1. Introduction
Transcription in bacteria and archaea is carried out by a single multimeric RNA polymerase, while three multimeric complexes (RNA pol I, II, and III) exist in most eukaryotes [
1,
2,
3]. Furthermore, plants contain two additional RNA pols (IV and V) that have evolved from RNA pol II [
4,
5,
6,
7,
8,
9,
10,
11,
12]. RNA pol I comprises 14 subunits and synthesizes precursor rRNA 45S (35S in yeast) of the three largest rRNAs [
3,
13,
14,
15]. RNA pol III contains 17 subunits and transcribes tRNAs, 5S rRNA and other non-coding RNAs [
13,
16,
17,
18]. RNA pol II is composed of 12 subunits and synthesizes mRNAs and some non-coding RNAs [
13,
19,
20,
21]. Plant-specific RNA pol IV and V, which have evolved from RNA pol II through duplication and functional divergence, also contain 12 subunits. These two enzymes influence epigenetic regulation and synthesize siRNAs, which play roles in transcriptional silencing via RNA-directed DNA methylation (RdDM), and the non-coding RNAs participating in plant growth, development, response to environmental changes or plant immunity [
4,
5,
6,
7,
8,
9,
10,
11,
12,
22,
23,
24].
RNA pol II, IV and V contain specific conserved subunits that may specialise some of their functions [
5,
22,
25]. This is the case of subunits NRPD1, NRPE1 and NRPB1 for RNA pols IV, V and II, respectively, in
Arabidopsis thaliana. In addition, other subunits are common to RNA pol IV and V, but are conserved to RNA pol II, like the subunits NRPDE2 and NRPB2, RNPDE4 and NRPB4, and NRPDE7 and NRPB7, which are shared by RNA pol IV and V and conserved with RNA pol II [
5,
22,
25,
26,
27]. Finally, several isoforms of the common subunit five, shared by all the RNA pols have been described, while a specific isoform, NRPE5, has been found for RNA pol V [
28]. In addition, several paralogues have been described for these and other subunits in different plants [
22,
25,
26,
28,
29,
30]. Based on the existence of these paralogues, it has been proposed that these may perform new functions or may have a different regulation, which is the case of the distinct isoforms of the shared subunits to the RNA pols in cultivated olive trees ‘Picual’ (
Olea europaea L. cv. Picual) [
28].
RNA pol IV and V have been reported to be involved in the biogenesis and functionality of 24-nt siRNA, which participates in RdDM [
9,
12,
25,
30]. RNA pol IV and V have also been proposed to participate in the transcription of long non-coding RNAs (lncRNAs) [
24,
31,
32,
33,
34,
35,
36,
37,
38]. Some of these lncRNAs are the intermediary of siRNA and found within intergenic regions [
34]. Although lncRNAs are also transcribed by RNA pol II, those synthesised by RNA pol IV and V contain some structural differences as regards the RNA pol II ones, such as lack of poly-A at the 3’ end region or lack of introns [
4]. lncRNAs transcribed by RNA pol IV and V are poorly characterized, in part because of their low expression and instability [
34,
39]. However, well-studied examples of non-polyA lncRNAs have been reported [
40,
41,
42]. Notably, the synthesis of non-polyA lncRNAs can be regulated by environmental conditions, which has been shown in
A. thaliana under abiotic stress [
43,
44,
45].
In this work we searched for genes that putatively code for specific subunits of RNA pol IV and V, and for those corresponding to RNA pol II, in the olive ‘Picual’ cultivar given its economic, agronomic and agro-ecological importance as one of the most important fruit trees in the Mediterranean Basin [
28,
46,
47,
48]. The paralogues for NRPD1, NRPE1 and NRPB1, NRPDE2 and NRPB2, NRPDE4 and NRPB4, NRPDE7 and NRPB7, and also for NRPB7-like, were identified, in addition to the putative pseudogenes, according to our transcriptomic analyses. The transcriptional studies from RNA-Seq data indicated a role of specific RNA pol IV and V during fruit development. Furthermore, according to the role of RNA pol IV and V in the transcription of lncRNAs, we studied the lncRNA transcriptome during fruit development, which revealed important changes in their expression and differed based on the analysed lncRNA type.
2. Results
A genome search for olive genes that are highly homologous to those that code for the specific subunits of RNA pol IV and V in A. thaliana was performed. Then their expression profile was studied by RNA-Seq in different plant organs/tissues in response to environmental stresses, and also during the fruit development process. As RNA pol IV and V have evolved from RNA pol II, the equivalent olive genes for the RNA pol II subunits were also identified. These genes coding for the RNA pol II subunits were used as references for the expression profiles and were compared to the RNA pol IV and V coding genes. An additional annotation and expression analysis of lncRNAs was also done.
2.1. Olive genes coding for RNA pol IV and V subunits
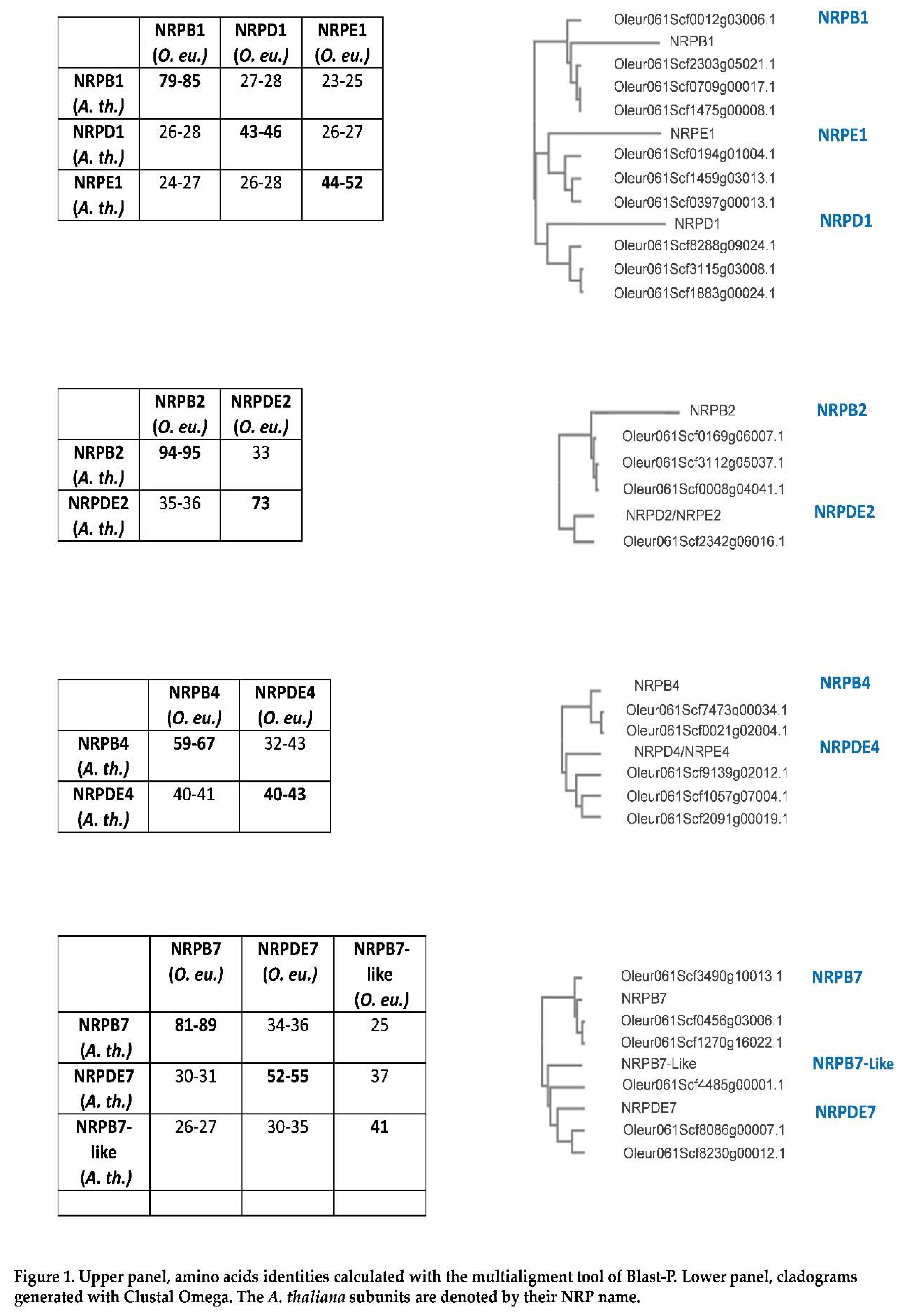
In order to search for the genes that putatively code for the specific subunits of RNA pol IV and V in olive, the
A. thaliana sequences for NRP1, NRP2, NRP4 and NRP7 of RNA pol II, IV and V were used as a query to find the corresponding homologues. Several paralogues were encountered: 16 for pol II and 12 for RNA pol IV and V (
Table 1 and Figure 1). This was not surprising because olive ancestors have quite probably undergone two whole genome duplication (WGD) events in the last 65 M years [
49,
50]. Furthermore, putative pseudogenes were found for RNA pol II as not expressed genes and containing inactivating mutations according to our RNA-Seq analyses under different conditions (see below).
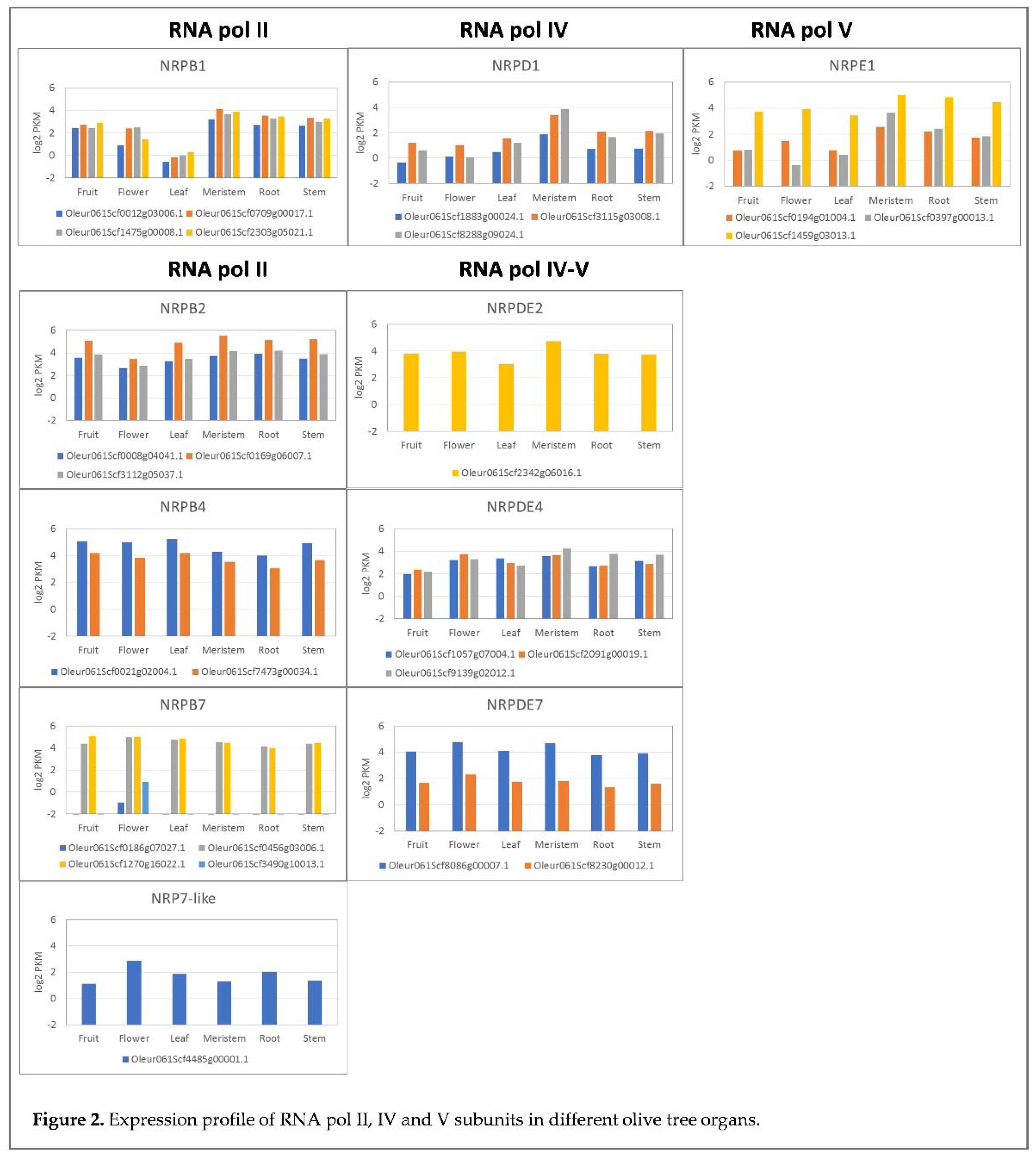
2.2. Gene expression profile in different plant organs/tissues
In a previous work, we found that those genes coding for subunits shared by RNA pols in ‘Picual’ cultivar [
28] were spatially and temporally regulated. In order to determine if the specific subunits of the RNA pol II, IV and V are spatially regulated in plant, a transcriptomic analysis was performed. Transcriptomic analyses were performed to determine if the specific subunits of RNA pol II, IV and V were spatially regulated in plants. The transcriptomic analysis of the genes coding for the specific subunits of RNA pol II, IV and V showed that some were regulated in the different analysed plant tissues (Figure 2). In line with this, the genes coding for RNA pol II subunits 1, 2, 4 and 7 generally had quite a homogeneous expression among the different analysed tissues, with some exceptions. For instance, the four genes coding for subunit 1 of RNA pol II (NRPB1) showed a lower expression level in leaves than in the other tissues. Similarly, two genes coding for subunit 7 (NRPB7), coding genes Oleur061Scf0186g07027.1 and Oleur061Scf3490g10013.1, seemed to be tissue-specific because they were expressed only in flowers. Furthermore, one of the genes coding for subunit 7 of RNA pols IV and V (Oleur061Scf8086g00007.1) showed the highest expression level for all tissues. The putative pseudogenes of the NRPB7 and NRPB7-like subunits were not expressed in any organ and tissue (not shown). For the RNA pol IV and V subunits, the general expression pattern was also quite homogeneous, except for the NRPD1 and NRPE1 subunits, which seemed to be expressed at variable levels depending on the plant tissue.
2.3. Expression profile in response to biotic and abiotic stresses
RNA pol IV and V might be involved in the response to stress stimuli to plants, according to reported data [
12,
51]. To examine this possibility, we studied the expression level of those genes coding for the different RNA pol II, IV and V subunits by analysing response to root injury,
V. dahliae infection and cold stress. As a result (Supplemental
Figure S1), the transcriptomic analysis of the genes coding for the specific RNA pol II, IV and V subunits showed no consistent expression pattern in response to any of the studied stresses. However for root injury stress, some of the genes coding for the RNA pol IV and V subunits seemed to slightly reduce its expression after injury, and the original expression level were recovered after a 7-day follow-up. This behaviour was not consistent in all the subunits or in all the paralogue genes of the same subunit.
In addition, no specific response to biotic stress produced by the induced infection by
V. dahliae was detected (Supplemental
Figure S2). In this case, minor changes were similar to the response to root injury, which is performed to induce
V. dahliae infection. Notably, some mayor changes were observed after 15 days post-inoculation. At the time of this follow-up, plants displayed clear severe disease symptoms. This fact could modify the general gene expression pattern, as previously described by [
52].
Regarding response to cold stress, changes in the expression for some genes of the RNA pol II, IV and V specific subunits were observed, but, once again, no consistent pattern was found (Supplemental
Figure S3).
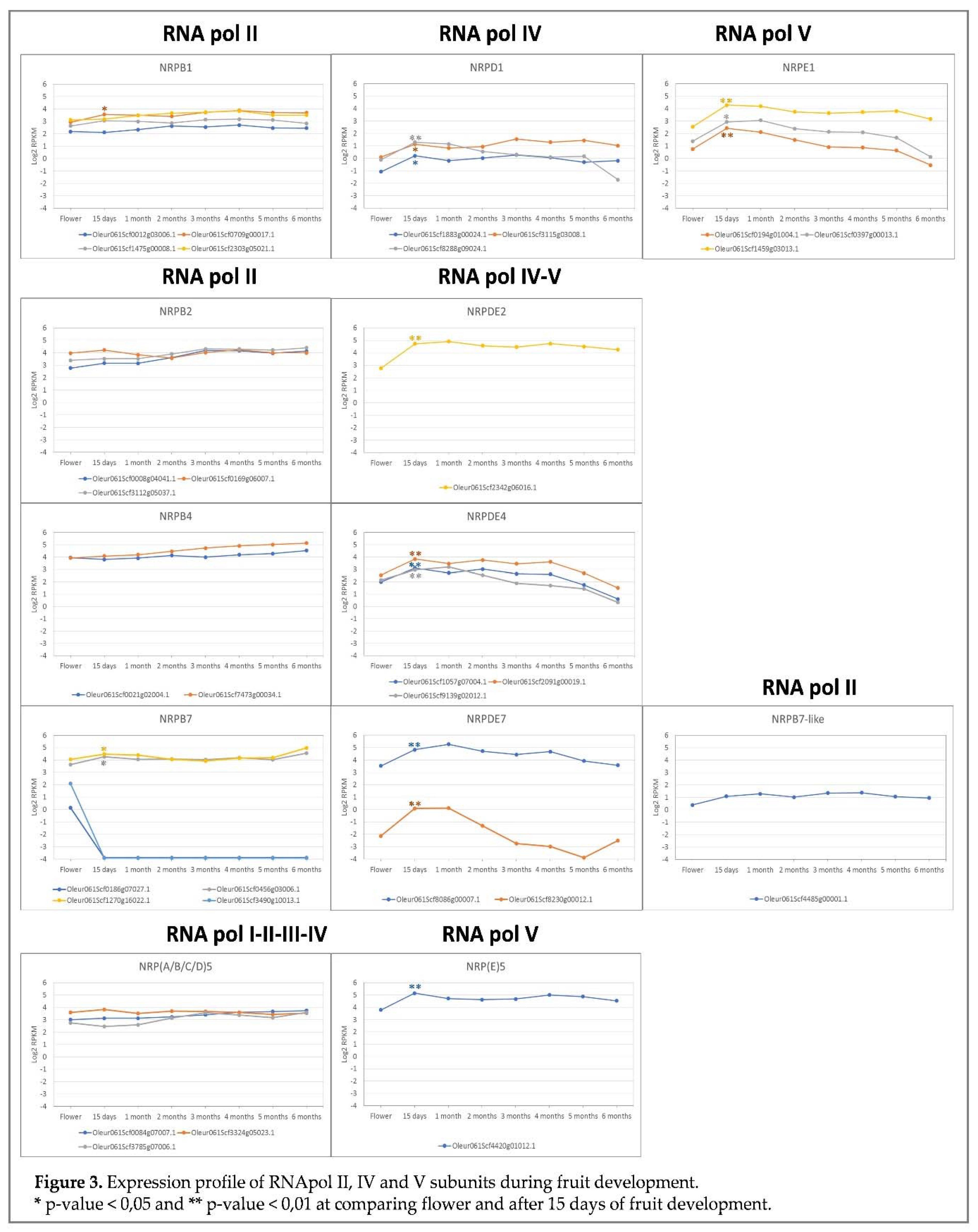
2.4. Expression profile during fruit development
In line with the reported role of RNA pol IV and V during plant development, we investigated whether this could be the case in olive trees. To investigate this, we performed a transcriptomic analysis of the genes that putatively code for the specific RNA polymerases II, IV and V subunits during fruit development. For this purpose, samples from three trees were analysed by RNA-Seq, which consisted of recently bloomed flowers and developing fruit at 15 days and every month from flowering to fully ripe fruit (
Figure S4) [
53].
Notably, all the RNA pol IV and V specific subunits showed a significant induction at 15 days after full bloom (AFB) (Figure 3), with an average fold change of 3.2 (1.8 – 8.0). However, this induction at 15 days AFB was observed only for three of the genes of RNA pol II (Oleur061Scf0709g00017.1 of subunit 1; Oleur061Scf1270g16022.1 and Oleur061Scf0456g03006.1 for subunit 7) with fold change from 1.3 to 1.6. Therefore, no relevant changes were observed in the gene expression of the genes of the specific RNA pol II subunits, except for the NRPB7 genes Oleur061Scf0186g07027.1 and Oleur061Scf3490g10013.1 which, according to the organ/tissue specificity proposed above (Figure 2), were rapidly repressed once fruit development began. In addition, the two putative pseudogenes identified for NRPB7 and NRPB7-like were not expressed during fruit development.
Olive cultivar ‘Picual’, like other plants, contains a gene that codes for the specific RNA pol V subunit NRPE5, which is an isoform of subunit NRP5, shared by all eukaryotic RNA polymerases [
28]. In line with this protein being specific for RNA pol V, for the corresponding gene (Oleur061Scf4420g01012.1), a significant induction was noted on the first 15 days of fruit development (Figure 3), which was not the case of the other NRP5 paralogues identified in olive tree.
Taken together, these data suggest that RNA pol IV and V could play a major role in early fruit development steps. As RNA pol IV and V have been proposed to participate in the transcription of lncRNAs [
24,
31,
32,
33,
34,
35,
36,
37,
38], we can speculate about a transcriptional response of these type of transcripts during this development process.
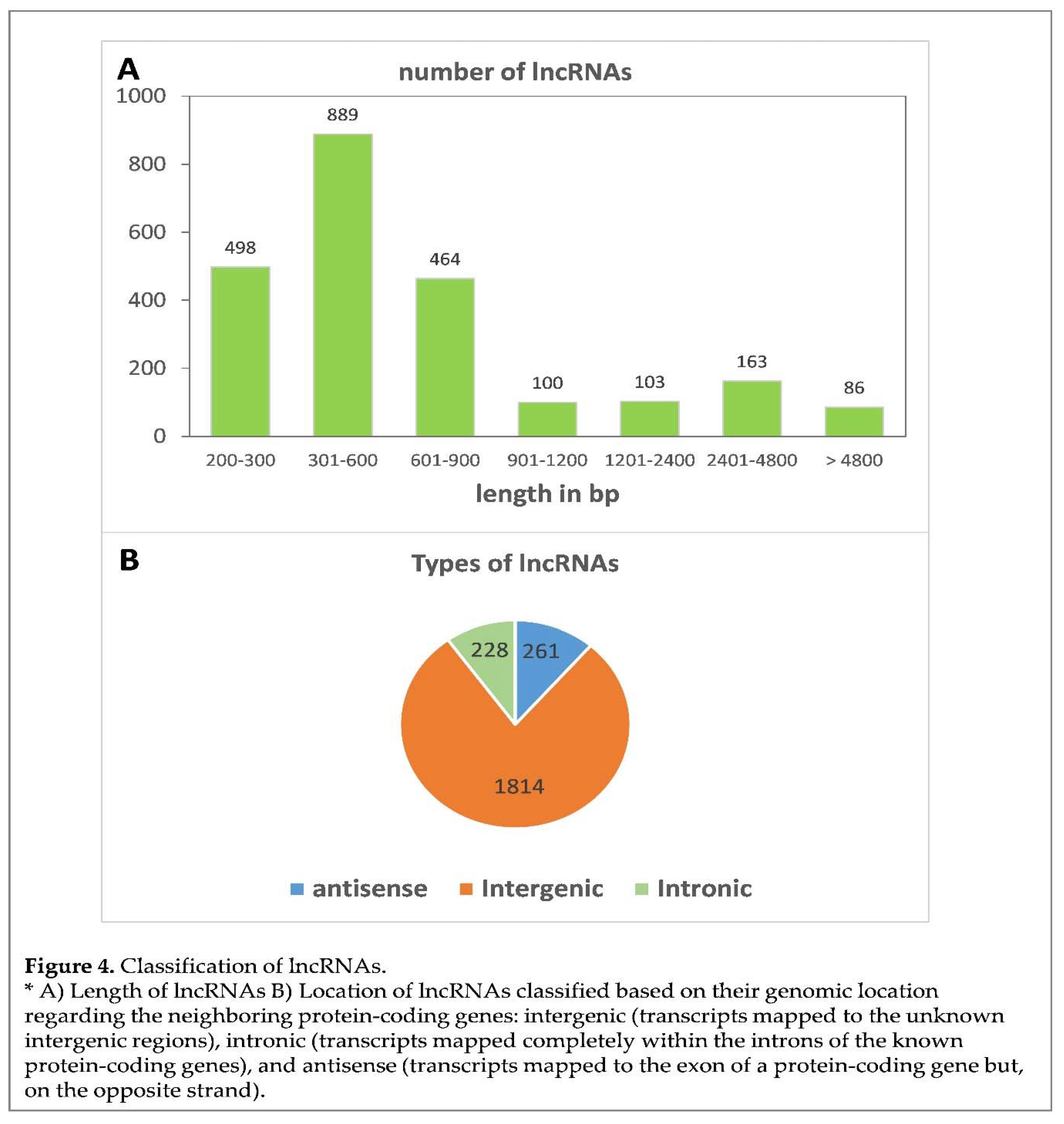
2.5. Annotation and expression of lncRNAs
In order to study the expression pattern of lncRNAs in olive, six strand-specific RNA-seq libraries were constructed using the total RNA of olive flower at full bloom and olive fruit at 15 days AFB, with three biological replicates each. An additional mix including different plant tissues was sequenced to obtain a broad representation of the lncRNA transcriptome in olive. From the total reads showing high quality (score > Q30), a final number of 744,825,368 clean reads was obtained from the seven libraries after trimming the adapters and reads that were shorter than 50bp (Supplemental
Table S1). These clean reads were aligned to the olive genome of the ‘Picual’ cultivar (
https://genomaolivar.dipujaen.es/db/downloads.php). Alignment rates appeared to range from 60.24% to 86.07% (Supplemental
Table S1). Subsequently, 120,670 total transcripts were assembled using Stringtie, and 79,654 transcripts resulted after filtering transcripts by size ≥ 200 bp. The length of lncRNAs ranged from 200 bp to 21,212 bp, although most were shorter than 900 pb (Figure 4A). These transcript sequences were analysed to identify putative lncRNAs. As a result, 3,603 total transcripts from both experimental conditions were selected as intergenic, intronic or antisense after annotation with GffCompare. No tRNAs were identified when applying tRNAscan-SE. Furthermore, 146 rRNAs and 370 coding transcripts were discarded by applying Barrnap and CPC2, respectively. Finally, 2,303 candidate non-poly-A lncRNAs were identified as non-coding RNAs by GreenNC, including 1,814 intergenic, 261 antisense and 228 intronic transcripts (Figure 4B).
The analysis allowed us to identify 1,899 lncRNAs (non-polyA lncRNAs) in flowers and 1,915 from 15 days AFB, which had 1,651 in common (Figure 5). In addition, changes in the lncRNAs expression pattern were observed. During the transition from flowering to 15 days AFB, 143 lncRNAs were found to be up-regulated and 273 down-regulated by using a false discovery rate (FDR) of 5% (
Table 2). However, no major changes in the average expression of lncRNAs was found between the flower and 15 days AFB samples, with 718.68 and 733.22 RPKMs, respectively (p-value = 0.8365), although some differences between the flower and the 15 days AFB samples were observed when discriminating by lncRNAs types (
Table 2). Specifically, a tendency towards increased expression in lncRNAs at 15 days AFB was observed in the intronic lncRNAs (234.96 RPKM in flowers to 336.53 RPKM at 15 days AFB with p-value = 0.0023) and intergenic lncRNAs (525.43 RPKM in flowers and 594.10 RPKM at 15 days AFB with p-value = 0.0399). However, no significant differences were found for the antisense lncRNAs (2,484.33 RPKM in flowers and 2,046.71 RPKM at 15 days AFB with p-value = 0.4484) (
Table 2).
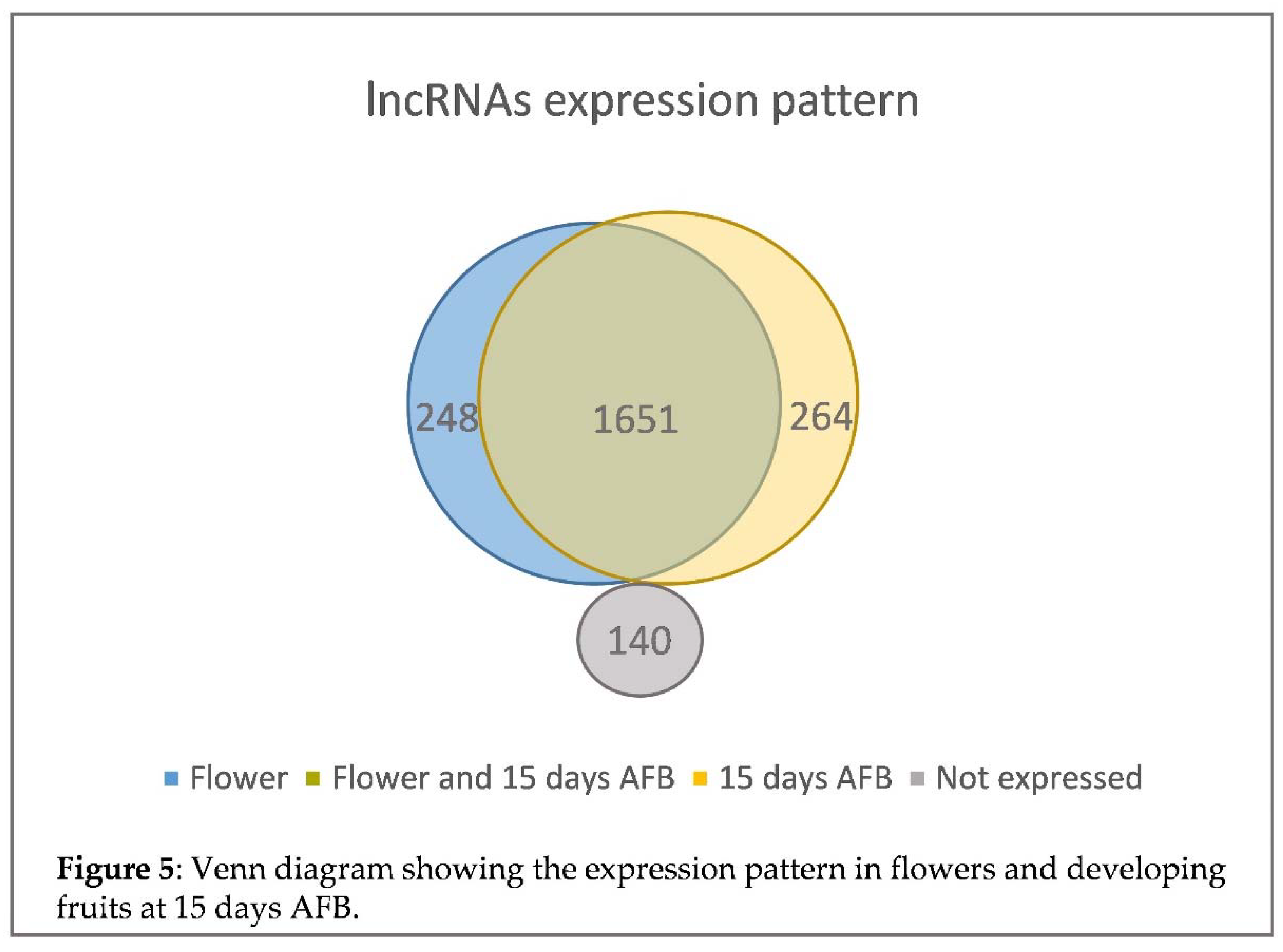
Together, these data indicate that the changes in the gene expression level for the specific RNA pol IV and V subunits during fruit development were accompanied by important changes in the expression of lncRNAs, and these changes differed depending on the type of analysed lncRNA.
3. Discussion
The role of RNA pol IV and V in plants is still being studied and is not fully understood. The olive tree is an important crop and a more complex one than the model plant A. thaliana. In this work, we searched for the genes that encode the specific subunits of RNA pol IV and V, as well as the RNA pol II from which they have evolved. A comprehensive analysis of the expression profile of these genes was also performed.
Several genes for the specific RNA pol IV and V subunits, and those corresponding to RNA pol II, were identified by a blast-p search with the corresponding
A. thaliana subunits [
7,
12,
22]. In line with this, for RNA pol IV and V subunits, protein identity with
A. thaliana homologues was variable, but within the 40-55% range, except for NRPDE2 with 73% identity (Figure 1). Identity was notably higher (79-95%) for the NRPB homologues, which indicated that they maintain greater conservation, while the NRPD and NRPE subunits have allowed major variation during evolution. RNA pol IV and V have evolved from RNA pol II [
4,
5,
22,
54,
55], and have apparently evolved more rapidly than the RNA pol II because their
A. thaliana and
O. europaea sequences have diverged more.
Several paralogues of the different specific RNA pol II, IV and V subunit genes were found for all the subunits, except for RNA pol IV/V subunit NRPDE2, which had only a single gene (
Table 1). This was not the only case for the NRP1, NRP2, NRP4 and NRP7 subunits identified in this work, but also for an additional specific RNA pol V subunit (the previously described NRPE5) with paralogues for additional common subunits for all the RNA polymerases [
28]. The presence of several paralogues is found for many genes in olive. One such case is the RNA pol subunits shared by the five RNA pols [
28]. These results are consistent with the olive cultivar genome that results from two independent whole-genome duplication (WGD) events, in addition to recent partial genome duplications [
49,
50]. In addition, several of the paralogues for RNA pols subunits have been identified in other organisms [
1,
5,
22,
25,
56].
It has been demonstrated that RNA pol IV and V play a role in silencing, plant growth, development, response to environmental changes or plant immunity [
4,
5,
6,
7,
8,
9,
10,
11,
12,
22,
23,
24]. Therefore, we can speculate that the regulation of the gene expression of these RNA pols could be expected in response to some growing conditions. The transcriptomic analysis run by several RNA-Seq experiments during stress or developing processes using the olive cultivar ‘Picual’ [
52,
57,
58,
59] has shown that all the genes coding for the specific subunits identified in this work are expressed under these conditions. Similarly, the genes corresponding to the RNA pol II subunits were also expressed. Although no common pattern for the changes in the expression of the RNA pol II, IV or V genes was observed, some cases of regulation by plant tissue appeared. Remarkably, two RNA pol II (NRPB7) genes presented strict organ specificity and were expressed only in flowers (Figure 2 and Figure 3). Furthermore, the response to biotic
V. dahliae infection [
52] or abiotics cold [
58] or root injury [
52] showed null or weak changes in the expression profile of most genes.
However, and notably, clear gene regulation occurred for the RNA pol IV and V subunits during fruit development. Indeed a consistent and significant overexpression of the RNA pol IV and V genes was observed at the beginning of fruit development, and this phenomenon did not occur for the RNA pol II subunits. This suggests a possible role of RNA pol IV and V during fruit development in agreement with the role of RNA pol IV and V during plant development and plant growth [
12]. According to the role of RNA pol IV and V during the synthesis of ncRNAs, relevant changes in the lncRNAs expression pattern were observed. In line with this, the synthesis of the non-polyA lncRNAs has been demonstrated to be regulated by environmental conditions in
A. thaliana under abiotic stress [
43,
44,
45]. Indeed we identified 2,303 lncRNAs (non-polyA) transcripts in flowers and fruit at 15 days AFB. Most were intergenic, while intronic and antisense lncRNAs were less frequent (Figure 4). Relevant changes in expression were found, and 284 lncRNA transcripts were expressed only in flowers and 264 only in fruit at 15 days AFB (Figure 5). Furthermore, a tendency toward an increasing expression was noted in the intronic and intergenic lncRNAs, but not in the antisense lncRNAs. Due to the role of RNA pol IV and V in the synthesis of the small ncRNAs that participate in silencing, we cannot rule out that some of these lncRNAs could be processed to small ncRNAs, which has been reported for other plants [
40,
60]. Indeed this fact has been observed for intergenic lncRNAs in
A. thaliana [
40] and, in our case, in 39 intergenic transcripts found in olive, which were also identified as putative siRNAs.
By way of conclusion, we identified the genes that code for the specific RNA IV and V subunits, and the corresponding ones in RNA pol II, in olive cultivar ‘Picual’. The expression analysis performed of different organs/tissues, biotic and abiotic stresses and a development process revealed that the expression of the RNA pol IV and V genes was induced in initial fruit development steps. This induction was accompanied by relevant changes in the expression of lncRNAs, and the expression of the intergenic and intronic lncRNAs tended to increase. These changes in the expression of lncRNAs may be important for controlling gene expression during fruit development. In addition, some intergenic transcripts are susceptible to be processed and to become siRNAs, which are known to play a role in the gene expression control. This reinforces the hypothesis that RNA pol IV and V may play a role in fruit development throughout the synthesis of lncRNAs.
4. Materials and Methods
4.1. Plant material
In order to analyse gene expression during fruit development in olive tree, flowers and fruit were collected from the ‘Picual’ olive cultivar growing in the experimental field of the University of Jaén (Jaén, Spain). Flower and fruit samples were collected from three different closely located trees and from south-facing branches to reduce environmental variability, as specified by [
53]. Therefore, three independent biological samples were collected at eight different times from full bloom (flowering) to fruit ripening 6 months later (15 day AFB), and monthly from 1 to 6 months AFB. These samples were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction.
4.2. Transcriptomic analysis
The total RNA from the triplicate samples of flowers and fruit at 15 days AFB was isolated using the Spectrum™ Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. PoliA+ RNA was purified and sequenced from the samples collected during fruit development as indicated by [
53]. Briefly, poliA+ RNA 150 bp x 2 paired-end Illumina sequences were obtained at Novogene (UK) and at least 50 M reads of Q30 sequences data were obtained from each biological replicate sample. The dataset is available at NCBI as BioProject: PRJNA870905.
For this work, an additional RNA-Seq of the total RNA was done. In this case, 150 bp x 2 paired-end Illumina sequences were obtained at Ascires (Spain) from the flower and 15 days AFB samples, as well as a mix of RNAs from flower, fruit, root, leaf, meristem and stem. For total RNA sequencing, at least 100 M reads were obtained per sample. The dataset is available at NCBI as BioProject: PRJNA989401.
Other RNA-Seq data were used as described in [
28]. Basically, a previous RNA-Seq from olive organ/tissues [
59] and several stresses, such as cold, injury or
Verticillium dahliae infection [
58], were analysed. The datasets are available at BioProject PRJNA556567 and at NCBI accession numbers SRR1525051, SRR1525052, SRR1524949, SRR1524950, SRR1524951, SRR1524952, SRR1525086, SRR1525087, SRR1525113, SRR1525114) SRR1525231, SRR1525237, SRR1524947, SRR1524948, SRR1525213, SRR1525114, SRR1525224, SRR1525226, SRR1525284, SRR1525285, SRR1525286, SRR1525287, SRR1525415, SRR1525416, SRR1525436, and SRR1525437.
The RNA-Seq analysis was performed with DNAstar (ArrayStar 17, Rockville, MD, USA) for the RNA-Seq analyses (
www.dnastar.com). Gene expression was carried out using a 95% false discovery rate (FDR).
4.3. Annotation of lncRNAs in olive
Assessments of raw sequence quality were first performed using the FastQC software (version 0.11.5,
http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Adapter sequences and reads shorter than 50 bp were trimmed with Fastq-mcf (EA-Utils version 1.04.759) (
http://expressionanalysis.github.io/ea-utils/). Next clean reads were mapped to the olive genome of the ‘Picual’ cultivar [
50] available in OliveTreeDB (
https://genomaolivar.dipujaen.es/db/downloads.php) using the HISAT2 software (v2.2.1) [
61]. Mapped reads were sorted and compressed by Samtools (v1.16.1) [
62], and then assembled and merged using StringTie v2.2.1 [
63]. The gffcompare tool (v0.12.6) [
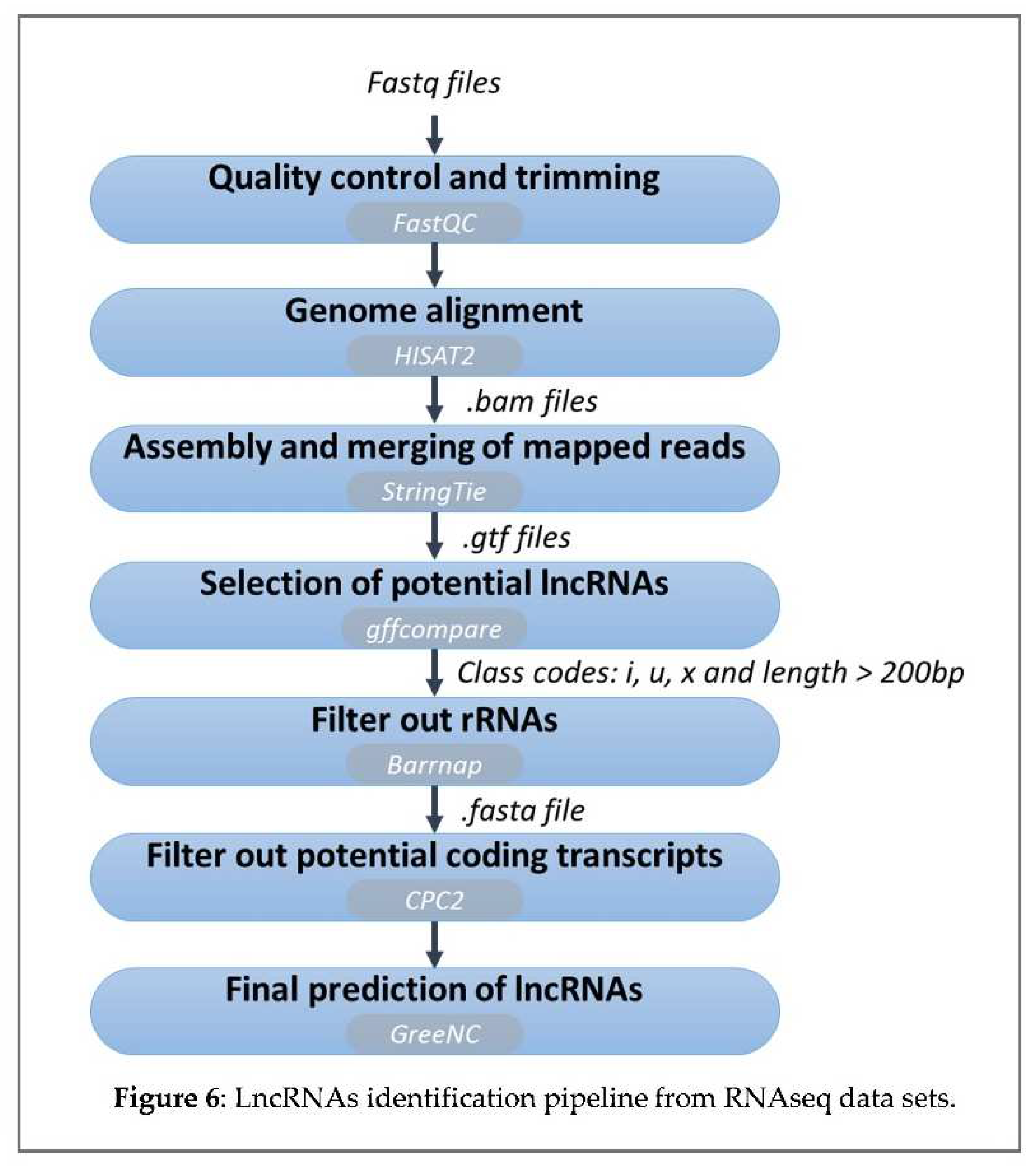
64] was used to identify the unannotated transcripts by comparing the assembled transcriptome to the reference ‘Picual’ transcriptome. Note that these unannotated transcripts corresponded to the non-polyA lncRNAs. Subsequently, the transcripts categorised as “u” (intergenic lncRNAs), “x” (antisense lncRNAs), “i” (intronic lncRNAs) and higher than 200 bp were selected as candidate lncRNAs.
However, as the selected transcripts could contain coding genes, they underwent another filtering process. The transfer RNAs were filtered using the tRNAscan-SE 2.0 tool [
65]. Barrnap tool v0.7 (
https://github.com/tseemann/barrnap) was applied to identify the ribosomal RNA genes and CPC2 software (
http://cpc2.cbi.pku.edu.cn) [
66] was applied to filter out those transcripts with coding ability. Finally, transcripts were analysed by the second script of GreeNC (
https://github.com/sequentiabiotech/GreeNC) to discriminate any other non-coding transcripts from lncRNAs and to identify any possible miRNA precursors Figure 6).
4.4. Analysis of the differentially expressed lncRNAs
The expression analysis was performed with DNAstar (ArrayStar 17, Rockville, MD, USA,
www.dnastar.com). Mapping was done with high-stringency parameters to differentiate between very similar paralogues, k-mer = 63 and 95% matches. Data were normalised based on reads per kilobase of transcript per million reads mapped (RPKM). A basal expression level of log2 RPKM = −2 was considered. Therefore, the genes with expression values above this threshold level were considered expressed, whereas those genes with expression values that equalled or were below the threshold level were considered not expressed. A comparison between samples was made using the parametric t-student test.
Supplementary Materials
The following supporting information can be downloaded at:
Preprints.org, Figure S1: title; Table S1: title; Video S1: title
Author Contributions
LncRNA analysis and writing, A.S.; investigation and data curation, M.M.; investigation, I.F.; lncRNAs supervision, A.B.; writing, conceptualization and original draft preparation, F.L. and F.N.; funding acquisition, F.L. and F.N.
Funding
work has been supported by grants from the Spanish Ministry of Science and Innovation (MCIN) and ERDF to F.N. and F.L. (PID2020-112853GB-C33 and PID2020-115853RR-C33, respectively), and the Junta de Andalucía (BIO258) and the University of Jaen to F.N.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
New RNA-Seq of the total RNA data is available at NCBI as BioProject: PRJNA989401.
Acknowledgments
The technical and human support provided by CICT of the Universidad de Jaén (UJA, MINECO, Junta de Andalucía, FEDER) is gratefully acknowledged. Alicia Serrano thanks the support received with a postdoctoral fellowship from the 2021–2023 grants programme for the requalification of the Spanish University system (modality Margarita Salas), funded by the European Union-NextGenerationEU.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ream, T. S.; Haag, J. R.; Pontvianne, F.; Nicora, C. D.; Norbeck, A. D.; Pasa-Tolic, L.; Pikaard, C. S. , Subunit compositions of Arabidopsis RNA polymerases I and III reveal Pol I- and Pol III-specific forms of the AC40 subunit and alternative forms of the C53 subunit. Nucleic Acids Res 2015, 43(8), 4163–78. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Bermúdez, A.; Martínez-Fernández, V.; Garrido-Godino, A. I.; Navarro, F. , Subunits common to RNA polymerases. In The Yeast Role in Medical Applications, Abdulkhair, W. M. H., Ed. IntechOpen: London, 2017; Vol. 1, pp 151-165.
- Werner, F.; Grohmann, D. , Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 2011, 9(2), 85–98. [Google Scholar] [CrossRef]
- Zhou, M.; Law, J. A. , RNA Pol IV and V in gene silencing: Rebel polymerases evolving away from Pol II's rules. Curr Opin Plant Biol 2015, 27, 154–64. [Google Scholar] [CrossRef] [PubMed]
- Tucker, S. L.; Reece, J.; Ream, T. S.; Pikaard, C. S. , Evolutionary history of plant multisubunit RNA polymerases IV and V: subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harb Symp Quant Biol 2010, 75, 285–97. [Google Scholar] [CrossRef] [PubMed]
- Lahmy, S.; Bies-Etheve, N.; Lagrange, T. , Plant-specific multisubunit RNA polymerase in gene silencing. Epigenetics 2010, 5(1), 4–8. [Google Scholar] [CrossRef]
- Haag, J. R.; Pikaard, C. S. , Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 2011, 12(8), 483–92. [Google Scholar] [CrossRef]
- Lopez, A.; Ramirez, V.; Garcia-Andrade, J.; Flors, V.; Vera, P. , The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genet 2011, 7(12), e1002434. [Google Scholar] [CrossRef]
- Ream, T.; Haag, J.; Pikaard, C. , Plant Multisubunit RNA Polymerases IV and V. In Nucleic Acid Polymerases, Trakselis, K. S. M. a. M. A., Ed. Springer-Verlag: Heidelberg, 2014; Vol. 30.
- Moo, L. d. R. C.; González, A. K.; Rodríguez-Zapata, L. C.; Suarez, V.; Castaño, E. , Expression of RNA polymerase IV and V in Oryza sativa. Electronic Journal of Biotechnology 2012, 15, 9–9. [Google Scholar]
- Huang, Y.; Kendall, T.; Forsythe, E. S.; Dorantes-Acosta, A.; Li, S.; Caballero-Perez, J.; Chen, X.; Arteaga-Vazquez, M.; Beilstein, M. A.; Mosher, R. A. , Ancient Origin and Recent Innovations of RNA Polymerase IV and V. Mol Biol Evol 2015, 32(7), 1788–99. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, X. Q.; Xie, H. T.; Zhao, S. S.; Wu, J. G. , Multifaceted roles of RNA polymerase IV in plant growth and development. J Exp Bot 2020, 71(19), 5725–5732. [Google Scholar] [CrossRef]
- Barba-Aliaga, M.; Alepuz, P.; Perez-Ortin, J. E. , Eukaryotic RNA Polymerases: The Many Ways to Transcribe a Gene. Front Mol Biosci 2021, 8, 663209. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.; Thuriaux, P.; Soutourina, J. , Structure-function analysis of RNA polymerases I and III. Curr Opin Struct Biol 2009, 19(6), 740–5. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Morcillo, M.; Taylor, N. M.; Gruene, T.; Legrand, P.; Rashid, U. J.; Ruiz, F. M.; Steuerwald, U.; Muller, C. W.; Fernandez-Tornero, C. , Solving the RNA polymerase I structural puzzle. Acta Crystallogr D Biol Crystallogr 2014, 70 Pt 10, 2570–82. [Google Scholar] [CrossRef] [PubMed]
- Moir, R. D.; Willis, I. M. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim Biophys Acta 2013, 1829(3-4), 361–375. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Tornero, C.; Bottcher, B.; Rashid, U. J.; Muller, C. W. , Analyzing RNA polymerase III by electron cryomicroscopy. RNA Biol 2011, 8(5), 760–5. [Google Scholar] [CrossRef] [PubMed]
- Turowski, T. W.; Boguta, M. , Specific Features of RNA Polymerases I and III: Structure and Assembly. Front Mol Biosci 2021, 8, 680090. [Google Scholar] [CrossRef] [PubMed]
- Armache, K. J.; Mitterweger, S.; Meinhart, A.; Cramer, P. , Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem 2005, 280(8), 7131–4. [Google Scholar] [CrossRef]
- Perez-Ortin, J. E.; Mena, A.; Barba-Aliaga, M.; Singh, A.; Chavez, S.; Garcia-Martinez, J. , Cell volume homeostatically controls the rDNA repeat copy number and rRNA synthesis rate in yeast. PLoS Genet 2021, 17(4), e1009520. [Google Scholar] [CrossRef]
- Tan, E. H.; Blevins, T.; Ream, T. S.; Pikaard, C. S. , Functional consequences of subunit diversity in RNA polymerases II and V. Cell Rep 2012, 1(3), 208–14. [Google Scholar] [CrossRef]
- Ream, T. S.; Haag, J. R.; Wierzbicki, A. T.; Nicora, C. D.; Norbeck, A. D.; Zhu, J. K.; Hagen, G.; Guilfoyle, T. J.; Pasa-Tolic, L.; Pikaard, C. S. , Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 2009, 33(2), 192–203. [Google Scholar] [CrossRef]
- Pikaard, C. S.; Tucker, S. , RNA-silencing enzymes Pol IV and Pol V in maize: more than one flavor? PLoS Genet 2009, 5(11), e1000736. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhou, W.; Zhang, X.; Li, Y. , Roles of long non-coding RNAs in plant immunity. PLoS Pathog 2023, 19(5), e1011340. [Google Scholar] [CrossRef] [PubMed]
- Haag, J. R.; Brower-Toland, B.; Krieger, E. K.; Sidorenko, L.; Nicora, C. D.; Norbeck, A. D.; Irsigler, A.; LaRue, H.; Brzeski, J.; McGinnis, K.; Ivashuta, S.; Pasa-Tolic, L.; Chandler, V. L.; Pikaard, C. S. , Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Rep 2014, 9, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Marcussen, T.; Oxelman, B.; Skog, A.; Jakobsen, K. S. , Evolution of plant RNA polymerase IV/V genes: evidence of subneofunctionalization of duplicated NRPD2/NRPE2-like paralogs in Viola (Violaceae). BMC Evol Biol 2010, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- He, X. J.; Hsu, Y. F.; Pontes, O.; Zhu, J.; Lu, J.; Bressan, R. A.; Pikaard, C.; Wang, C. S.; Zhu, J. K. , NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev 2009, 23(3), 318–30. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Parras, I.; Ramirez-Tejero, J. A.; Luque, F.; Navarro, F. Several Isoforms for Each Subunit Shared by RNA Polymerases are Differentially Expressed in the Cultivated Olive Tree (Olea europaea L.). Front Mol Biosci 2021, 8, 679292. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, J. T.; Seetharam, A. S.; Hufford, M. B.; Beilstein, M. A.; Mosher, R. A. , Evidence for a Unique DNA-Dependent RNA Polymerase in Cereal Crops. Mol Biol Evol 2018, 35(10), 2454–2462. [Google Scholar] [CrossRef]
- Chakraborty, T.; Trujillo, J. T.; Kendall, T.; Mosher, R. A. , A null allele of the pol IV second subunit impacts stature and reproductive development in Oryza sativa. Plant J 2022, 111(3), 748–755. [Google Scholar] [CrossRef]
- Bohmdorfer, G.; Rowley, M. J.; Kucinski, J.; Zhu, Y.; Amies, I.; Wierzbicki, A. T. , RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J 2014, 79(2), 181–91. [Google Scholar] [CrossRef]
- Wierzbicki, A. T.; Haag, J. R.; Pikaard, C. S. , Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 2008, 135(4), 635–48. [Google Scholar] [CrossRef]
- Liu, X.; Hao, L.; Li, D.; Zhu, L.; Hu, S. , Long non-coding RNAs and their biological roles in plants. Genomics Proteomics Bioinformatics 2015, 13(3), 137–47. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Vandivier, L. E.; Tu, B.; Gao, L.; Won, S. Y.; Zheng, B.; Gregory, B. D.; Chen, X. , Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res 2015, 25(2), 235–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. V.; Chekanova, J. A. , Long Noncoding RNAs in Plants. Adv Exp Med Biol 2017, 1008, 133–154. [Google Scholar] [PubMed]
- Chen, L.; Zhu, Q. H.; Kaufmann, K. , Long non-coding RNAs in plants: emerging modulators of gene activity in development and stress responses. Planta 2020, 252(5), 92. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Kaya, S. B.; Cagirici, H. B. , Long Non-coding RNA in Plants in the Era of Reference Sequences. Front Plant Sci 2020, 11, 276. [Google Scholar] [CrossRef]
- Wierzbicki, A. T.; Blevins, T.; Swiezewski, S. , Long Noncoding RNAs in Plants. Annu Rev Plant Biol 2021, 72, 245–271. [Google Scholar] [CrossRef] [PubMed]
- Rai, M. I.; Alam, M.; Lightfoot, D. A.; Gurha, P.; Afzal, A. J. , Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111(5), 997–1005. [Google Scholar] [CrossRef]
- Heo, J. B.; Sung, S. , Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331(6013), 76–9. [Google Scholar] [CrossRef]
- Shin, J. H.; Chekanova, J. A. , Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis. PLoS Genet 2014, 10(9), e1004612. [Google Scholar] [CrossRef]
- Kim, D. H.; Sung, S. Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev Cell 2017, 40(3), 302–312 e4. [Google Scholar] [CrossRef]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Zhang, T.; Qi, Y.; Gerstein, M. B.; Guo, Y.; Lu, Z. J. , Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J 2014, 80(5), 848–61. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Zhang, Y.; Dong, J.; Sun, Y.; Lim, B. L.; Liu, D.; Lu, Z. J. , Systematic characterization of novel lncRNAs responding to phosphate starvation in Arabidopsis thaliana. BMC Genomics 2016, 17, 655. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yamada, M.; Han, X.; Ohler, U.; Benfey, P. N. , High-Resolution Expression Map of the Arabidopsis Root Reveals Alternative Splicing and lincRNA Regulation. Dev Cell 2016, 39(4), 508–522. [Google Scholar] [CrossRef]
- Gutiérrez, F.; Arnaud, T.; Garrido, A. , Contribution of polyphenols to the oxidative stability of virgin olive oil. Journal of the Science of Food and Agriculture 2001, 81(15), 1463–1470. [Google Scholar] [CrossRef]
- Donaire, L.; Pedrola, L.; Rosa Rde, L.; Llave, C. , High-throughput sequencing of RNA silencing-associated small RNAs in olive (Olea europaea L.). PLoS One 2011, 6(11), e27916. [Google Scholar] [CrossRef] [PubMed]
- Conde, C.; Delrot, S.; Geros, H. , Physiological, biochemical and molecular changes occurring during olive development and ripening. J Plant Physiol 2008, 165(15), 1545–62. [Google Scholar] [CrossRef]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F. J.; Llorens, C.; Roig, F. J.; Parmaksiz, I.; Dundar, E.; Xie, F.; Zhang, B.; Ipek, A.; Uranbey, S.; Erayman, M.; Ilhan, E.; Badad, O.; Ghazal, H.; Lightfoot, D. A.; Kasarla, P.; Colantonio, V.; Tombuloglu, H.; Hernandez, P.; Mete, N.; Cetin, O.; Van Montagu, M.; Yang, H.; Gao, Q.; Dorado, G.; Van de Peer, Y. , Genome of wild olive and the evolution of oil biosynthesis. Proc Natl Acad Sci U S A 2017, 114(44), E9413–E9422. [Google Scholar] [CrossRef]
- Jiménez-Ruiz, J.; Ramírez Tejero, J.; Fernández Pozo, N.; Leyva-Pérez, M. D. L. O.; Yan, H.; de la Rosa, R.; Belaj, A.; Montes, E.; Rodríguez-Ariza, M.; Navarro, F.; Barroso, J.; Beuzón, C.; Valpuesta, V.; Bombarely, A.; Luque, F. , Transposon activation is a major driver in the genome evolution of cultivated olive trees ( Olea europaea L.). The Plant Genome 2020, e20010. [Google Scholar] [CrossRef]
- Popova, O. V.; Dinh, H. Q.; Aufsatz, W.; Jonak, C. , The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol Plant 2013, 6(2), 396–410. [Google Scholar] [CrossRef]
- Jiménez-Ruiz, J.; Leyva-Pérez, M. d. l. O.; Schilirò, E.; Barroso, J. B.; Bombarely, A.; Mueller, L.; Mercado-Blanco, J.; Luque, F. , Transcriptomic Analysis of Olea europaea L. Roots during the Verticillium dahliae Early Infection Process. The Plant Genome 2017, 10(1), 1–15. [Google Scholar] [CrossRef]
- Moret, M.; Ramirez-Tejero, J. A.; Serrano, A.; Ramirez-Yera, E.; Cueva-Lopez, M. D.; Belaj, A.; Leon, L.; de la Rosa, R.; Bombarely, A.; Luque, F. , Identification of Genetic Markers and Genes Putatively Involved in Determining Olive Fruit Weight. Plants (Basel) 2022, 12(1). [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Hall, B. D. , A multistep process gave rise to RNA polymerase IV of land plants. J Mol Evol 2007, 64(1), 101–12. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Jones, A. M.; Searle, I.; Patel, K.; Vogler, H.; Hubner, N. C.; Baulcombe, D. C. , An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol 2009, 16(1), 91–3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, H. , Step-wise and lineage-specific diversification of plant RNA polymerase genes and origin of the largest plant-specific subunits. New Phytol 2015, 207(4), 1198–212. [Google Scholar] [CrossRef]
- Leyva-Pérez, M. O.; Jiménez-Ruiz, J.; Gómez-Lama Cabanas, C.; Valverde-Corredor, A.; Barroso, J. B.; Luque, F.; Mercado-Blanco, J. , Tolerance of olive (Olea europaea) cv Frantoio to Verticillium dahliae relies on both basal and pathogen-induced differential transcriptomic responses. New Phytol 2018, 217(2), 671–686. [Google Scholar] [CrossRef]
- Leyva-Pérez, M. O.; Valverde-Corredor, A.; Valderrama, R.; Jiménez-Ruiz, J.; Muñoz-Mérida, A.; Trelles, O.; Barroso, J. B.; Mercado-Blanco, J.; Luque, F. , Early and delayed long-term transcriptional changes and short-term transient responses during cold acclimation in olive leaves. DNA Res 2015, 22(1), 1–11. [Google Scholar] [CrossRef]
- Ramírez-Tejero, J. A.; Jiménez-Ruiz, J.; Leyva-Pérez, M. O.; Barroso, J. B.; Luque, F. , Gene Expression Pattern in Olive Tree Organs (Olea europaea L.). Genes (Basel) 2020, 11(5). [Google Scholar] [CrossRef]
- Chekanova, J. A.; Shaw, R. J.; Belostotsky, D. A. , Analysis of an essential requirement for the poly(A) binding protein function using cross-species complementation. Curr Biol 2001, 11(15), 1207–14. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J. M.; Park, C.; Bennett, C.; Salzberg, S. L. , Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019, 37(8), 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. , The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25(16), 2078–9. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G. M.; Leek, J. T.; Salzberg, S. L. , Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016, 11(9), 1650–67. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Res 2020, 9. [Google Scholar] [CrossRef]
- Chan, P. P.; Lin, B. Y.; Mak, A. J.; Lowe, T. M. , tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res 2021, 49(16), 9077–9096. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y. J.; Yang, D. C.; Kong, L.; Hou, M.; Meng, Y. Q.; Wei, L.; Gao, G. , CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res 2017, 45(W1), W12–W16. [Google Scholar] [CrossRef]
Table 1.
Coding genes for RNA pol II, IV and V subunits in olive (Olea europaea L. cv. Picual).
Table 1.
Coding genes for RNA pol II, IV and V subunits in olive (Olea europaea L. cv. Picual).
| RNA pol II |
RNA pol IV |
RNA pol V |
| NRPB1 |
Oleur061Scf2303g05021.1 |
NRPD1 |
Oleur061Scf8288g09024.1
Oleur061Scf3115g03008.1
Oleur061Scf1883g00024.1 |
NRPE1 |
Oleur061Scf1459g03013.1
Oleur061Scf0397g00013.1
Oleur061Scf0194g01004.1 |
| Oleur061Scf0709g00017.1 |
| Oleur061Scf1475g00008.1 |
| Oleur061Scf0012g03006.1 |
| NRPB2 |
Oleur061Scf0169g06007.1
Oleur061Scf0008g04041.1
Oleur061Scf3112g05037.1 |
NRPDE2 |
Oleur061Scf2342g06016.1 |
| NRPB4 |
Oleur061Scf7473g00034.1
Oleur061Scf0021g02004.1 |
NRPDE4 |
Oleur061Scf9139g02012.1 |
| Oleur061Scf1057g07004.1 |
| Oleur061Scf2091g00019.1 |
| NRPB7 |
Oleur061Scf0456g03006.1 |
NRPDE7 |
Oleur061Scf8086g00007.1
Oleur061Scf8230g00012.1 |
| Oleur061Scf1270g16022.1 |
| Oleur061Scf3490g10013.1 |
| Oleur061Scf0186g07027.1 |
| Oleur061Scf0397g02002.1 |
| NRPB7-like |
Oleur061Scf4485g00001.1 |
|
|
|
|
| Oleur061Scf7934g03011.1 |
|
|
|
|
Table 2.
lncRNA expression analysis in flowers and at 15 days AFB.
Table 2.
lncRNA expression analysis in flowers and at 15 days AFB.
| LncRNA type |
lncRNAs |
Average RPKMs |
| |
up regulated |
down regulated |
flower |
15 days AFB |
p-value |
| Intergenic |
106 |
237 |
525.43 |
594.10 |
0.0399 |
| Intronic |
13 |
22 |
234.96 |
336.53 |
0.0023 |
| Antisense |
24 |
14 |
2484.33 |
2046.71 |
0.4484 |
| Total |
143 |
273 |
718.68 |
733.22 |
0.8365 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).











