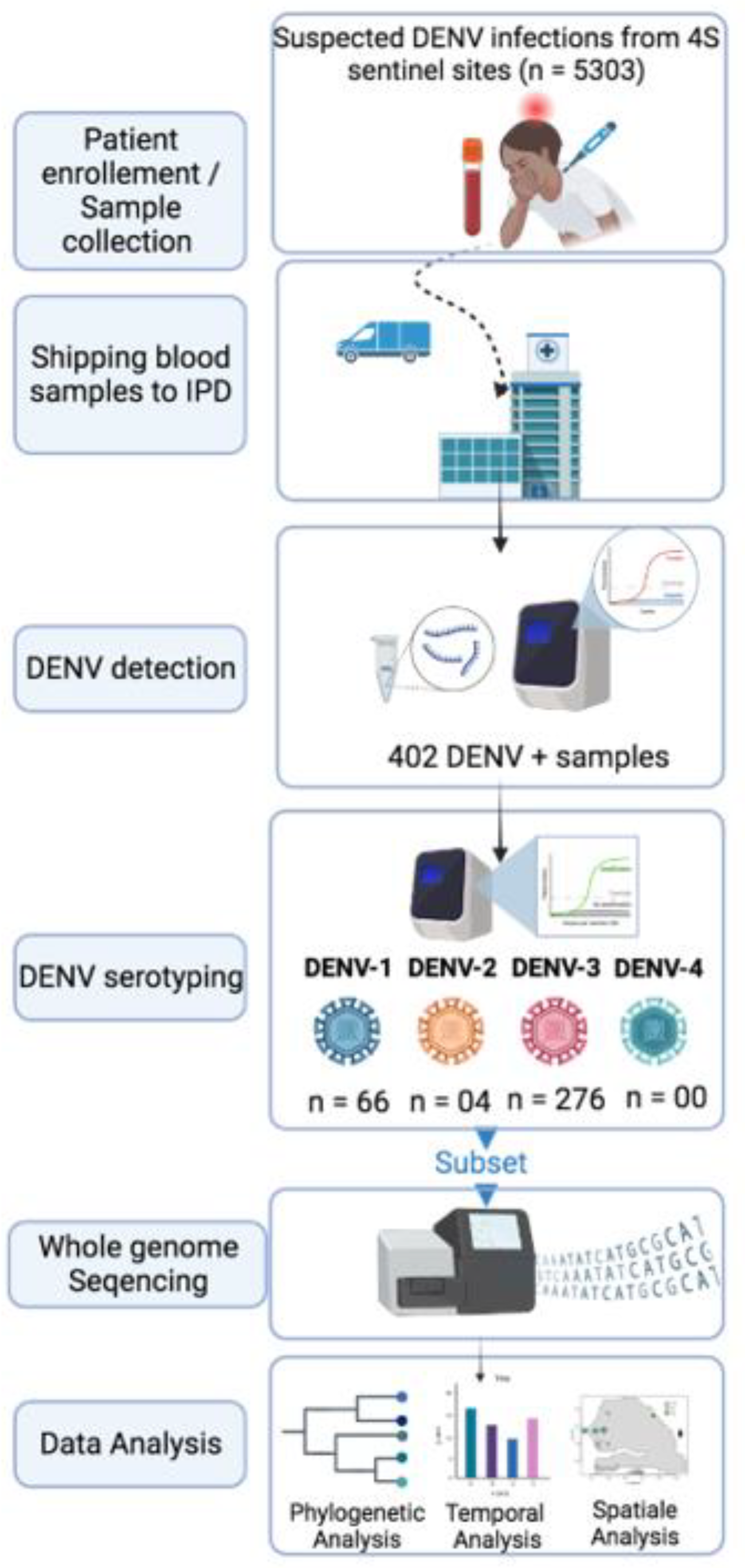
1. Introduction
Dengue fever (DF) is recognized as the most widespread arboviral disease globally (1). Its transmission occurs when infected mosquito vectors from the Aedes genus bite humans (2). Astonishingly, over one-third of the world's population is at risk of DENV infection (3). Many factors including climate changes , increasing travel and trades and urbanization (2) participate to the disease spread worldwide. According to WHO estimates approximately 3.6 billion people worldwide are at risk of dengue infections (4), and reported cases range from 50 to 100 million annually (1).Tragically, the disease results in an estimated 10,000 deaths each year (3). Infection with dengue virus (DENV), the etiological agent of DF, can manifest in various clinical forms, ranging from a self-limited disease known as DF to the life-threatening severe dengue (5).
DF is prevalent in tropical and subtropical areas ; the virus epidemiology is well known in Asia and America (6).
In Africa the virus was thought to be rare for long but detection from returning travelers (7,8) and recently reported outbreaks highlight the virus circulation in the continent (9,10). Due to the lack of sufficient diagnostic tools and effective surveillance, the true burden of DENV infection is likely to be underestimated (8,11).
In Senegal the first dengue case was reported in 1970 and was mainly dominated by the occurrence of sylvatic cycle up to years 2000s (12). The first urban dengue epidemic took place in 2009 and was caused by DENV-3 ; following years yearly outbreak affecting different region of the countries (13–15) and linked to different serotypes were noticed (13,16,17). Despite the recurrent reports few studies assessed the virus diversity in Senegal exist in the literature (16).
DENV belonging to the Flavivirus genus within the Flaviviridae family [Kraemer, 2015]. DENV is enveloped and possess a single-stranded RNA genome with positive polarity, approximately 10.6 kilobases in length. The DENV genomic RNA encodes three structural proteins namely: capsid (C), pre-membrane /membrane (prM), and envelope (E) proteins, along with seven nonstructural (NS) proteins: NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 [Chambers, 1990].
Antigenically DENVs are categorized into four distinct serotypes, namely DENV-1, DENV-2, DENV-3, and DENV-4, each inducing only limited cross-protection immunity (18). Serotypes share 65-70 % in amino acid sequence (19). Furthermore, each serotype is subsequently subdivided into various genotypes which follow a marked geographic distribution (18). Within each serotypes genotypes are defined by group of virus sharing less than 6% nucleotide divergence (19).Antigenic differences among serotypes and genotypes play a crucial role in dengue epidemiology and pose challenges for vaccine development and disease control strategies (19,20).
Therefore, it is essential to engage in monitoring strains that are prevalent in a specific region. This surveillance will inform the choice of suitable prophylactic and preventive actions (10). Additionally, diverse DENV serotypes or genotypes have been observed to elicit distinct immune responses. This variance influences their capacity to infect particular target cells , more severe manifestations of dengue or infect mosquitoes (21–23).
Although reports of dengue circulation in Africa exist, there are only a few studies that have examined the genetic make up of the prevalent strains, focusing on both the serotype and/or genotype levels (24–26).
Dengue strains in Africa remain poorly characterized with African sequences representing < 1% of global sequences data (Phillipe Selhorst., 2023). Existing sequences data are mainly obtained during outbreak periods (Burkina, Senegal) or returning travelers (8). In contrast many studies in Asia (27–30) and America (31) were focused on the genetic diversity of circulating DENV strains.
Due to the unprecedented and growing numbers of confirmed DENV cases in Senegal (Multifoci) and the co-circulation of different viral serotypes (29) continuous monitoring of circulating virus variants (serotypes/genotypes) and genomic surveillance of viral strains appears to be pivotal to anticipate worsen situations. Herein to get insight about the circulating DENV serotypes/genotypes and understand their spatial and temporal distribution across the Senegal we combine epidemiology, RT-qPCR and genome sequencing to uncover the virus genetic diversity in from January 2019 to March 2023.
2. Materials and Methods
2.1. The 2.1 Febrile illnesses surveillance system
The increasing threat of emerging pathogens to public health requires a reliable surveillance system to control their spread. In Senegal, the Institut Pasteur de Dakar partnered with the Senegalese Ministry of Health and the WHO country office to implement a nationwide Syndromic Sentinel Surveillance System called the 4S network in 2011 (32). Initially, the surveillance was limited to virologic surveillance of Influenza-like Illness (ILI). In 2015, it was expanded to include a wider range of pathogens associated with public health priority syndromes such as malaria, dengue-like syndromes, and diarrheal syndromes. This syndromic approach enables the early detection of unexpected and/or unusual occurrences of specific symptoms to monitor the evolution of the diseases under surveillance, investigate outbreaks, and implement appropriate response actions. The network has up to 20 sentinel sites distributed across the 14 administrative regions of Senegal, selected based on the WHO-recommended attributes (33). The DENV suspected samples tested during this study were collected throughout the 4S network system.
2.2. Samples shipping to WHOCC
At a weekly basis suspected samples collected from sentinel sites were shipped with clinical and demographic forms at the virology department at the Institut Pasteur de Dakar. At IPD samples were subjected to molecular screening for the detection of 07 medically important arboviruses including Dengue, Zika, Yellow Fever, Chikungunya, Rift valley fever, West Nile and Crimee Congo Hemorrhagic fever virus.
2.3. RNA extraction
Blood sample from suspected dengue cases were subjected to centrifugation at 2000 rpm for 5 mn to obtain sera which were harvested and aliquoted on 2ml cryotubes for immediate use and further biobanking. RNA extraction was performed using 140 µl of sera using Qiagen viral RNA mini kit according to the manufactures’ recommendation. RNA was eluted to a final volume of 60µl and conserved to - 80 until further use.
2.4. RT-qPCR DENV detection
DENV RNA presence on extracted RNA was assessed using RT-qPCR with sets of primers targeting 3’-UTR region of all dengue serotypes (34). Reaction were performed on CFX machine (14); following temperature profile were used during reaction : 50°C – 10mn, 40 cycles of 95° C – 1mn ; 95° C – 15 secondes and 95 ° C – 30 secondes. All sample with a Ct values below the fixed cut of value of 32 were considered as DENV+.
2.5. DENV serotyping
Serotypes of DENV+ samples were assessed by RT-qPCR according to a protocol previously described by Dieng and colleagues (35). Briefly CDC dengue typing kit (36) were used according to the manufactures recommendations. The system allow the simultaneous detection of DENV serotypes from 5µl of input RNA. Each of DENV serotypes can be read in different dye channels.
2.6. cDNA synthesis and amplicons generation
To maximize yield and genome coverage, a subset of DENV+ RNA samples with Ct values < 32 was chosen for sequencing. For selected samples cDNA synthesis was carried out using the Luna Script RT SuperMix (5X) from New England Biolab, Ipswich, MA, USA. In brief, 8µl of RNA was mixed with 2µl of master mix, pipetted up and down up to ten times, and briefly centrifuged. The mixture was then incubated at 25°C for 2 minutes, 55°C for 20 minutes, and 95°C for 2 minutes, and finally placed directly on ice until further use. Then according to the serotype a specific whole genome multiplex PCR was conducted in order to amplify the entire coding region of DENV using two primer pools (1 & 2) in separated tubes. Reactions conditions were previously described by Dieng and colleagues (37). Amplification success was checked at the end of the reaction by agarose gel based electrophoresis.
2.7. Library preparation and Sequencing
Obtained amplicons were purified using 1X Ampure XP Beads (Beckman Coulter Inc.), and cleaned-up concentrations of each PCR product were measured using a Qubit dsDNA HS Assay kit (Thermo Fisher Scientific) on a Qubit fluorimeter (Thermo Fisher Scientific). Targeted whole-genome sequencing of DENV 1-3 was undertaken for each sample; equal concentrations of pool 1 and pool 2 amplicons were pooled per sample before library preparation using Illumina DNA preparation kit Nextera DNA flex (Illumina Inc.) according to manufacturer's recommendations; whole-genome sequencing was performed with paired-end reads using Illumina MiSeq reagent kit V3 (300 cycles) on an Illumina MiSeq instrument. Consensus sequences of around 10 Kb (corresponding to the full CDS) were generated by de-novo assembling using Genome Detective (
https://www.genomedetective.com/app/ accessed on February 15, 2023).
2.8. Datasets construction and phylogenetic analysis
DENV serotypes/genotypes/lineages identification was performed using dual procedures:
- i)
by using the genome detective dengue typing tool and ii) by using a maximum likelihood (ML) phylogenetic analysis to put newly sequenced DENV strains in a global context and explore the relationship with others available global sequences.
For this purpose we retrieved from US National Institutes of Health National Institute of Allergy and Infectious Diseases Virus Pathogen Database and Analysis Re source (
http://www.viprbrc.org) representative sequences of described dengue genotypes for each serotype. Downloaded dataset for each serotype (DENV-1, n = 202; DENV-2, n = 257 , DENV-3, n = 133 ) containing all genomes from Africa and ≈10% of the remaining genomes. Newly generated sequences were aligned with downloaded datasets , full details of used sequences can be found in (
Table S1). Multiple sequence alignment was performed using MAFFT (ref) and then manually curated to remove artefacts with AliView (38). Maximum Likelihood (ML) trees were generated using IQ-TREE (39), under appropriate models which were inferred as best fit models for DENV 1-3 by ModelFinder application implemented in IQ-TREE software ((40). Tree topologies robustness was determined using 1000 replicates. Tree visualization was performed using Figtree (
http://tree.bio.ed.ac.uk) and R software (41).
2.8. Temporal trend and Spatial mapping of detected serotypes
According the RT-qPCR serotyping results temporal and spatial mapping of detected DENV serotypes were performed using epidemiological week of sampling and informations related to latitude and longitude of region where sentinels sites from which samples were collected. Temporal trends was represented using barplot, spatial distribution was represented by a maps made using maplots package within R ; pie chart representing the proportion of each detected serotypes at a given region were represented.
Figure 0.
Summary of used workflow during this study
Figure 0.
Summary of used workflow during this study
3. Results
From January 2019 to February 2023 5303 suspected arboviral cases were collected from 4S sentinel sites and shipped to WHOCC for arboviruses and haemorrhagic fever viruses where they are subjected to molecular testing for the detection of DENV RNA ; 402 among them where panDENV positives by RT-qPCR.
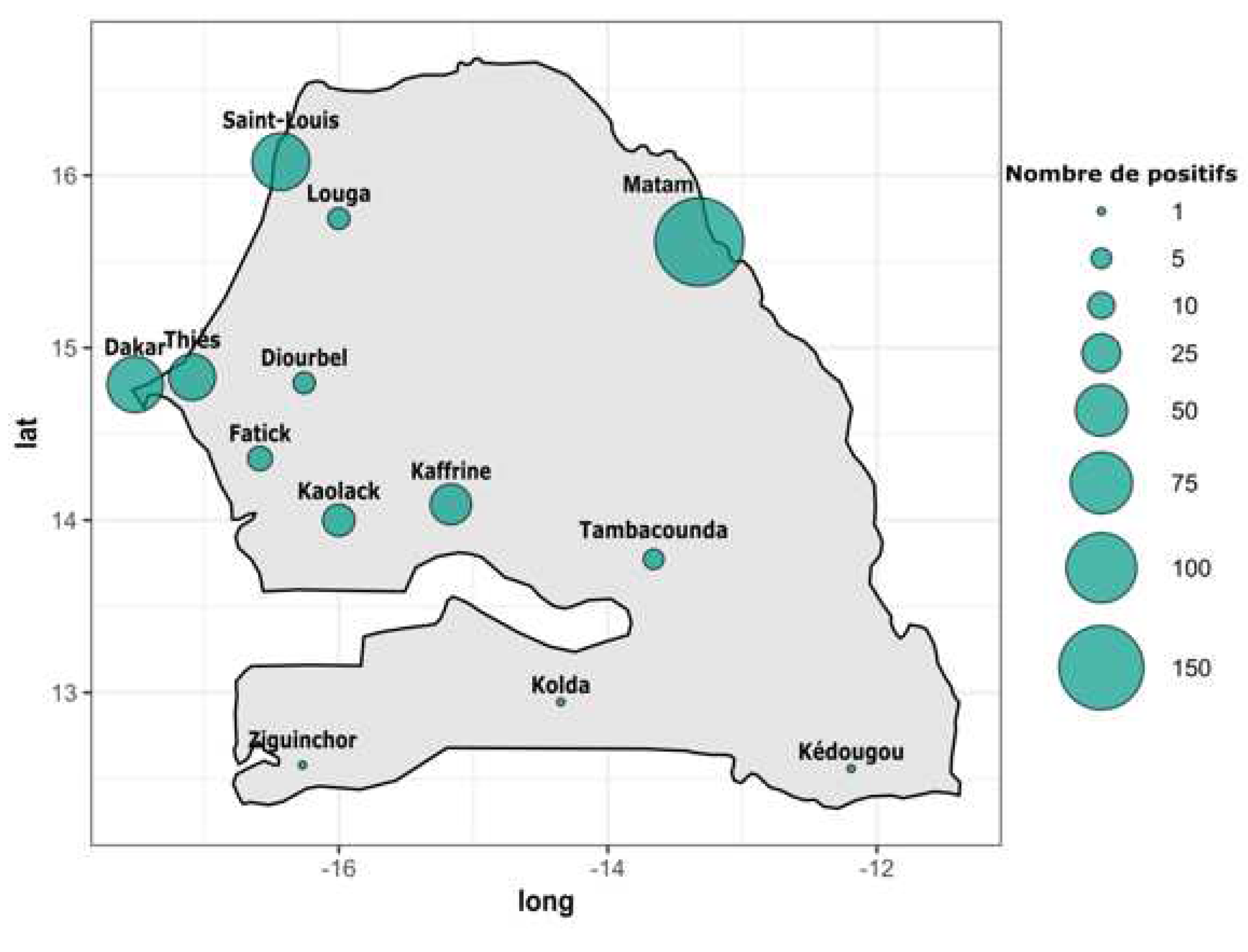
According to the region of provenance RT-qPCR DENV+ samples were collected from twelve out of fourteen administrative regions of Senegal. In term of occurrence the highest number of confirmed DENV+ cases were recorded in Matam (n = 166), Saint louis (n = 64), Dakar (n = 60) , Thies (n = 39), Kaffrine (n = 29) and Kaolack (n = 16). Remaining regions including Diourbel, Fatick, Kedougou, Kolda, Louga, Tambacounda and Ziguinchor all recorded less than ten confirmed DENV+ cases (
Figure 1 and
Table S1).
According to the year of collection the table summarise the number of suspected and confirmed DENV+ samples according to the year of collection. Briefly, the highest number of confirmed dengue cases was obtained in 2022, the same year we recorded the highest confirmed DENV+ cases (n = 216). The lowest number of suspected (n = 109 ) and confirmed cases (n = 15) were obtained during the year 2023.
Table 1.
Repartition of suspected and confirmed DENV cases according to the year of sampling.
Table 1.
Repartition of suspected and confirmed DENV cases according to the year of sampling.
| Year of collection |
Number of suspected cases |
Number of confirmed DENV cases |
| 2019 |
890 |
19 |
| 2020 |
862 |
20 |
| 2021 |
1353 |
131 |
| 2022 |
2089 |
216 |
| 2023 |
109 |
15 |
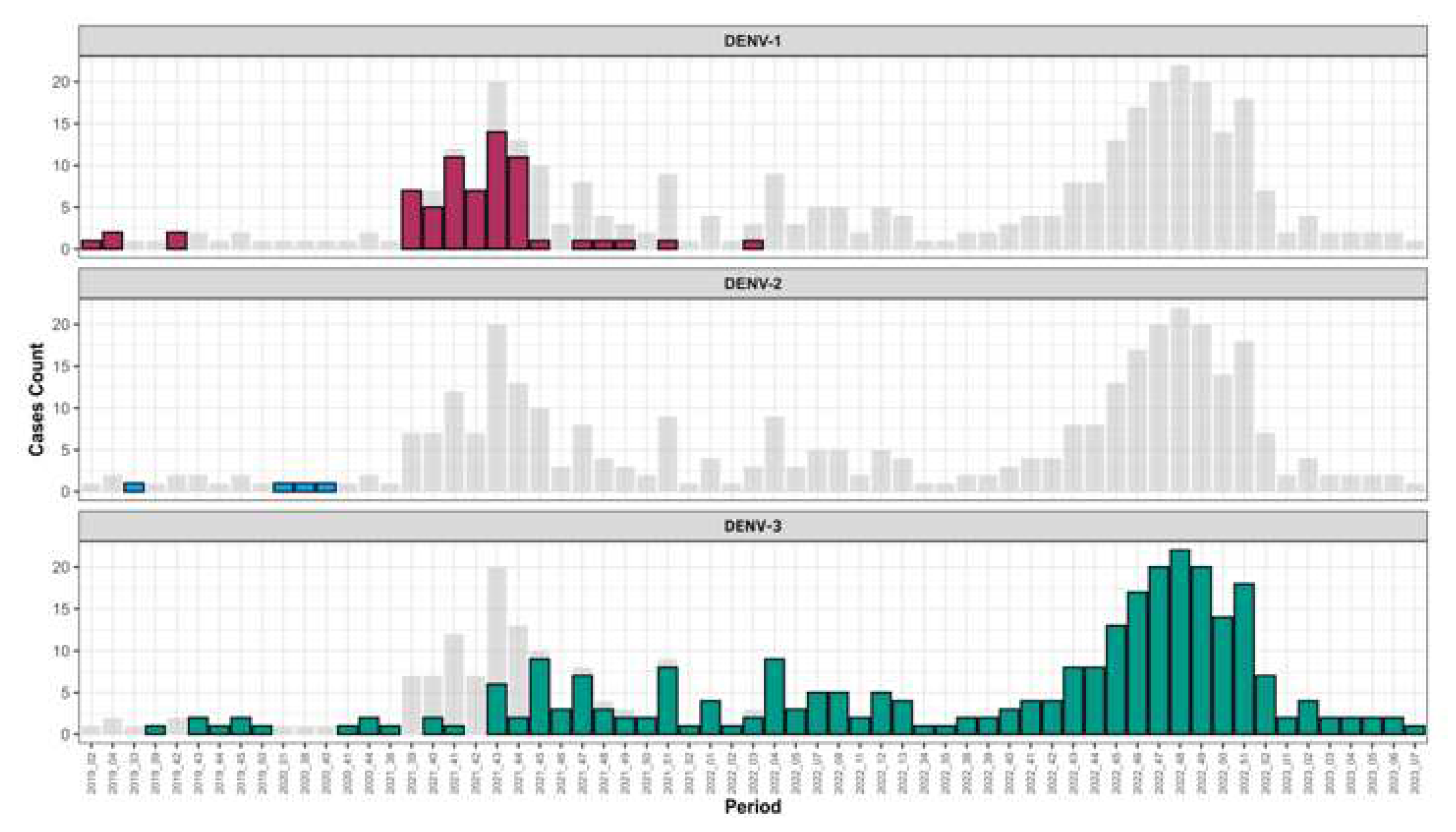
Serotyping results by RT-qPCR of 346 out of 402 DENV+ samples shows the circulation of three DENV serotypes.
In term of occurrence DENV-3 is the most prevalent serotype (n = 276) followed by DENV-1 (n = 66) ; the less prevalent serotype among screened DENV+ samples was DENV-2 (n = 04) (
Table S2). Interestingly temporal distribution of circulating serotype during study period show that between February 2019 to March 2022 the three serotypes co-circulated after March 2022 DENV 1-2 were no longer detected and only DENV-3 circulated till March 2023 (
Figure 2 and
Table S3 &
Table S4 ).
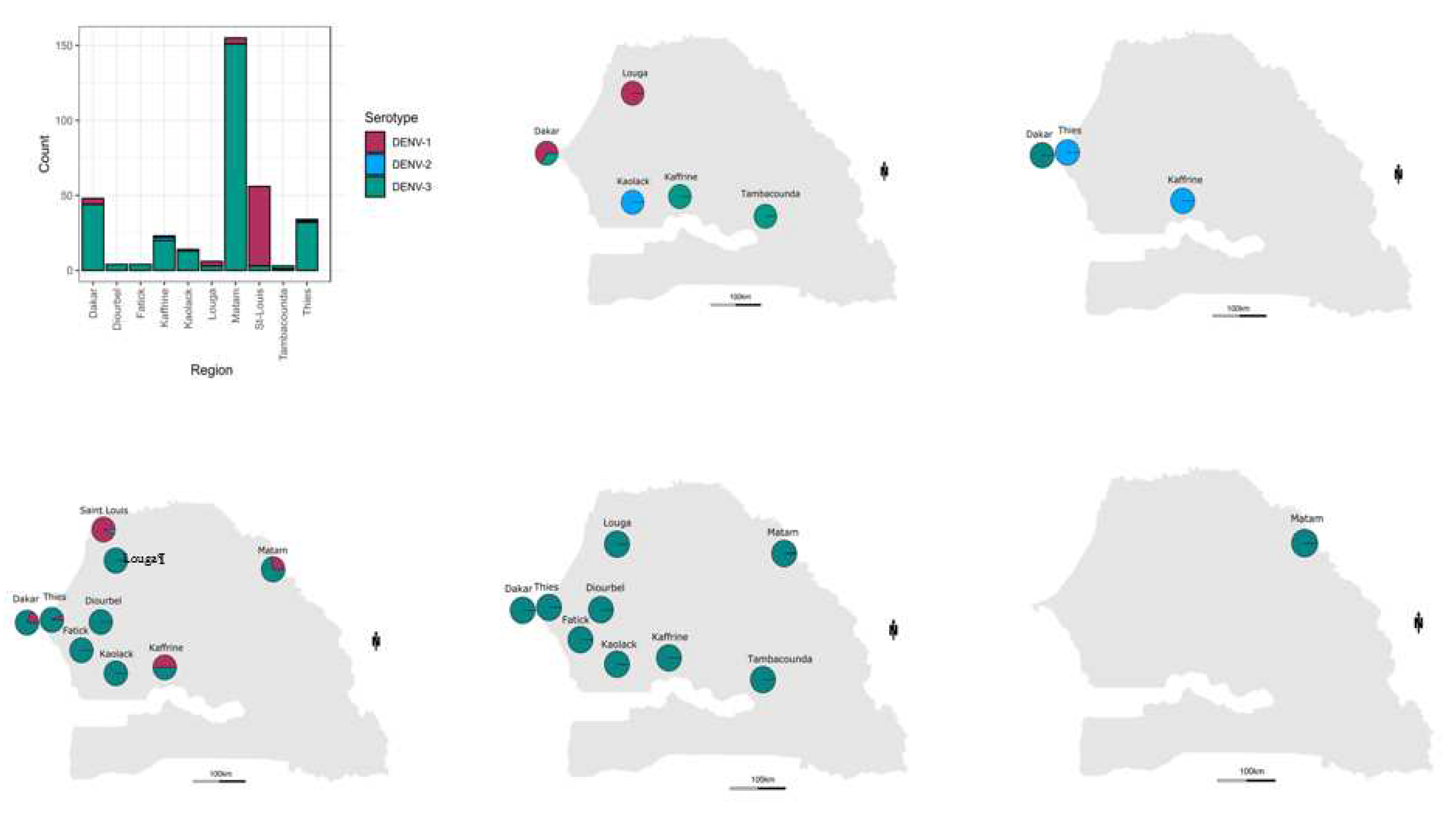
Geographically the most widely distributed serotype was DENV-3 found in ten out of ten regions were serotypes samples were retrieved ; the highest number of DENV+ cases associated to this serotype was found in Matam (n = 151), Dakar (n = 44) and Thies (n = 32). DENV-1 was detected in four regions including Saint-Louis (n = 53), Dakar (n = 04, Matam (n = 04), ), Louga (n = 03), Kaffrine (n = 01) and Thies (n = 01). Finally DENV-2 is the less detected and spreaded serotype with the detection of only 04 cases in Kaffrine (n = 2), Kaolack (n = 1) and Thies (n = 01) (
Figure 2 ;
Table S2)
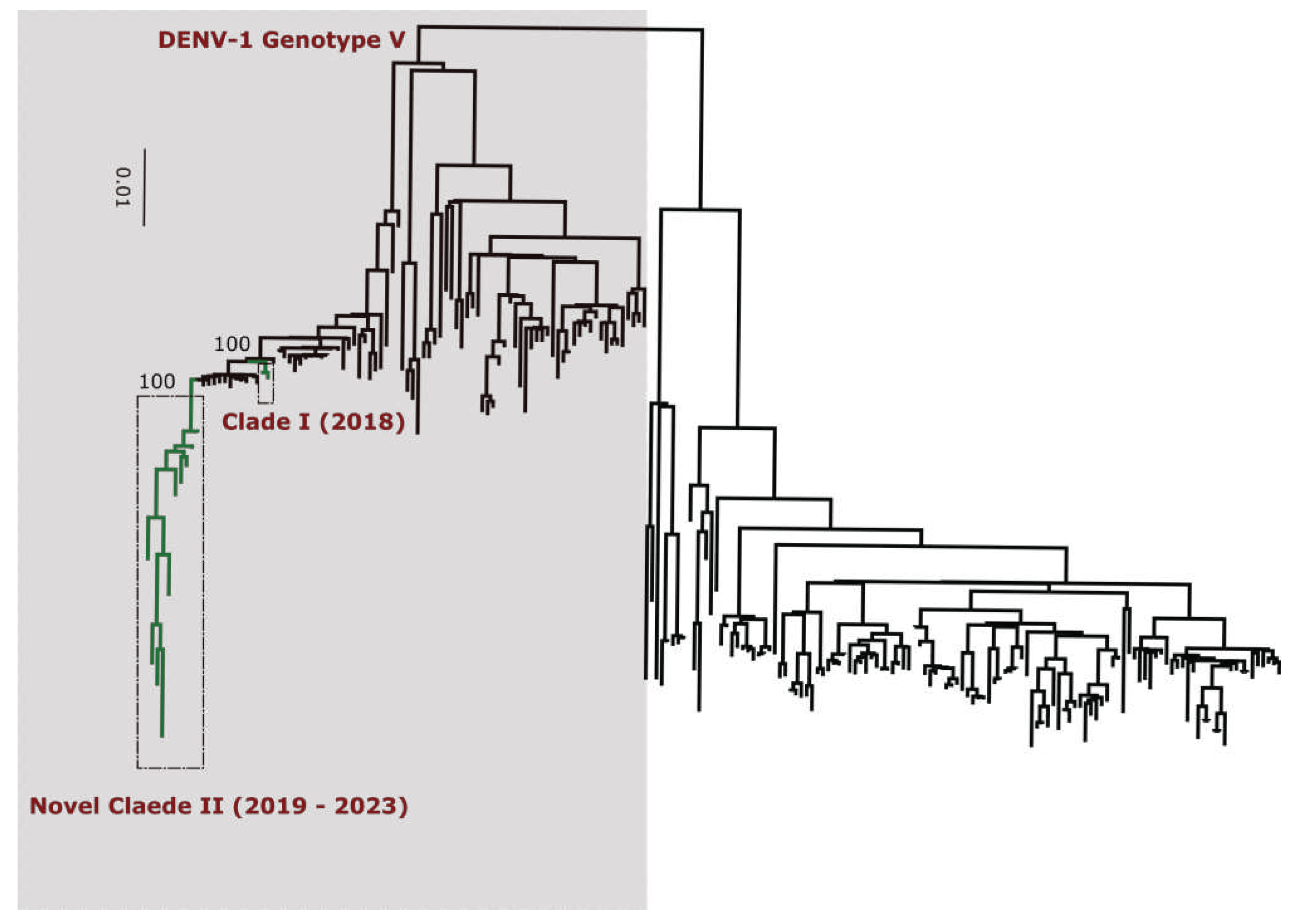
DENV typing tool results (
Table S7) classify all DENV-1 sequences into genotype V (
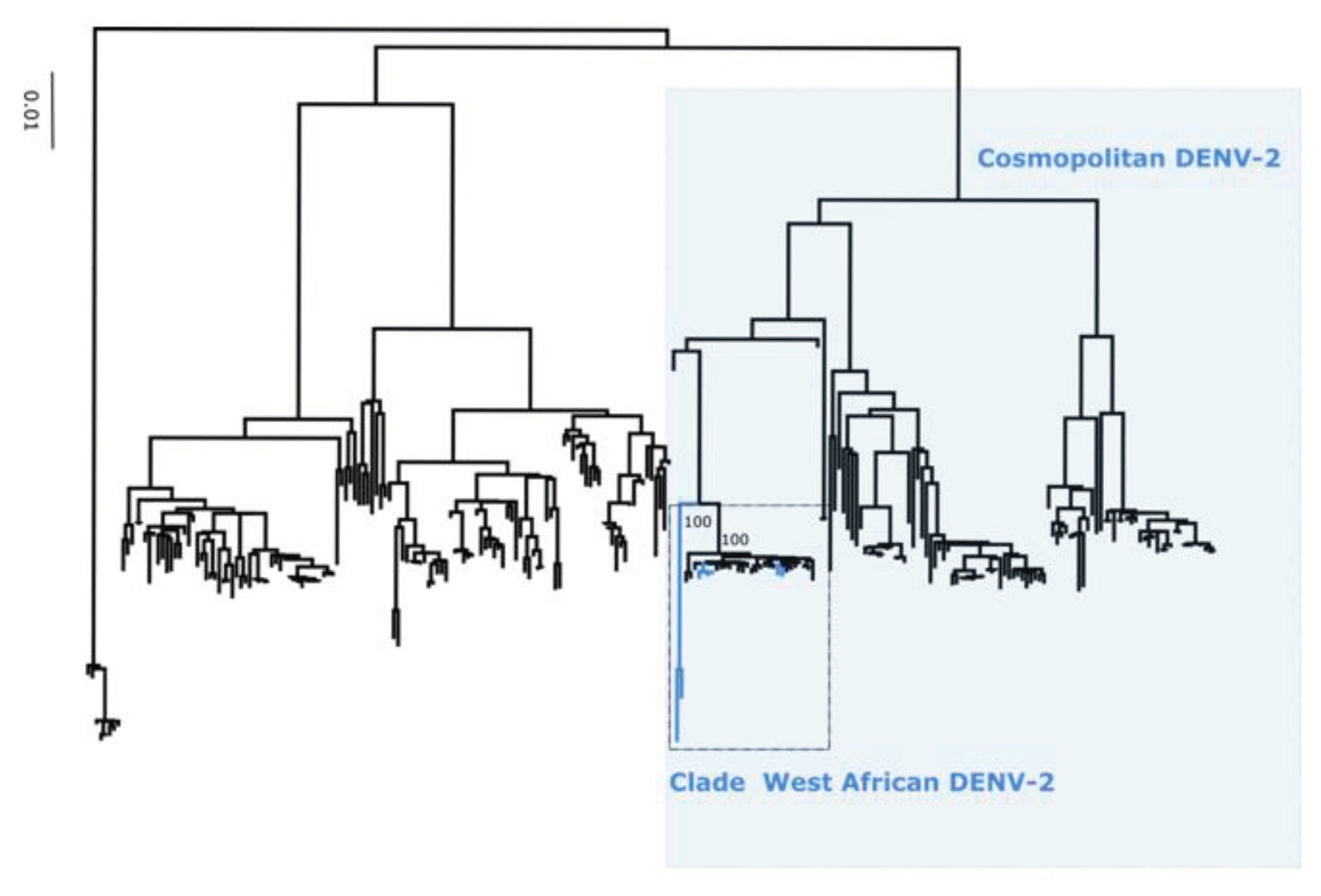
Figure 3), DENV-2 into cosmopolitan genotype (
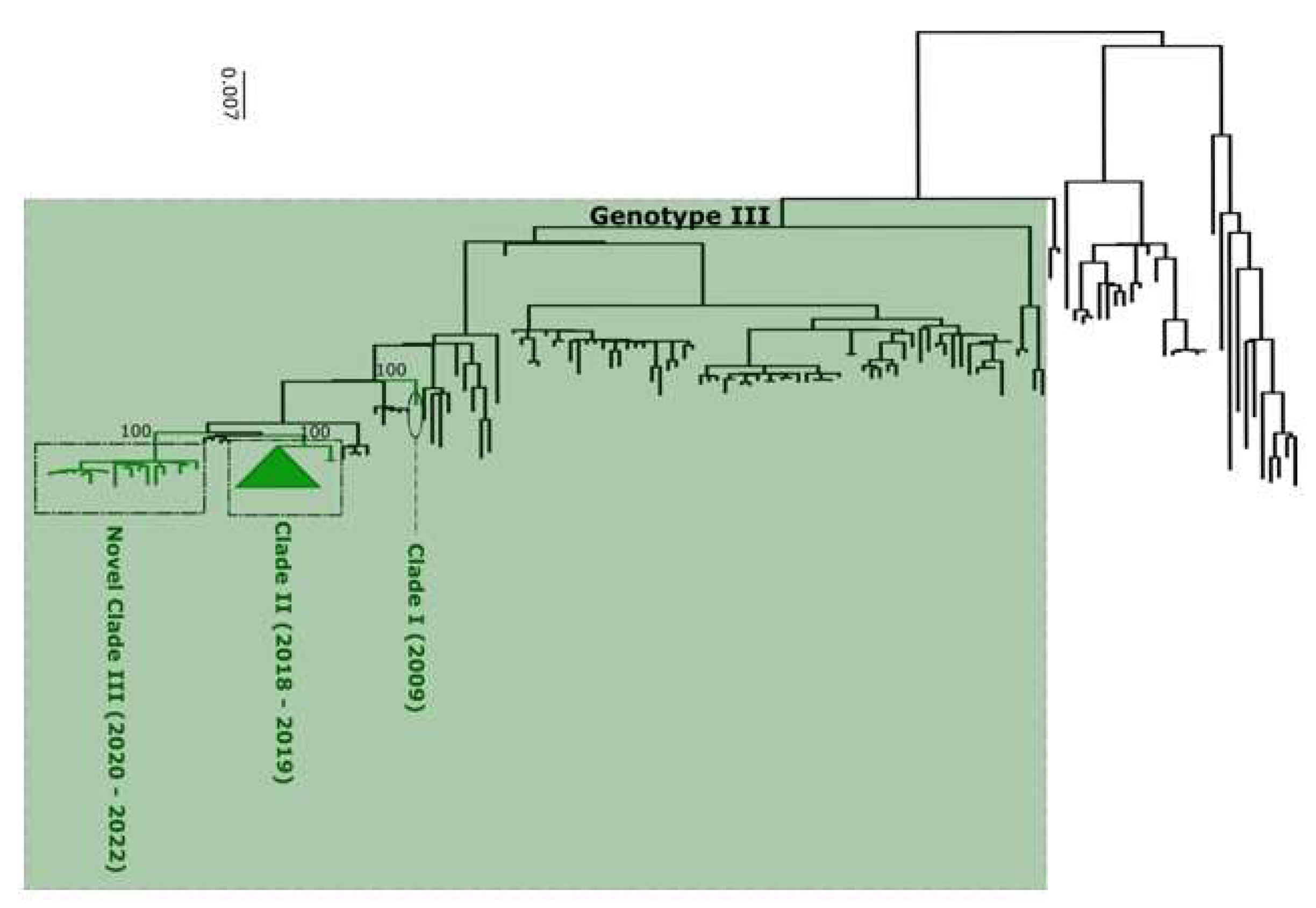
Figure 4) and finally DENV-3 as genotype III (
Figure 5).
Table 2.
Summary of sequenced DENV samples during this study.
Table 2.
Summary of sequenced DENV samples during this study.
| ID |
Virus type |
Genotype |
Region |
Isolate |
Sample type |
Collection Date |
| SH 377553 |
DENV-1 |
Genotype V |
Saint-Louis |
Human |
Serum |
29-10-2021 |
| SH 377552 |
DENV-1 |
Genotype V |
Saint-Louis |
Human |
Serum |
29-10-2021 |
| SH 377551 |
DENV-1 |
Genotype V |
Saint-Louis |
Human |
Serum |
28-10-2021 |
| SH 377538 |
DENV-1 |
Genotype V |
Saint-Louis |
Human |
Serum |
01-11-2021 |
| SH 377555 |
DENV-1 |
Genotype V |
Saint-Louis |
Human |
Serum |
29-10-2021 |
| SH 377554 |
DENV-1 |
Genotype V |
Saint-Louis |
Human |
Serum |
29-10-2021 |
| SH 322838 |
DENV-1 |
Genotype V |
Dakar |
Human |
Serum |
21-10-2019 |
| SH 322765 |
DENV-1 |
Genotype V |
Dakar |
Human |
Serum |
15-10-2021 |
| SH 318479 |
DENV-1 |
Genotype V |
Louga |
Human |
Serum |
24-01-2019 |
| SH 318478 |
DENV-1 |
Genotype V |
Louga |
Human |
Serum |
24-01-2019 |
| SH 318267 |
DENV-1 |
Genotype V |
Louga |
Human |
Serum |
10-01-2019 |
| SH 326097 |
DENV-2 |
Genotype II |
Kaffrine |
Human |
Serum |
30-09-2020 |
| SH 323592 |
DENV-2 |
Genotype II |
Thies |
Human |
Serum |
05-01-2020 |
| SH 377545 |
DENV-3 |
Genotype III |
Saint-Louis |
Human |
Serum |
27-10-2021 |
| SH 377540 |
DENV-3 |
Genotype III |
Saint-Louis |
Human |
Serum |
26-10-2021 |
| SH 377524 |
DENV-3 |
Genotype III |
Thies |
Human |
Serum |
28-10-2021 |
| SH 377522 |
DENV-3 |
Genotype III |
Thies |
Human |
Serum |
28-10-2021 |
| SH 392244 |
DENV-3 |
Genotype III |
NA |
Human |
Serum |
NA |
| SH 402404 |
DENV-3 |
Genotype III |
Fatick |
Human |
Serum |
22-08-2022 |
| SH 392518 |
DENV-3 |
Genotype III |
Tambacounda |
Human |
Serum |
19-03-2022 |
| SH 392408 |
DENV-3 |
Genotype III |
Matam |
Human |
Serum |
15-03-2022 |
| SH 392406 |
DENV-3 |
Genotype III |
Matam |
Human |
Serum |
15-03-2022 |
| SH 392265 |
DENV-3 |
Genotype III |
Matam |
Human |
Serum |
25-02-2022 |
| SH 392263 |
DENV-3 |
Genotype III |
Matam |
Human |
Serum |
25-02-2022 |
| SH 392260 |
DENV-3 |
Genotype III |
Matam |
Human |
Serum |
23-02-2022 |
| SH 392169 |
DENV-3 |
Genotype III |
Matam |
Human |
Serum |
22-02-2022 |
| SH 392168 |
DENV-3 |
Genotype III |
Matam |
Human |
Serum |
21-02-2022 |
| SH 330006 |
DENV-3 |
Genotype III |
Dakar |
Human |
Serum |
03-11-2020 |
| SH 330004 |
DENV-3 |
Genotype III |
Dakar |
Human |
Serum |
03-11-2020 |
| SH 329067 |
DENV-3 |
Genotype III |
NA |
Human |
Serum |
NA |
| SH 327002 |
DENV-3 |
Genotype III |
Dakar |
Human |
Serum |
12-10-2020 |
| SH 392572 |
DENV-3 |
Genotype III |
Matam |
Human |
Serum |
21-03-2022 |
| SH 392571 |
DENV-3 |
Genotype III |
Matam |
Human |
Serum |
22-03-2022 |
| SH 323342 |
DENV-3 |
Genotype III |
Kaffrine |
Human |
Serum |
10-12-2019 |
| SH 322872 |
DENV-3 |
Genotype III |
Kaffrine |
Human |
Serum |
28-10-2019 |
All this assignment were confirmed by phylogenetic analysis (
Figure 1,
Figure 2 and
Figure 3). Additionnally, phylogenetic analysis shows that in comparison to limited previously available full genome sequences sequences from Senegal DENV-1 and DENV-3 sequences clustered in different clades (hereafter namely as Novel Clades (2019 - 2023) for DENV-1 and Novel Clade III (2020 - 2022)) ; DENV-2 sequences are closely related to DENV-2 cosmopolitan detected in West Africa.
4. Discussion
Senegal is a West African country with a reliable and efficient syndromic surveillance system as exemplified by previous early detection of epidemic prone disease and subsequent organization of appropriate response (13,42,43).
This system allowed the notification of many DENV outbreak in Senegal (13,14,17) ; despite the recurrent occurrence of dengue epidemics and/or sporadic cases studies focusing on the circulating serotypes/genotypes and their associated spatial and temporal distribution are limited (16). This present study aimed to address this concern by investigating the circulating dengue variants in Senegal between 2019 to 2023 through the syndromic sentinel surveillance network of Senegal (4S network). To the best of our knowledge this study represent the first multiyear countrywide study focusing on temporal and spatial distribution of dengue virus serotype/genotype and viral genetic diversity using full genome sequences.
Among collected suspected dengue samples (n = 5303), 402 were DENV RNA positive samples (
Figure 0). Interestingly the confirmed cases were distributed around twelve out of fourteen administrative regions of Senegal vs seven regions during 2017 -2018 study (Multifoci), with a DENV RNA positivity rate of 7.58 %. The highest number of dengue positive cases were recorded in Matam region with 166 cases followed by Saint-Louis (n = 64), Dakar (n = 60), Kaffrine (n = 29) ; others regions where dengue was detected recorded a number of cases below twenty (
Table S1). Interestingly, the Matam regions was never been associated to any dengue outbreaks or recurrent cases notifications in the past supporting studies highlighting that the introduction of new groups of viruses to populations lacking prior exposure (serological naivety) has the potential to trigger unprecedented outbreaks and can be linked to more severe manifestations of dengue. (27,44).
Compared to previous Senegalese study (Multifoci) the highest number of suspected DENV samples were enrolled thus probably lead to the highest number of confirmed cases recorded during the present study. In contrast a study on genetic diversity of dengue virus in Bangkok yield a highest prevalence of DENV positivity (25.09 %) compared to our study (29) while a DENV RNA prevalence of 38.24 % was obtained during single year study in India (45). This discrepancy is probably do the fact that compared to Senegal dengue is highly endemic in Thailand and India. Indeed Bangkok , the capital city of Thailand, is located in the center of the country and serve as a transportation hub and dengue is known to circulate there since 1950s (46,47). In India studies report that DENV is reported every year and the size, severity, duration of outbreaks are increasing (45,48).
All together findings highlight the rapid spread of arboviral disease between neighboring regions thanks travel and trades activities (49,50). It is well known that frequent reintroductions of pathogens pose a significant challenge to elimination campaigns, especially in areas experiencing substantial regional and international travel. This is because humans serve as the reservoir host for both epidemic dengue and chikungunya (51). None DENV+ RNA sample was recorded in Sedhiou region in Southern Senegal. This is probably due to the fact that the sentinel site in this region was implemented recently and issues on proper samples transportation du IPD were noticed (Samba Sagn personnal communication). According to the year of collection the highest number of confirmed DENV cases was noticed in 2022 ; the trends follow the number of collected samples during the same year which is higher compared to other years (
Table 1). This is probably due to the fact that between 2019 to 2021 most of the surveillance effort were focused on the covid-19 pandemic.
For assignment of DENV serotype/genotype the only nationwide dengue spatial mapping study based on partial CprM gene used limited number of samples collected between 2017 to 2018. To get more insight and up to date genetic diversity of circulating dengue strains at the serotype/genotype levels using RT-qPCR we serotyped 347 out of 402 dengue positives samples and generated 34 nearly complete DENV genomes (
Table 2 ). Serotyping using RT-qPCR shows that detected dengue virus strains in Senegal during the study period belong to DENV 1-3 ; the most represented serotype was DENV-3 (n = 276), followed by DENV=1 (n = 66) and finally DENV-2 (n = 04). None of the DENV+ samples was linked to DENV-4 (
Table S2). This finding corroborate those obtained by Dieng and colleagues which showed the co-circulation of DENV 1-3 in Senegal between 2017 to 2018 (16).
Any marked spatial distribution pattern of serotype was observed compared to previous study in Senegal (16). In contrast a study performed in India show a regional diversity of DENV serotypes (45).
But based on observations DENV-3 was the dominant serotype in Dakar, Thies, Fatick, Diourbel, Kaolack, Kaffrine, Tambacounda and Matam. DENV-1 was dominant serotype in Saint-Louis. DENV-2 and DENV-3 were co-dominant in Louga (
Figure 2 A ;
Table S2). Multiple serotype infections were more prominent in samples tested in Thies and Kaffrine where DENV 1-3 were noticed. In Africa limited countries as Gabon, Burkina Faso reported the co-circulation of at least three DENV serotypes (10,52). This is probably linked to the fact that dengue surveillance and awareness is lacking in the continent (8). Limited availability of data about DENV circulating serotype/genotypes is mainly to the limited availability of national research institutions in many areas (48).
In contrast studies in South America reveals the co-circulation in high frequency of dengue viruses serotypes (53). In another hand the hyperendemic behaviour of DENV virus epidemiology is well known and documented in Asian countries as India (27,28,54) and in China, Malaysia , Thailand (29).
Interestingly temporal trends of serotype’s circulation shows that from the third week of year 2022 DENV-1 and DENV-2 serotypes were no longer circulating among confirmed DENV cases but only DENV-3 was noticed (
Figure 2). This serotype shift was associated with a widespread and increased frequency of cases related to this serotype. Findings corroborate those of Suzuki and Colleagues in a study performed in Japan (55). The fact that only DENV-3 was detected in Senegal up to may be due to an increased viral fitness of this serotype compared to DENV 1-2 (56). In is well know that different virus serotypes can be associated to different phenotypic traits (57). For instance, a research conducted in Colombia examined the replicative capability of DENV within C6/36 mosquito cells and populations of A. aegypti. This was done using a distinct viral strain for each DENV serotype, revealing varying degrees of fitness among the serotypes (57). Beside intra-serotypic genetic diversification other parameters as cross-protective immunity between serotypes may explain observed DENV serotype replacement phenomenon (58). All up mentioned hypothesis about differential viral fitness should be confirmed by in vitro and in vivo studies since vector-driven selection may contributed to viral replacement phenomenon as described previously in New Caledonia (23).
Performed phylogenetic analysis as well as genotyping using genomedetective dengue typing tools show that the genotype diversity of detected DENV serotypes was relatively low during our study. Indeed, each of all detected serotype consisted of a single genotype ; DENV-1 belong to genotype V, DENV-2 to genotype cosmopolitan and finally DENV-3 to genotype III. This trends is comparable of genotypic dengue virus make up found in Africa (25). However the genetic diversity of Senegalese DENV strains was more pronounced within each genotype. Indeed in each genotype of characterized DENV sequences fall into different clades ; with the exception of DENV-2 sequences which falls into one clade composed by virus detected in West Africa as previously described by Dieng and Colleagues (37). DENV-1 sequences were distributed in two separate clade namely clade I 2018 composed by viruses sampled during 2018 outbreak in Thies regions (15) and Clade 2019 – 2022 including principally strains associated to outbreak in Rosso in 2021 (17) in addition to sporadic cases collected in late 2019 and beginning of year 2022. The same trend was observed for DENV-3 with the observations of the occurrence circulation of viruses belonging to two clades : Clade II 2018 -2019 shared with strains circulating in Thies 2018 (15), in Senegal 2019 in addition to a newly identified Clade III 2020 – 2022 which is closely related to virus sampled in Burkina Faso 2017 and Ethiopia 2019. The occurrence of viral strains belonging to different clusters is a hallmark of different origin of transmission and call for in depth phylogeographic studies to elucidate origin and dispersal patterns.
In general, the observed presence of various strains from different serotypes/genotypes/clades could potentially account for the consistent reports of dengue outbreaks within the country since 2017. Collectively, these findings underscore the vital importance of maintaining an ongoing genomic surveillance of DENV in Senegal. The data generated through this surveillance can be utilized to support public health laboratories in monitoring the diversity of the virus, which is essential for implementing effective control measures.
5. Conclusions
In summary, it is essential to maintain ongoing monitoring of the circulating DENV serotypes/genotypes in Senegal. This ongoing surveillance will provide crucial information to guide proactive and well-informed public health interventions. By remaining vigilant and adaptable in the face of viral variants, we can effectively navigate and respond to emerging waves of infections, minimizing their impact and safeguarding the health of the population. Consistent genomic surveillance, coupled with real-time data analysis, offers invaluable insights into the evolutionary dynamics of the virus. This, in turn, aids in making informed decisions for public health responses. Comprehending the patterns of circulation of these DENV variants contributes to a comprehensive understanding of the virus's current status at a local/regional context. Such understanding enables authorities to implement appropriate measures, including refining testing strategies and enhancing contact tracing efforts. These measures effectively mitigate the impact of new infection waves and prevent rapid spread within communities.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.Table S1: Number of DENV positives samples recorded per regions from 2019 to 2023 through the 4S network ; Table S2 : Summary of the number of serotyped samples at each monitoring region ; Table S3: Reparation of detected DENV RNA positive samples per Year_Week from 2019 to 2023 ; Table S4: Reparation of detected DENV serotypes per Year_Week from 2019 to 2023
Author Contributions
Conceptualization, Idrissa Dieng and Oumar Faye; Data curation, Idrissa Dieng; Formal analysis, Idrissa Dieng, Diamilatou Balde and Mignane Ndiaye; Funding acquisition, Boly Diop, Amadou Sall, Ousmane Faye and Cheikh Loucoubar; Investigation, Cheikh Talla, Mamadou Barry, Samba Sagne and Boly Diop; Methodology, Idrissa Dieng, Diamilatou Balde, Mignane Ndiaye and Mouhamed Kane; Project administration, Cheikh Talla, Mamadou Barry, Boly Diop, Amadou Sall, Ousmane Faye, Gamou Fall, Cheikh Loucoubar and Oumar Faye; Resources, Cheikh Talla, Mouhamed Kane, Moussa DIAGNE and Oumar Faye; Software, Idrissa Dieng and Aboubacry Gaye; Supervision, Cheikh Talla, Mamadou Barry, Samba Sagne, Boubacar Diallo, Abdourahmane Sow, Gamou Fall, Cheikh Loucoubar and Oumar Faye; Validation, Idrissa Dieng, Gamou Fall and Oumar Faye; Visualization, Idrissa Dieng and Aboubacry Gaye; Writing – original draft, Idrissa Dieng; Writing – review & editing, Idrissa Dieng and Aboubacry Gaye.
Funding
Not special funding were received
Informed Consent Statement
Not applicable
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request
Acknowledgments
We would like to convey special thanks to the virology lab workers at the Institut Pasteur de Dakar
Conflicts of Interest
The authors declare no conflict of interest
References
- WHO. Global strategy for dengue prevention and control, 2012-2020. [Internet]. Geneva, Switzerland: World Health Organization; 2012 [cité 12 sept 2020]. Disponible sur: http://apps.who.int/iris/bitstream/10665/75303/1/9789241504034_eng.pdf.
- Messina JP, Brady OJ, Scott TW, Zou C, Pigott DM, Duda KA, et al. Global spread of dengue virus types: mapping the 70 year history. Trends in Microbiology. mars 2014;22(3):138-46. [CrossRef]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 25 avr 2013;496(7446):504-7. [CrossRef]
- WHO. World health statistics 2012. Geneva, Switzerland: World Health Organization; 2012.
- WHO/TDR, éditeur. Dengue: guidelines for diagnosis, treatment, prevention, and control. New ed. Geneva: TDR : World Health Organization; 2009. 147 p.
- Guzman MG, Harris E. Dengue. The Lancet. janv 2015;385(9966):453-65.
- Fourié T, El Bara A, Dubot-Pérès A, Grard G, Briolant S, Basco LK, et al. Emergence of dengue virus serotype 2 in Mauritania and molecular characterization of its circulation in West Africa. PLoS Negl Trop Dis. oct 2021;15(10):e0009829.
- Amarasinghe A, Kuritsky JN, Letson GW, Margolis HS. Dengue Virus Infection in Africa. Emerg Infect Dis. août 2011;17(8):1349-54.
- Tarnagda Z, Cissé A, Bicaba BW, Diagbouga S, Sagna T, Ilboudo AK, et al. Dengue Fever in Burkina Faso, 2016. Emerg Infect Dis. janv 2018;24(1):170-2. [CrossRef]
- Letizia AG, Pratt CB, Wiley MR, Fox AT, Mosore M, Agbodzi B, et al. Retrospective Genomic Characterization of a 2017 Dengue Virus Outbreak, Burkina Faso. Emerg Infect Dis [Internet]. juin 2022 [cité 15 sept 2022];28(6). Disponible sur: https://wwwnc.cdc.gov/eid/article/28/6/21-2491_article.htm. [CrossRef]
- Were F. The dengue situation in Africa. Paediatr Int Child Health. mai 2012;32(s1):18-21. [CrossRef]
- Diallo M, Ba Y, Sall AA, Diop OM, Ndione JA, Mondo M, et al. Amplification of the Sylvatic Cycle of Dengue Virus Type 2, Senegal, 1999–2000: Entomologic Findings and Epidemiologic Considerations. Emerg Infect Dis. mars 2003;9(3):362-7.
- Dieng I, Diarra M, Diagne MM, Faye M, Dior Ndione MH, Ba Y, et al. Field Deployment of a Mobile Biosafety Laboratory Reveals the Co-Circulation of Dengue Viruses Serotype 1 and Serotype 2 in Louga City, Senegal, 2017. J Trop Med. 2021;2021:8817987. [CrossRef]
- Dieng I, Fall C, Barry MA, Gaye A, Dia N, Ndione MHD, et al. Re-Emergence of Dengue Serotype 3 in the Context of a Large Religious Gathering Event in Touba, Senegal. IJERPH. 16 déc 2022;19(24):16912. [CrossRef]
- Gaye A, Ndiaye T, Sy M, Deme AB, Thiaw AB, Sene A, et al. Genomic investigation of a dengue virus outbreak in Thiès, Senegal, in 2018. Sci Rep. 14 mai 2021;11(1):10321.
- Dieng I, Ndione MHD, Fall C, Diagne MM, Diop M, Gaye A, et al. Multifoci and multiserotypes circulation of dengue virus in Senegal between 2017 and 2018. BMC Infect Dis. 24 août 2021;21(1):867.
- Dieng I, Ndiaye M, Ndione MH, Sankhe S, Diagne MM, Sagne SN, et al. Molecular Characterization of Circulating DENV-2 During Outbreak in Northern Senegal, Rosso 2018 [Internet]. LIFE SCIENCES; 2021 déc [cité 26 janv 2022]. Disponible sur: https://www.preprints.org/manuscript/202112.0270/v1.
- Harapan H, Michie A, Sasmono RT, Imrie A. Dengue: A Minireview. Viruses. 30 juill 2020;12(8):829.
- Holmes E, Twiddy S. The origin, emergence and evolutionary genetics of dengue virus. Infection, Genetics and Evolution. mai 2003;3(1):19-28. [CrossRef]
- Usme-Ciro JA, Méndez JA, Laiton KD, Páez A. The relevance of dengue virus genotypes surveillance at country level before vaccine approval. Human Vaccines & Immunotherapeutics. 2 sept 2014;10(9):2674-8. [CrossRef]
- OhAinle M, Balmaseda A, Macalalad AR, Tellez Y, Zody MC, Saborío S, et al. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci Transl Med. 21 déc 2011;3(114):114ra128. [CrossRef]
- Fried JR, Gibbons RV, Kalayanarooj S, Thomas SJ, Srikiatkhachorn A, Yoon IK, et al. Serotype-specific differences in the risk of dengue hemorrhagic fever: an analysis of data collected in Bangkok, Thailand from 1994 to 2006. PLoS Negl Trop Dis. 2 mars 2010;4(3):e617.
- O’Connor O, Ou TP, Aubry F, Dabo S, Russet S, Girault D, et al. Potential role of vector-mediated natural selection in dengue virus genotype/lineage replacements in two epidemiologically contrasted settings. Emerging Microbes & Infections. 1 janv 2021;10(1):1346-57. [CrossRef]
- Amoako N, Duodu S, Dennis FE, Bonney JHK, Asante KP, Ameh J, et al. Detection of Dengue Virus among Children with Suspected Malaria, Accra, Ghana. Emerg Infect Dis. août 2018;24(8):1544-7. [CrossRef]
- Ayolabi CI, Olusola BA, Ibemgbo SA, Okonkwo GO. Detection of Dengue viruses among febrile patients in Lagos, Nigeria and phylogenetics of circulating Dengue serotypes in Africa. Infection, Genetics and Evolution. nov 2019;75:103947. [CrossRef]
- Yamashita A, Sakamoto T, Sekizuka T, Kato K, Takasaki T, Kuroda M. DGV: Dengue Genographic Viewer. Front Microbiol [Internet]. 7 juin 2016 [cité 30 déc 2020];7. Disponible sur: http://journal.frontiersin.org/Article/10.3389/fmicb.2016.00875/abstract.
- Shrivastava S, Tiraki D, Diwan A, Lalwani SK, Modak M, Mishra AC, et al. Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 season. Ansari AA, éditeur. PLoS ONE. 22 févr 2018;13(2):e0192672. [CrossRef]
- Jagtap S, Pattabiraman C, Sankaradoss A, Krishna S, Roy R. Evolutionary dynamics of dengue virus in India. Katzelnick L, éditeur. PLoS Pathog. 3 avr 2023;19(4):e1010862. [CrossRef]
- Poltep K, Phadungsombat J, Nakayama EE, Kosoltanapiwat N, Hanboonkunupakarn B, Wiriyarat W, et al. Genetic Diversity of Dengue Virus in Clinical Specimens from Bangkok, Thailand, during 2018–2020: Co-Circulation of All Four Serotypes with Multiple Genotypes and/or Clades. TropicalMed. 4 sept 2021;6(3):162. [CrossRef]
- De Simone TS, Nogueira RMR, Araújo ESM, Guimarães FR, Santos FB, Schatzmayr HG, et al. Dengue virus surveillance: the co-circulation of DENV-1, DENV-2 and DENV-3 in the State of Rio de Janeiro, Brazil. Transactions of the Royal Society of Tropical Medicine and Hygiene. sept 2004;98(9):553-62.
- Lim JK, Carabali M, Camacho E, Velez DC, Trujillo A, Egurrola J, et al. Epidemiology and genetic diversity of circulating dengue viruses in Medellin, Colombia: a fever surveillance study. BMC Infect Dis. déc 2020;20(1):466. [CrossRef]
- Dia N, Diene Sarr F, Thiam D, Faye Sarr T, Espié E, OmarBa I, et al. Influenza-Like Illnesses in Senegal: Not Only Focus on Influenza Viruses. PLoS One [Internet]. 27 mars 2014 [cité 24 oct 2019];9(3). Disponible sur: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3968133/. [CrossRef]
- Bob NS, Barry MA, Diagne MM, Faye M, Ndione MHD, Diallo A, et al. Detection of Rift Valley Fever Virus Lineage H From South Africa Through the Syndromic Sentinel Surveillance Network in Senegal. Open Forum Infect Dis. mars 2022;9(3):ofab655. [CrossRef]
- Wagner D, de With K, Huzly D, Hufert F, Weidmann M, Breisinger S, et al. Nosocomial Acquisition of Dengue. Emerg Infect Dis. oct 2004;10(10):1872-3. [CrossRef]
- Dieng I, Cunha MDP, Diagne MM, Sembène PM, Zanotto PMDA, Faye O, et al. Origin and Spread of the Dengue Virus Type 1, Genotype V in Senegal, 2015–2019. Viruses. 4 janv 2021;13(1):57. [CrossRef]
- Santiago GA, Vergne E, Quiles Y, Cosme J, Vazquez J, Medina JF, et al. Analytical and Clinical Performance of the CDC Real Time RT-PCR Assay for Detection and Typing of Dengue Virus. Harris E, éditeur. PLoS Negl Trop Dis. 11 juill 2013;7(7):e2311.
- Dieng I, Diallo A, Ndiaye M, Mhamadi M, Diagne MM, Sankhe S, et al. Full genome analysis of circulating DENV-2 in Senegal reveals a regional diversification into separate clades. Journal of Medical Virology. 5 août 2022;jmv.28027. [CrossRef]
- Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 15 nov 2014;30(22):3276-8. [CrossRef]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol Biol Evol. janv 2015;32(1):268-74. [CrossRef]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. juin 2017;14(6):587-9. [CrossRef]
- R Core Team. R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021.
- Fall A, Dieng I, Touré CT, Mhamadi M, Sadio BD, Ndione MHD, et al. Institut Pasteur Dakar Mobile Lab: Part of the Solution to Tackle COVID Pandemic in Senegal, a Model to Be Exploited. COVID. 20 oct 2022;2(10):1509-17. [CrossRef]
- Diagne MM, Ndione MHD, Gaye A, Barry MA, Diallo D, Diallo A, et al. Yellow Fever Outbreak in Eastern Senegal, 2020–2021. Viruses. 28 juill 2021;13(8):1475. [CrossRef]
- Vu D, Mutai N, Heath C, Ndenga B, Labeaud AD. Dengue Viremia in Kenyan children With Acute Febrile Illness. Open Forum Infectious Diseases. 1 déc 2016;3(suppl_1):597. [CrossRef]
- Alagarasu K, Patil JA, Kakade MB, More AM, Yogesh B, Newase P, et al. Serotype and genotype diversity of dengue viruses circulating in India: a multi-centre retrospective study involving the Virus Research Diagnostic Laboratory Network in 2018. International Journal of Infectious Diseases. oct 2021;111:242-52. [CrossRef]
- Hammon WMcD. DENGUE HEMORRHAGIC FEVER—DO WE KNOW ITS CAUSE? *. The American Journal of Tropical Medicine and Hygiene. 1 janv 1973;22(1):82-91.
- Halstead SB. Immune enhancement of viral infection. Prog Allergy. 1982;31:301-64.
- Cecilia D, Patil JA, Kakade MB, Walimbe A, Alagarasu K, Anukumar B, et al. Emergence of the Asian genotype of DENV-1 in South India. Virology. oct 2017;510:40-5. [CrossRef]
- Brunette GW, Nemhauser JB. Travel-Related Infectious Diseases. In: CDC Yellow Book 2020 [Internet]. Oxford University Press; 2019 [cité 22 août 2023]. p. 169-394. Disponible sur: https://academic.oup.com/book/25289/chapter/191511526.
- Diagne CT, Barry MA, Ba Y, Faye O, Sall AA. Dengue epidemic in Touba, Senegal: implications for the Grand Magal Pilgrimage for travellers. Journal of Travel Medicine. 14 oct 2019;26(7):tay123. [CrossRef]
- Moncayo AC, Fernandez Z, Ortiz D, Diallo M, Sall A, Hartman S, et al. Dengue Emergence and Adaptation to Peridomestic Mosquitoes. Emerg Infect Dis. oct 2004;10(10):1790-6. [CrossRef]
- Caron M, Grard G, Paupy C, Mombo IM, Bikie Bi Nso B, Kassa Kassa FR, et al. First Evidence of Simultaneous Circulation of Three Different Dengue Virus Serotypes in Africa. PLoS One [Internet]. 21 oct 2013 [cité 27 oct 2019];8(10). Disponible sur: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3804462/. [CrossRef]
- Fritsch H, Moreno K, Lima IAB, Santos CS, Costa BGG, De Almeida BL, et al. Phylogenetic Reconstructions Reveal the Circulation of a Novel Dengue Virus-1V Clade and the Persistence of a Dengue Virus-2 III Genotype in Northeast Brazil. Viruses. 28 avr 2023;15(5):1073. [CrossRef]
- Racherla RG, Pamireddy ML, Mohan A, Mudhigeti N, Mahalakshmi PA, Nallapireddy U, et al. Co-circulation of four dengue serotypes at South Eastern Andhra Pradesh, India: A prospective study. Indian J Med Microbiol. juin 2018;36(2):236-40. [CrossRef]
- Suzuki K, Phadungsombat J, Nakayama EE, Saito A, Egawa A, Sato T, et al. Genotype replacement of dengue virus type 3 and clade replacement of dengue virus type 2 genotype Cosmopolitan in Dhaka, Bangladesh in 2017. Infection, Genetics and Evolution. nov 2019;75:103977. [CrossRef]
- Ritchie SA, Pyke AT, Hall-Mendelin S, Day A, Mores CN, Christofferson RC, et al. An Explosive Epidemic of DENV-3 in Cairns, Australia. Vasilakis N, éditeur. PLoS ONE. 16 juill 2013;8(7):e68137.
- Quintero-Gil DC, Uribe-Yepes A, Ospina M, Díaz FJ, Martinez-Gutierrez M. Differences in the replicative capacities of clinical isolates of dengue virus in C6/36 cells and in urban populations of Aedes aegypti from Colombia, South America. The Brazilian Journal of Infectious Diseases. juill 2018;22(4):257-72.
- Adams B, Holmes EC, Zhang C, Mammen MP, Nimmannitya S, Kalayanarooj S, et al. Cross-protective immunity can account for the alternating epidemic pattern of dengue virus serotypes circulating in Bangkok. Proceedings of the National Academy of Sciences. 19 sept 2006;103(38):14234-9. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).