Submitted:
05 October 2023
Posted:
09 October 2023
You are already at the latest version
Abstract
Keywords:
Introduction
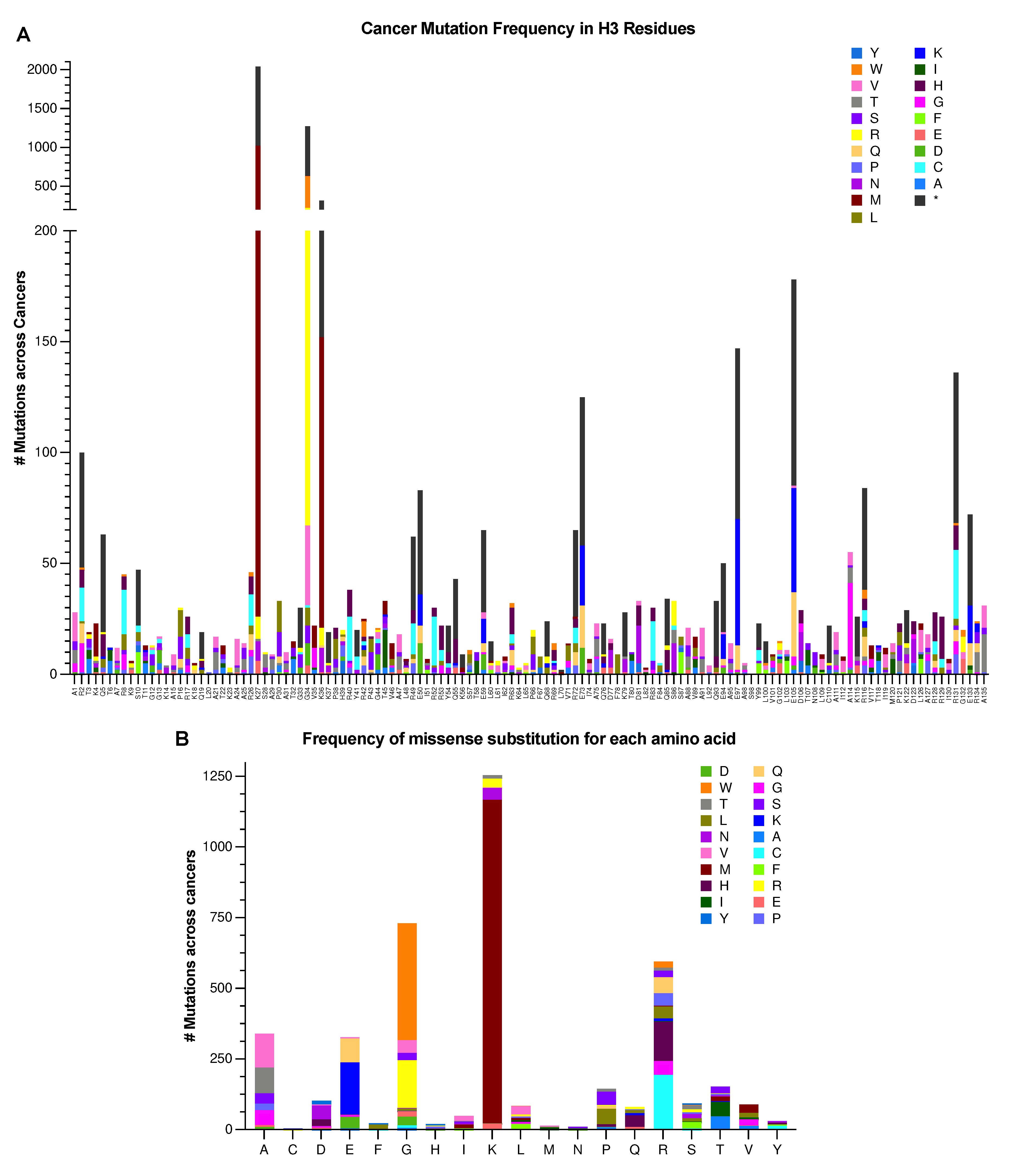
Oncohistones are Novel, Cancer-Causing Histone Mutants
Prior Work Investigating Histone Function in Yeast
Advantages of Using Yeast Models to Investigate Oncohistones
Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xu, J.; Murphy, S.L.; Kochanek, K.D.; Arias, E. Mortality in the United States, 2021. NCHS Data Brief 2022, 1–8. [Google Scholar]
- Hartwell, L.H. Yeast and cancer. Biosci Rep 2004, 24, 523–544. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.M. Histone structure and function. Curr Opin Cell Biol 1991, 3, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Millar, C.B.; Grunstein, M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol 2006, 7, 657–666. [Google Scholar] [CrossRef]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Luger, K.; Dechassa, M.L.; Tremethick, D.J. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 2012, 13, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Stützer, A.; Liokatis, S.; Kiesel, A.; Schwarzer, D.; Sprangers, R.; Söding, J.; Selenko, P.; Fischle, W. Modulations of DNA Contacts by Linker Histones and Post-translational Modifications Determine the Mobility and Modifiability of Nucleosomal H3 Tails. Mol Cell 2016, 61, 247–259. [Google Scholar] [CrossRef]
- Nacev, B.A.; Feng, L.; Bagert, J.D.; Lemiesz, A.E.; Gao, J.; Soshnev, A.A.; Kundra, R.; Schultz, N.; Muir, T.W.; Allis, C.D. The expanding landscape of 'oncohistone' mutations in human cancers. Nature 2019, 567, 473–478. [Google Scholar] [CrossRef]
- Albers, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P.; Wilson, J.; Hunt, T. Molecular Biology of the Cell, 5th edn.(Garland Science, New York, 2008).
- Flaus, A.; Downs, J.A.; Owen-Hughes, T. Histone isoforms and the oncohistone code. Curr Opin Genet Dev 2021, 67, 61–66. [Google Scholar] [CrossRef]
- Seal, R.L.; Denny, P.; Bruford, E.A.; Gribkova, A.K.; Landsman, D.; Marzluff, W.F.; McAndrews, M.; Panchenko, A.R.; Shaytan, A.K.; Talbert, P.B. A standardized nomenclature for mammalian histone genes. Epigenetics Chromatin 2022, 15, 34. [Google Scholar] [CrossRef]
- Singh, R.; Bassett, E.; Chakravarti, A.; Parthun, M.R. Replication-dependent histone isoforms: a new source of complexity in chromatin structure and function. Nucleic Acids Res 2018, 46, 8665–8678. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.C.; Denu, J.M. Chemical mechanisms of histone lysine and arginine modifications. Biochim Biophys Acta 2009, 1789, 45–57. [Google Scholar] [CrossRef]
- Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352. [Google Scholar] [CrossRef]
- Thorne, A.W.; Kmiciek, D.; Mitchelson, K.; Sautiere, P.; Crane-Robinson, C. Patterns of histone acetylation. Eur J Biochem 1990, 193, 701–713. [Google Scholar] [CrossRef]
- Tjeertes, J.V.; Miller, K.M.; Jackson, S.P. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. Embo j 2009, 28, 1878–1889. [Google Scholar] [CrossRef]
- Schneider, J.; Bajwa, P.; Johnson, F.C.; Bhaumik, S.R.; Shilatifard, A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem 2006, 281, 37270–37274. [Google Scholar] [CrossRef]
- Bernier, M.; Luo, Y.; Nwokelo, K.C.; Goodwin, M.; Dreher, S.J.; Zhang, P.; Parthun, M.R.; Fondufe-Mittendorf, Y.; Ottesen, J.J.; Poirier, M.G. Linker histone H1 and H3K56 acetylation are antagonistic regulators of nucleosome dynamics. Nat Commun 2015, 6, 10152. [Google Scholar] [CrossRef]
- Hong, L.; Schroth, G.P.; Matthews, H.R.; Yau, P.; Bradbury, E.M. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 "tail" to DNA. J Biol Chem 1993, 268, 305–314. [Google Scholar] [CrossRef]
- Clayton, A.L.; Hazzalin, C.A.; Mahadevan, L.C. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell 2006, 23, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.M.; O'Neill, L.P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol 1995, 6, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.S.; Yue, W.W.; Oppermann, U.; Klose, R.J. Dynamic protein methylation in chromatin biology. Cell Mol Life Sci 2009, 66, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 2012, 48, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Miller, K.M. Histone methylation and the DNA damage response. Mutat Res Rev Mutat Res 2019, 780, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Grant, P.A.; Briggs, S.D.; Sun, Z.W.; Bone, J.R.; Caldwell, J.A.; Mollah, S.; Cook, R.G.; Shabanowitz, J.; Hunt, D.F.; et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 2002, 22, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Gogol, M.; Carey, M.; Pattenden, S.G.; Seidel, C.; Workman, J.L. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 2007, 21, 1422–1430. [Google Scholar] [CrossRef]
- Shilatifard, A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 2006, 75, 243–269. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Humphrey, E.L.; Erlich, R.L.; Schneider, R.; Bouman, P.; Liu, J.S.; Kouzarides, T.; Schreiber, S.L. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A 2002, 99, 8695–8700. [Google Scholar] [CrossRef]
- Shi, X.; Hong, T.; Walter, K.L.; Ewalt, M.; Michishita, E.; Hung, T.; Carney, D.; Peña, P.; Lan, F.; Kaadige, M.R.; et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006, 442, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Lam, U.T.F.; Tan, B.K.Y.; Poh, J.J.X.; Chen, E.S. Structural and functional specificity of H3K36 methylation. Epigenetics Chromatin 2022, 15, 17. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.K.; Nguyen, T.T.T.; Li, A.Y.; Yeo, Y.P.; Chen, E.S. Histone H3 lysine 36 methyltransferase mobilizes NER factors to regulate tolerance against alkylation damage in fission yeast. Nucleic Acids Res 2018, 46, 5061–5074. [Google Scholar] [CrossRef] [PubMed]
- Pai, C.C.; Deegan, R.S.; Subramanian, L.; Gal, C.; Sarkar, S.; Blaikley, E.J.; Walker, C.; Hulme, L.; Bernhard, E.; Codlin, S.; et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat Commun 2014, 5, 4091. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, H.; Fong, N.; Erickson, B.; Bentley, D.L. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci U S A 2011, 108, 13564–13569. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, M.R.; Jha, D.K.; Ucles, S.A.; Flood, D.M.; Strahl, B.D.; Stevens, S.W.; Kress, T.L. Histone H3K36 methylation regulates pre-mRNA splicing in Saccharomyces cerevisiae. RNA Biol 2016, 13, 412–426. [Google Scholar] [CrossRef]
- Yuan, H.; Li, N.; Fu, D.; Ren, J.; Hui, J.; Peng, J.; Liu, Y.; Qiu, T.; Jiang, M.; Pan, Q.; et al. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J Clin Invest 2017, 127, 3375–3391. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.H.; Cheung, W.L.; Hsu, J.Y.; Diaz, R.L.; Smith, M.M.; Allis, C.D. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 2005, 120, 25–36. [Google Scholar] [CrossRef]
- Sierra, F.; Lichtler, A.; Marashi, F.; Rickles, R.; Van Dyke, T.; Clark, S.; Wells, J.; Stein, G.; Stein, J. Organization of human histone genes. Proc Natl Acad Sci U S A 1982, 79, 1795–1799. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: from mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R. Sophisticated Conversations between Chromatin and Chromatin Remodelers, and Dissonances in Cancer. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- West, A.C.; Johnstone, R.W. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014, 124, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.Y.; Jones, D.T.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.A.; Tönjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Broniscer, A.; McEachron, T.A.; Lu, C.; Paugh, B.S.; Becksfort, J.; Qu, C.; Ding, L.; Huether, R.; Parker, M.; et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 2012, 44, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Behjati, S.; Tarpey, P.S.; Presneau, N.; Scheipl, S.; Pillay, N.; Van Loo, P.; Wedge, D.C.; Cooke, S.L.; Gundem, G.; Davies, H.; et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 2013, 45, 1479–1482. [Google Scholar] [CrossRef]
- Papillon-Cavanagh, S.; Lu, C.; Gayden, T.; Mikael, L.G.; Bechet, D.; Karamboulas, C.; Ailles, L.; Karamchandani, J.; Marchione, D.M.; Garcia, B.A.; et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet 2017, 49, 180–185. [Google Scholar] [CrossRef]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef]
- Szenker, E.; Ray-Gallet, D.; Almouzni, G. The double face of the histone variant H3.3. Cell Res 2011, 21, 421–434. [Google Scholar] [CrossRef]
- Lewis, P.W.; Müller, M.M.; Koletsky, M.S.; Cordero, F.; Lin, S.; Banaszynski, L.A.; Garcia, B.A.; Muir, T.W.; Becher, O.J.; Allis, C.D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013, 340, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Jain, S.U.; Hoelper, D.; Bechet, D.; Molden, R.C.; Ran, L.; Murphy, D.; Venneti, S.; Hameed, M.; Pawel, B.R.; et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 2016, 352, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Huang, Y.; Mao, G.; Yang, S.; Rennert, G.; Gu, L.; Li, H.; Li, G.M. Cancer-driving H3G34V/R/D mutations block H3K36 methylation and H3K36me3-MutSα interaction. Proc Natl Acad Sci U S A 2018, 115, 9598–9603. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shan, C.M.; Wang, J.; Bao, K.; Tong, L.; Jia, S. Molecular basis for the role of oncogenic histone mutations in modulating H3K36 methylation. Sci Rep 2017, 7, 43906. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Shi, J.; Shi, X.; Li, W.; Wen, H. Histone H3.3 G34 Mutations Alter Histone H3K36 and H3K27 Methylation In Cis. J Mol Biol 2018, 430, 1562–1565. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Liu, X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 2015, 350, aac4383. [Google Scholar] [CrossRef]
- Justin, N.; Zhang, Y.; Tarricone, C.; Martin, S.R.; Chen, S.; Underwood, E.; De Marco, V.; Haire, L.F.; Walker, P.A.; Reinberg, D.; et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun 2016, 7, 11316. [Google Scholar] [CrossRef]
- Jain, S.U.; Khazaei, S.; Marchione, D.M.; Lundgren, S.M.; Wang, X.; Weinberg, D.N.; Deshmukh, S.; Juretic, N.; Lu, C.; Allis, C.D.; et al. Histone H3.3 G34 mutations promote aberrant PRC2 activity and drive tumor progression. Proc Natl Acad Sci U S A 2020, 117, 27354–27364. [Google Scholar] [CrossRef]
- Mitchener, M.M.; Muir, T.W. Oncohistones: Exposing the nuances and vulnerabilities of epigenetic regulation. Mol Cell 2022, 82, 2925–2938. [Google Scholar] [CrossRef]
- Bennett, R.L.; Bele, A.; Small, E.C.; Will, C.M.; Nabet, B.; Oyer, J.A.; Huang, X.; Ghosh, R.P.; Grzybowski, A.T.; Yu, T.; et al. A Mutation in Histone H2B Represents a New Class of Oncogenic Driver. Cancer Discov 2019, 9, 1438–1451. [Google Scholar] [CrossRef]
- Fang, D.; Gan, H.; Lee, J.H.; Han, J.; Wang, Z.; Riester, S.M.; Jin, L.; Chen, J.; Zhou, H.; Wang, J.; et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 2016, 352, 1344–1348. [Google Scholar] [CrossRef] [PubMed]
- Lowe, B.R.; Yadav, R.K.; Henry, R.A.; Schreiner, P.; Matsuda, A.; Fernandez, A.G.; Finkelstein, D.; Campbell, M.; Kallappagoudar, S.; Jablonowski, C.M.; et al. Surprising phenotypic diversity of cancer-associated mutations of Gly 34 in the histone H3 tail. Elife 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Silveira, A.B.; Kasper, L.H.; Fan, Y.; Jin, H.; Wu, G.; Shaw, T.I.; Zhu, X.; Larson, J.D.; Easton, J.; Shao, Y.; et al. H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol 2019, 137, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Y. Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair. J Biol Chem 2017, 292, 11951–11959. [Google Scholar] [CrossRef] [PubMed]
- Fnu, S.; Williamson, E.A.; De Haro, L.P.; Brenneman, M.; Wray, J.; Shaheen, M.; Radhakrishnan, K.; Lee, S.H.; Nickoloff, J.A.; Hromas, R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A 2011, 108, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Hu, X.; Jing, Q.; Zeng, X.; Chan, K.M.; Han, J. Mechanism of cancer: Oncohistones in action. J Genet Genomics 2018, 45, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Bonner, E.R.; Dawood, A.; Gordish-Dressman, H.; Eze, A.; Bhattacharya, S.; Yadavilli, S.; Mueller, S.; Waszak, S.M.; Nazarian, J. Pan-cancer atlas of somatic core and linker histone mutations. NPJ Genom Med 2023, 8, 23. [Google Scholar] [CrossRef]
- Okosun, J.; Bödör, C.; Wang, J.; Araf, S.; Yang, C.Y.; Pan, C.; Boller, S.; Cittaro, D.; Bozek, M.; Iqbal, S.; et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 2014, 46, 176–181. [Google Scholar] [CrossRef]
- Kang, T.Z.E.; Zhu, L.; Yang, D.; Ding, D.; Zhu, X.; Wan, Y.C.E.; Liu, J.; Ramakrishnan, S.; Chan, L.L.; Chan, S.Y.; et al. The elevated transcription of ADAM19 by the oncohistone H2BE76K contributes to oncogenic properties in breast cancer. J Biol Chem 2021, 296, 100374. [Google Scholar] [CrossRef]
- McBurney, K.L.; Leung, A.; Choi, J.K.; Martin, B.J.; Irwin, N.A.; Bartke, T.; Nelson, C.J.; Howe, L.J. Divergent Residues Within Histone H3 Dictate a Unique Chromatin Structure in Saccharomyces cerevisiae. Genetics 2016, 202, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Libuda, D.E.; Winston, F. Amplification of histone genes by circular chromosome formation in Saccharomyces cerevisiae. Nature 2006, 443, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, P.R.; Ganguli, D.; Nagarajavel, V.; Clark, D.J. Regulation of histone gene expression in budding yeast. Genetics 2012, 191, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.M.; Murray, K. Yeast H3 and H4 histone messenger RNAs are transcribed from two non-allelic gene sets. J Mol Biol 1983, 169, 641–661. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Liu, Y.; Wang, A.; Qin, Y.; Luo, M.; Wu, Q.; Boeke, J.D.; Dai, J. Construction of Comprehensive Dosage-Matching Core Histone Mutant Libraries for Saccharomyces cerevisiae. Genetics 2017, 207, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.L.; Smith, M.M. Comparison of the structure and cell cycle expression of mRNAs encoded by two histone H3-H4 loci in Saccharomyces cerevisiae. Mol Cell Biol 1988, 8, 945–954. [Google Scholar] [CrossRef]
- Liang, D.; Burkhart, S.L.; Singh, R.K.; Kabbaj, M.H.; Gunjan, A. Histone dosage regulates DNA damage sensitivity in a checkpoint-independent manner by the homologous recombination pathway. Nucleic Acids Res 2012, 40, 9604–9620. [Google Scholar] [CrossRef]
- Hereford, L.; Fahrner, K.; Woolford, J., Jr.; Rosbash, M.; Kaback, D.B. Isolation of yeast histone genes H2A and H2B. Cell 1979, 18, 1261–1271. [Google Scholar] [CrossRef]
- Norris, D.; Osley, M.A. The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol Cell Biol 1987, 7, 3473–3481. [Google Scholar] [CrossRef]
- Vidal, M.; Gaber, R.F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol 1991, 11, 6317–6327. [Google Scholar] [CrossRef]
- Taunton, J.; Hassig, C.A.; Schreiber, S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996, 272, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulos, T.; Thireos, G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. Embo j 1992, 11, 4145–4152. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.H.; Brownell, J.E.; Sobel, R.E.; Ranalli, T.A.; Cook, R.G.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 1996, 383, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Seol, J.H.; Kim, H.J.; Yoo, J.K.; Park, H.J.; Cho, E.J. Analysis of Saccharomyces cerevisiae histone H3 mutants reveals the role of the alphaN helix in nucleosome function. Biochem Biophys Res Commun 2008, 374, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Ngubo, M.; Reid, J.L.; Patterton, H.G. Distinct structural groups of histone H3 and H4 residues have divergent effects on chronological lifespan in Saccharomyces cerevisiae. PLoS One 2022, 17, e0268760. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Srinivasan, M.; Nakanishi, S.; Leatherwood, J.; Shilatifard, A.; Sternglanz, R. A conserved patch near the C terminus of histone H4 is required for genome stability in budding yeast. Mol Cell Biol 2011, 31, 2311–2325. [Google Scholar] [CrossRef] [PubMed]
- Sankar, A.; Mohammad, F.; Sundaramurthy, A.K.; Wang, H.; Lerdrup, M.; Tatar, T.; Helin, K. Histone editing elucidates the functional roles of H3K27 methylation and acetylation in mammals. Nat Genet 2022, 54, 754–760. [Google Scholar] [CrossRef]
- White, C.L.; Suto, R.K.; Luger, K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. Embo j 2001, 20, 5207–5218. [Google Scholar] [CrossRef]
- Doyon, Y.; Selleck, W.; Lane, W.S.; Tan, S.; Côté, J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol 2004, 24, 1884–1896. [Google Scholar] [CrossRef]
- Spedale, G.; Timmers, H.T.; Pijnappel, W.W. ATAC-king the complexity of SAGA during evolution. Genes Dev 2012, 26, 527–541. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Xue, H.; Cao, M.; Bai, G.; Mu, Z.; Yao, Y.; Sun, S.; Fang, D.; Huang, J. Cryo-EM structure of SETD2/Set2 methyltransferase bound to a nucleosome containing oncohistone mutations. Cell Discov 2021, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Lachner, M.; Sengupta, R.; Schotta, G.; Jenuwein, T. Trilogies of histone lysine methylation as epigenetic landmarks of the eukaryotic genome. Cold Spring Harb Symp Quant Biol 2004, 69, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Duina, A.A.; Miller, M.E.; Keeney, J.B. Budding yeast for budding geneticists: a primer on the Saccharomyces cerevisiae model system. Genetics 2014, 197, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Hyland, E.M.; Yuan, D.S.; Huang, H.; Bader, J.S.; Boeke, J.D. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 2008, 134, 1066–1078. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Maertens, A.M.; Hyland, E.M.; Dai, J.; Norris, A.; Boeke, J.D.; Bader, J.S. HistoneHits: a database for histone mutations and their phenotypes. Genome Res 2009, 19, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, S.; Sanderson, B.W.; Delventhal, K.M.; Bradford, W.D.; Staehling-Hampton, K.; Shilatifard, A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol 2008, 15, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Collord, G.; Martincorena, I.; Young, M.D.; Foroni, L.; Bolli, N.; Stratton, M.R.; Vassiliou, G.S.; Campbell, P.J.; Behjati, S. Recurrent histone mutations in T-cell acute lymphoblastic leukaemia. Br J Haematol 2019, 184, 676–679. [Google Scholar] [CrossRef]
- Lemon, L.D.; Kannan, S.; Mo, K.W.; Adams, M.; Choi, H.G.; Gulka, A.O.D.; Withers, E.S.; Nurelegne, H.T.; Gomez, V.; Ambrocio, R.E.; et al. A Saccharomyces cerevisiae model and screen to define the functional consequences of oncogenic histone missense mutations. G3 (Bethesda) 2022, 12. [Google Scholar] [CrossRef]
- Smith, E.R.; Eisen, A.; Gu, W.; Sattah, M.; Pannuti, A.; Zhou, J.; Cook, R.G.; Lucchesi, J.C.; Allis, C.D. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A 1998, 95, 3561–3565. [Google Scholar] [CrossRef]
- Cooke, S.L.; Soares, B.L.; Müller, C.A.; Nieduszynski, C.A.; Bastos de Oliveira, F.M.; de Bruin, R.A.M. Tos4 mediates gene expression homeostasis through interaction with HDAC complexes independently of H3K56 acetylation. J Biol Chem 2021, 296, 100533. [Google Scholar] [CrossRef]
- Yadav, R.K.; Jablonowski, C.M.; Fernandez, A.G.; Lowe, B.R.; Henry, R.A.; Finkelstein, D.; Barnum, K.J.; Pidoux, A.L.; Kuo, Y.M.; Huang, J.; et al. Histone H3G34R mutation causes replication stress, homologous recombination defects and genomic instability in S. pombe. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- D'Arcy, S.; Luger, K. Understanding histone acetyltransferase Rtt109 structure and function: how many chaperones does it take? Curr Opin Struct Biol 2011, 21, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Bagert, J.D.; Mitchener, M.M.; Patriotis, A.L.; Dul, B.E.; Wojcik, F.; Nacev, B.A.; Feng, L.; Allis, C.D.; Muir, T.W. Oncohistone mutations enhance chromatin remodeling and alter cell fates. Nat Chem Biol 2021, 17, 403–411. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




