1. Introduction
To date, 86 distinct viruses belonging to 17 families and 34 genera were demonstrated to infect grapevine (
Vitis spp.), the largest number of viruses detected in a particular plant species [
1,
2,
3,
4]. The occurrence of so many different viruses in grapevine could be due to the long history of grapevine domestication and exposure to the coexisting viruses, or high incidence of persistently infecting viruses. This list can be extended to include extensive exchange of grapevine germplasm among grape growing countries or regions, broad use of grafting as an essential viticulture practice [
5] and to a substantial research effort toward identifying viruses infecting this high cash value crop using new detection technologies such as next generation sequencing (NGS) for virus identification [
3,
6]. Grapevines are commonly infected with multiple viruses, likely because unregulated grafting has permitted the broad spread of viruses. Because of these ubiquitous co-infections, it is difficult to establish exact etiological roles of a given virus in a disease. All grapevine viruses are efficiently disseminated through infected propagating materials and grafting among varieties of scions and rootstocks [
7,
8] while many of these viruses can also be spread by different invertebrate vectors.
Currently, majority of documented grapevine viruses are classified into four major groups based on the diseases with which they are associated. Grapevine leafroll disease (GLRD) is inarguably the most important virus disease in grapevine economically, comparable with those caused by fungal pathogens. GLRD complex occurs in all major grape-growing regions worldwide, causing significant losses to the grape and wine industry. Grapevine infections with GLRD result in major reductions in yield and fruit quality and shorten the productive lifespan of vineyards. Although GLRD is associated with six species of grapevine leafroll-associated viruses (GLRaVs) belonging to three genera of the
Closteroviridae family,
Grapevine leafroll-associated virus 3 (GLRaV-3) from the
Ampelovirus genus is regarded as the most economically destructive of the GLRaVs due to its global distribution, high prevalence and disease severity [
8,
9,
10,
11,
12].
To investigate the molecular or cellular aspects of RNA virus infection or virus gene functions, it is essential to generate experimentally amenable, biologically active cDNA clones of the virus genomes. These cDNA clones can initiate virus infection either upon mechanical inoculation of the susceptible plants or upon agro-infiltration [
13]. The latter approach is especially critical for the non-mechanically transmissible viruses. Recently, several cDNA clones of the grapevine viruses have been developed and utilized as basic tools in molecular virology [
14,
15,
16,
17,
18,
19,
20]. In addition to application of the reverse genetics for studying virus and host functional genomics, virus cDNA clones are critical for fulfilling Koch’s postulates for the virus diseases.
Unlike many herbaceous plants, that can be readily infected via mechanical inoculation with cloned virus cDNA, grapevine and other woody perennials are recalcitrant to mechanical inoculation. A choice alternative to mechanical inoculation is agro-infection that relies on the natural ability of
Agrobacterium tumefaciens to deliver target DNA into the nucleus of plant cells. The agro-infection can be achieved through four available inoculation methods, depending on the virus-host combination. In the first method, an agrobacterium suspension is directly rubbed on the leaf surface or injected into the leaves using a needless syringe. This method is suitable for the viruses of herbaceous hosts [
17,
21,
22,
23,
24]. The second method is agro-drenching, which has been effective for the
Solanaceae species through drenching of soil with agrobacterium suspensions [
25]. Agro-drenching does also work for some grapevine viruses such as
Grapevine virus A (GVA) and
Grapevine rupestris stem pitting-associated virus (GRSPaV) [
14,
16]. However, this method requires a long time for establishing virus infection and has low infection rate. The third method is agro-pricking which was previously described and used by Yepes et al. [
18] in agro-infection of grapevine plantlets by using the infectious clones of
Grapevine red blotch virus (GRBV) and based on their report this method worked with good infection rate. The fourth, more efficient method is vacuum-mediated agro-infiltration developed for woody plant viruses such as
Grapevine leafroll-associated virus-2(GLRaV-2),
Apple chlorotic leaf spot virus (ACLSV), GRBV and
Grapevine Pinot gris virus (GPGV) [
15,
18,
20,
26].
Despite substantial progress in developing methods for the delivery of cDNA clones, further improvements of these methods remain to be crucial for empowering research and practical applications of the viruses of grapevine and other woody plants. Accordingly, the main objective of this study was to optimize an experimental system for launching grapevine infections with virus cDNA clones in general and GLRaV-3 in particular. We first compared the performance of four mentioned inoculation methods in initiating grapevine infection with GLRaV-3cDNA clones and found that vacuum-mediated agro-infiltration is the most effective of these. We further tested a series of experimental conditions impacting the survival of the infiltrated plantlets and infectivity rate and developed the optimized protocol for entire process of vacuum agro-infiltration and following plant recovery. This protocol has a strong potential for broad applications in woody plant virology, including the fulfilment of Koch’s postulates, virus-host interactions including pathogenesis, as well as virus and plant functional genomics using virus vectors for gene expression virus-induced RNA interference.
2. Material and Methods
2.1. Establishment of Grapevine Tissue Culture
The first step was to generate grapevine plantlets from certified virus-free grapevine mother plants through tissue culture. In this study, certified cuttings from different varieties including Cabernet franc, Syrah, and Chardonnay, kindly provided by Foundation Plant Services US-Davis, were rooted and potted and later used as mother plants to establish the tissue culture system. Plants established in the greenhouse were first tested for the most common grapevine viruses involved in important diseases in commercial grapes, such as leafroll, rugose wood, infectious degeneration and decline, and newly identified viruses such as red blotch. To reach this goal, the plants were monitored for disease symptoms and tested by multiplex RT-PCR with primers targeting 17 grapevine viruses as described in Xiao et al. [
27]. Once established, grapevine plants in the greenhouse produced a good number of shoots. The new shoots were cut and treated for growing on agar medium. For this reason, a method of semi-sterilized tissue culture for rapid propagation of grapevines used by Shan and Seaton [
28] was adopted with some changes. First, all leaves were removed and shoot tips, roughly 7-10 cm in length, with one node on each were cut. These cuttings were rinsed for one hour in running tap water and then surface sterilized for 6 minutes in 5% bleach solution while stirring. From this point onward, all plant work was conducted in the laminar flow hood. After sterilization, all the single-node cuttings were rinsed three times in sterile deionized water for 10 minutes each. Subsequently, all the cuttings were drained on sterile tissue paper. Finally, bases of single-node cuttings were cut off with a sterile blade at a 45-degree angle and planted into glass tubes with OH medium, the shoot initiation agar medium containing 0.2 gr/L cefotaxime (
Table S1). One single-node cutting per tube was maintained in the growth chamber at a constant temperature of 22°C with a 16-h photoperiod at light intensity of 50µmol/m
2/s. These tubes were periodically checked for contamination. After about 2 weeks, once the new shoots started growing, GS1 liquid medium containing 0.2 g/L cefotaxime was added into the glass tubes (
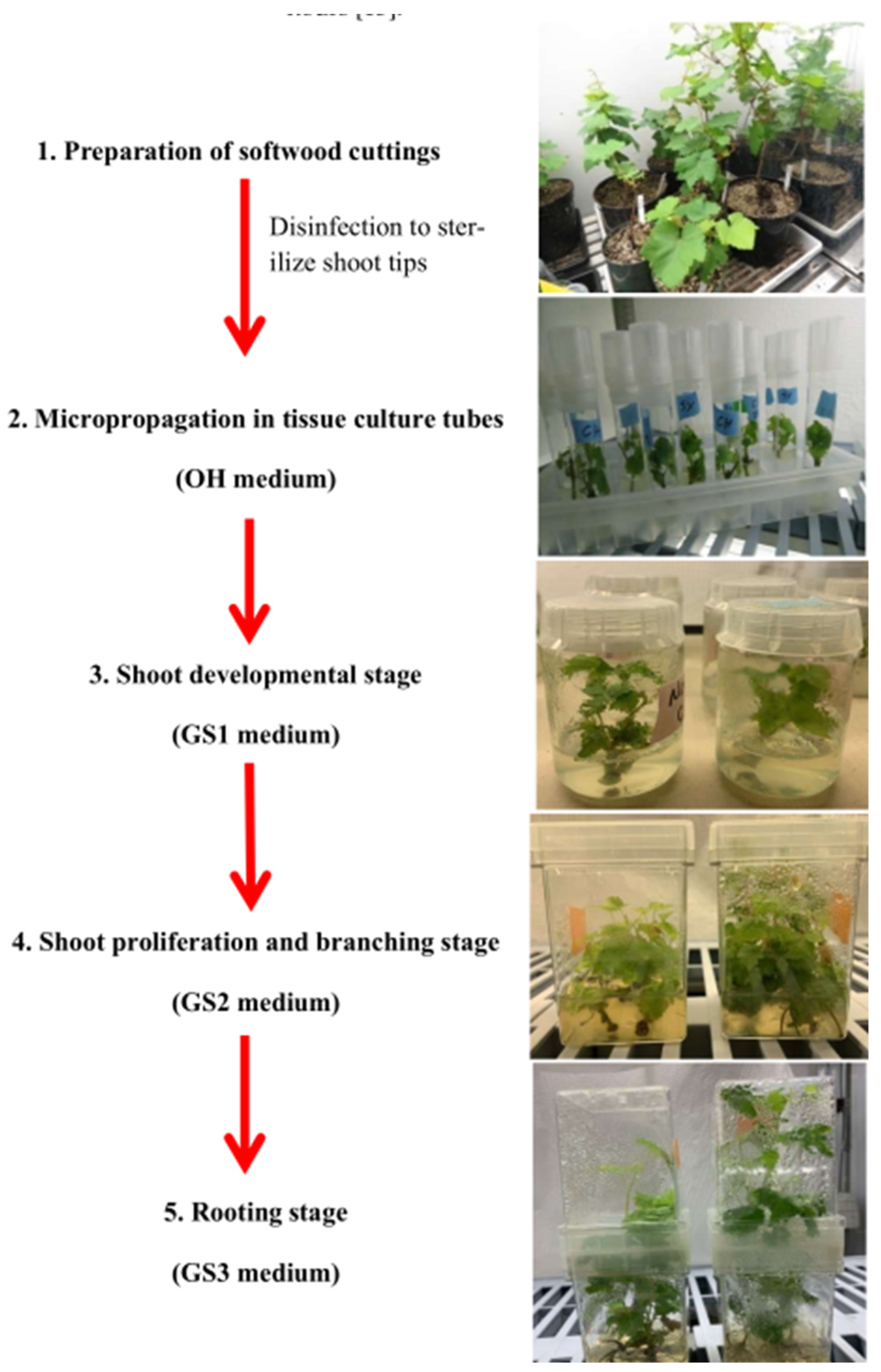
Table S1). About 2-3 weeks later, the new shoots were removed from the stem and transferred into new containers with GS1 agar medium. New shoots established on agar media moved through the three typical grape stages– GS1, GS2 and GS3 (
Table S1). While GS1 medium allows plant growth without promoting shoot or root elongation, GS2 medium promotes shoot proliferation and elongation and GS3 medium promotes root development (
Figure 1).
2.2. Agrobacterium Preparation
Agro-infiltration was conducted by using
A. tumefaciens (EHA105) containing the binary vector pCB301 carrying full-length GLRaV-3 cDNA clone or its GFP-tagged variant which were recently constructed in our lab (data not published) and vLR2-p24 (GLRaV-2 RNA silencing suppressor) (RSS), kindly provided by Dr. Valerian Dolja, have been used [
15]. Briefly, after streaking glycerol stocks of the agrobacterium on LB agar plates containing kanamycin (50 μg/ml) and rifampicin (25 μg/ml) and incubating at 30°C for 2 days, a single colony was cultured in 5 ml of LB containing the same antibiotics and 10 mM morpholine ethanesulfonic acid (MES) (pH 5.85) and 20 μM acetosyringone at 30°C with shaking at 250 rpm. The overnight cultures were sub-cultured and grown at 30°C with shaking in a larger volume of the same LB medium until reaching optical density at 600 nm (OD
600) of 1.0. Then agrobacterial cultures were centrifuged for 10 min at 5000 rpm and the resultant pellets were re-suspended and washed in infiltration buffer (10 mM MES, pH 5.85, 10 mM MgCl
2 and 150 μM acetosyringone). A second round of centrifugation was carried out and the pellet was re-suspended in an appropriate amount of infiltration buffer. The final OD
600 of agrobacterial suspension was adjusted to 2.0 for infectious viral clones or to 0.5 for vLR2-24, followed by induction at room temperature for two hours [
15].
2.3. Four Methods Applied in Agro-Inoculation
Four different techniques including vacuum-based agro-infiltration, agro-pricking, agro-drenching and agro-injection were tested and compared in this study. In all methods, 8–11-week-old, healthy-looking, tissue-cultured grapevine plantlets were used. To prepare them for agro-inoculation, plantlets were trimmed with a pair of scissors to remove most of the leaves and side shoots, especially near the bottom of the plantlets. In addition, some of the roots were trimmed to one third of their length. In vacuum infiltration and pricking methods, trimmed plantlets were poked on their stems and roots with a 31-gauge needle (
Figure 2a-c). The reason behind trimming and pricking was to create minor wounds to facilitate entry of agrobacterium into plant tissue. Trimming would also reduce the number of leaf and branches to allow better recovery from the shock produced by the infiltration process and rapid loss of humidity after transplanting them in soil after infiltration.
2.3.1. Vacuum-Based Agro-Infiltration
To examine the infectivity of our GLRaV-3 cDNA clones we followed the protocol of Kurth et al. [
15] with minor modifications. Agrobacterium preparation was done, as mentioned earlier, for all agrobacterium-containing viral clones including pLR3, pLR3-GFP and p24 as an RSS. In each beaker, 300 ml of each virus-genome containing agrobacterial suspension were prepared at the OD
600 of 2.0 for the viral clone and OD
600 of 0.5 for RSS p24. Plantlets were submerged in the beaker containing the bacterial suspension, which was placed in a nucerite desiccator connected to a vacuum pump (Edwards-Lab-2 Single-Phase) (
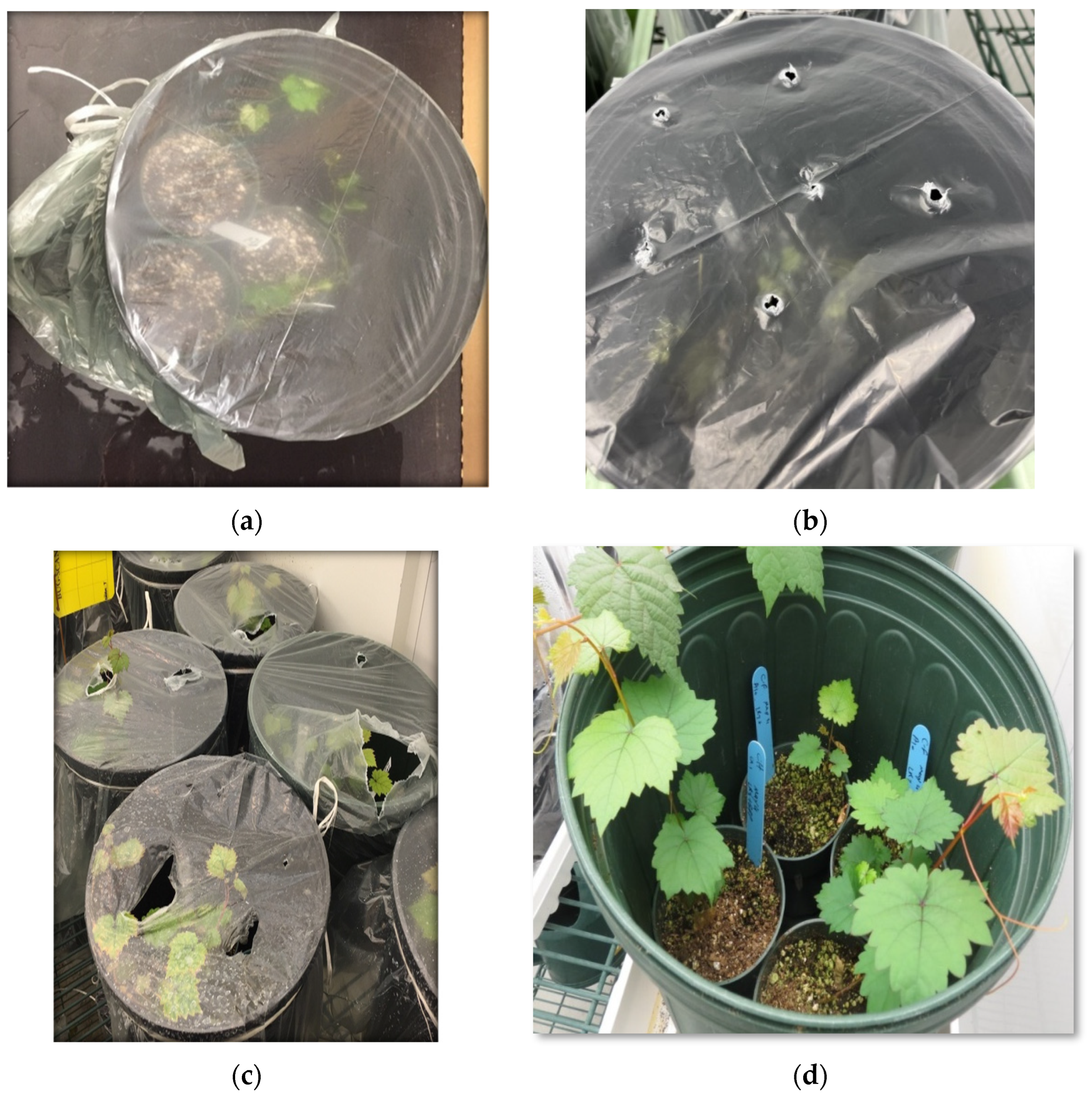
Figure 2d-f).
Plantlets after trimming were placed in a 500 mL beaker containing 300 mL of agrobacterial suspension carrying viral full-length clone (d) which was subsequently placed in a nucerite desiccator (e) that was connected to a vacuum pump (f). Finally, vacuum was applied for a desired length of time, followed by a quick vacuum release to allow entry of agrobacterial suspension into the stem and roots through stomata.
The infiltration procedure comprised of three cycles, each including applying the vacuum for 10 minutes followed by a quick vacuum release. After the infiltration, plantlets were rinsed with tap water and potted gently in 2.5-inch round plastic nursery pots and maintained in the growth room. For maintaining the humidity, the pots containing infiltrated plantlets were placed in a 3-gallon plastic pot covered with clear plastic sheets and allowed to recover in a growth chamber for one month at a temperature of 21-22°C with 16-hour photoperiod at light intensity of 50 µmol/m
2/s (
Figure 3a).
2.3.2. Agro-Pricking
In the second method, the protocol described by Yepes et al. [
18] with some modifications was applied. After trimming the plantlets, stems and roots were again gently wounded by pricking with a 31-gauge needle dipped in agar culture of
A. tumefaciens containing viral infectious clones, followed submerging them for 30 min in a beaker with 300 ml of each viral-containing agrobacterial cells as described for method 1. Afterwards, they were washed, potted, and kept in the condition described previously.
2.3.3. Agro-Drenching
Agro-drenching was conducted as described by Ryu et al. [
25] with minor changes. First, grapevine plantlets were trimmed, poked with a 31-gauge needle, as described above, and planted in 2.5-inch round plastic nursery pots. Afterwards, 5-10 ml of the agrobacterial suspension at the same optical density, as described above, was poured into the soil close to the crown of each plantlet. These mini pots were kept in the growth chamber with the same conditions described earlier.
2.3.4. Agro-Injection
Similar to the other methods, 8–11-week-old tissue cultured plantlets were first trimmed, followed by injection with agrobacterial suspension containing viral clone alone or mixture of agrobacteria containing both viral clone and the construct expressing p24. For this procedure, the bacterial inoculum was diluted with infiltration buffer to reach the final OD
600 of 1.0. The bacterial suspensions were injected gently by using a 1-ml needleless syringe through the stomata of the lower epidermis [
29].
It should be noted that in all four treatments, grapevine plantlets were mock-infiltrated using the respective method with agrobacterium containing the p24 expression plasmid only.
2.4. The Effect of Various Factors on the Survival and Infectivity Rate in Vacuum-Based Agro-Infiltration Technique
After conducting all four methods and comparing the preliminary results of nested-RT-PCR and RT-qPCR, based on the infectibility percentage and virus expression level in plantlets, we found that vacuum-based agro-infiltration gave the best performance. Therefore, we decided to focus only on this technique to test different factors and conditions to optimize this experimental system to launch infection of grapevine using infectious viral clones. It is important to note that in each experiment, only one factor was varied at a time and all other conditions were kept constant.
2.4.1. Age and the Cultivar of Tissue Cultured Plantlets
To study the impacts of age of plantlets on the survival and infection rate, three groups of plantlets were chosen. The first group was 5-7 weeks old, the second group 8-11 weeks old and the third group 12-16 weeks old. Three grapevine cultivars were tested (Cabernet franc, Syrah and Chardonnay) to find out which cultivar is more conducive to the vacuum infiltration procedure as judged by both survival and infection rate. In this experiment, pLR-3 containing agrobacterium at OD600 of 2.0 and p24 at OD600 of 0.5 were infiltrated into the plantlets with three vacuum cycles of 10 minutes for each followed by a quick vacuum release. Infiltrated plantlets were monitored for survival and assayed for infectivity at 2 months post infection.
2.4.2. Impacts of Humidity Level
Humidity level is one of the most important factors in the plant growth chamber, as it affects the rate of transpiration and nutrient absorption. To study the impacts of humidity level on the survival and infection rates of grapevine plantlets post agro-infiltration the plastic cover of the pots was removed from pots containing infiltrated plantlets at three different times in a gradual or instant manner. Here, pLR-3 containing agrobacterium at OD
600 of 2.0 and p24 at the OD
600 of 0.5 were infiltrated into the Syrah and Cabernet franc plantlets following the standard procedure as described previously. After infiltration and potting the plantlets in 2.5-inch round plastic nursery pots, these small pots were placed in 3-gallon plastic pots which were covered with plastic sheets and kept in a growth chamber and one week later, the plastic covers were removed instantly or gradually (
Figure 3a & b). In the gradual manner, after one week post infiltration, several holes with the diameter of 4-5 mm were made by poking the plastic cover. These holes were made gradually bigger until the covers were removed (
Figure 3c & d). In other pots, the covers were removed after 2 or 3weeks either gradually or instantly to observe the effects of humidity on both the survival and the infection rates.
2.4.3. Effects of Vacuum Duration and Agrobacterial Density (OD600) on Plantlet Survival and Infectivity
As the vacuum is an important step in this infiltration procedure, the effect of various vacuum durations on the survival and infection rates was tested. Three-time durations were evaluated: 5, 10 and 15 minutes, each for three cycles. For example, consider the 5-minute duration level. Plantlets that were submerged in agrobacterial suspension in a beaker were subjected to vacuum for 5 minutes, followed by a quick release. The same procedure was repeated twice. In the second part of this experiment, various OD600 for pLR3-containing bacterium including 1.0, 2.0 and 3.0 and different OD600 for supplemental bacterium containing p24 RSS such as 0.5 and 1.0 were infiltrated into 8–11-week-old grapevine plantlets with three vacuum cycles of 10 min each and quick pressure release after each cycle.
2.4.4. Effects of Various RSSs on Infectivity
To investigate the impacts of viral RNA silencing suppressors (RSSs) on the infection rates, several such suppressors derived from different viruses including p24 of GLRaV-2, p19 of Tomato bushy stunt virus, p21 of Beet yellows virus, HC-Pro of Turnip mosaic virus and TCV-CP of Turnip crinkle viruswere tested. As negative controls, plantlets were vacuum infiltrated with agrobacterium containing only GLRaV-3 cDNA clones without any RSS. In this experiment, cDNA clone-containing agrobacterium of OD600 of 2.0 and different supplemental agrobacteria containing RSS at OD600 of 1.0 were infiltrated into 8–11-week-old grapevine plantlets with vacuum duration of 10 minutes for each cycle.
2.4.5. Impact of Dormancy Treatment on the Infection Rates and Virus Titer
To study the influence of dormancy on infectivity rate, virus titer, and symptom development, 8-10 months after agro-infiltration, grapevine plants that were inoculated via agro-infiltration 8-10 months earlier were subjected to either one or two cycles of dormancy. As negative controls, grapevines derived from the same agro-infiltration procedure were kept under the normal growth conditions. Each dormancy treatment comprised of 50 grapevines, 25 of which tested positive whereas the remaining 25 plants tested negative for GLRaV-3based on nested RT-PCR. Dormancy treatment was repeated once for all experiments. Before inducing dormancy, each plant was trimmed such that only three to four leaves were left. In the single dormancy treatment, 50 grapevine plantlets were subjected to an incremental drop in temperature over a 3-week period. In week one, the temperature was reduced from 22 to 16°C; in week two, the temperature was further dropped to 10°C; from the third week and forward, the plants were kept at 4°C for 2 months. At the end of the cold treatment cycle, the temperature was returned to 22°C using the same increment but in reverse order.
The double dormancy treatment was simply comprised of two cycles of single dormancy treatment that were interrupted by a 2-month break period between them. In other words, at the completion of the first dormancy cycle, plants were returned to 22°C for 2 months to recover and grow followed by a second cycle of dormancy.
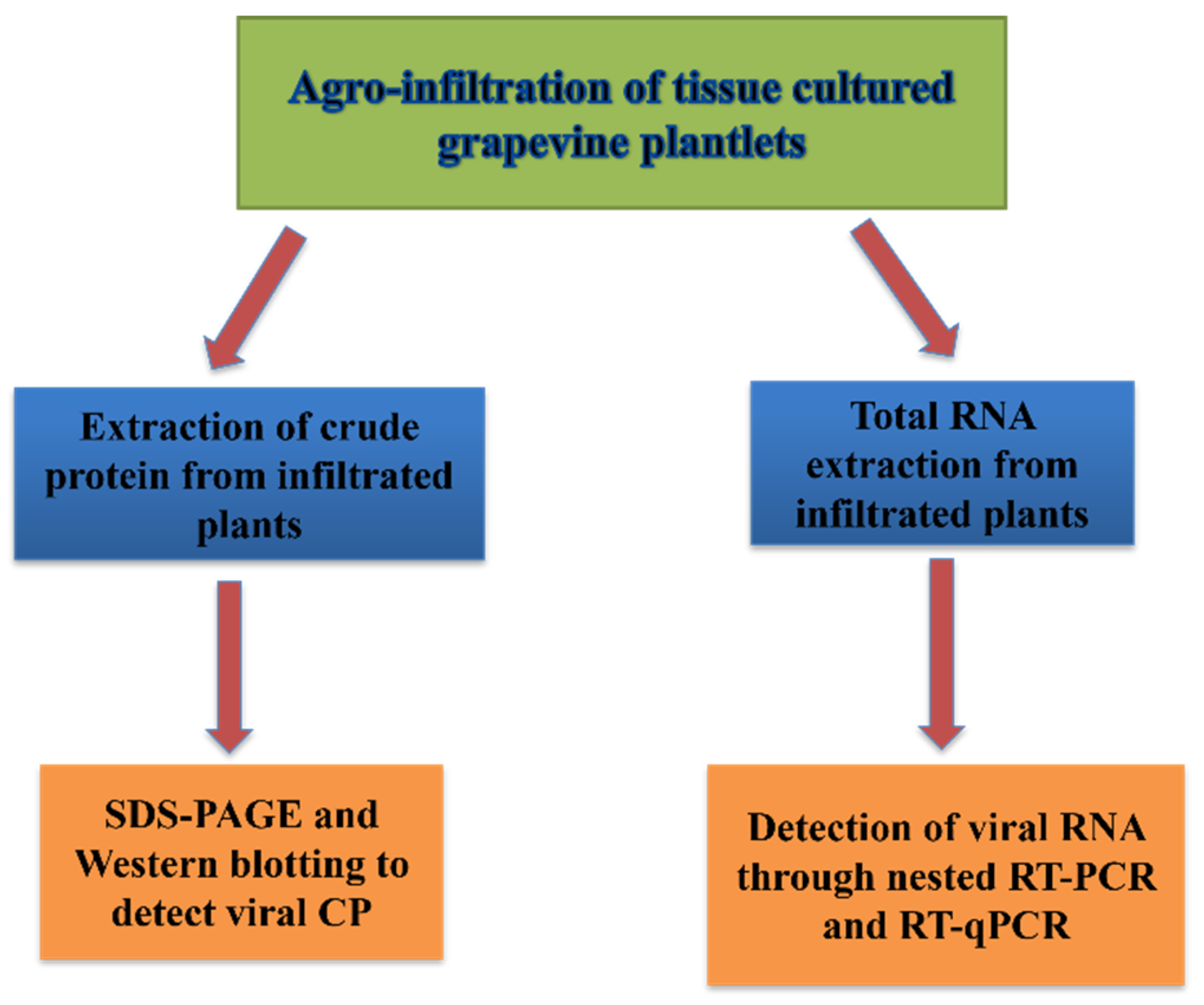
2.5. Infectivity Assays
Various ways and methods have been used for detecting the virus after infiltration. The best and more sensitive method, especially at the early months post infiltration, was based on the detection of virus RNA, such as nested-RT-PCR and qRT-PCR. Other assays such as sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting, which is based on the expression of the virus capsid protein was also helpful for testing the infectivity success but were a bit less sensitive. A schematic of these methods is shown in (
Figure 4).
2.5.1. Total RNA Isolation, RT-PCR, Nested RT-PCR and RT-qPCR
To test for systemic infection of the virus resulting from agro-infiltration new and non-infiltrated leaves of grapevines were collected at two months post infiltration (mpi). Subsequently, the veins and petioles were obtained from leaf samples and ground to fine powder in liquid nitrogen using a mortar and pestle and stored at -80
oC. Total RNA was extracted from 50 mg of ground tissue samples following the protocol as described in Xiao et al. [
30]. The quality and concentration of RNA preps were measured with a Nano Drop (ND-1000, Thermo Fisher) and stored at -80
oC until further use. First-strand cDNA synthesis was primed with random hexamers using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies) essentially following the protocol of Shabanian et al. [
31].
Conventional RT-PCR was conducted by using virus-specific primers following Shabanian et al. [
31] with a minor change in the cDNA used in PCR. Because of the extremely low level of viral RNA present in infiltrated plants, in each PCR reaction 2-3 μl of cDNA (instead of 1μl in regular PCR reaction, was used. Furthermore, nested RT-PCR and RT-qPCR were used for detecting the low level of virus RNA. Here, two sets of primers with the same reverse primer were used to amplify a region at 3ʹ end of GLRaV-3genome followed by electrophoresis on agarose gels. The first round of nested RT-PCR reaction mix (25 µl) contained 2-3 µl of cDNA, 2.5 µl 10XPCR buffer (containing 2.0 mM MgCl
2), 0.2 mM dNTPs, 0.2 µM of each primer (F1_16289 and R_17240) and 1.0 unit of Taq DNA polymerase (Gene DireX) to amplify a 952 bp DNA. PCR conditions included an initial denaturation step at 94ºCfor 5 min, then 35 cycles at 94ºC for 30 s, 53ºC for 30 s and 72ºC for 1 min, followed by a final extension at 72ºC for 7 min. For the second round PCR, everything remained the same as the first round PCR except the following: 1) 2 µl of the amplification product from the first round PCR was used as a template instead of cDNA; 2) an internal forward primer (F2_16896) and the same reverse primer were used to amplify a 345 bp DNA; and 3) PCR was conducted for 20 cycles instead of 35 cycles. RT-qPCR was conducted following the method of Shabanian et al. [
31]. Primers used for these assays are shown in
Table S2.
2.5.2. Western Blotting
Western blot was performed following the protocol of Shabanian et al. [
31] with some modifications. As was mentioned earlier, the viral titer in infiltrated plants was extremely low, so to have more reliable results, petioles and leaf-midribs were cut by using a single edged razor blade and ground to fine powder in liquid nitrogen. Subsequently, 0.5 g of the tissue powder was homogenized in four volume of Bradly buffer and then all steps were done as prescribed by Shabanian et al. [
31]. In this experiment, the primary antibody generated in rat against recombinant CP of GLRaV-3 at the dilution of 1:5,000 and secondary antibody (goat anti-rat IgG) conjugated with horseradish peroxidase (Sigma) at the dilution of 1:5,000 followed by incubation with SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Fisher Scientific).
3. Results
3.1. Vacuum-Based Agro-Infiltration is the Best Approach for Agro-Infection
Four inoculation methods were tested and compared to assess their efficacy in launching infection of grapevine plantlets using agrobacteria containing full-length cDNA clones of GLRaV-3 (summarized in
Table 1). The vacuum-based method gave the highest levels of infection, followed by pricking, while agro-drenching and injection failed to produce infection. A total of 39 of the 50 Syrah plantlets survived the vacuum-based inoculation procedure, with 22 plants (56%) testing positive for GLRaV-3 at 6 months post inoculation (
Table 1). For Cabernet franc, 38 of the 50 plantlets survived, and 20 (53% of survivors) tested positive for GLRaV-3 at 6 mpi. On the other hand, inoculation via the pricking method yielded a higher rate of survival but a much lower rate of infection. For example, 43 of the 50 Syrah plantlets survived with only 7 testing positive (16% of survivors). Similarly, 42 of the 50 Cabernet franc plantlets survived, and only six of them (14% of survivors) were infected with GLRaV-3 (
Table 1).
3.2. Impacts of Age and the Cultivar of Grapevine Plantlets on Survival and Infectivity Rates
Both the age and specific cultivars of grapevine plantlets had considerable impacts on survival and infection rates. Survival rate was positively correlated with the age of plantlets (
Table 2). For example, in the 5–7-weeks-old group, 18-25% of the plantlets survived after agro-infiltration while in the 8–11-weeks-old group, 65-82% of plantlets survived and finally in the 12–16-weeks-old group, the survival rate reached 76% for Chardonnay, 88% for Cabernet franc, and 92% for Syrah. Based on the differences observed in survival rate among the three cultivars, Syrah had the highest survival rate for all three age groups and Chardonnay had the lowest (
Table 2). On the other hand, the infectivity rate did not follow the same trend as for the survival rate. The highest infection rate was consistently observed for plantlets of the 8–11-weeks-old group for all three cultivars. It is important to note that infection rate varied considerably among the three cultivars, with Syrah being the highest at 63%, followed by Cabernet franc at 58%, and Chardonnay the lowest at 44% (
Table 2).
3.3. The Effects of Humidity Control on Plantlet Survival and Infectivity
Results from an initial trial of agro-infiltration showed that low humidity level had drastic negative effects on the survival rate and consequently on the infection rate (
Table 3). If the plastic covers were removed at one- or two-weeks post-infiltration (wpi), almost none of the plantlets survived. However, if the removals were done step by step by poking the cover after the second week post infiltration, the number of survivors and consequently the infection rate increased. Therefore, in subsequent experiments all agro-infiltrated plantlets were kept under high relative humidity conditions for at least 3 weeks.
3.4. Effects of Vacuum Treatment Duration and Density of Agrobacterium on Infectivity Rates
To find out the optimal duration of vacuum application on the survival and infection, three different experimental series were tested. These included three cycles of vacuum for 5, 10 and 15 minutes, followed by a quick vacuum release for each cycle. The survival rate at 4 mpi was negatively correlated with the duration of vacuum, as expected (Table 4a). For instance, with the three 5 min cycles of vacuum application, 84% and 82% of Syrah and Cabernet franc plantlets survived, respectively, and in the case of using three cycles of 10 minutes vacuum setting, 78% and 74% of Syrah and Cabernet franc plantlets survived, respectively. The lowest survival rate was observed among plantlets that were subjected to the three cycles of 15 minutes treatment where only 42% of Syrah plantlets and 36% of the Cabernet franc plantlets were viable (Table 4a). Interestingly, the infection rate did not follow the same trend. The three 10 min vacuum cycles gave the best results, with 66% of the infiltrated Syrah plantlets and 64% of the Cabernet franc plantlets being positive for GLRaV-3. We thus concluded that the best vacuum cycle to initiate viral infection of grapevine plantlets derived from tissue culture is three 10 min vacuum cycles.
Table 4.
a. Effects of vacuum duration on survival and infection rates. Grapevine plantlets that were co-infiltrated with agrobacterium containing GLRaV-3 infectious clone and the suppressor of RNA silencing p24 were tested by nested RT-PCR at 4 mpi for the presence of GLRaV-3.
Table 4.
a. Effects of vacuum duration on survival and infection rates. Grapevine plantlets that were co-infiltrated with agrobacterium containing GLRaV-3 infectious clone and the suppressor of RNA silencing p24 were tested by nested RT-PCR at 4 mpi for the presence of GLRaV-3.
| Vacuum duration |
5 Minutes |
10 Minutes |
15 Minutes |
| Cultivar of plantlets |
SY |
CF |
SY |
CF |
SY |
CF |
| No. of infiltrated plants |
50 |
50 |
50 |
50 |
50 |
50 |
| No. of survived plants |
42 |
41 |
39 |
37 |
21 |
18 |
| No. of infected plants |
8 |
5 |
26 |
24 |
12 |
11 |
| Percent of infection of survivor plants |
19% |
12% |
66% |
64% |
57% |
61% |
The influence of cell density of agrobacteria containing cDNA clones of GLRaV-3 and those containing the plasmid for the RSS p24 on survival and infection rate also were tested. The best cell density or OD600 for agrobacterium containing GLRaV-3 clone is 2.0 regardless of whether p24 was used at either OD600 of 0.5 or 1.0 (Table 4b). All other combinations gave a lower percentage of infections.
Table 4.
b. The effects of agrobacterial cell density (OD600) on infection and survival rates. Status of infection was based on nested RT-PCR at 4 mpi.
Table 4.
b. The effects of agrobacterial cell density (OD600) on infection and survival rates. Status of infection was based on nested RT-PCR at 4 mpi.
OD 600
|
GLRaV-3 |
1 |
2 |
3 |
| P24 |
0.5 |
1 |
0.5 |
1 |
0.5 |
1 |
| Cultivar of plantlets |
SY |
SY |
SY |
SY |
SY |
SY |
| No. of infiltrated plants |
50 |
50 |
50 |
50 |
50 |
50 |
| No. of survived plants |
42 |
39 |
38 |
34 |
23 |
18 |
| No. of infected plants |
5 |
9 |
26 |
24 |
15 |
11 |
| Percent of infection of survivor plants |
12% |
23% |
68% |
70% |
65% |
61% |
3.5. Effects of Co-Infiltration with Virus RSSs on Infectivity
In a preliminary experiment, it was found that co-infiltration with p24, an RSS encoded by GLRaV-2, was essential for launching infection with the GLRaV-3 infectious cDNA clone. To find out the most effective RSS for this purpose, the following five RSS in co-infiltration experiments were tested: HC-Pro, p19, p21, TCV-CP and p24. As negative controls, grapevine plantlets were infiltrated with only GLRaV-3 cDNA clone. All RSSs had major positive impacts on the infection rate, albeit to different degrees and depending on the cultivars used (Table 5a). For example, p19 increased the rate of infection by 3.7-fold for Syrah and 2.6-fold for Cabernet franc when compared to the no RSS control. For both cultivars, the top three RSSs were p19, HC-Pro, and TCV-CP. Interestingly, p24 ranked the 4th for Syrah and tied for 4th place with p21 where Cabernet franc was concerned (Table 5b).
Table 5.
a. Effects of co-infiltration with RNA silencing suppressors on infectivity percentage. 8-11 weeks old grapevine plantlets were infiltrated with agrobacterium containing infectious GLRaV-3 clone or GLRaV-3 clone and one of the following RNA silencing suppressors: HC-Pro, p19, p24, TCV CP and p21. The status of GLRaV-3 infection was assessed based on nested RT-PCR at 4 mpi.
Table 5.
a. Effects of co-infiltration with RNA silencing suppressors on infectivity percentage. 8-11 weeks old grapevine plantlets were infiltrated with agrobacterium containing infectious GLRaV-3 clone or GLRaV-3 clone and one of the following RNA silencing suppressors: HC-Pro, p19, p24, TCV CP and p21. The status of GLRaV-3 infection was assessed based on nested RT-PCR at 4 mpi.
| RNA silencing suppressors |
No SRS |
HC-Pro |
P24 |
P19 |
TCV |
P21 |
| SY |
CF |
SY |
CF |
SY |
CF |
SY |
CF |
SY |
CF |
SY |
CF |
| No. of infiltrated ones |
50 |
50 |
50 |
50 |
50 |
50 |
50 |
50 |
50 |
50 |
50 |
50 |
| No. of survived ones |
44 |
40 |
42 |
37 |
40 |
39 |
44 |
38 |
39 |
36 |
43 |
38 |
| No. of infected plants |
8 |
9 |
22 |
19 |
20 |
18 |
26 |
23 |
21 |
23 |
17 |
18 |
| Percentage of infection of survivor plants |
18% |
23% |
52% |
51% |
50% |
51% |
59% |
60% |
54% |
64% |
40% |
47% |
Table 5.
b. Ranking table of different RNA silencing suppressors based on the impacts of them on the infectivity percentage.
Table 5.
b. Ranking table of different RNA silencing suppressors based on the impacts of them on the infectivity percentage.
Rank
|
Based on infection % of all plants |
Based on infection % of survivor plants |
| SY |
CF |
SY |
CF |
| 1 |
P19 (52%) |
TCV (46%) |
P19 (59%) |
TCV (64%) |
| 2 |
HC-Pro (44%) |
P19 (46%) |
TCV (54%) |
P19 (60%) |
| 3 |
TCV (42%) |
HC-Pro (1%) |
HC-Pro (52%) |
HC-Pro (51%) |
| 4 |
P24 (40%) |
P24 (36%) |
P24 (50%) |
P24 (51%) |
| 5 |
P21 (34%) |
P21 (36%) |
P21 (40%) |
P21 (47%) |
| No RSS |
16% |
18% |
18% |
23% |
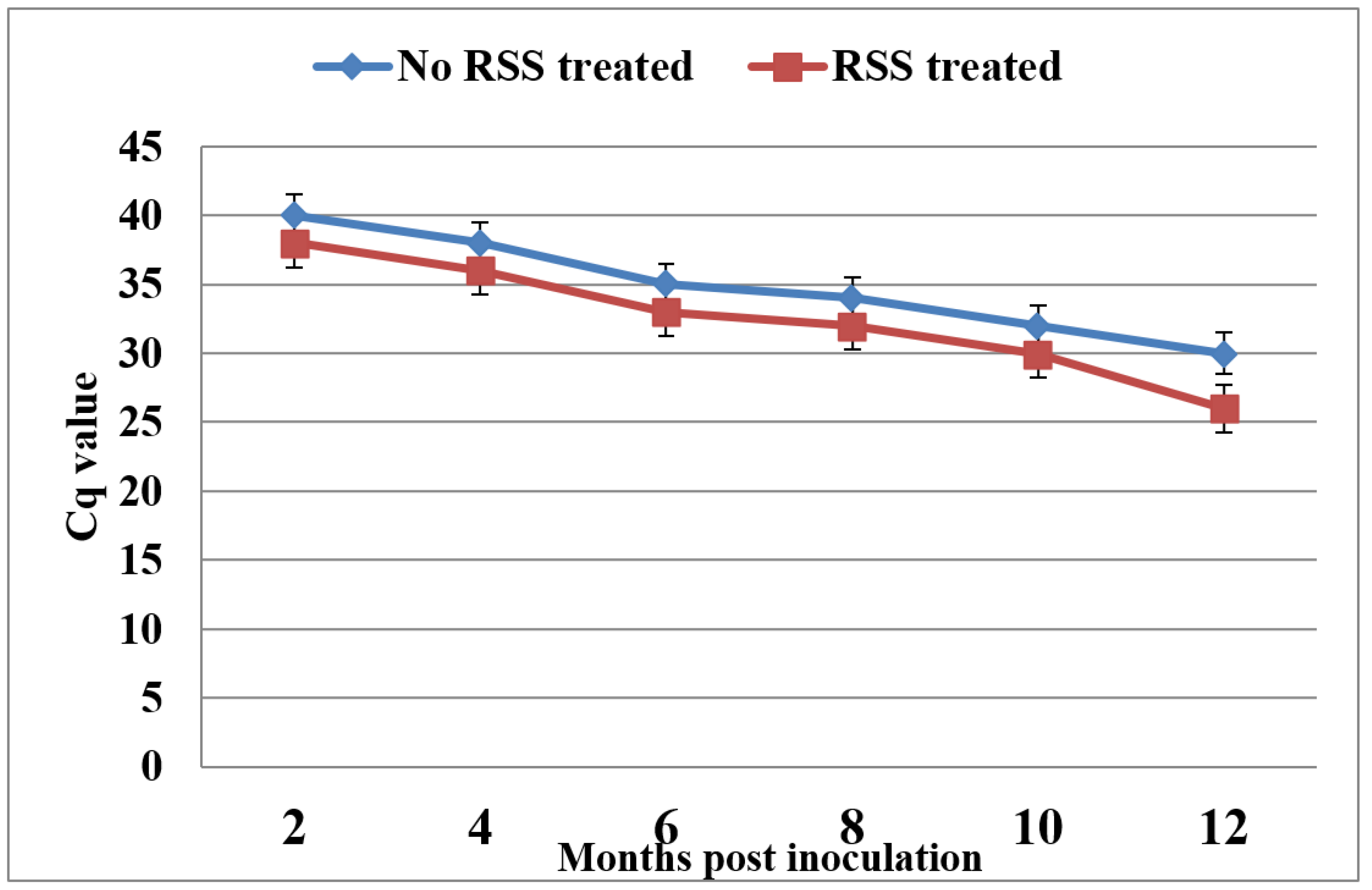
To test if the increase in infection rate of grapevine plantlets in the presence of RSSs is due to enhanced virus replication, a time course RT-qPCR analysis was conducted for three Syrah plants that were co-infiltrated with GLRaV-3 cDNA clone and p24 at 2, 4, 6, 8, 10 and 12 mpi (
Figure 5). It should be noted that the Cq values for the no RSS treated samples were consistently higher than the corresponding values for the RSS treatment reflecting the lower levels of virus RNA compared with RSS-treated samples. Two conclusions can be drawn from these data: 1) a positive signal was first detected at 2mpi for the grapevine samples that were co-infiltrated with GLRaV-3 full-length clone and the RSS and with 2 months delay in the grapevine samples without using RSS; 2) as time progressed, the Cq values decreased for both and the difference in the Cq between the two groups of plants increased considerably at 12 mpi.
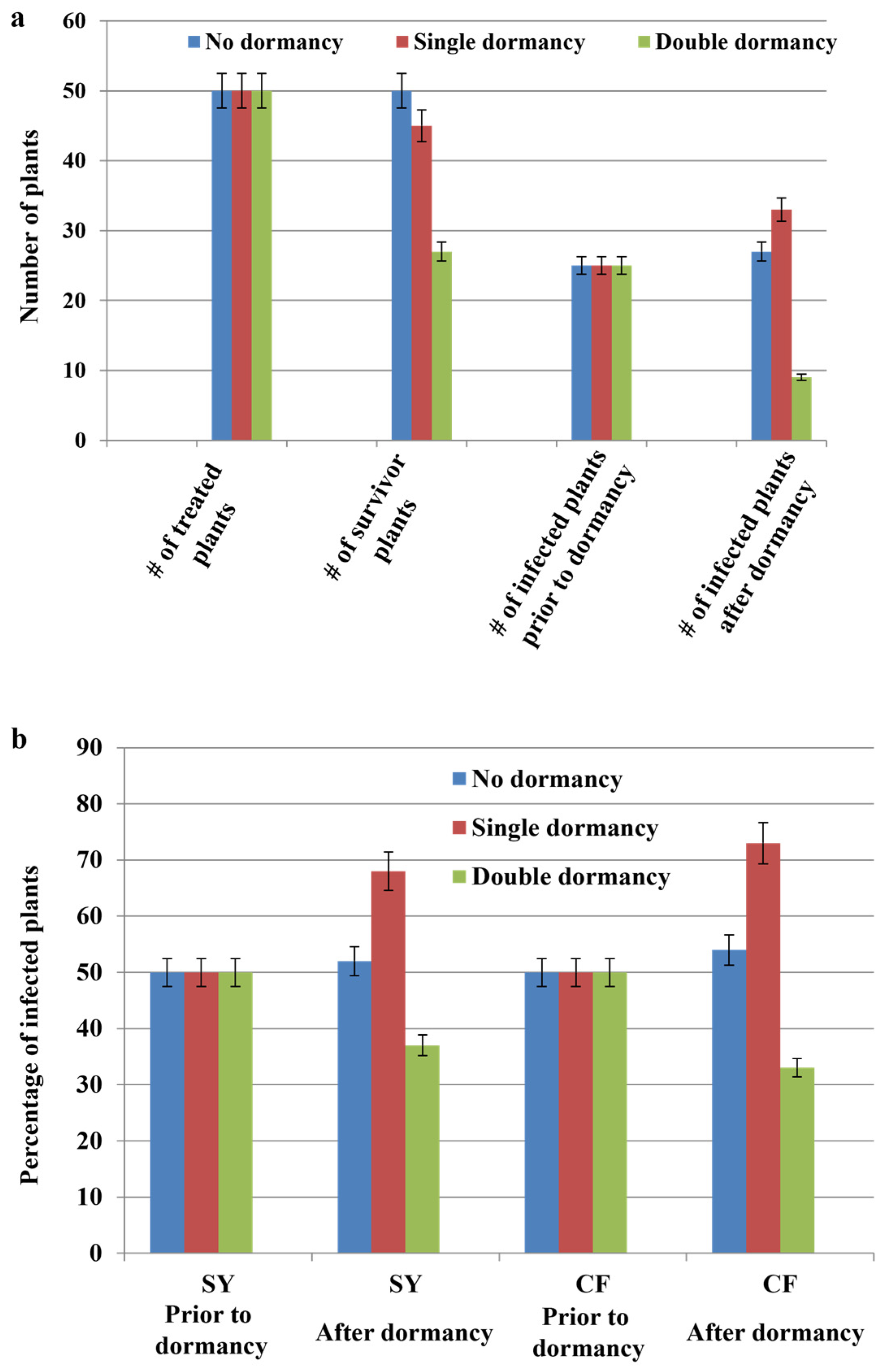
2.6. Effects of Dormancy Treatment on Infection Rate
The single cycle dormancy treatment had a major positive impact on the infection rates among the infiltrated plantlets. For example, among the 47 Syrah plants that survived dormancy treatment, 32 tested positive for GLRaV-3 reflecting a 28% increase in the number of infected plants (
Table 6). A similar trend was observed for the Cabernet franc plants: 33 of the 45 plants that survived the single cycle dormancy treatment tested positive for GLRaV-3 (
Table 6). In contrast, the double cycle dormancy treatment was detrimental to plant survival, which in turn resulted in a reduction in the number of plants that tested positive for GLRaV-3 at the completion of the treatment cycle (
Figure 6a & b). For example, 21 Syrah and 23 Cabernet franc plants of 50 died from going through the two cycles of dormancy. Of the survivors, only 11 Syrah and 9 Cabernet franc plants tested positive for the virus (
Table 6).
Although systemic infection was detectable after 4 mpi by nested RT-PCR and RT-qPCR, a single round of PCR amplification failed to detect any positives prior to dormancy treatment (
Figure 7a &b). In contrast, after one dormancy cycle, even conventional RT-PCR was able to identify GLRaV-3 positives. As shown in (
Figure 7c & d), the titer of the virus at 12 mpi was high enough for some of the samples to test positive after the first round of nested-RT-PCR. The intensity of PCR products further increased for those tested positive in the first round or became visible for those that did not show positive amplification in the first round PCR.
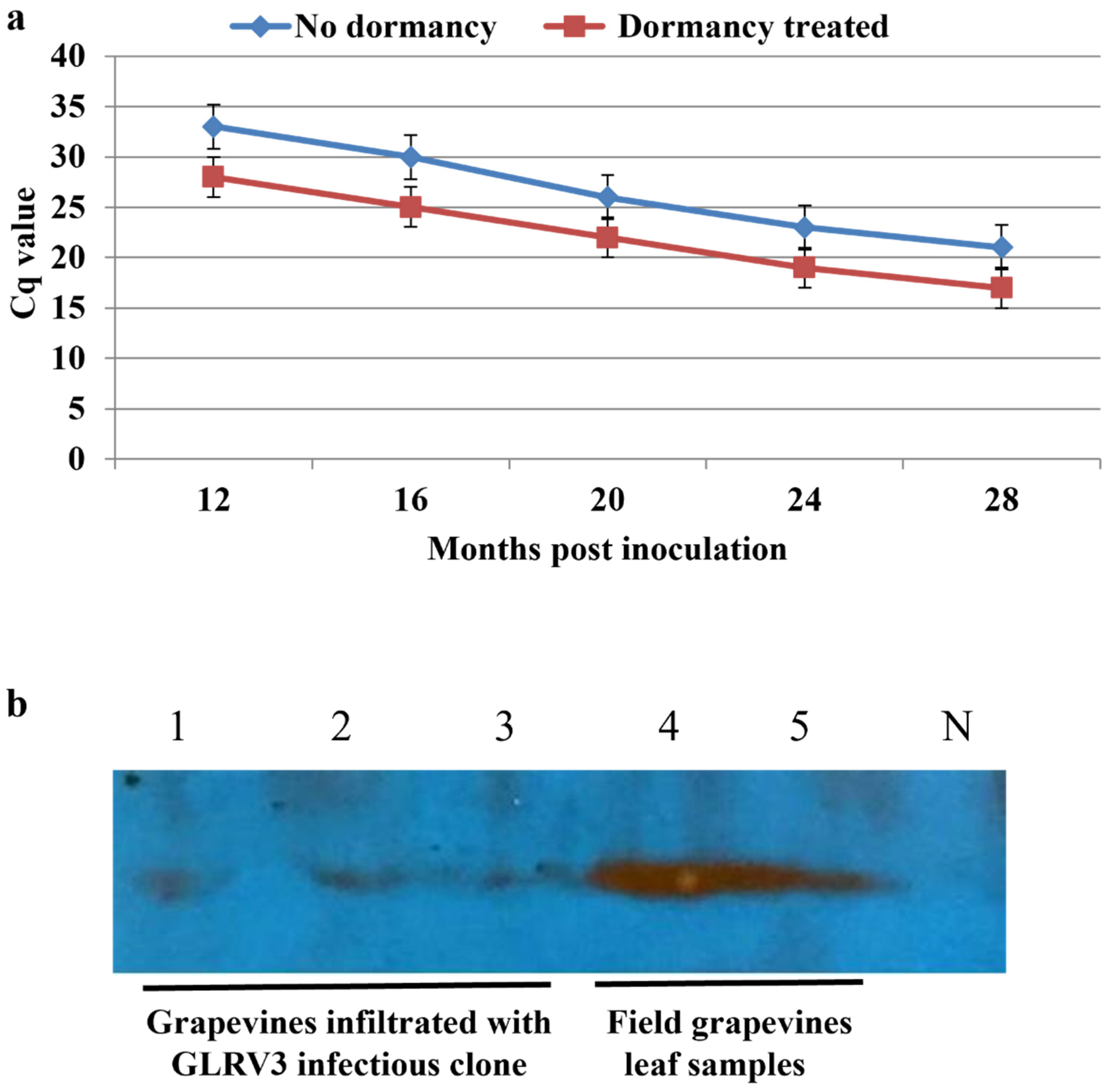
The RT-qPCR results also confirmed those from nested RT-PCR, which revealed that at 4 mpi and before dormancy treatment, the Cq value was too high (35-38) to be confidently called as a positive. However, after a single dormancy cycle the Cq value reduced to 25-30 at 12 mpi. Taken together, these data demonstrate that dormancy had a considerable positive impact on virus titer (
Figure 8a). No virus capsid protein was detected by Western blotting prior to the one-year mark in contrast to field samples used as positive controls (
Figure 8b, lanes 4-6). However, 14 mpi and after the dormancy treatment the capsid protein could be confidently detected in vines infiltrated with pLR-3 (
Figure 8b, lanes 1-3).
4. Discussion
The state of research on viruses of woody perennial plants including grapevine lags far behind those that infect herbaceous plants due to the compound effect of several factors. First, grapevines are commonly infected with a mixture of distinct viruses and strains complicating elucidation of the etiological contribution of individual viruses in a disease. Second, many pathogenic viruses of woody plants do not infect herbaceous plants which are incomparably more amenable for investigating all aspects of virus infection. In this respect, GLRaV-2 seems to be a rare exception in being readily agro-inoculated into
Nicotiana benthamiana plants [
32]. Third, unlike most viruses of herbaceous plants, grapevine viruses cannot be transmitted among vines through mechanical inoculation although in some studies dodders (
Cuscuta spp.) have shown different level of success in transferring some viruses to woody plants [
33,
34]. Therefore, the preferred way to initiate infection with a virus in grapevine is the delivery of infectious clones using agrobacterium. This procedure, dubbed ‘agro-infection’, was invented by Grimsley et al. [
35] to launch infection of turnip with
Cauliflower mosaic virus. Since then, agrobacterium-based inoculation has been widely used in the plant virology community. In fact, this approach has become the method of choice when it comes to studies of viruses that infect grapevines and other woody perennials.
Drenching was the first agrobacterium-based inoculation method that was developed to launch grapevine infection using micro-propagated plantlets with an infectious DNA clone of GVA [
14]. This method appears to be effective in launching grapevine infection with GVA and Grapevine Pinot gris virus cDNA clones [
14,
20]. Unfortunately, it failed to initiate systemic infection for GRSPaV at the practically acceptable level [
16].
In 2012, a breakthrough was achieved through development of a vacuum-based agro-infiltration method to launch infection of micro-propagated grapevine plantlets with a GFP-tagged cDNA clone of GLRaV-2 [
15]. The authors tested 15 grape cultivars and found that Syrah and Cabernet franc were most susceptible, Zinfandel less so, whereas the remaining 12 cultivars failed to be infected. It was also shown that p24, an RNAi suppressor encoded by GLRaV-2 significantly improved the infection rate [
15,
36] opening the possibility of method application for other grapevine-infecting viruses. The utility of this new technology was tested by Yepes et al. [
18] to launch infection with an infectious clone of GRBV, a single-stranded DNA virus of the family
Geminiviridae responsible for an emerging grapevine red blotch disease [
37]. In the latter study, the effectiveness of vacuum infiltration in launching GRBV infection in grapevine plantlets derived from seven
V. vinifera cultivars and four rootstocks has been tested. It was concluded that: 1) both vacuum agro-infiltration and agro-pricking worked equally well; 2) 4-6 weeks-old plantlets were superior to those 8-12 weeks-old; 3) none of the three silencing suppressors (GLRaV-2 p24, TBSV p19, and CMV 2b) was necessary for initiating GRBV infection; and 4) the duration and level of vacuum were specific to the grapevine cultivars tested [
18]. The achieved rate of infection varied significantly among the grape cultivars. Thus, Syrah performed the best, with an average of 55% of the plantlets testing positive for GRBV, followed by Pinot noir (22%), Cabernet franc and Chardonnay (20%). In contrast, the infection rates in rootstocks were lower, averaging at 16% among the four rootstocks [
18].
In this study, a systematic approach to test a set of factors that may influence the survival and infection rate of grapevine plantlets has been used. First, four inoculation methods of the grapevine plantlets with a cDNA clone of GLRaV-3 (unpublished data) were applied. Among those the vacuum agro-infiltration was the most effective followed by pricking, whereas soil drenching and direct infiltration with a needleless syringe failed to produce infection. We have demonstrated that plantlets of 8-11 weeks of age gave the best performance both in terms of survival and infectivity. We compared five different silencing suppressors in agro-infiltration and demonstrated that four of them were highly effective as each substantially increased the infection rate between 40-50 %. In line with previous research [
15,
18], maintaining plantlets at high humidity during the first several weeks after infiltration was critical for plantlets survival and hence infectivity. Lastly, we have shown that a single cycle of dormancy treatment for two months at 4 ºC significantly increased virus titer and enhanced symptom development.
It was also reconfirmed that the grapevine cultivar and the age of plantlets have direct impacts on the survival and infection rates. The 8-11 weeks old plantlets showed the best outcome in survival (65-82%) and infectivity (44-63%) rates, while the older or younger plantlets resulted in either reduced infectivity or survival rates similar to the findings by Yepes et al. [
18]. Also like the reports by Kurth et al. [
15] and Yepes et al. [
18], Syrah and Cabernet franc showed higher survival and infection rates compared with Chardonnay.
We have also found that the application of virus RNA silencing suppressors is an important factor in the virus infection rates and virus titer in agreement with the results of Kurth et al. [
15] but in contrast to the findings by Yepes et al. [
18]. This difference could be due to differences in genome type, genome size and structure, expression strategies, and infection process between GRBV and GLRaV-3. Indeed, GRBV possesses single-strand DNA genome of only ~3.2 kb, whereas positive-strand RNA genome of GLRaV-3 has a much larger positive sense ssRNA of nearly 19 kb.
As mentioned above, the GLRaV-3 titer and agro-infection rates exhibited a sharp variation before and after cold treatment
. According to RT-qPCR and nested-RT-PCR analysis before and after dormancy, the cold treatment increased the titer and infection rates of 16% and 19% in Syrah and Cabernet franc respectively, similar to work by Yepes et al. [
18] with GRBV cDNA clone.
5. Conclusions
To our knowledge, this is the most comprehensive report on the optimization of agro-inoculation methods for launching grapevine infections with a viral cDNA clone. In particular, we established that the best material for inoculation is the 2-4 months-old micro-propagated Syrah or Cabernet franc plantlets. We also found that co-infiltration of the plantlets with a strong suppressor of RNA silencing, such as HC-Pro, p19, or p24 significantly increases the infection rate. Upon agro-infiltration, plantlets must be protected from loss of humidity during the first 3 weeks. Finally, we showed that a single cycle of dormancy treatment of lignified plantlets at 4ºC for 2 months significantly increases virus titer.
This work is certain to facilitate overall progress in the field of grapevine virology by providing an optimized protocol of grapevine agro-infection. This protocol constitutes a critical tool for investigations into the etiological roles of distinct viruses in grapevine disease complexes often caused by several viruses, as well as into advanced studies on the molecular and cellular aspects of virus-interactions in grapevine. Furthermore, methodical approaches developed in our work has clear potential applications for studying challenging but important pathogenic viruses of other woody crop plants such as citrus, stone fruits and pome trees.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
MS designed and performed the experiments, analyzed the data, wrote the initial drafts of the manuscript. CL constructed the infectious cDNA clones of GLRaV-3 and assisted with agro-infiltration experiments, and monitoring symptom development. AE assisted with agro-infiltration experiments, performed infectivity assays, designed and executed dormancy treatment, and monitoring disease symptoms. VD provided the protocol of tissue culture, vacuum-mediate agro-infiltration, and essential reagents including the various RNA silencing suppressors and revised the manuscript. BM obtained the funding, conceived and designed the project, and revised and finalized the manuscript.
Funding
This project was funded by the Discovery Grant Program of the Natural Sciences Engineering Research Council of Canada, projects RGPIN-2014-05306 and RGPIN-2020-04718 was awarded to BM.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We thank M. Mucci for the help with the greenhouse and growth chambers facility, Foundation Plant Services at UC-Davis California for providing virus-free grapevine cuttings to establish our tissue culture plantlets.
Conflicts of Interest
No conflict of interest in any form exists in relation to this publication.
Consent for publication
All authors have consented for the publication of this manuscript.
References
- Martelli, G.P. An overview on grapevine viruses, viroids, and the diseases they cause. Grapevine viruses: molecular biology, diagnostics and management edited by Meng, B., Martelli, G.P., Golino, D.A., & Fuchs, M. Springer International Publishing AG, Cham, Switzerland. 2017, Pages 31-46. [CrossRef]
- Martelli, G.P. Where Grapevine Virology Is Heading To. In Proceedings of the 19th Congress of ICVG, Santiago, Chile. 2018, Pages. 10–15.
- Fuchs, M. Grapevine viruses: A multitude of diverse species with simple but overall, poorly adopted management solutions in the vineyard. J. Plant Pathol. 2020, 102, 643–653. [Google Scholar] [CrossRef]
- Crnogorac, A.; Panno, S.; Mandić, A.; Gasˇpar, M.; Caruso, A.G.; Noris, E.; Davino, S.; Maltic, S. Survey of five major grapevine viruses infecting Blatina and Zˇilavka cultivars in Bosnia and Herzegovina. PLoS ONE, 2021, 16(1): e0245959. [CrossRef]
- Reynolds, A. The Grapevine, Viticulture, and Winemaking: A Brief Introduction. Grapevine viruses: molecular biology, diagnostics and management edited by Meng, B., Martelli, G.P., Golino, D.A., & Fuchs, M. Springer International Publishing AG, Cham, Switzerland. 2017, Pages 3-31. [CrossRef]
- Saldarelli, P.; Giampetruzzi, A.; Maree, H.J.; Rwahnih, A. High throughput sequencing: advantages beyond virus identification edited by Meng, B., Martelli, G.P., Golino, D.A., & Fuchs, M. Springer International Publishing AG, Cham, Switzerland. 2017, Pages 625-642. [CrossRef]
- Prosser, S.W.; Goszczynski, D.E.; Meng, B. Molecular analysis of double-stranded RNAs reveals complex infection of grapevines with multiple viruses. Virus Res. 2007, 124, 151–159. [Google Scholar] [CrossRef]
- Martelli, G.P. Directory of virus and virus-like diseases of the grapevine and their agents. J. Plant Pathol. 2014, 96, 1–136. [Google Scholar]
- Dolja, V.V.; Kreuze, K.F.; Valkonen, J.P.T. Comparative and functional genomics of closteroviruses. Virus Res. 2006, 117(1), 38–51. [Google Scholar] [CrossRef]
- Maree, H.J.; Almeida, R.P.P.; Bester, R.; Chooi, K.M.; Cohen, D.; Dolja, V.V.; Fuchs, M.F.; Golino, D.A.; Jooste, A.E.C.; Martelli, G.P.; Naidu, R.A.; Rowhani, A.; Saldarelli, P.; Burger, J.T. Grapevine leafroll-associated virus-3. Front. Microbiol. 2013, 4, 1–21. [Google Scholar] [CrossRef]
- Naidu, R.; Rowhani, A.; Fuchs, M.; Golino, D.; Martelli, G.P. Grapevine leafroll: A complex viral disease affecting a high-value fruit crop. Plant Dis. 2014, 98, 1172–1185. [Google Scholar] [CrossRef] [PubMed]
- Maliogka, V.I.; Martelli, G.P.; Fuchs, M.; Katis, N.I. Control of viruses infecting grapevine. Adv. Virus Res. 2015, 91, 175–227. [Google Scholar] [CrossRef]
- Pugachev, K.V.; Abernathy, E.S.; Frey, T.K. Improvement of the specific infectivity of the Rubella virus (RUB) infectious clone: Determinants of cytopathogenicity induced by RUB map to the nonstructural proteins. J. Virol. 1997, 71, 562–568. [Google Scholar] [CrossRef]
- Muruganantham, M.; Moskovitz, Y.; Haviv, S.; Horesh, T.; Fenigstein, A.; Preez, J.; Stephan, D.; Burger, J.T.; Mawassi, M. Grapevine virus A-mediated gene silencing in Nicotiana benthamiana and Vitis vinifera. J. Virol. Methods. 2009, 155, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Kurth, E.G.; Peremyslov, V.V.; Prokhnevsky, A.I.; Kasschau, K.D.; Miller, M.; Carrington, J.C.; Dolja, V.V. Virus-derived gene expression and RNA interference vector for grapevine. Virol. 2012, 86, 6002–6009. [Google Scholar] [CrossRef]
- Meng, B.; Venkataraman, S.; Li, C.; Wanga, W.; Dayan-Glick, C.; Mawassi, M. Construction and biological activities of the first infectious cDNA clones of the genus Foveavirus. Virol. 2013, 435, 453–462. [Google Scholar] [CrossRef]
- Lovato, A.; Faoro, F.; Gambino, G.; Maffi, D.; Bracale, M.; Polverari, A.; Santi, L. Construction of a synthetic infectious cDNA clone of Grapevine Algerian latent virus (GALV-Nf) and its biological activity in Nicotiana benthamiana and grapevine plants. Virol. J. 2014, 11, 186. [Google Scholar] [CrossRef] [PubMed]
- Yepes, L.M.; Cieniewicz, E.; Krenz, B.; McLane, H.; Thompson, J.R.; Perry, K.L.; Fuchs, M. Causative Role of Grapevine Red Blotch Virus in Red Blotch Disease. Phythopathol. 2018, 108(7), 902–909. [Google Scholar] [CrossRef]
- Jarugula, S.; Gowda, S.; Dawson, W.O.; Naidu, R. Development of infectious cDNA clones of Grapevine leafroll-associated virus 3 and analyses of the 5′ non-translated region for replication and virion formation. Virol. 2018, 523, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Tarquini, G.; Zaina, G.; Ermacora, P.; Amicis, F.D.; Franco-Orozco, B.; Loi, N.; Martini, M.; Bianchi, G.L.; Pagliari, L.; Firrao, G.; Paoli, E.; Musetti, R. Agroinoculation of Grapevine Pinot gris virus in tobacco and grapevine provides insights on viral pathogenesis. PLOS ONE. 2019. [Google Scholar] [CrossRef]
- Turpen, T.H.; Turpen, A.M.; Weinzettl, N.; Kumagai, M.H.; Dawson, W.O. Transfection of whole plants from wounds inoculated with Agrobacterium tumefaciens containing cDNA of Tobacco mosaic virus. J. Virol. Methods. 1993, 42, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, H.B. Rapid delivery of foreign genes into plants by direct rub-inoculation wh intact plasmid DNA of a Tomato bushy stunt virus gene vector. J. Virol. 1999, 73, 7823–7829. [Google Scholar] [CrossRef]
- Ratcliff, F.; Martin-Hernandez, A.M.; Baulcombe, D.C. Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001, 25, 237–245. [Google Scholar]
- Du, J.; Rietman, H.; Vleeshouwers, V.G. Agroinfiltration and PVX Agroinfection in Potato and Nicotiana benthamiana. J. Vis. Exp. 2014, 83, e50971. [Google Scholar] [CrossRef]
- Ryu, C.M.; Anand, A.; Kang, L.; Mysore, K.S. Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J. 2004, 40, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jelkmann, W. Construction of Full-length Infectious cDNA Clones of Apple chlorotic leaf spot virus and Their Agroinoculation to Woody Plants by a Novel Method of Vacuum Infiltration. Plant Dis. 2017, 101, 2110–2115. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Shabanian, M.; Moore, C.; Li, C.; Meng, B. Survey for major viruses in commercial Vitis vinifera wine grapes in Ontario. Virol. J. 2018, 15, 127. [Google Scholar] [CrossRef] [PubMed]
- Shan, F.; and Seaton, K. Semi-sterilized tissue culture for rapid propagation of grapevines (Vitis vinifera L.) using immature cuttings. Hortscience 2014, 49(7), 949–954. [Google Scholar] [CrossRef]
- Zottini, M.; Barizza, E.; Costa, A.; Formentin, E.; Ruberti, C.; Carimi, F.; Lo Schiavo, F. Agroinfiltration of grapevine leaves for fast transient assays of gene expression and for long-term production of stable transformed cells. Plant Cell Rep. 2008, 27, 845–85. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Kim, W.S.; Meng, B. A highly effective and versatile technology for the isolation of RNAs from grapevines and other woody perennials for use in virus diagnostics. Virol. J. 2015a, 12, 1–15. [Google Scholar] [CrossRef]
- Shabanian, M.; Xiao, H.; Meng, B. Seasonal dynamics and tissue distribution of two major viruses associated with grapevine leafroll under cool climate conditions. Eur. J. Plant Pathol. 2020. [Google Scholar] [CrossRef]
- Liu, Y.P.; Peremyslov, V.V.; Medina, V.; and Dolja, V.V. Tandem leader proteases of grapevine leafroll-associated virus-2: host-specific functions in the infection cycle. Virol. 2009, 383, 291–299. [Google Scholar] [CrossRef]
- Mikona, C.; Jelkmann, W. Replication of Grapevine leafroll-associated virus-7 (GLRaV-7) by Cuscuta species and its transmission to herbaceous plants. Plant Dis. 2010, 94(4), 471–6. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.G.; Fuchs, M. Herbaceous plant hosts as supermodels for grapevine viruses: a historical perspective. J. Plant Pathol. 2022, 27, 1–30. [Google Scholar] [CrossRef]
- Grimsley, N.; Hohn, B.; Hohn, T.; Walden, R. “Agroinfection,” an alternative route for viral infection of plants by using the Ti plasmid. Proc. Natl. Acad. Sci. U.S.A. 1986, 83, 3282–3286. [Google Scholar] [CrossRef] [PubMed]
- Chiba, M.; Reed, J.C.; Prokhnevsky, A.I.; Chapman, E.J.; Mawassi, M.; Koonin, E.V.; Carrington, J.C.; Dolja, V.V. Diverse suppressors of RNA silencing enhance agroinfection by a viral replicon. Virol. 2006, 346(1), 7–14. [Google Scholar] [CrossRef]
- Krenz, B.; Thompson, J.R.; McLane, H.L.; Fuchs, M.; Perry, K.L. Grapevine red blotch-associated virus is widespread in the United States. Phytopathol. 2012, 104(11), 1232–40. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Establishment of grapevine tissue culture from grapevine cuttings. This involves five stages: 1) preparation of softwood cuttings; 2) micro-propagation of shoot tips in glass tubes; 3) shoot development; 4) shoot proliferation and elongation; and 5) rooting. The entire process takes about 4 months.
Figure 1.
Establishment of grapevine tissue culture from grapevine cuttings. This involves five stages: 1) preparation of softwood cuttings; 2) micro-propagation of shoot tips in glass tubes; 3) shoot development; 4) shoot proliferation and elongation; and 5) rooting. The entire process takes about 4 months.
Figure 2.
Preparation of grapevine plantlets and vacuum-mediated agro-infiltration. Grapevine plantlets, 8-11-week-old, generated from tissue culture were removed from magenta boxes and placed on a bench (a), trimmed with a pair of scissors to remove some of the leaves and roots (b), and pierced with a 31-gauge needle to create minor wounds along the stem and major roots (c).
Figure 2.
Preparation of grapevine plantlets and vacuum-mediated agro-infiltration. Grapevine plantlets, 8-11-week-old, generated from tissue culture were removed from magenta boxes and placed on a bench (a), trimmed with a pair of scissors to remove some of the leaves and roots (b), and pierced with a 31-gauge needle to create minor wounds along the stem and major roots (c).
Figure 3.
Transplantation, recovery and establishment of grapevine plantlets in soil. Infiltrated plantlets were gently transplanted individually in 2.5-inch round plastic pots, several of which were then placed in a 3-gallon plastic pot followed by covering with transparent plastic sheets (a). These covered pots were kept in a growth room at 21-22° C with 16/8 hrs photo period for 3-4 weeks. To gradually expose the plantlets to the conditions in the growth room, small holes were poked using a pen on each plastic cover after 2-3 weeks post infiltration (b). These holes were gradually enlarged to allow more air exchange (c). When the plantlets were fully established in soil, plastic covers were removed (d).
Figure 3.
Transplantation, recovery and establishment of grapevine plantlets in soil. Infiltrated plantlets were gently transplanted individually in 2.5-inch round plastic pots, several of which were then placed in a 3-gallon plastic pot followed by covering with transparent plastic sheets (a). These covered pots were kept in a growth room at 21-22° C with 16/8 hrs photo period for 3-4 weeks. To gradually expose the plantlets to the conditions in the growth room, small holes were poked using a pen on each plastic cover after 2-3 weeks post infiltration (b). These holes were gradually enlarged to allow more air exchange (c). When the plantlets were fully established in soil, plastic covers were removed (d).
Figure 4.
A schematic overview of different methods that were used to demonstrate infectivity in grapevines after infiltration with viral infectious clone. CP: viral capsid protein; RT-PCR: reverse transcription-polymerase chain reaction; qPCR: quantitative PCR; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Figure 4.
A schematic overview of different methods that were used to demonstrate infectivity in grapevines after infiltration with viral infectious clone. CP: viral capsid protein; RT-PCR: reverse transcription-polymerase chain reaction; qPCR: quantitative PCR; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Figure 5.
Effects of RNA silencing suppressor (p24) on infection rate and viral titer in grapevines after inoculation with GLRaV-3 full-length clone through agro-infiltration. Results of RT-qPCR using primers F1-14117 & R-14327 of grapevines infiltrated with GLRV-3 clone alone (blue line) or co-infiltrated with GLRaV-3 clone and RNA silencing suppressor p24 (red line).
Figure 5.
Effects of RNA silencing suppressor (p24) on infection rate and viral titer in grapevines after inoculation with GLRaV-3 full-length clone through agro-infiltration. Results of RT-qPCR using primers F1-14117 & R-14327 of grapevines infiltrated with GLRV-3 clone alone (blue line) or co-infiltrated with GLRaV-3 clone and RNA silencing suppressor p24 (red line).
Figure 6.
Effects of dormancy treatment on plantlets survival and infection rate. (a) The number of plantlets that survived or tested positive for GLRaV-3 after going through a single or two cycles of dormancy treatment at 4 oC as compared to those in the no dormancy treatment group. (b) percentage of Syrah or Cabernet franc plants that tested positive for GLRaV-3 prior to dormancy treatment as compared to after dormancy treatment. Note, a single dormancy treatment resulted in a significant increase in infectivity rate while double dormancy treatment decreased the percentage of plants infected by GLRaV-3. .
Figure 6.
Effects of dormancy treatment on plantlets survival and infection rate. (a) The number of plantlets that survived or tested positive for GLRaV-3 after going through a single or two cycles of dormancy treatment at 4 oC as compared to those in the no dormancy treatment group. (b) percentage of Syrah or Cabernet franc plants that tested positive for GLRaV-3 prior to dormancy treatment as compared to after dormancy treatment. Note, a single dormancy treatment resulted in a significant increase in infectivity rate while double dormancy treatment decreased the percentage of plants infected by GLRaV-3. .
Figure 7.
Effects of dormancy treatments on infection rate and viral titer in grapevines after inoculation with GLRaV-3 full-length clone through agro-infiltration. Results of first round PCR amplification using external primers F1-16289 & R-17240 (a) and second round PCR with internal primer set F1-16896 & R-17240 (b) at 4 mpi of grapevine plantlets infiltrated with both GLRV-3 cDNA clone and p24 prior to dormancy. M: molecular size marker; P1: agrobacterial cells containing GLRaV-3 full-length cDNA clone; P2: Plasmid DNA containing full-length cDNA clone of GLRaV-3; N: grapevine plantlet mock-infiltrated with buffer; Lanes 1-9: samples collected from individual infiltrated grapevines. (c) and (d): Detection of GLRaV-3 using nested RT-PCR in grapevines that were co-infiltrated with GLRaV-3 full-length clone and p24. Grapevine plants were subjected to a single cycle dormancy treatment. (c): Results of first round RT-PCR using primers F1-16289 & R-17240; (d): results of second round PCR using primers F1-16896 & R-17240. M: molecular size marker; P1: agrobacterial cells containing GLRaV-3 full-length cDNA clone; P2: Plasmid DNA containing full-length cDNA clone of GLRaV-3; N: Cabernet franc vine mock-infiltrated with buffer; Lanes 1-8: samples collected from individual infiltrated grapevine of Cabernet franc at a given time.
Figure 7.
Effects of dormancy treatments on infection rate and viral titer in grapevines after inoculation with GLRaV-3 full-length clone through agro-infiltration. Results of first round PCR amplification using external primers F1-16289 & R-17240 (a) and second round PCR with internal primer set F1-16896 & R-17240 (b) at 4 mpi of grapevine plantlets infiltrated with both GLRV-3 cDNA clone and p24 prior to dormancy. M: molecular size marker; P1: agrobacterial cells containing GLRaV-3 full-length cDNA clone; P2: Plasmid DNA containing full-length cDNA clone of GLRaV-3; N: grapevine plantlet mock-infiltrated with buffer; Lanes 1-9: samples collected from individual infiltrated grapevines. (c) and (d): Detection of GLRaV-3 using nested RT-PCR in grapevines that were co-infiltrated with GLRaV-3 full-length clone and p24. Grapevine plants were subjected to a single cycle dormancy treatment. (c): Results of first round RT-PCR using primers F1-16289 & R-17240; (d): results of second round PCR using primers F1-16896 & R-17240. M: molecular size marker; P1: agrobacterial cells containing GLRaV-3 full-length cDNA clone; P2: Plasmid DNA containing full-length cDNA clone of GLRaV-3; N: Cabernet franc vine mock-infiltrated with buffer; Lanes 1-8: samples collected from individual infiltrated grapevine of Cabernet franc at a given time.
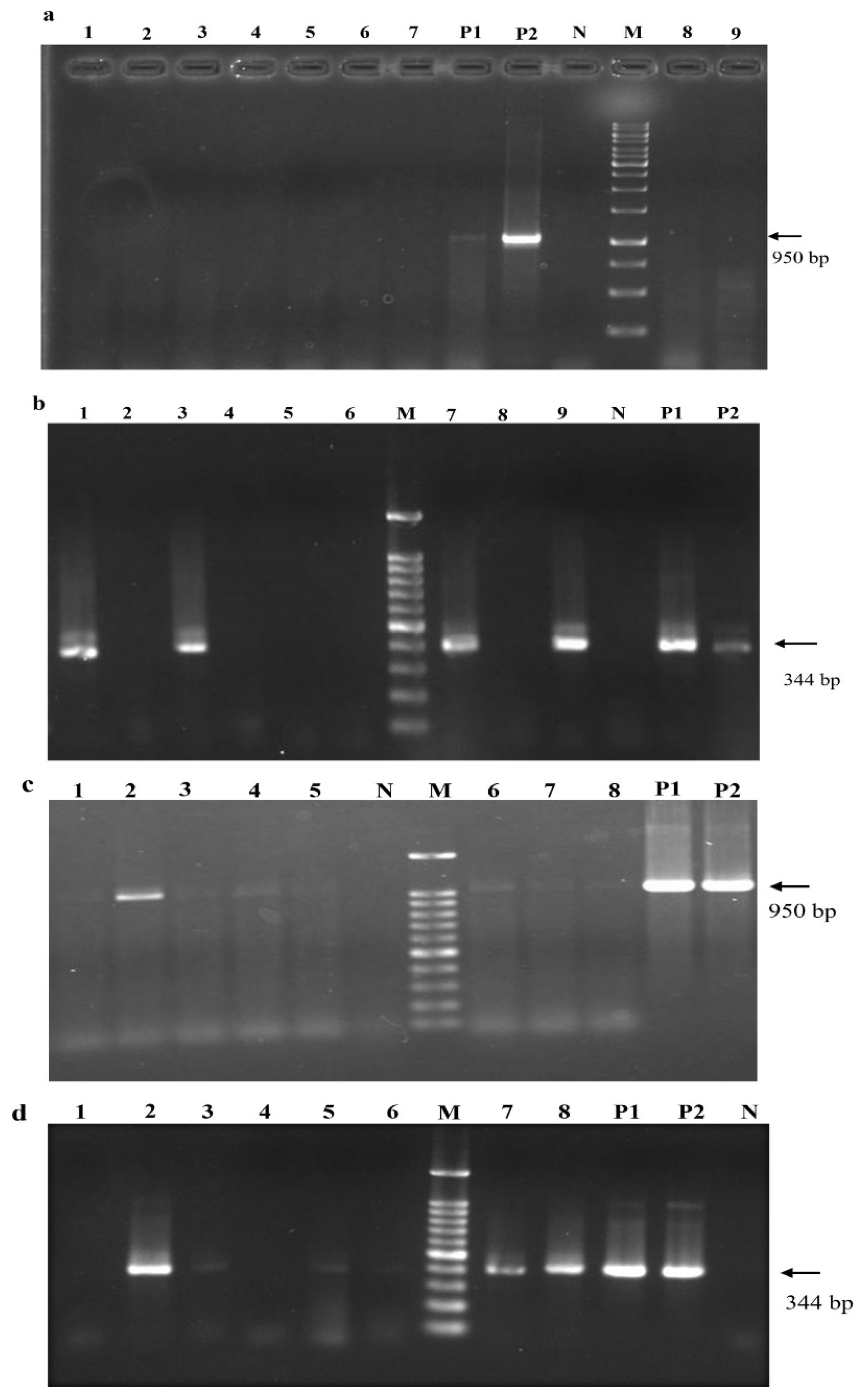
Figure 8.
(a): Trends in GLRaV-3 titer as judged by RT-qPCR testing grapevine co-infiltrated with viral infectious clone and p24 that were subjected to a single cycle dormancy treatment (red line) as compared to those that did not go through dormancy treatment (blue line). Primers F1-14117 & R-14327 were used in RT-qPCR. Note that at 12 mpi, the titer of GLRaV-3 was significantly higher after two months dormancy.
(b): The results of Western blotting for the capsid protein in leaf samples of Cabernet franc grapevines. Lanes 1, 2, and 3: leaf samples from grapevines infiltrated with pLR-3 full-length cDNA clone which tested positive for GLRaV-3 nested-RT-PCR. Lanes 4 & 5: leaf samples collected in October from field grapevines infected with GLRaV-3. These field samples, which were used previously to study the seasonal dynamics of GLRaV-2 and GLRaV-3 [
31], were included as positive controls. N: Grapevine infiltrated with agrobacteria containing p24 as a negative control.
Figure 8.
(a): Trends in GLRaV-3 titer as judged by RT-qPCR testing grapevine co-infiltrated with viral infectious clone and p24 that were subjected to a single cycle dormancy treatment (red line) as compared to those that did not go through dormancy treatment (blue line). Primers F1-14117 & R-14327 were used in RT-qPCR. Note that at 12 mpi, the titer of GLRaV-3 was significantly higher after two months dormancy.
(b): The results of Western blotting for the capsid protein in leaf samples of Cabernet franc grapevines. Lanes 1, 2, and 3: leaf samples from grapevines infiltrated with pLR-3 full-length cDNA clone which tested positive for GLRaV-3 nested-RT-PCR. Lanes 4 & 5: leaf samples collected in October from field grapevines infected with GLRaV-3. These field samples, which were used previously to study the seasonal dynamics of GLRaV-2 and GLRaV-3 [
31], were included as positive controls. N: Grapevine infiltrated with agrobacteria containing p24 as a negative control.
Table 1.
Effects of different agro-infiltration methods on grapevine plantlets survival and infection rates. The status of infection was based on nested RT-PCR at 6months post inoculation (mpi). SY: Syrah; CF: Cabernet franc.
Table 1.
Effects of different agro-infiltration methods on grapevine plantlets survival and infection rates. The status of infection was based on nested RT-PCR at 6months post inoculation (mpi). SY: Syrah; CF: Cabernet franc.
Inoculation
methods
|
Vacuum-based
agro-infiltration
|
Agro-pricking |
Agro-drenching |
Agro-injection |
| Cultivar of plantlets |
SY |
CF |
SY |
CF |
SY |
CF |
SY |
CF |
| No. of infiltrated plants |
50 |
50 |
50 |
50 |
50 |
50 |
50 |
50 |
| No. of survived ones |
39 |
38 |
43 |
42 |
46 |
45 |
44 |
43 |
| No. of infected plantlets |
22 |
20 |
7 |
6 |
0 |
0 |
0 |
0 |
Percent of infection of
survivor plants |
56% |
52% |
16% |
14% |
0% |
0% |
0% |
0% |
Table 2.
Effects of age and cultivars of grapevine plantlets on survival and infection rates. The status of infection was based on nested RT-PCR at 4mpi. SY: Syrah, CF: Cabernet franc; CH: Chardonnay.
Table 2.
Effects of age and cultivars of grapevine plantlets on survival and infection rates. The status of infection was based on nested RT-PCR at 4mpi. SY: Syrah, CF: Cabernet franc; CH: Chardonnay.
| Age of plantlets |
5-7 weeks old |
8-11 weeks old |
12-16 weeks old |
| Cultivar of plantlets |
SY |
CF |
CH |
SY |
CF |
CH |
SY |
CF |
CH |
| No. of infiltrated plants |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
| No. of survived ones |
25 |
21 |
18 |
82 |
78 |
65 |
92 |
88 |
76 |
| No. of infected plantlets |
14 |
11 |
8 |
52 |
46 |
29 |
13 |
10 |
5 |
| Percent of infection of survivor plants |
56 |
52 |
44 |
63 |
58 |
44 |
14 |
11 |
7 |
Table 3.
Effects of humidity control on survival and infection rates of Syrah plantlets after vacuum-based agro-infiltration. Status of infection was assessed by using nested RT-PCR at 4 mpi.
Table 3.
Effects of humidity control on survival and infection rates of Syrah plantlets after vacuum-based agro-infiltration. Status of infection was assessed by using nested RT-PCR at 4 mpi.
| Time of cover removal |
1 week |
2 weeks |
3 weeks |
| Manner of cover removal |
Instant |
Gradual |
Instant |
Gradual |
Instant |
Gradual |
| No. of infiltrated plants |
100 |
100 |
100 |
100 |
100 |
100 |
| No. of survived plants |
0 |
6 |
2 |
18 |
36 |
78 |
| No. of infected plants |
0 |
0 |
0 |
4 |
11 |
51 |
| Percent of infection of survivor plants |
0% |
0% |
0% |
22% |
30% |
65% |
Table 6.
Effects of dormancy treatment on survival and infection rates of grapevine plantlets that were infiltrated with full-length infectious clone of GLRaV-3. Plantlets were subjected to either a single dormancy treatment for two months or two cycles of dormancy treatment each of two months. Status of GLRaV-3 infection was assessed by nested-RT-PCR.
Table 6.
Effects of dormancy treatment on survival and infection rates of grapevine plantlets that were infiltrated with full-length infectious clone of GLRaV-3. Plantlets were subjected to either a single dormancy treatment for two months or two cycles of dormancy treatment each of two months. Status of GLRaV-3 infection was assessed by nested-RT-PCR.
|
Duration of dormancy at 4° C
|
No dormancy treatment |
Single dormancy |
Double dormancy |
| Cultivar of plantlets |
SY |
CF |
SY |
CF |
SY |
CF |
| Total no. of infiltrated plants |
50 |
50 |
50 |
50 |
50 |
50 |
| No. of plants that survived dormancy treatment |
50 |
50 |
47 |
45 |
29 |
27 |
| No. of infected plants prior to dormancy |
25 |
25 |
25 |
25 |
25 |
25 |
| No. of plants tested positive after dormancy |
26 |
27 |
32 |
33 |
11 |
9 |
| Percentage of infection of survivor plants |
52% |
54% |
68% |
73% |
37% |
33% |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).