Submitted:
05 October 2023
Posted:
06 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
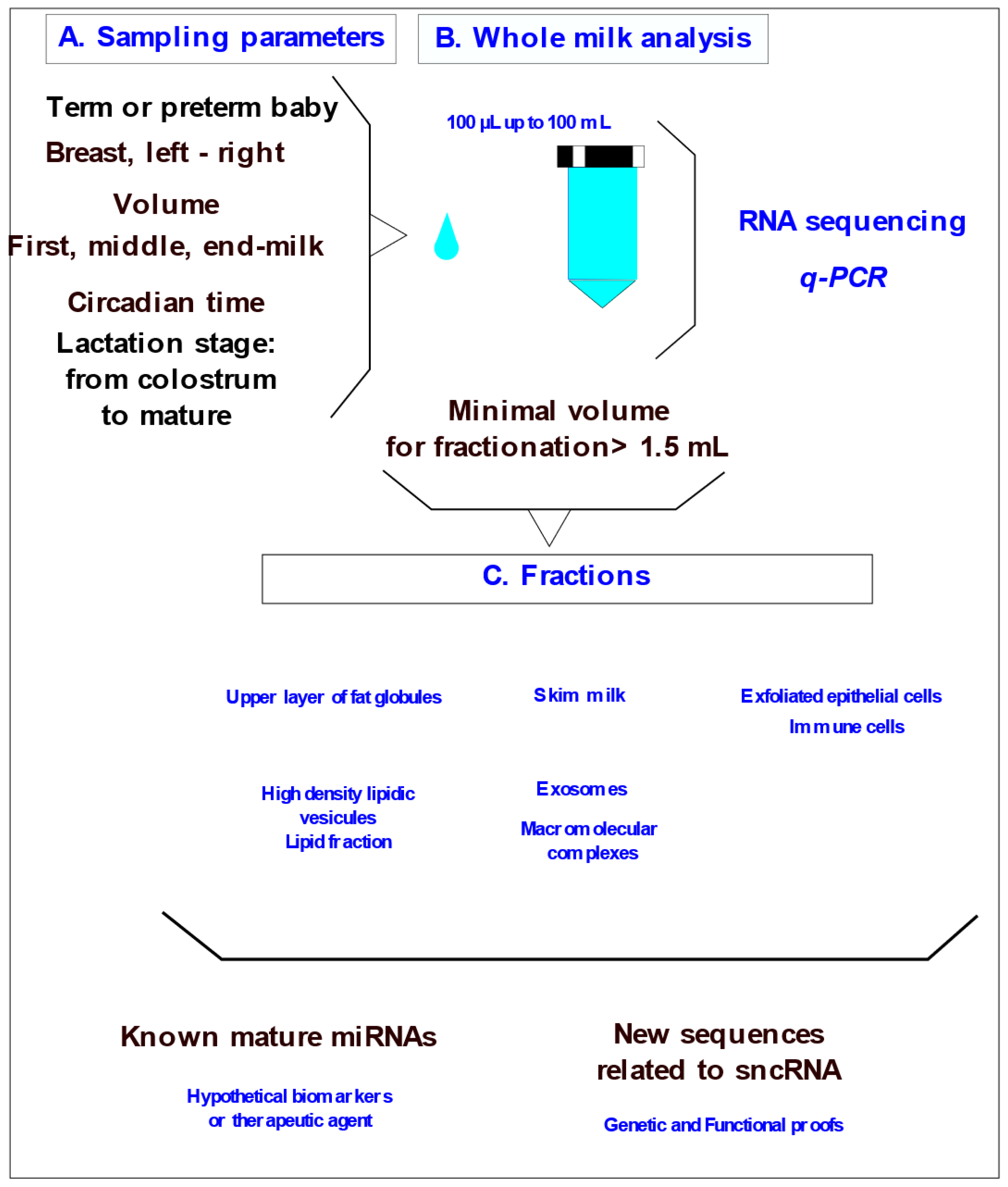
1. RNA content and mature miRNA diversity
2. Bioavailability of mature miRNAs in the digestive system
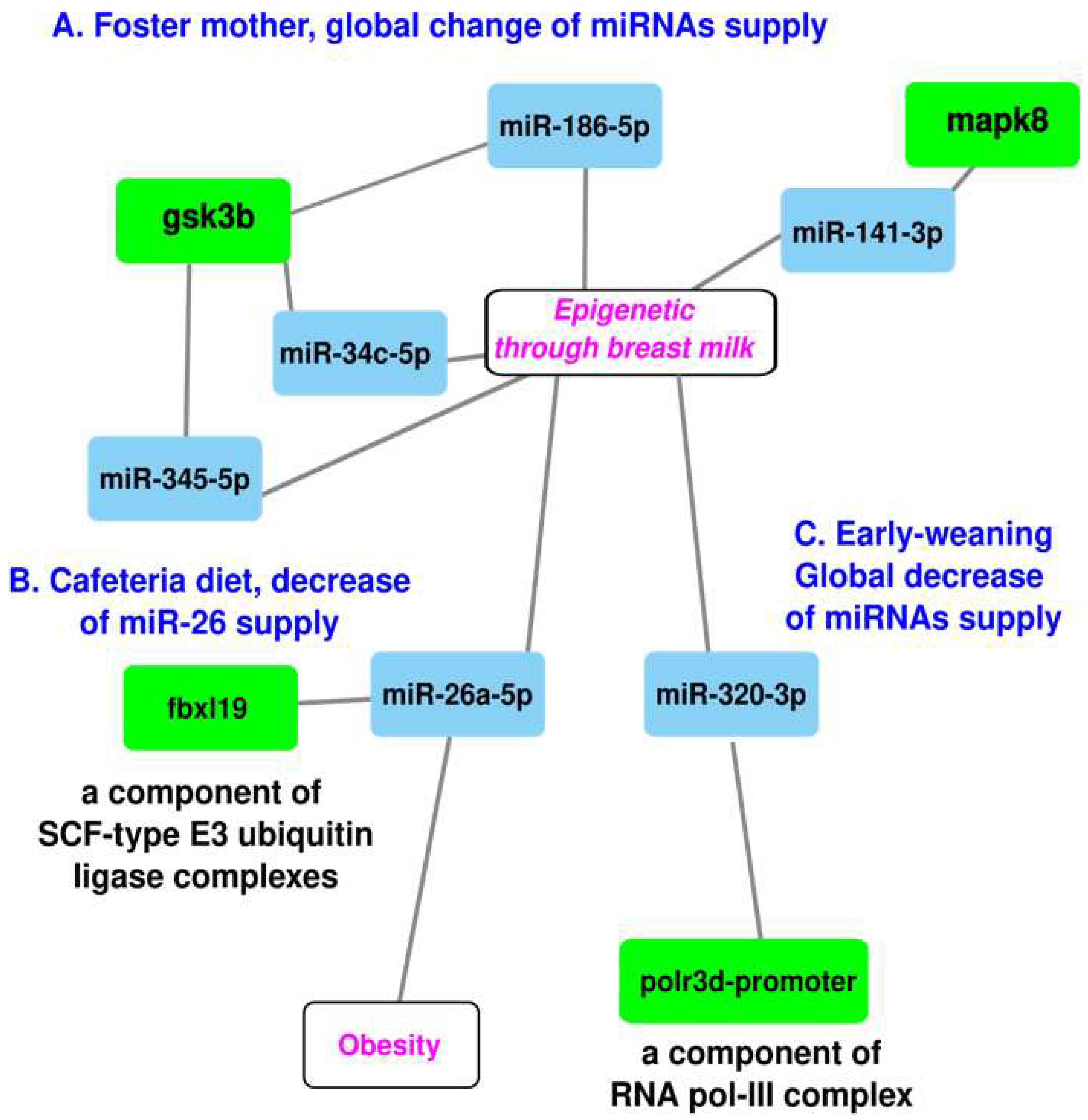
3. PotentialsofmiRNAsforinvivotreatmentofoffspring
Conclusions and Perspectives
Funding
Acknowledgments
References
- Playford RJ, Macdonald CE, Johnson WS. Colostrum and milk-derived peptide growth factors for the treatment of gastrointestinal disorders. Am J Clin Nutr. 2000 Jul;72(1):5-14. PMID: 10871554. [CrossRef]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T. A uniform system for microRNA annotation. RNA. 2003 Mar;9(3):277-9. PMID: 12592000; PMCID: PMC1370393. [CrossRef]
- Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu Rev Genet. 2015;49:213-42Epub 2015 Oct 14. PMID: 26473382; PMCID: PMC4743252. [CrossRef]
- Alles J, Fehlmann T, Fischer U, Backes C, Galata V, Minet M, Hart M, Abu-Halima M, Grässer FA, Lenhof HP, Keller A, Meese E. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019 Apr 23;47(7):3353-3364. PMID: 30820533; PMCID: PMC6468295. [CrossRef]
- Bartel DP. 2018. Metazoan MicroRNAs. Cell 173: 20–51. [CrossRef]
- Li, Z. F., Liang, Y. M., Lau, P. N., Shen, W., Wang, D. K., Cheung, W. T., et al. (2013). Dynamic localisation of mature microRNAs in human nucleoli is influenced by exogenous genetic materials. PLoS One 8:e70869. [CrossRef]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010; 56: 1733–41. PMID: 20847327. [CrossRef]
- Melnik BC, John SM, Schmitz G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr J. 2013 Jul 25;12:103. Review. PubMed PMID: 23883112; PubMed Central PMCID: PMC3725179. [CrossRef]
- Melnik B C, G. Smitz G. (2017b). DNA methyltransferase 1-targeting miRNA-148a of dairy milk: a potential bioactive modifier of the human epigenome. Funct. Foods Health Dis. 7, 671–687. [CrossRef]
- Melnik BC, Schmitz G. MicroRNAs: Milk's epigenetic regulators. Best Pract Res Clin Endocrinol Metab. 2017a Aug;31(4):427-442. Epub 2017 Oct 20. PMID: 29221571. [CrossRef]
- Melnik BC. Milk disrupts p53 and DNMT1, the guardians of the genome: implications for acne vulgaris and prostate cancer. Nutr Metab (Lond). 2017 Aug 15;14:55. PMID: 28814964; PMCID: PMC5556685. [CrossRef]
- Zhao S, Fung-Leung WP, Bittner A, et al. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells[Comparative Study]. PloS one. 2014;9(1):e78644. PubMed PMID:24454679; PubMed Central PMCID: PMC3894192. eng.
- Songia P, Chiesa M, Valerio V, Moschetta D, Myasoedova VA, D’Alessandra Y, Poggio P. Direct screening of plasma circulating microRNAs. RNA Biol 2018;15:1268–72.
- Raymond F, Lefebvre G, Texari L, Pruvost S, Metairon S, Cottenet G, Zollinger A, Mateescu B, Billeaud C, Picaud JC, Silva-Zolezzi I, Descombes P, Bosco N. Longitudinal Human Milk miRNA Composition over the First 3 mo of Lactation in a Cohort of Healthy Mothers Delivering Term Infants. J Nutr. 2022 Jan 11;152(1):94-106. PMID: 34510208. [CrossRef]
- Hicks SD, Carney MC, Tarasiuk A, DiAngelo SL, Birch LL, Paul IM. Breastmilk microRNAs are Stable Throughout Feeding and Correlate With Maternal Weight. Transl Genet Genom (2017) 5:1–8. [CrossRef]
- Alsaweed M, Lai CT, Hartmann PE, Geddes DT, Kakulas F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci Rep 2016c;6:20680.
- Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, Zhou X, Wang X, Gao X, Li X. (2012) Immune-related MicroRNAs are Abundant in Breast Milk Exosomes. Int J Biol Sci 8(1):118-123. [CrossRef]
- Floris I, Billard H, Boquien CY, Joram-Gauvard E, Simon L, Legrand A, Boscher C, Rozé JC, Bolaños-Jiménez F, Kaeffer B. MiRNA Analysis by Quantitative PCR in Preterm Human Breast Milk Reveals Daily Fluctuations of hsa-miR-16-5p. PLoS One. 2015 Oct 16;10(10):e0140488. ECollection 2015. PubMed PMID: 26474056; PubMed Central PMCID: PMC4608744. [CrossRef]
- Holzhausen EA, Kupsco A, Chalifour BN, Patterson WB, Schmidt KA, Mokhtari P, Baccarelli AA, Goran MI, Alderete TL. Influence of technical and maternal-infant factors on the measurement and expression of extracellular miRNA in human milk. Front Immunol. 2023 Jul 10;14:1151870. PMID: 37492577; PMCID: PMC10363855. [CrossRef]
- Alexandre-Gouabau M-C, Le Dréan G, Kaeffer B, Abderrahlan A, De Coppet P, Bobin P, Croyal M, De Luca A, Hankard R, Robitaille J. Gestational Diabetes Meletis modifies human breast milk content in insulin sensitivity regulators. DOHAD, 2022, Vancouver, Canada.
- Lukasik, A.; Brzozowska, I.; Zielenkiewicz, U.; Zielenkiewicz, P. Detection of Plant miRNAs Abundance in Human Breast Milk. Int. J. Mol. Sci. 2017, 19, 37.
- Benmoussa A, Laugier J, Beauparlant CJ, Lambert M, Droit A, Provost P. Complexity of the microRNA transcriptome of cow milk and milk-derived extracellular vesicles isolated via differential ultracentrifugation. J Dairy Sci. 2020 Jan;103(1):16-29. Epub 2019 Oct 31. PMID: 31677838. [CrossRef]
- Karlsson O, Rodosthenous RS, Jara C, Brennan KJ, Wright RO, Baccarelli AA, Wright RJ. Detection of long non-coding RNAs in human breastmilk extracellular vesicles: Implications for early child development. Epigenetics. 2016 Oct 2;11(10):721-729. Epub 2016 Nov 1. PMID: 27494402; PMCID: PMC5094628. [CrossRef]
- Mourtzi N, Siahanidou T, Tsifintaris M, Karamichali E, Tasiopoulou A, Sertedaki A, Pesmatzoglou M, Kapetanaki A, Liosis G, Baltatzis G, Vlachakis D, Chrousos GP, Giannakakis A. lncRNA NORAD is consistently detected in breastmilk exosomes and its expression is downregulated in mothers of preterm infants. Int J Mol Med. 2021 Dec;48(6):216. Epub 2021 Oct 15. PMID: 34651660; PMCID: PMC8559700. [CrossRef]
- Alsaweed M, Lai CT, Hartmann PE, Geddes DT, Kakulas F. Human milk cells and lipids conserve numerous known and novel miRNAs, some of which are differentially expressed during lactation. PLoS One 2016a;11:e0152610.
- Alsaweed M, Lai CT, Hartmann PE, Geddes DT, Kakulas F. Human milk cells contain numerous miRNAs that may change with milk removal and regulate multiple physiological processes. Int J Mol Sci 2016b;17:995.
- Munch EM, Harris RA, Mohammad M, Benham AL, Pejerrey SM, et al. (2013) Transcriptome Profiling of microRNA by Next-Gen Deep Sequencing Reveals Known and Novel miRNA Species in the Lipid Fraction of Human Breast Milk. PLoS ONE 8(2): e50564. [CrossRef]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005 May;6(5):376-85. PMID: 15852042. [CrossRef]
- Desvignes, T.; Batzel, P.; Berezikov, E.; Eilbeck, K.; Eppig, J.T.; McAndrews, M.S.; Singer, A.; Postlethwait, J.H. miRNA Nomenclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends Genet. 2015, 31, 613–626.
- Zonneveld MI, Brisson AR, van Herwijnen MJC, Tan S, van de Lest CHA, Redegeld FA, Garssen J, Wauben MHM and Nolte-’t Hoen ENM. Recovery of extracellular vesicles from human breast milk is influenced by sample collection and vesicle isolation procedure. (2014) Journal of Extracellular Vesicles 2014, 3: 24215 -. [CrossRef]
- Zheng Z, Mo J, Lin F, Wang J, Chen J, Luo H, Liu Y, Su C, Gu X, Xiong F, Zha L. Milk Exosomes from Gestational Diabetes Mellitus (GDM) and Healthy Parturient Exhibit Differential miRNAs Profiles and Distinct Regulatory Bioactivity on Hepatocyte Proliferation. Mol Nutr Food Res. 2023 Jun 25:e2300005. Epub ahead of print. PMID: 37357556. [CrossRef]
- Wu F, Zhi X, Xu R, Liang Z, Wang F, Li X, Li Y, Sun B. Exploration of microRNA profiles in human colostrum. Ann Transl Med. 2020 Sep;8(18):1170. PMID: 33241019; PMCID: PMC7576086. [CrossRef]
- Ishibashi O, Ohkuchi A, Ali MM, Kurashina R, Luo SS, Ishikawa T, Takizawa T, Hirashima C, Takahashi K, Migita M, Ishikawa G, Yoneyama K, Asakura H, Izumi A, Matsubara S, Takeshita T, Takizawa T. Hydroxysteroid (17-β) dehydrogenase 1 is dysregulated by miR-210 and miR-518c that are aberrantly expressed in preeclamptic placentas: a novel marker for predicting preeclampsia. Hypertension. 2012 Feb;59(2):265-73. Epub 2011 Dec 27. PMID: 22203747. [CrossRef]
- Patuleia SIS, van Gils CH, Oneto Cao AM, Bakker MF, van Diest PJ, van der Wall E, Moelans CB. The Physiological MicroRNA Landscape in Nipple Aspirate Fluid: Differences and Similarities with Breast Tissue, Breast Milk, Plasma and Serum. Int J Mol Sci. 2020 Nov 11;21(22):8466. PMID: 33187146; PMCID: PMC7696615. [CrossRef]
- Ahlberg E, Al-Kaabawi A, Thune R, Simpson MR, Pedersen SA, Cione E, Jenmalm MC, Tingö L. Breast milk microRNAs: Potential players in oral tolerance development. Front Immunol. 2023 Mar 14;14:1154211. PMID: 36999032; PMCID: PMC10045994. [CrossRef]
- Chiba T, Kooka A, Kowatari K, Yoshizawa M, Chiba N, Takaguri A, Fukushi Y, Hongo F, Sato H, Wada S. Expression profiles of hsa-miR-148a-3p and hsa-miR-125b-5p in human breast milk and infant formulae. Int Breastfeed J. 2022 Jan 3;17(1):1. PMID: 34980190; PMCID: PMC8725387. [CrossRef]
- Li A, Li Y, Zhang X, Zhang C, Li T, Zhang J, Li C. The human milk oligosaccharide 2'-fucosyllactose attenuates β-lactoglobulin-induced food allergy through the miR-146a-mediated toll-like receptor 4/nuclear factor-κB signaling pathway. J Dairy Sci. 2021 Oct;104(10):10473-10484. Epub 2021 Jul 30. PMID: 34334202. [CrossRef]
- Kosaka N, Izumi H, Sekine K, Ochiya T (2010) microRNA as a new immune-regulatory agent in breast milk. Silence 1: 7.
- Leiferman A, Shu J, Upadhyaya B, Cui J, Zempleni J. Storage of Extracellular Vesicles in Human Milk, and MicroRNA Profiles in Human Milk Exosomes and Infant Formulas. J Pediatr Gastroenterol Nutr. 2019 Aug;69(2):235-238. PMID: 31169664; PMCID: PMC6658346. [CrossRef]
- Elsarraj HS, Hong Y, Valdez K, Carletti M, Salah SM, Raimo M, Taverna D, Prochasson P, Bharadwaj U, Tweardy DJ, Christenson LK, Behbod F. A novel role of microRNA146b in promoting mammary alveolar progenitor cell maintenance. J Cell Sci. 2013 Jun 1;126(Pt 11):2446-58. Epub 2013 Apr 9. PMID: 23572509; PMCID: PMC3679487. [CrossRef]
- Mirza AH, Kaur S, Nielsen LB, Størling J, Yarani R, Roursgaard M, Mathiesen ER, Damm P, Svare J, Mortensen HB and Pociot F (2019) Breast Milk-Derived Extracellular Vesicles Enriched in Exosomes From Mothers With Type 1 Diabetes Contain Aberrant Levels of microRNAs. Front. Immunol. 10:2543. [CrossRef]
- Agarwal V, Bell GW, Nam JW, Bartel DP: Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4. [CrossRef]
- Benmoussa, A. & Provost, P. Milk microRNAs in health and disease. Comprehensive Reviews in food science and food safety 18, 703–722 (2019).
- Shah KB, Fields DA, Pezant NP, Kharoud HK, Gulati S, Jacobs K, Gale CA, Kharbanda EO, Nagel EM, Demerath EW, Tryggestad JB. Gestational Diabetes Mellitus Is Associated with Altered Abundance of Exosomal MicroRNAs in Human Milk. Clin Ther. 2022 Feb;44(2):172-185.e1. Epub 2022 Jan 26. Erratum in: Clin Ther. 2022 Jul;44(7):1034. PMID: 35090750; PMCID: PMC9089438. [CrossRef]
- Golan-Gerstl R, Elbaum Shiff Y, Moshayoff V, Schecter D, Leshkowitz D, Reif S. Characterization and biological function of milk-derived miRNAs. Mol Nutr Food Res. 2017 Oct;61(10). Epub 2017 Jul 31. PMID: 28643865. [CrossRef]
- Xi Y, Jiang X, Li R, Chen M, Song W, Li X. The levels of human milk microRNAs and their association with maternal weight characteristics. Eur J Clin Nutr. 2016 Apr;70(4):445-9. Epub 2015 Oct 21. PMID: 26486303. [CrossRef]
- Zamanillo R, Sánchez J, Serra F, Palou A. Breast milk supply of microRNA associated with leptin and adiponectin is affected by maternal overweight/obesity and influences infancy BMI. Nutrients. 2019;11(11):2589. [CrossRef]
- Karbiener M, Pisani DF, Frontini A, et al. MicroRNA-¬26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells. 2014;32(6):1578-¬1590. [CrossRef]
- Kupsco A, Prada D, Valvi D, Hu L, Petersen MS, Coull B, Grandjean P, Weihe P, Baccarelli AA. Human milk extracellular vesicle miRNA expression and associations with maternal characteristics in a population-based cohort from the Faroe Islands. Sci Rep. 2021 Mar 12;11(1):5840. PMID: 33712635; PMCID: PMC7970999. [CrossRef]
- Shiff YE, Reif S, Marom R, Shiff K, Reifen R, Golan-Gerstl R. MiRNA-320a Is Less Expressed and miRNA-148a More Expressed in Preterm Human Milk Compared to Term Human Milk. J Funct Food (2019) 57:68–74. [CrossRef]
- Chen Q, et al,. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res. 2021 PMID: 32801357.
- Title AC, Denzler R, Stoffel M. Uptake and Function Studies of Maternal Milk-derived MicroRNAs. J Biol Chem. 2015 Sep 25;290(39):23680-91. Epub 2015 Aug 3. PMID: 26240150; PMCID: PMC4583031. [CrossRef]
- Laubier J, Castille J, Le Guillou S, Le Provost F. No effect of an elevated miR-30b level in mouse milk on its level in pup tissues. RNA Biol. 2015;12(1):26-9. PMID: 25763824; PMCID: PMC4615842. [CrossRef]
- Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012 Jul 1;9(8):840-6. PMID: 22751203; PMCID: PMC3518396. [CrossRef]
- Beuzelin D, Pitard B, Kaeffer B. Oral Delivery of miRNA With Lipidic Aminoglycoside Derivatives in the Breastfed Rat. Front Physiol. 2019 Aug 13;10:1037. PMID: 31456698; PMCID: PMC6700720. [CrossRef]
- Tavares GA, Torres A, Le Drean G, Queignec M, Castellano B, Tesson L, Remy S, Anegon I, Pitard B, Kaeffer B. Oral Delivery of miR-320-3p with Lipidic Aminoglycoside Derivatives at Mid-Lactation Alters miR-320-3p Endogenous Levels in the Gut and Brain of Adult Rats According to Early or Regular Weaning. International Journal of Molecular Sciences, 2023, 24 (1), pp.191. ⟨10.3390/ijms24010191⟩.
- Liao Y, Du X, Li J, Lönnerdal B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol Nutr Food Res. 2017 Nov;61(11). Epub 2017 Aug 15. PMID: 28688106. [CrossRef]
- Manca S, Upadhyaya B, Mutai E, Desaulniers AT, Cederberg RA, White BR, Zempleni J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci Rep. 2018 Jul 27;8(1):11321. PMID: 30054561; PMCID: PMC6063888. [CrossRef]
- Riquelme I, Tapia O, Leal P, Sandoval A, Varga MG, Letelier P, Buchegger K, Bizama C, Espinoza JA, Peek RM, Araya JC, Roa JC. miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in gastric cancer through regulation of the PI3K/AKT/mTOR pathway. Cell Oncol (Dordr). 2016 Feb;39(1):23-33. Epub 2015 Oct 12. PMID: 26458815; PMCID: PMC4751587. [CrossRef]
- Horns F, Martinez JA, Fan C, Haque M, Linton JM, Tobin V, Santat L, Maggiolo AO, Bjorkman PJ, Lois C, Elowitz MB. Engineering RNA export for measurement and manipulation of living cells. Cell. 2023 Aug 17;186(17):3642-3658.e32. Epub 2023 Jul 11. PMID: 37437570. [CrossRef]
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Patel, S.K.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022, 601, 446–451.
- Ghoshal B, Bertrand E, Bhattacharyya SN. Non-canonical argonaute loading of extracellular vesicle-derived exogenous single-stranded miRNA in recipient cells. J Cell Sci. 2021 May 1;134(9):jcs253914. Epub 2021 May 17. PMID: 33785534. [CrossRef]
- Beuzelin, D.; Kaeffer, B. Exosomes and miRNA-Loaded Biomimetic Nanovehicles, a Focus on Their Potentials Preventing Type-2 Diabetes Linked to Metabolic Syndrome. Front. Immunol. 2018, 9, 2711.
- Liu J, Wang P, Zhang P, Zhang X, Du H, Liu Q, Huang B, Qian C, Zhang S, Zhu W, Yang X, Xiao Y, Liu Z, Luo D. An integrative bioinformatics analysis identified miR-375 as a candidate key regulator of malignant breast cancer. J Appl Genet. 2019 Nov;60(3-4):335-346. Epub 2019 Aug 1. PMID: 31372832. [CrossRef]
- Hicks SD, Beheshti R, Chandran D, Warren K, Confair A. Infant consumption of microRNA miR-375 in human milk lipids is associated with protection from atopy. Am J Clin Nutr (2022) 116(6):1654–62. [CrossRef]
- Simpson M, Brede G, Johansen J et al (2015) Human breast milk miRNA, maternal probiotic supplementation and atopic dermatitis in offspring. PLoS One 10:e0143496.
- Liu H, Geng Z, Su J. Engineered mammalian and bacterial extracellular vesicles as promising nanocarriers for targeted therapy. Extracell Vesicles Circ Nucleic Acids 2022;3:63-86. [CrossRef]
- Afrin H, Geetha Bai R, Kumar R, Ahmad SS, Agarwal SK, Nurunnabi M. Oral delivery of RNAi for cancer therapy. Cancer Metastasis Rev. 2023 Mar 27:1–26. Epub ahead of print. PMID: 36971908; PMCID: PMC10040933. [CrossRef]
- Gao F, Wang F, Cao H, Chen Y, Diao Y, Kapranov P. Evidence for Existence of Multiple Functional Human Small RNAs Derived from Transcripts of Protein-Coding Genes. Int J Mol Sci. 2023 Feb 19;24(4):4163. PMID: 36835575; PMCID: PMC9959880. [CrossRef]
- Melnik BC, Schmitz G. Milk Exosomal microRNAs: Postnatal Promoters of β Cell Proliferation but Potential Inducers of β Cell De-Differentiation in Adult Life. Int J Mol Sci. 2022 Sep 29;23(19):11503. PMID: 36232796; PMCID: PMC9569743. [CrossRef]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366-82. PMID: 17947979; PMCID: PMC3685583. [CrossRef]
- Ma RCW, Popkin BM (2017) Intergenerational diabetes and obesity—A cycle to break? PLoS Med 14(10): e1002415. [CrossRef]
- Armstrong J, Reilly JJ, Team CHI. Breastfeeding and lowering the risk of childhood obesity. Lancet. 2002;359(9322):2003-2004. [CrossRef]
- Ozkan H, Tuzun F, Taheri S, Korhan P, Akokay P, Yılmaz O, Duman N, Özer E, Tufan E, Kumral A, Özkul Y. Epigenetic Programming Through Breast Milk and Its Impact on Milk-Siblings Mating. Front Genet. 2020 Oct 2;11:569232. PMID: 33133155; PMCID: PMC7565666. [CrossRef]
- Pomar CA, Serra F, Palou A, Sánchez J. Lower miR-26a levels in breastmilk affect gene expression in adipose tissue of offspring. FASEB J. 2021 Oct;35(10):e21924. PMID: 34582059. [CrossRef]
- Arslanoglu, S., Ziegler, E. E., and Moro, G. E. (2010). World association of perinatal medicine working group on N. Donor human milk in preterm infant feeding: evidence and recommendations. J. Perinat. Med. 38, 347–351.
- Kakimoto, Y.; Matsushima, Y.; Tanaka, M.; Hayashi, H.; Wang, T.; Yokoyama, K.; Ochiai, E.; Osawa, M. MicroRNA Profiling of Gastric Content From Breast-Fed and Formula-Fed Infants to Estimate Last Feeding: A Pilot Study. Int. J. Leg. Med. 2019, 134, 903–909.
- Carrillo-Lozano E, Sebastián-Valles F, Knott-Torcal C. Circulating microRNAs in Breast Milk and Their Potential Impact on the Infant. Nutrients. 2020 Oct 8;12(10):3066. PMID: 33049923; PMCID: PMC7601398. [CrossRef]
- Acharya A, Berry DC, Zhang H, et al. miR-¬26 suppresses adipocyte progenitor differentiation and fat production by targeting. Genes Dev. 2019;33(19--20):1367-¬1380. [CrossRef]
- Babiarz, J.E. et al. (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 22, 2773–2785.
- Kim DH, Saetrom P, Snøve O Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008 Oct 21;105(42):16230-5. Epub 2008 Oct 13. PMID: 18852463; PMCID: PMC2571020. [CrossRef]
- Yeganeh M, Hernandez N. RNA polymerase III transcription as a disease factor. Genes Dev. 2020 Jul 1;34(13-14):865-882. PMID: 32611613; PMCID: PMC7328520. [CrossRef]
- Kulaberoglu Y, Malik Y, Borland G, Selman C, Alic N, Tullet JMA. Corrigendum: RNA Polymerase III, Ageing and Longevity. Front Genet. 2021 Sep 2;12:758135. Erratum for: Front Genet. 2021 Jul 06;12:705122. PMID: 34539762; PMCID: PMC8444630. [CrossRef]
- Shenderov, B. A., and Midtvedt, T. (2014). Epigenomic programing: a future way to health? Microb. Ecol. Health Dis. 25:10.3402/mehd.v25.24145.
- Yu AM, Choi YH, Tu MJ. RNA Drugs and RNA targets for small molecules. Principles, progress, and challenges. Pharmacol Rev (2020) 72(4):862–98. [CrossRef]
- Torri A, Jaeger J, Pradeu T, Saleh MC. The origin of RNA interference: Adaptive or neutral evolution? PloS Biol (2022) 20(6):e3001715. [CrossRef]
- Denzler R, Agarwal V, Stefano J, Bartel D, and Stoffel M. Assessing the ceRNA Hypothesis with Quantitative Measurements of miRNA and Target Abundance. Molecular Cell 2014 54, 766–776, June 5, 2014.
- Harman, J. C., Guidry, J. J., and Gidday, J. M. (2020). Intermittent hypoxia promotes functional neuroprotection from retinal ischemia in untreated first-generation offspring: proteomic mechanistic insights. Invest. Ophthalmol. Vis. Sci. 61:15. [CrossRef]
- Ip S, Chung M, Raman G, Chew P, Magula N, DeVine D, Trikalinos T, Lau J: Breastfeeding and maternal and infant health outcomes in developed countries. Evid RepTechnol Assess (Full Rep) 2007, 153:1–186.
- Huang S. Towards a unification of the 2 meanings of "epigenetics". PLoS Biol. 2022 Dec 27;20(12):e3001944. PMID: 36574409; PMCID: PMC9829431. [CrossRef]
- Smythies, J., Edelstein, L., and Ramachandran, V. (2014). Molecular mechanisms for the inheritance of acquired characteristics-exosomes, microRNA shuttling, fear and stress: lamarck resurrected? Front. Genet. 5:133. [CrossRef]
- Mezo-González C E, García Santillán J A, Kaeffer B, Gourdel M, Croyal M, Bolaños-Jiménez F. Both maternal and paternal obesity in the rat induce learning deficits in offspring linked to impaired brain glutamatergic signaling. 2023. Acta Physiologica, in press.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




