Submitted:
09 October 2023
Posted:
11 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Parental materials and F1 progeny used to generate the F1DH population
2.2. Development of the F1DH population using microspore culture
2.3. Flow cytometry analysis
2.4. Plant materials for genetic map construction and DNA extraction
2.5. Sequencing library construction and high-throughput resequencing
2.6. SNP identification and genotyping
2.7. Genetic map construction and evaluation
2.8. Relationship between the genetic and physical maps
3. Results
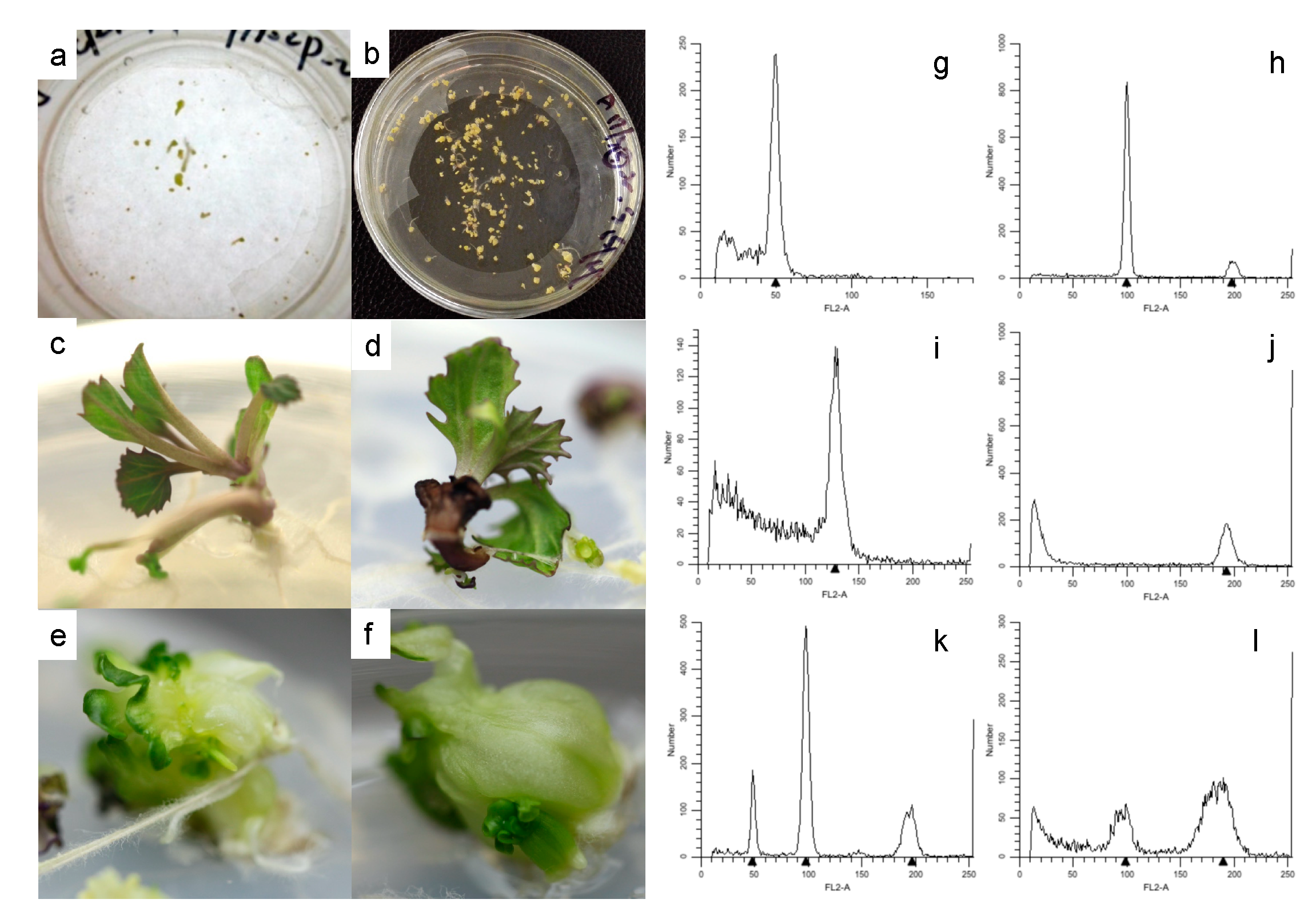
3.1. Microspore embryogenesis of ornamental kale F1 progeny
3.2. Ploidy level and spontaneous chromosome doubling
3.3. Construction of the F1DH mapping population
3.4. Whole-genome resequencing of the F1DH mapping population and two parents
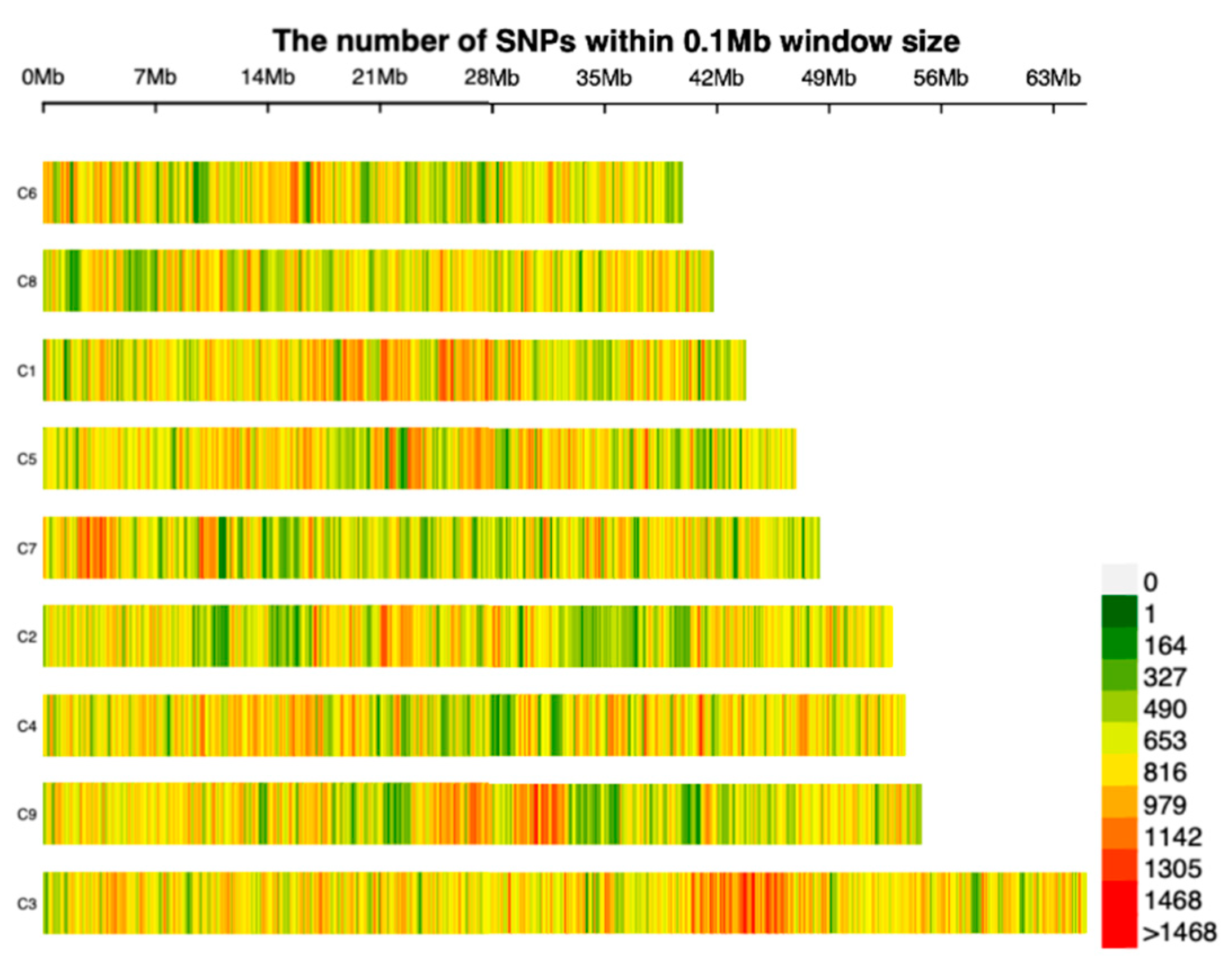
3.5. SNP marker identification
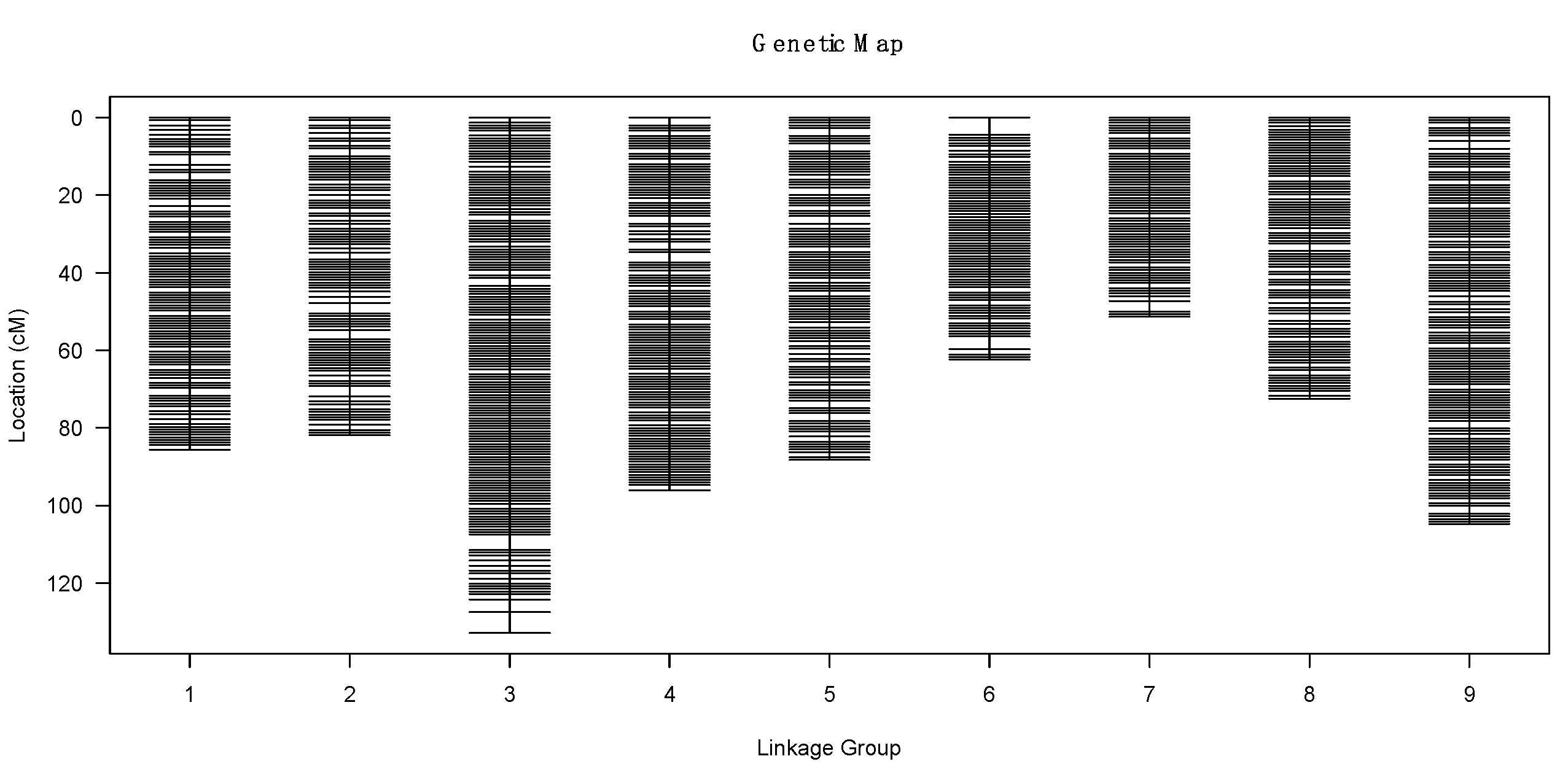
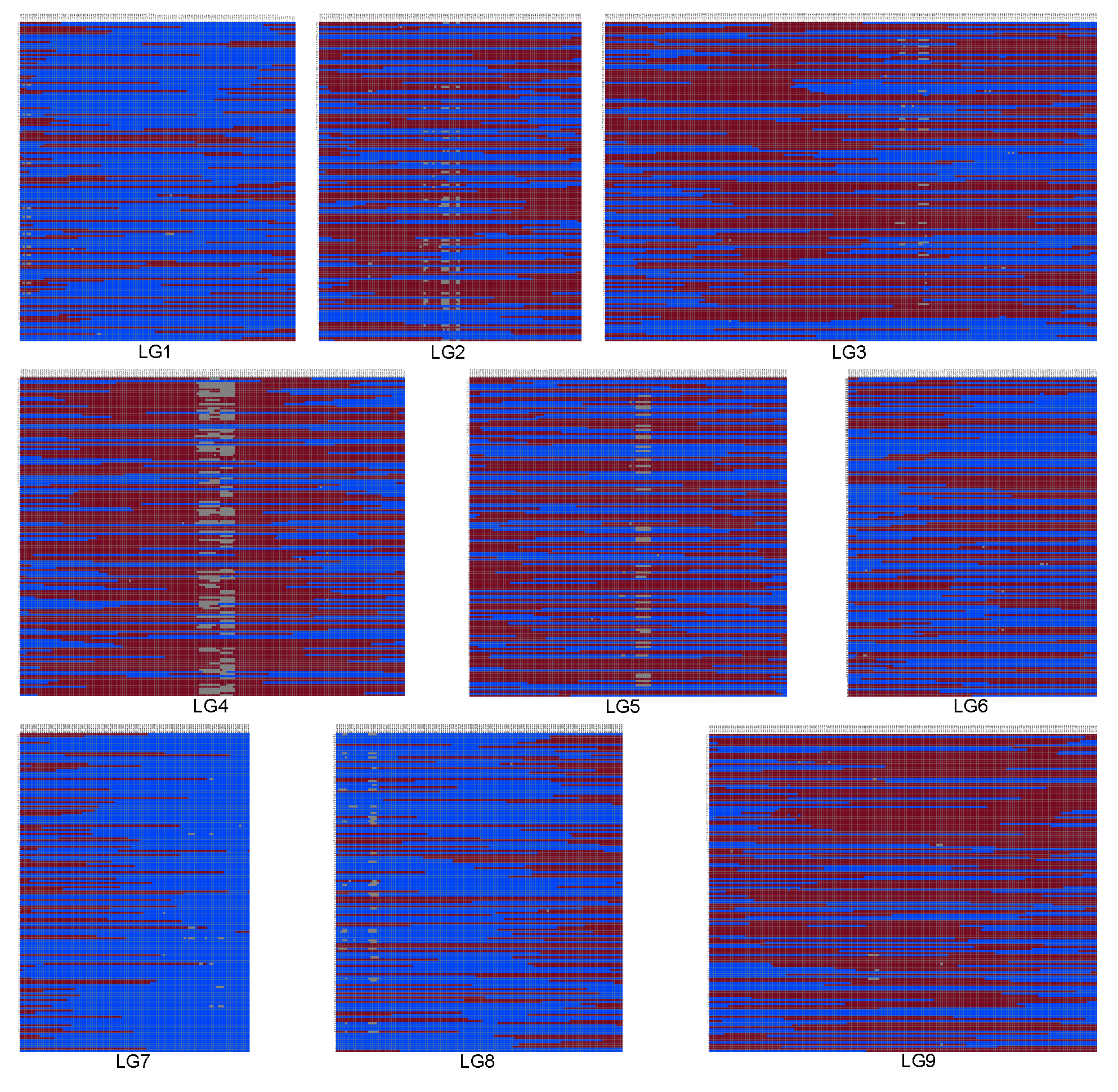
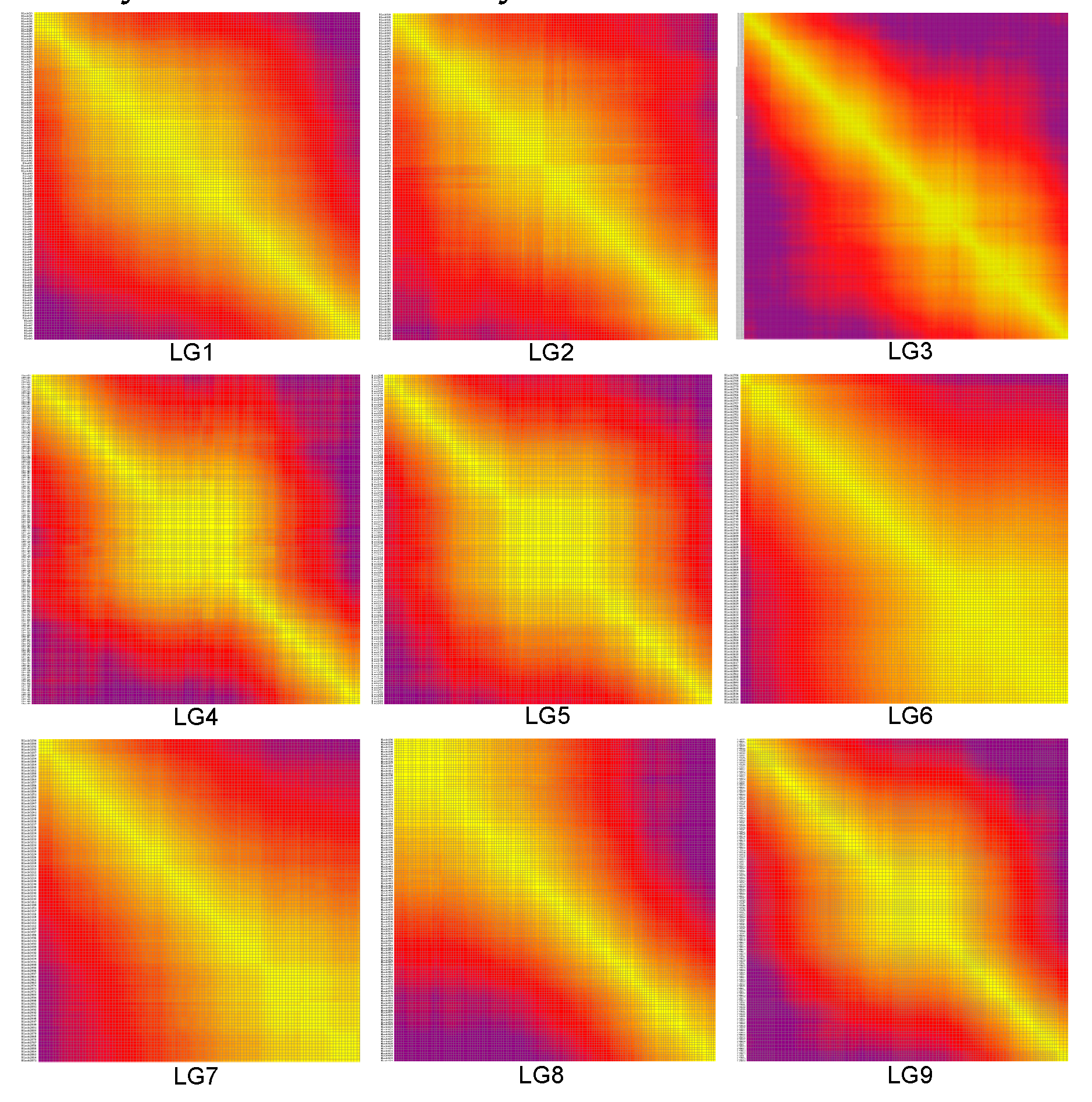
3.6. Construction of the high-density bin genetic linkage map
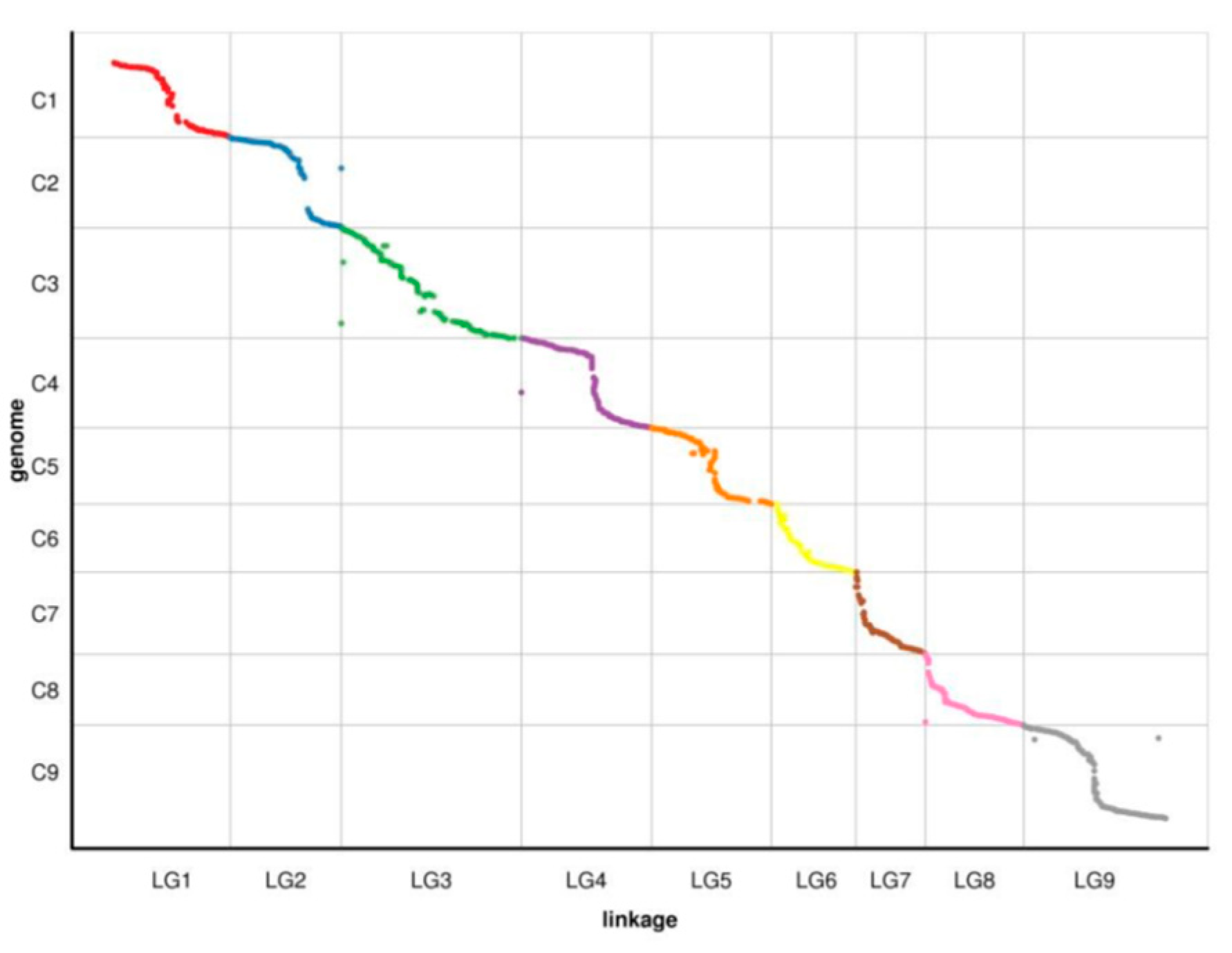
3.7. Collinearity of the genetic and physical maps
4. Discussion
4.1. Development of DH lines is an effective method for germplasm innovation of ornamental kale
4.2. The F1DH population will play important roles in genetic analysis of ornamental kale
4.3. Features of the high-density genetic map of ornamental kale
4.4. The application prospects of the high-density genetic map of ornamental kale
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.; Yu, X. Pollination with laser-irradiated pollens breaks cross-incompatibility between zicaitai (Brassica campestris var. purpurea) and ornamental kale (Brassica oleracea var. acephala) to produce hybrids with the aid of ovule culture. Scientia horticulturae 2006, 108, 397–402. [Google Scholar]
- Forster, B.P.; Heberle-Bors, E.; Kasha, K.J.; Touraev, A. The resurgence of haploids in higher plants. Trends in plant science 2007, 12, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M. Haploids in flowering plants: origins and exploitation. Plant biotechnology journal 2010, 8, 377–424. [Google Scholar] [CrossRef] [PubMed]
- Forster, B.P.; Thomas, W.T. Doubled haploids in genetics and plant breeding. Plant breeding reviews 2010, 25, 57–88. [Google Scholar]
- Ferrie, A.; Caswell, K. Isolated microspore culture techniques and recent progress for haploid and doubled haploid plant production. Plant Cell, Tissue and Organ Culture (PCTOC) 2011, 104, 301–309. [Google Scholar] [CrossRef]
- Zhang, W.; Fu, Q.; Dai, X.; Bao, M. The culture of isolated microspores of ornamental kale (Brassica oleracea var. acephala) and the importance of genotype to embryo regeneration. Scientia Horticulturae 2008, 117, 69–72. [Google Scholar]
- Yuan, S.; Liu, Y.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Sun, P. Effect of combined cold pretreatment and heat shock on microspore cultures in broccoli. Plant Breeding 2011, 130, 80–85. [Google Scholar] [CrossRef]
- Lv, H.; Wang, Q.; Yang, L.; Fang, Z.; Liu, Y.; Zhuang, M.; Zhang, Y.; Yang, Y.; Xie, B.; Wang, X. :. Breeding of cabbage (Brassica oleracea L. var. capitata) with fusarium wilt resistance based on microspore culture and marker-assisted selection. Euphytica 2014, 200, 465–473. [Google Scholar]
- Lou, P.; Zhao, J.; He, H.; Hanhart, C.; Pino Del Carpio, D.; Verkerk, R.; Custers, J.; Koornneef, M.; Bonnema, G. Quantitative trait loci for glucosinolate accumulation in Brassica rapa leaves. New phytologist 2008, 179, 1017–1032. [Google Scholar] [CrossRef]
- Hirani, A.H.; Li, G. Genetic Mapping, Quantitative Trait Analysis, and Gene Cloning in Brassica oleracea. In: The Brassica oleracea Genome. Springer; 2021: 7-22.
- Gao, M.; Li, G.; Yang, B.; Qiu, D.; Farnham, M.; Quiros, C. High-density Brassica oleracea linkage map: identification of useful new linkages. Theoretical and applied genetics 2007, 115, 277–287. [Google Scholar] [CrossRef]
- Wang, W.; Huang, S.; Liu, Y.; Fang, Z.; Yang, L.; Hua, W.; Yuan, S.; Liu, S.; Sun, J.; Zhuang, M. Construction and analysis of a high-density genetic linkage map in cabbage (Brassica oleracea L. var. capitata). BMC genomics 2012, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jaganathan, D.; Bohra, A.; Thudi, M.; Varshney, R.K. Fine mapping and gene cloning in the post-NGS era: advances and prospects. Theoretical and Applied Genetics 2020, 133, 1791–1810. [Google Scholar] [CrossRef] [PubMed]
- Parkin, I.A.; Koh, C.; Tang, H.; Robinson, S.J.; Kagale, S.; Clarke, W.E.; Town, C.D.; Nixon, J.; Krishnakumar, V.; Bidwell, S.L. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome biology 2014, 15, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 2014, 5, 3930. [Google Scholar] [CrossRef]
- Cai, X.; Wu, J.; Liang, J.; Lin, R.; Zhang, K.; Cheng, F.; Wang, X. Improved Brassica oleracea JZS assembly reveals significant changing of LTR-RT dynamics in different morphotypes. Theoretical and Applied Genetics 2020, 133, 3187–3199. [Google Scholar] [CrossRef]
- Lv, H.; Wang, Y.; Han, F.; Ji, J.; Fang, Z.; Zhuang, M.; Li, Z.; Zhang, Y.; Yang, L. A high-quality reference genome for cabbage obtained with SMRT reveals novel genomic features and evolutionary characteristics. Scientific reports 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Guo, N.; Wang, S.; Gao, L.; Liu, Y.; Wang, X.; Lai, E.; Duan, M.; Wang, G.; Li, J.; Yang, M. Genome sequencing sheds light on the contribution of structural variants to Brassica oleracea diversification. BMC biology 2021, 19, 1–15. [Google Scholar] [CrossRef]
- Sun, D.; Wang, C.; Zhang, X.; Zhang, W.; Jiang, H.; Yao, X.; Liu, L.; Wen, Z.; Niu, G.; Shan, X. Draft genome sequence of cauliflower (Brassica oleracea L. var. botrytis) provides new insights into the C genome in Brassica species. Horticulture research 2019, 6. [Google Scholar] [CrossRef]
- Belser, C.; Istace, B.; Denis, E.; Dubarry, M.; Baurens, F.-C.; Falentin, C.; Genete, M.; Berrabah, W.; Chèvre, A.-M.; Delourme, R. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nature plants 2018, 4, 879–887. [Google Scholar] [CrossRef]
- Lee, J.; Izzah, N.K.; Choi, B.-S.; Joh, H.J.; Lee, S.-C.; Perumal, S.; Seo, J.; Ahn, K.; Jo, E.J.; Choi, G.J. Genotyping-by-sequencing map permits identification of clubroot resistance QTLs and revision of the reference genome assembly in cabbage (Brassica oleracea L.). DNA research 2016, 23, 29–41. [Google Scholar] [CrossRef]
- Zhao, Z.; Gu, H.; Sheng, X.; Yu, H.; Wang, J.; Huang, L.; Wang, D. Genome-wide single-nucleotide polymorphisms discovery and high-density genetic map construction in cauliflower using specific-locus amplified fragment sequencing. Frontiers in plant science 2016, 7, 334. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wang, J.; Zhao, Z.; Sheng, X.; Shen, Y.; Branca, F.; Gu, H. Construction of a high-density genetic map and identification of loci related to hollow stem trait in broccoli (Brassic oleracea L. italica). Frontiers in plant science 2019, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Custers, J.; Cordewener, J.; Fiers, M.; Maassen, B.; van Lookeren Campagne, M.; Liu, C. Androgenesis in Brassica: a model system to study the initiation of plant embryogenesis. Current trends in the embryology of angiosperms 2001, 451–470. [Google Scholar]
- Lv H-h Wang Q-b Yang L-m Fang Z-y Liu Y-m Zhuang, M.; Zhang Y-y Yang Y-h Xie B-y Wang, X.-w. Breeding of cabbage (Brassica oleracea L. var. capitata) with fusarium wilt resistance based on microspore culture and marker-assisted selection. Euphytica 2014, 200, 465–473. [Google Scholar]
- Han, S.; Guo, N.; Zhang, Y.; Zong, M.; Wang, G.; Liu, F. Researches on the double haploid breeding of ornamental kale (Brassica oleracea var. acephala). Journal of Agricultural Biotechnology 2018, 26, 521–529. [Google Scholar]
- Dpooležel, J.; Binarová, P.; Lcretti, S. Analysis of nuclear DNA content in plant cells by flow cytometry. Biologia plantarum 1989, 31, 113–120. [Google Scholar] [CrossRef]
- Murray, M.; Thompson, W. Rapid isolation of high molecular weight plant DNA. Nucleic acids research 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Reumers, J.; De Rijk, P.; Zhao, H.; Liekens, A.; Smeets, D.; Cleary, J.; Van Loo, P.; Van Den Bossche, M.; Catthoor, K.; Sabbe, B. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nature biotechnology 2012, 30, 61–68. [Google Scholar] [CrossRef]
- Liu, D.; Ma, C.; Hong, W.; Huang, L.; Liu, M.; Liu, H.; Zeng, H.; Deng, D.; Xin, H.; Song, J. Construction and analysis of high-density linkage map using high-throughput sequencing data. Plos one 2014, 9, e98855. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Feng, Q.; Qian, Q.; Zhao, Q.; Wang, L.; Wang, A.; Guan, J.; Fan, D.; Weng, Q.; Huang, T. High-throughput genotyping by whole-genome resequencing. Genome research 2009, 19, 1068–1076. [Google Scholar] [CrossRef]
- Van Os, H.; Stam, P.; Visser, R.G.; van Eck, H.J. SMOOTH: a statistical method for successful removal of genotyping errors from high-density genetic linkage data. Theoretical and applied genetics 2005, 112, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Zhang, X.; Miao, C.; Zhang, J.; Ming, R.; Schnable, J.C.; Schnable, P.S.; Lyons, E.; Lu, J. ALLMAPS: robust scaffold ordering based on multiple maps. Genome biology 2015, 16, 1–15. [Google Scholar] [CrossRef]
- Li, B.; Lu, X.; Dou, J.; Aslam, A.; Gao, L.; Zhao, S.; He, N.; Liu, W. Construction of a high-density genetic map and mapping of fruit traits in watermelon (Citrullus lanatus L.) based on whole-genome resequencing. International journal of molecular sciences 2018, 19, 3268. [Google Scholar] [CrossRef] [PubMed]
- Ferrie, A.; Caswell, K. Isolated microspore culture techniques and recent progress for haploid and doubled haploid plant production. Plant Cell, Tissue and Organ Culture (PCTOC) 2011, 104, 301–309. [Google Scholar] [CrossRef]
- Demo, B.K. Development of intervarietal substitution lines in Brassica napus L. using marker assisted selection and mapping of QTL for agronomically important traits. Göttingen, Univ., Diss 2007, 2007. [Google Scholar]
- Yadava, S.K.; Ramchiary, N. Molecular Linkage Mapping in Brassica juncea: Founding the Basis for Marker-Assisted Selection. The Brassica juncea Genome 2022, 197–219. [Google Scholar]
- Boopathi, N.M.; Boopathi, N.M. Linkage map construction. Genetic Mapping and Marker Assisted Selection: Basics, Practice and Benefits 2020, 179–227. [Google Scholar]
- Yu, H.; Wang, J.; Sheng, X.; Zhao, Z.; Shen, Y.; Branca, F.; Gu, H. Construction of a high-density genetic map and identification of loci controlling purple sepal trait of flower head in Brassica oleracea L. italica. BMC plant biology 2019, 19, 1–8. [Google Scholar] [CrossRef]
- MPerez-de-Castro, A.; Vilanova, S.; Cañizares, J.; Pascual, L.; MBlanca, J.; JDiez, M.; Prohens, J.; Picó, B. Application of genomic tools in plant breeding. Current genomics 2012, 13, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.-H.; Yang, H.-J.; Jung, K.-H.; Yoo, S.-C.; Paek, N.-C. Quantitative trait locus mapping and candidate gene analysis for plant architecture traits using whole genome re-sequencing in rice. Molecules and cells 2014, 37, 149. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zeng, L.; Tao, Y.; Vuong, T.; Wan, J.; Boerma, R.; Noe, J.; Li, Z.; Finnerty, S.; Pathan, S.M. Pinpointing genes underlying the quantitative trait loci for root-knot nematode resistance in palaeopolyploid soybean by whole genome resequencing. Proceedings of the National Academy of Sciences 2013, 110, 13469–13474. [Google Scholar] [CrossRef] [PubMed]
- Scheben, A.; Batley, J.; Edwards, D. Genotyping-by-sequencing approaches to characterize crop genomes: choosing the right tool for the right application. Plant biotechnology journal 2017, 15, 149–161. [Google Scholar] [CrossRef]
- Torkamaneh, D.; Boyle, B.; Belzile, F. Efficient genome-wide genotyping strategies and data integration in crop plants. Theoretical and Applied Genetics 2018, 131, 499–511. [Google Scholar] [CrossRef]
- Le Nguyen, K.; Grondin, A.; Courtois, B.; Gantet, P. Next-generation sequencing accelerates crop gene discovery. Trends in plant science 2019, 24, 263–274. [Google Scholar] [CrossRef]

| Sample | Total Clean Reads | Total Clean Bases | Q30 Percentage (%) | GC Percentage (%) |
|---|---|---|---|---|
| 05-DH-65 | 145,119,251 | 43,453,720,350 | 91.45 | 37.59 |
| 06-DH-71 | 72,896,717 | 21,830,253,446 | 85.59 | 36.74 |
| F1DH offspring | 694,710,016 | 208,094,995,584 | 90.88 | 37.07 |
| Total | 912,725,984 | 273,378,969,380 | 90.84 | 37.07 |
| Sample | Clean Reads | Mapped (%) | Properly mapped (%) |
|---|---|---|---|
| 05-DH-65 | 145,119,251 | 94.12 | 80.85 |
| 06-DH-71 | 72,896,717 | 92.59 | 78.45 |
| F1DH offspring (average) | 9,199,180 | 93.55 | 81.41 |
| LG ID | Total Bin Marker | Total Distance (cM) | Average Distance (cM) | Max Gap (cM) | Gaps<5cM (%) |
|---|---|---|---|---|---|
| LG1 | 156 | 85.73 | 0.55 | 2.67 | 100 |
| LG2 | 164 | 81.89 | 0.5 | 2.73 | 98.36 |
| LG3 | 272 | 132.88 | 0.49 | 5.35 | 99.73 |
| LG4 | 221 | 96.06 | 0.43 | 2.67 | 98.89 |
| LG5 | 179 | 88.25 | 0.49 | 2 | 100 |
| LG6 | 170 | 62.4 | 0.37 | 3.34 | 100 |
| LG7 | 126 | 51.34 | 0.41 | 2.67 | 100 |
| LG8 | 180 | 72.45 | 0.4 | 2 | 100 |
| LG9 | 228 | 104.81 | 0.46 | 0.02 | 100 |
| Total | 1696 | 775.81 | 0.46 | 99.66 |
| LG ID | Spearman |
|---|---|
| LG1 | 0.9987 |
| LG2 | 0.9926 |
| LG3 | 0.9778 |
| LG4 | 0.9816 |
| LG5 | 0.9842 |
| LG6 | 0.9976 |
| LG7 | 0.9981 |
| LG8 | 0.9641 |
| LG9 | 0.9803 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





