, on the other hand, is developed using the PHP language. Its main function is to act as a bridge between the front-end tier and the back-end tier. This tier receives requests from the client side, processes them, and then communicates with the database. The back-end tier is the database itself resides. It is responsible for storing, managing, and retrieving data from the database. We use the database management system MySQL to store all the lily-db data.
3. Result and Discussion
3.1. Content of the Lily-db
3.1.1. Germplasm Information:
To develop the Lilium species germplasm information database, morphological characters of 141 cultivars maintained at various research institutes in South Korea were collected from published manuals and books. The morphological and taxonomical information of these cultivars was then compiled and stored in a user-friendly database. To easily navigate the phenotypic data (
Figure S1), two search options were incorporated: cultivar name and lily taxonomic group. Users can search for specific cultivars by name or browse through different taxonomic groups to access information from this database.
3.1.2. Molecular Markers Developments and Database Features:
In the lily-database, three distinct types of molecular markers, namely SSR, SNP, and COS, have been introduced. SSR markers were mined from seven datasets, and their summarized results are presented in
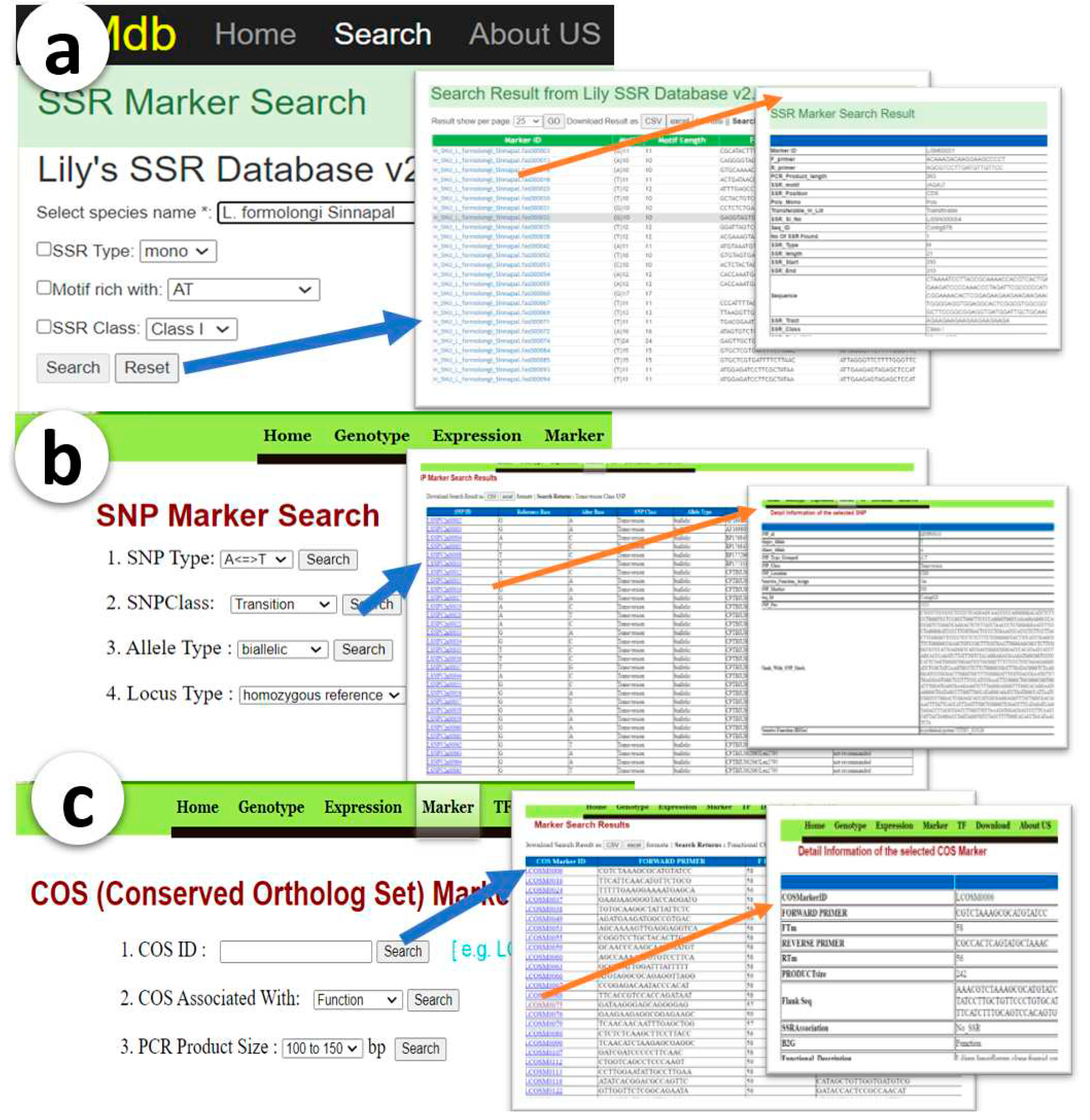
Table 2. The result reveals that, approximately 12% of the analyzed RNA sequences contain SSR motifs, with an average of one SSR found in 2793 bp of sequences in Lily species. As expected, Class II SSR types (SSR loci less than 20 nucleotides) dominate over Class I SSR types (SSR loci greater than 20 nucleotides). Additionally, AT-rich motifs are more abundant than GC-rich motifs, and tri-nucleotide repeats predominate among all other SSR repeats in this study. A total 47,863 primers pairs were design and integrated into the SSR marker database. This database offers a sophisticated search interface with three search criteria, that can be use single or in different combinations. The search results are visualized on a HTML page, providing users with quick access to detailed information about each marker (
Figure 1a). Each SSR markers contains 41 attributes that are unique compared to other SSR marker databases. Most of the published SSR marker databases only present the forward and reverse primer sets, while in some cases, genomic coordinates and flanking sequences are presented. In the Lily SSR maker datasets, all 41 attributes were obtained from the in-silico characterization of the markers. This information will help in selecting the best primer sets for research interests.
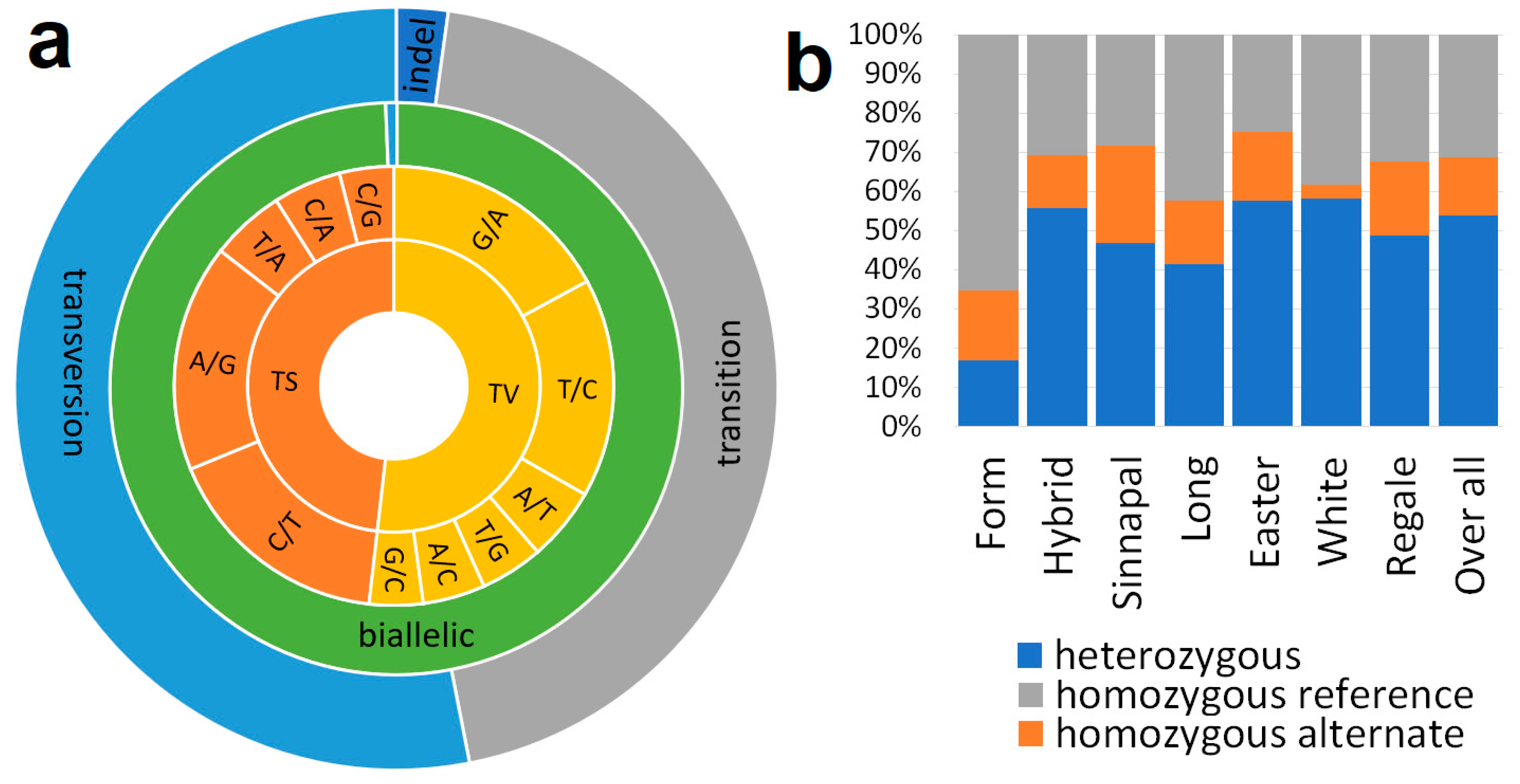
A comprehensive summary of the SNP mining results is presented in
Figure 2. Our findings revealed that the majority of these SNPs can be classified into two primary categories: Transitions and Transversions, with Transitions being slightly more abundant (9,363 vs. 11,109 SNPs). Within the Transition category, we observed distinct allele combinations, including A/G, C/A, C/G, C/T, and T/A. Similarly, Transversions exhibited a diverse array of allele combinations, such as A/C, A/T, G/A, G/C, T/C, and T/G. Notably, a significant proportion of these SNPs were found to be biallelic, accounting for 99.36% of the total. Furthermore, we conducted an analysis of allele distribution among various lily species. Our observations indicated a prevalence of heterozygous conditions, with heterozygotes being more abundant than homozygotes. These results provide valuable insights into the genetic diversity and allelic distribution within the lily species, offering essential information for further genetic research and breeding programs.
A total of 20,929 SNPs were successfully identified and stored in a dedicated database. The SNP-databse search interface enable users search SNP markers with three search criteria options, including SNP type, SNP class, and SNP position, which can be easily used to filter and retrieve relevant data. The interface (
Figure 1b) was designed to provide an intuitive and user-friendly experience. Users can simply build their search criteria from the dropdown list and submit their queries. The search results are displayed in a user-friendly format, allowing users to easily navigate and analyze the data. A total of 27 attributes are displayed for each SNP, providing valuable insights that can be used to identify the best markers for further analysis.
In this study, a total of 179,988, 85,647, and 90,115 transcriptome sequences from
L. longiflorum Easter,
L. longiflorum White, and
L. formolongi Sinnapal were reciprocally compared, resulting in the identification of 12,695 common sequences that were conserved among the three lily species (
Figure S2). Subsequently, we developed 1,213 COS markers, representing 9.65% of the conserved sequences (
Table S2), for potential marker development. These markers have been stored in a searchable database for future use. The search interface for COS (Conserved Ortholog Sequences) markers offers users with an efficient and effective way to search for COS marker sets with 10 attributes. The user-friendly interface permits users to search the entire COS dataset based on three individual search criteria (
Figure 1c). Once the user has selected their desired search criteria, they can submit their query and the results will appear in a table format.
3.1.3. Transcription Factors Genes:
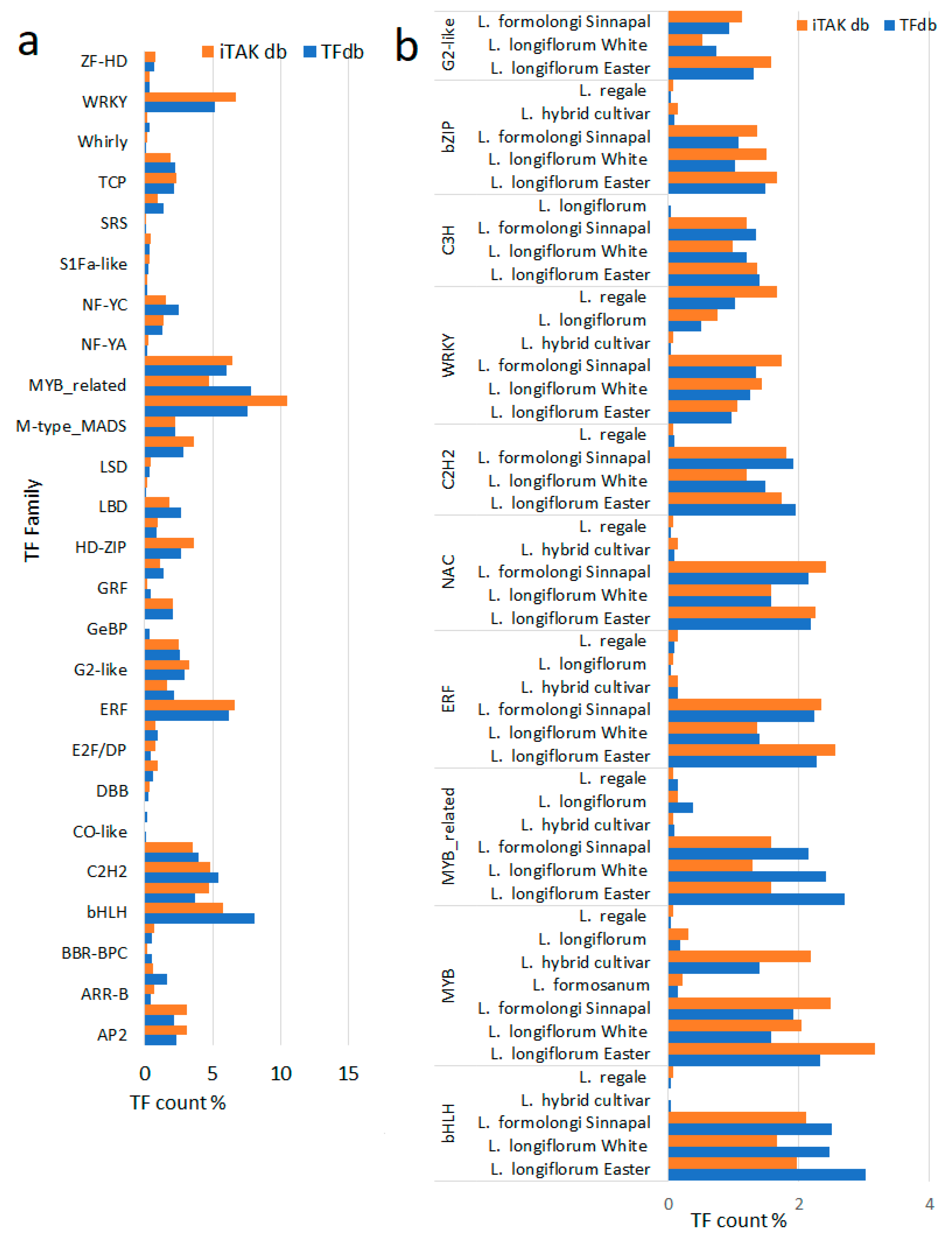
A comprehensive search for putative transcription factor (TF) genes was conducted on transcriptome sequences. Initially, all sequences were subjected to a BLAST search against the plant TF-database v5, resulting in the identification of 2151 TF genes. Subsequently, these genes underwent further analysis using iTAKdb tools, leading to the annotation of 1327 TF genes. These annotations accounted for approximately 1% of all genes within the transcriptomes (
Table S3). These TF genes were subsequently categorized into 46 to 49 families based on their distinctive DNA binding domains, as detailed in
Table S4,
Table S5 and
Figure 2. Notably, the MYB superfamily emerged as the largest family, followed by the bHLH, ERF, NAC, C2H2, WRKY, C3H, bZIP, and G2-like families. Interestingly, the copy numbers of each family varied, with the identification of between 1 and 65 copies per family. To enhance the utility of these TF genes, a user-friendly search page was developed, organized by TF family. All identified families are presented in a tabulated format, accompanied by links to the TF database. Users can easily access comprehensive details for each TF family by clicking on the respective links. This search page (
Figure S3) facilitates efficient exploration of plant regulatory networks, allowing users to identify specific factors involved in crucial responses in a streamlined manner.
3.1.4. Gene Expression Data:
The gene expression database is a comprehensive repository that highlights the details of gene regulation. Transcriptome data from 22 cDNA libraries were analyzed to calculate the FPKM value for each identified gene, and the data were grouped based on the experiments, including treatments and cultivars. These experiments focused on three specific lily species: Sinnapal Lily (data from L. formolongi cv. Sinnapal Lily transcriptome of 6 cDNA libraries), White tower Lily (data from L. longiflorum White tower Lily transcriptome of 10 cDNA libraries), and Easter Lily (data from L. longiflorum Easter Lily transcriptome). This valuable resource provides crucial insights into the gene expression patterns of these lily species.
To facilitate easy accessibility and data exploration, the database has been precisely organized into groups according to the specific experiments conducted (
Figure S4 to S6). Users can navigate through the vast dataset using various search criteria, such as Unige ID, gene status, gene name, and Gene Ontology (GO) ID, enabling them to pinpoint and analyze gene expression profiles relevant to their research interests. Search results are presented in tabular form, allowing users to copy or download the data in XLS and CSV formats for further use.
This database serves as an indispensable tool for the scientific community, fostering a deeper understanding of gene expression dynamics and paving the way for innovative discoveries in the realm of molecular biology and genetics. It holds the potential to accelerate research efforts and contribute to significant advancements in our knowledge of gene regulation and its implications in various biological processes.
3.2. Applications, Limitations and Future Directions
The current version of Lily-db contains a wide range of data that can be applied in several ways to improve Lilium species breeding programs. One notable example is the morphological data of the cultivars, which can be utilized by researchers to select and characterize germplasm. Additionally, molecular markers found within the database can be employed to distinguish between different genotypes. This is particularly useful in situations where the morphology of the cultivars is similar or when the cultivars have mixed identities. Biswas et al [
1] use 46 SSR makers from this database for genetic diversity, population structure, and phylogenetic studies of Korean Lily germplasm. Expression and transcriptomes data present in this database help to understand gene expression in Lilium species during biotic stress.
In the first version of Lily-db only contains the data generated by our research group, recently many other transcriptomes data are now available in the public domain we will update the Lily database incorporation with public data. More features will be included in the next version of this database.
4. Conclusions
Lily-db is a compressive database that contains various types of data such as morphological, molecular markers, gene expression, and transcription factor genes. This comprehensive resource offers researchers a broad range of information that can be used in numerous research applications, making it an essential tool for those studying Lilium species in the fields of genetics, genomics, and breeding programs.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1. Lily Germplasm Search User Interface: This is designed to facilitate easy access to the diverse germplasm resources available in this database. The user interface offers a user-friendly and efficient platform to explore and retrieve information related to Lily germplasm. Search returns a list of the germplasm with morphological information and images of the cultivars/variety of the germplasm. Figure S2. Vine diagram for conserved ortholog sequences identification from the RNA-seq data of Lilium species. Figure S3. TF data search user interface: This figure illustrates the User Interface of the TF Data Search platform. The user interface offers an intuitive and user-friendly experience, enabling researchers to efficiently access valuable information related to TFs. Figure S4. Expression data search user interface for Sinnapal Lily cultivar. Figure S5. Expression data search user interface for White tower Lily cultivar. Figure S6. Expression data search user interface for Easter Lily cultivar. Table S1. List of the RNA-sequencing libraries and read information. Table S2. Lily Conserved Orthologous Sequenced based (COS) marker development and characterization. Table S3. Summary of the TF analysis. Table S4. Distribution of Lily TF genes among the TF family. Table S5. Distribution of Lily TF family genes in different family and species level.
Author Contributions
The work presented here was carried out in collaboration among all authors. M.K.B. was involved in transcriptome assembly and annotation, SSR mining, database development, and drafted the manuscript; S.K.N and D.B. wrote and managed the server to host the database; J.H. and P.K. grew the plant materials and maintained the greenhouse, extract RNA and quality control; MKB and I.S.N. conceived and designed the experiments. All authors have read and approved the final manuscript.
Acknowledgments
This study was financially supported by the Golden Seed Project (Center for Horticultural Seed Development, No. 213007-05-2-CG100) of the Ministry of Agriculture, Food and Rural Affairs (MAFRA), South Korea.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Biswas, M.K.; Bagchi, M.; Nath, U.K.; Biswas, D.; Natarajan, S.; Jesse, D.M.I.; Park, J.; Nou, I. Transcriptome wide SSR discovery cross-taxa transferability and development of marker database for studying genetic diversity population structure of Lilium species. Scientific Reports 2020, 10, 18621. [Google Scholar] [CrossRef] [PubMed]
- Angiosperm Phylogeny Group; Chase, M.W.; Christenhusz, M.J.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 2016, 181, 1–20. [Google Scholar]
- Biswas, M.K.; Nath, U.K.; Howlader, J.; Bagchi, M.; Natarajan, S.; Kayum, M.A.; Kim, H.; Park, J.; Kang, J.; Nou, I. Exploration and exploitation of novel SSR markers for candidate transcription factor genes in Lilium species. Genes 2018, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Buschman, J. Globalisation-flower-flower bulbs-bulb flowers, IX International Symposium on Flower Bulbs. 2004, 673, 27–33. [Google Scholar]
- Wilford, R.; Gardens, K.R.B. The Kew Gardener s Guide to Growing Bulbs: The art and science to grow your own bulbs; White Lion Publishing, 2019. [Google Scholar]
- Li, Y.; Zhang, L.; Wang, T.; Zhang, C.; Wang, R.; Zhang, D.; Xie, Y.; Zhou, N.; Wang, W.; Zhang, H. The complete chloroplast genome sequences of three lilies: Genome structure, comparative genomic and phylogenetic analyses. J Plant Res 2022, 1–15. [Google Scholar] [CrossRef]
- Du, Y.; Bi, Y.; Yang, F.; Zhang, M.; Chen, X.; Xue, J.; Zhang, X. Complete chloroplast genome sequences of Lilium: insights into evolutionary dynamics and phylogenetic analyses. Scientific reports 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Liu, H.; Yu, Y.; Deng, Y.; Li, J.; Huang, Z.; Zhou, S. The chloroplast genome of Lilium henrici: genome structure and comparative analysis. Molecules 2018, 23, 1276. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Wang, T.; Zhang, C.; Wang, R.; Zhang, D.; Xie, Y.; Zhou, N.; Wang, W.; Zhang, H. The complete chloroplast genome sequences of three lilies: Genome structure, comparative genomic and phylogenetic analyses. J Plant Res 2022, 1–15. [Google Scholar] [CrossRef]
- Howlader, J.; Robin, A.H.K.; Natarajan, S.; Biswas, M.K.; Sumi, K.R.; Song, C.Y.; Park, J.; Nou, I. Transcriptome analysis by rna–seq reveals genes related to plant height in two sets of parent-hybrid combinations in easter lily (Lilium longiflorum). Scientific Reports 2020, 10, 9082. [Google Scholar] [CrossRef]
- Du, F.; Wu, Y.; Zhang, L.; Li, X.; Zhao, X.; Wang, W.; Gao, Z.; Xia, Y. De novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium Oriental hybrid ‘Sorbonne’. Plant Mol Biol Rep 2015, 33, 281–293. [Google Scholar] [CrossRef]
- Sun, M.; Zhao, Y.; Shao, X.; Ge, J.; Tang, X.; Zhu, P.; Wang, J.; Zhao, T. EST–SSR Marker Development and Full-Length Transcriptome Sequence Analysis of Tiger Lily (Lilium lancifolium Thunb). Applied Bionics and Biomechanics 2022, 2022. [Google Scholar] [CrossRef] [PubMed]
- Droc, G.; Lariviere, D.; Guignon, V.; Yahiaoui, N.; This, D.; Garsmeur, O.; Dereeper, A.; Hamelin, C.; Argout, X.; Dufayard, J. The banana genome hub. Database 2013, 2013, bat035. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H. The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res 2015, 43, D1036–D1041. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3. 0: an upgraded Brassicaceae database. Nucleic Acids Res 2022, 50, D1432–D1441. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Lee, T.; Cheng, C.; Zheng, P.; Bubble, K.; Crabb, J.; Gasic, K.; Yu, J.; Humann, J.; Hough, H. Resources for peach genomics, genetics and breeding research in GDR, the Genome Database for Rosaceae, X International Peach Symposium. 2022, 1352, 149–156. [Google Scholar]
- Liu, H.; Wang, X.; Liu, S.; Huang, Y.; Guo, Y.; Xie, W.; Liu, H.; ul Qamar, M.T.; Xu, Q.; Chen, L. Citrus Pan-Genome to Breeding Database (CPBD): A comprehensive genome database for citrus breeding. Molecular Plant 2022, 15, 1503–1505. [Google Scholar] [CrossRef]
- Lee, S.; Nguyen, X.T.; Kim, J.; Kim, N. Genetic diversity and structure analyses on the natural populations of diploids and triploids of tiger lily, Lilium lancifolium Thunb., from Korea, China, and Japan. Genes & Genomics 2016, 38, 467–477. [Google Scholar]
- Wen, C.S.; Hsiao, J.Y. Altitudinal genetic differentiation and diversity of Taiwan lily (Lilium longiflorum var. formosanum; Liliaceae) using RAPD markers and morphological characters. Int J Plant Sci 2001, 162, 287–295. [Google Scholar] [CrossRef]
- Shahin, A.; Smulders, M.J.; van Tuyl, J.M.; Arens, P.; Bakker, F.T. Using multi-locus allelic sequence data to estimate genetic divergence among four Lilium (Liliaceae) cultivars. Frontiers in plant science 2014, 5, 567. [Google Scholar] [CrossRef]
- Yuan, S.; Ge, L.; Liu, C.; Ming, J. The development of EST-SSR markers in Lilium regale and their cross-amplification in related species. Euphytica 2013, 189, 393–419. [Google Scholar] [CrossRef]
- Varshney, A.; Sharma, M.P.; Adholeya, A.; Dhawan, V.; Srivastava, P.S. Enhanced growth of micropropagated bulblets of Lilium sp. inoculated with arbuscular mycorrhizal fungi at different P fertility levels in an alfisol. The journal of horticultural science and biotechnology 2002, 77, 258–263. [Google Scholar] [CrossRef]
- Xi, M.; Sun, L.; Qiu, S.; Liu, J.; Xu, J.; Shi, J. In vitro mutagenesis and identification of mutants via ISSR in lily (Lilium longiflorum). Plant Cell Rep 2012, 31, 1043–1051. [Google Scholar] [CrossRef]
- Yin, Z.; Zhao, B.; Bi, W.; Chen, L.; Wang, Q. Direct shoot regeneration from basal leaf segments of Lilium and assessment of genetic stability in regenerants by ISSR and AFLP markers. , 49In Vitro Cellular & Developmental Biology-Plant 2013, 49, 333–342. [Google Scholar]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Hancock, B. Trinity v3, a DDoS tool, hits the streets. Comput Secur 2000, 19, 574. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 1–16. [Google Scholar] [CrossRef]
- Huang, H.; Wang, L.; Tak, B.C.; Wang, L.; Tang, C. Cap3: A cloud auto-provisioning framework for parallel processing using on-demand and spot instances, 2013 IEEE Sixth International Conference on Cloud Computing. IEEE, 2013; pp. 228–235. [Google Scholar]
- Biswas, M.K.; Natarajan, S.; Biswas, D.; Nath, U.K.; Park, J.; Nou, I. LSAT: Liliaceae Simple Sequences Analysis Tool, a web server. Bioinformation 2018, 14, 181. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.stylefix |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).