Submitted:
29 November 2023
Posted:
30 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Transcriptomic Analysis
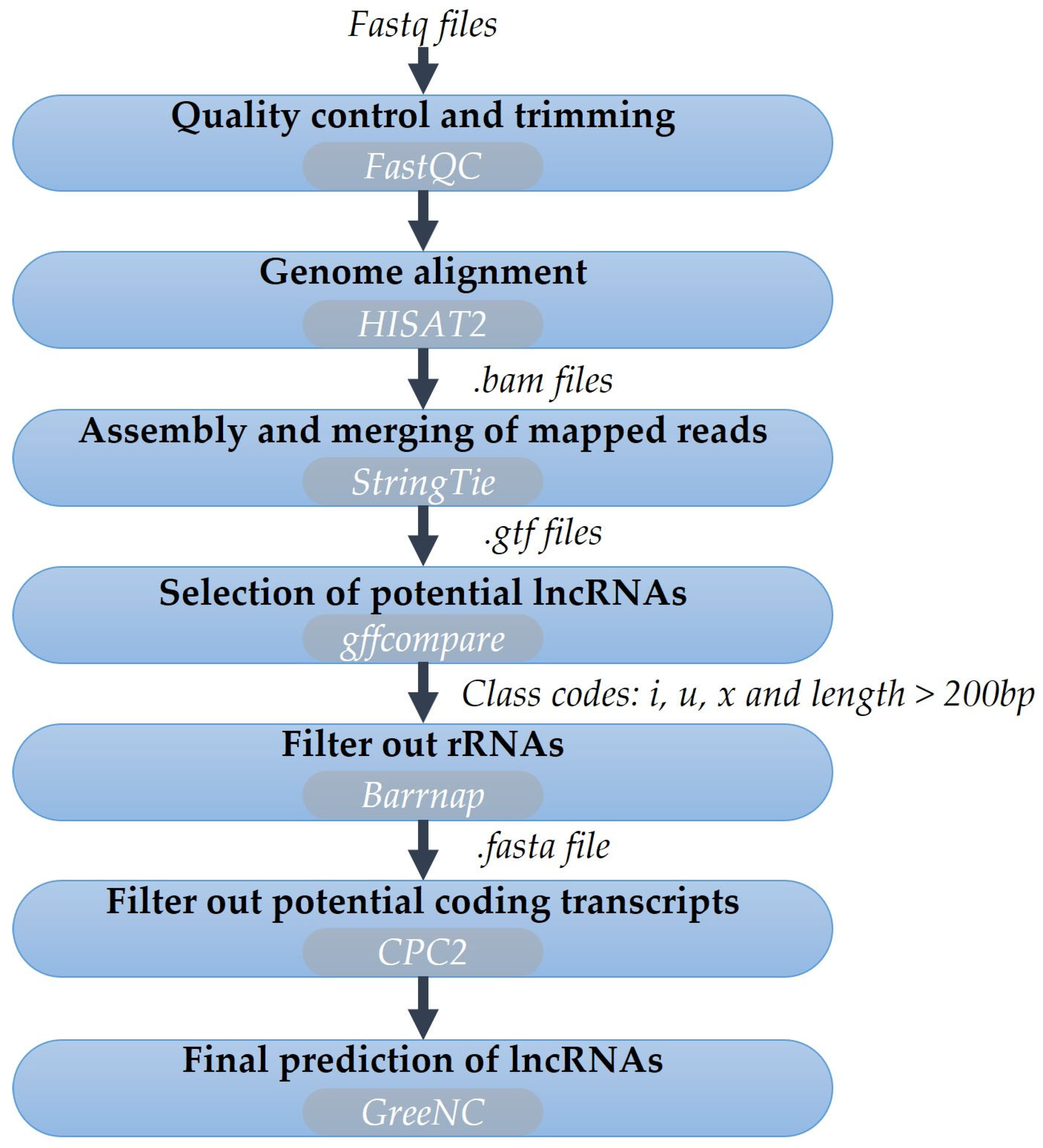
2.3. Annotation of lncRNAs in Olive
2.4. Analysis of the Differentially Expressed lncRNAs
3. Results
3.1. Olive Genes Coding for RNA pol IV and V Subunits
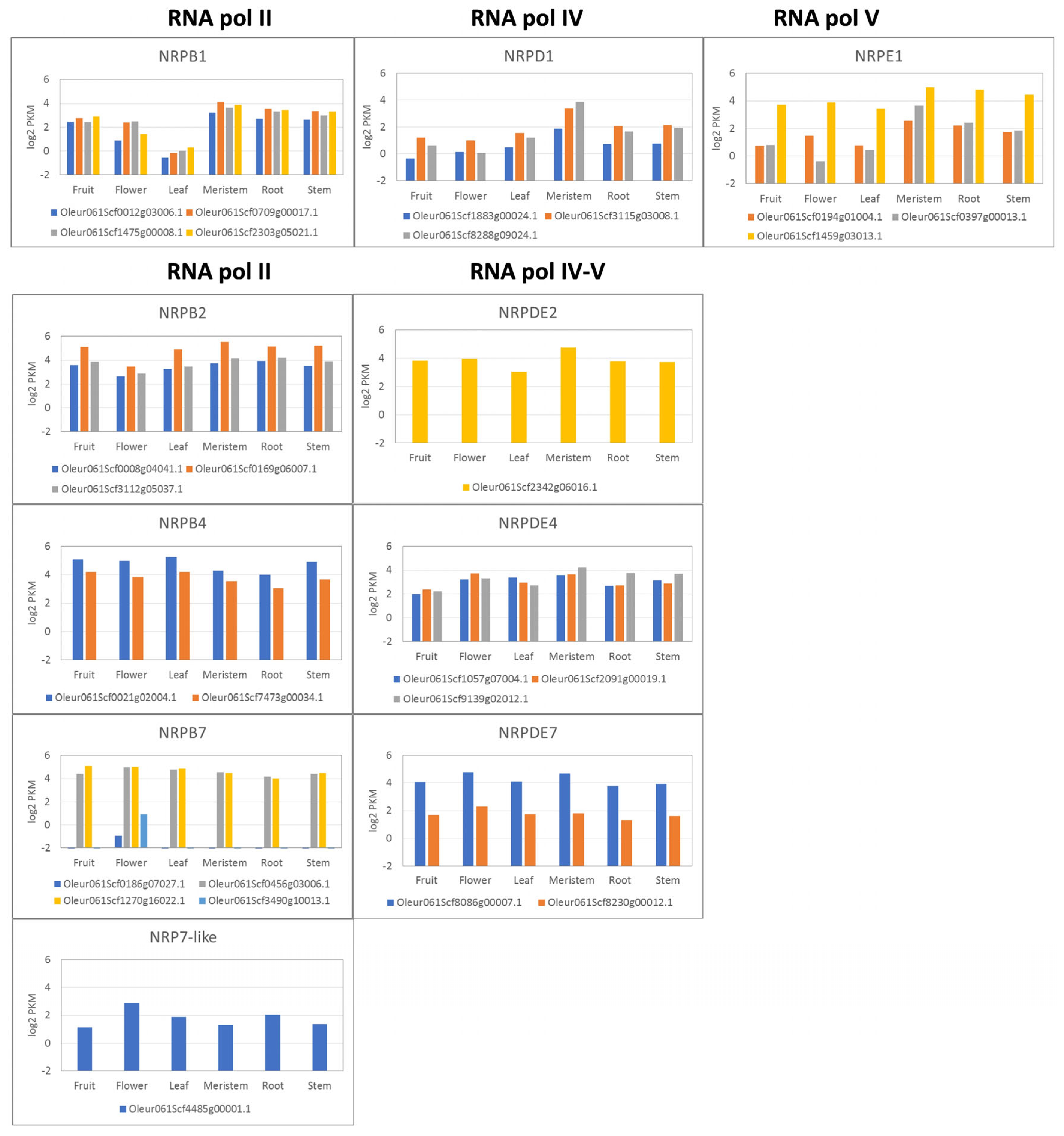
3.2. Gene Expression Profile in Different Plant Organs/Tissues
3.3. Expression Profile in Response to Biotic and Abiotic Stresses
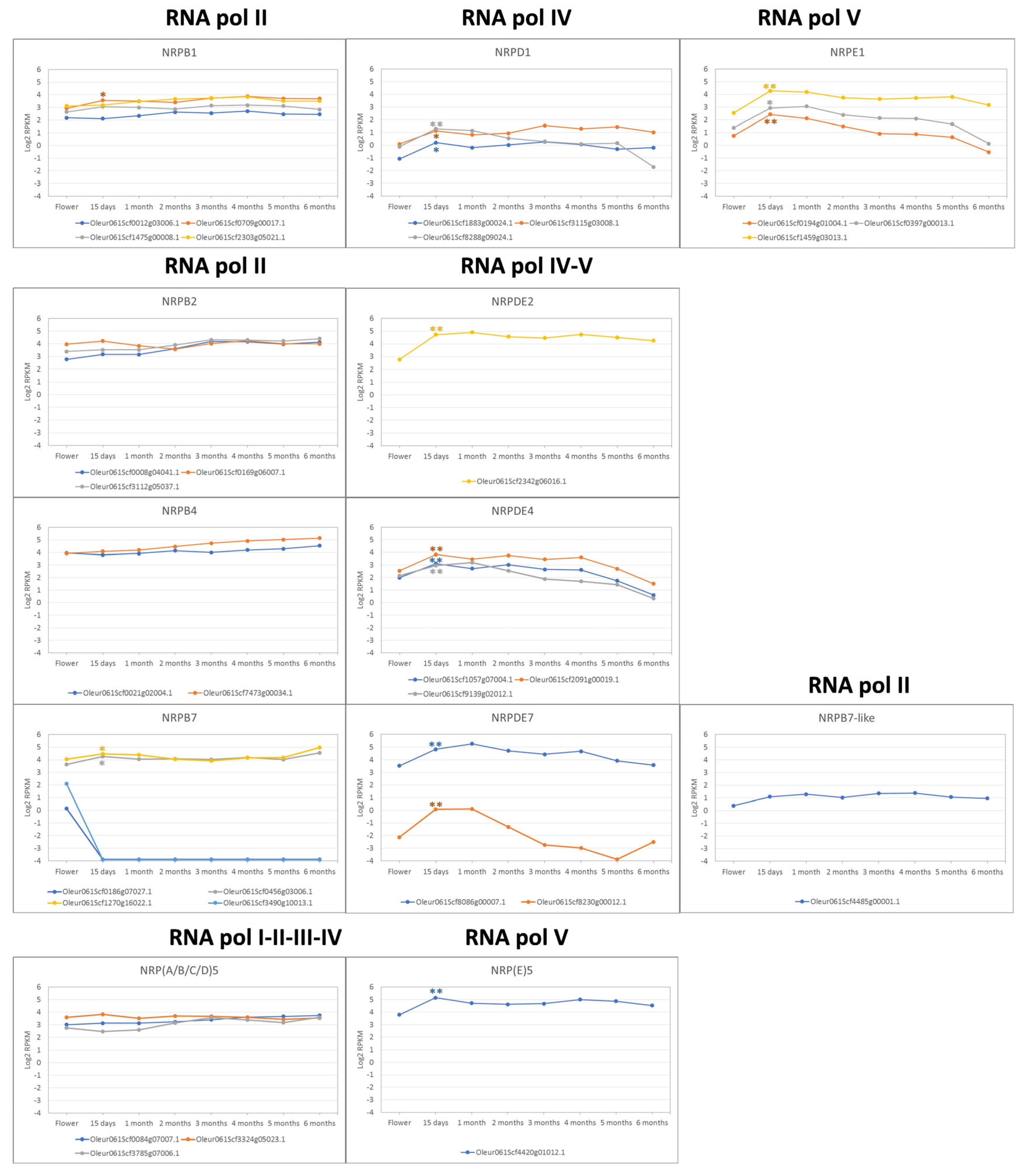
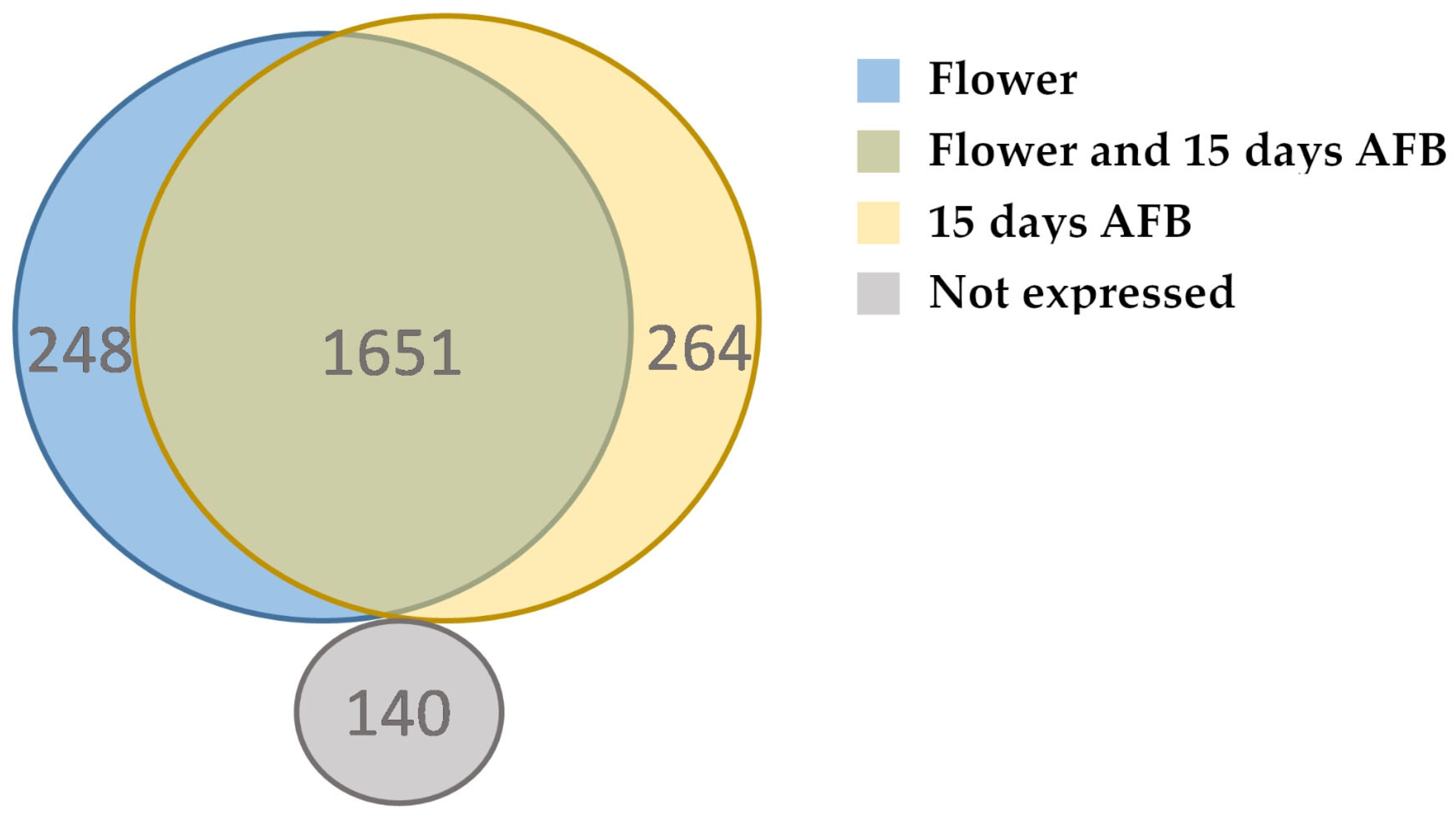
3.4. Expression Profile during Fruit Development
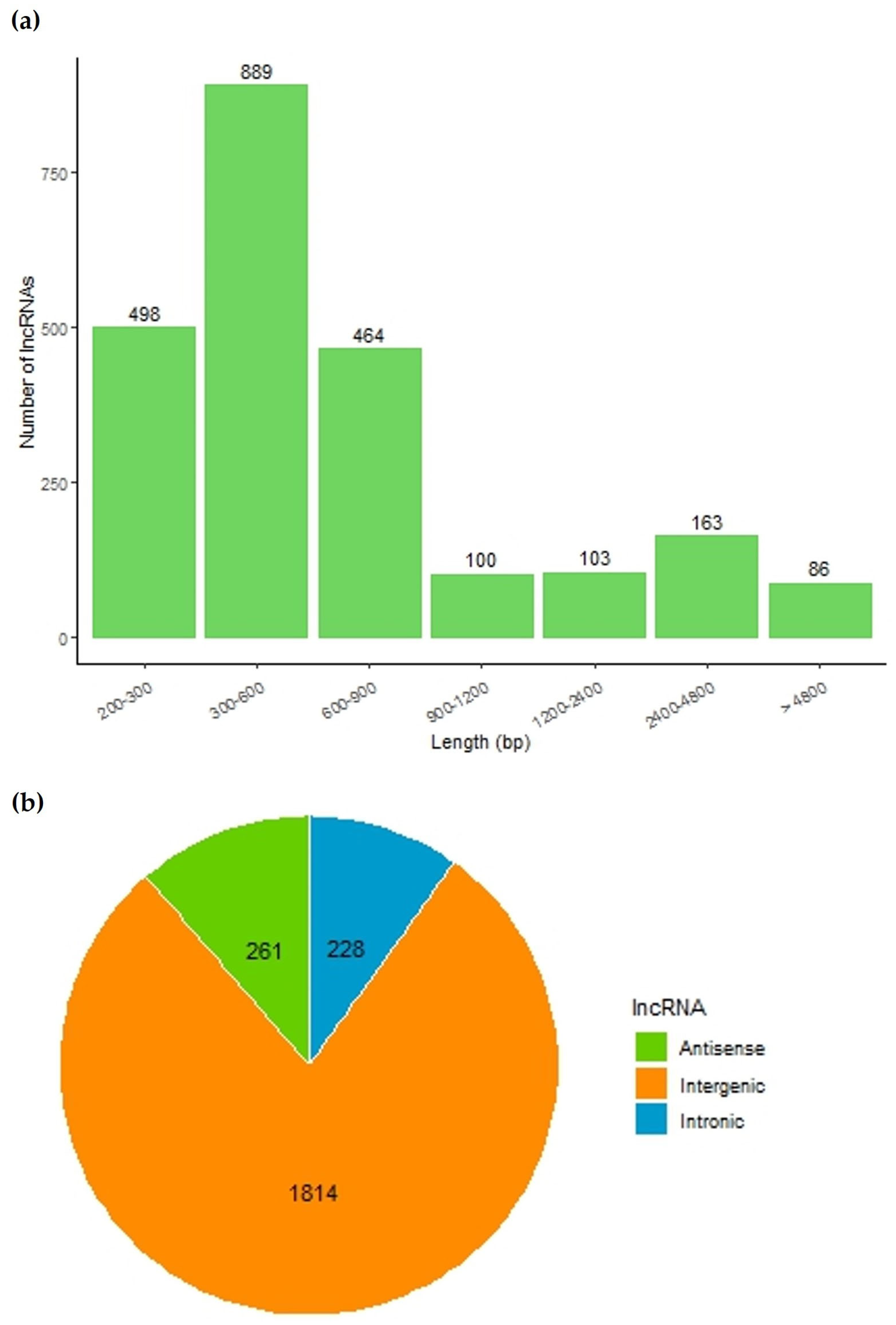
3.5. Annotation and Expression of lncRNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ream, T. S.; Haag, J. R.; Pontvianne, F.; Nicora, C. D.; Norbeck, A. D.; Pasa-Tolic, L.; Pikaard, C. S. Subunit compositions of Arabidopsis RNA polymerases I and III reveal Pol I- and Pol III-specific forms of the AC40 subunit and alternative forms of the C53 subunit. Nucleic Acids Res 2015, 43, 4163–4178. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Bermúdez, A.; Martínez-Fernández, V.; Garrido-Godino, A. I.; Navarro, F. Subunits common to RNA polymerases. In The Yeast Role in Medical Applications; Abdulkhair, W. M. H., Ed.; IntechOpen: London, 2017; Volume 1, pp. 151–165. [Google Scholar]
- Werner, F.; Grohmann, D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 2011, 9, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Law, J. A. RNA Pol IV and V in gene silencing: Rebel polymerases evolving away from Pol II's rules. Curr Opin Plant Biol 2015, 27, 154–164. [Google Scholar] [CrossRef]
- Tucker, S. L.; Reece, J.; Ream, T. S.; Pikaard, C. S. Evolutionary history of plant multisubunit RNA polymerases IV and V: subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harb Symp Quant Biol 2010, 75, 285–297. [Google Scholar] [CrossRef]
- Lahmy, S.; Bies-Etheve, N.; Lagrange, T. Plant-specific multisubunit RNA polymerase in gene silencing. Epigenetics 2010, 5, 4–8. [Google Scholar] [CrossRef]
- Haag, J. R.; Pikaard, C. S. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 2011, 12, 483–492. [Google Scholar] [CrossRef]
- Lopez, A.; Ramirez, V.; Garcia-Andrade, J.; Flors, V.; Vera, P. The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genet 2011, 7, e1002434. [Google Scholar] [CrossRef] [PubMed]
- Ream, T.; Haag, J.; Pikaard, C. Plant Multisubunit RNA Polymerases IV and V. In Nucleic Acid Polymerases; Trakselis, K. S. M. a. M. A., Ed.; Springer-Verlag: Heidelberg, 2014; Volume 30. [Google Scholar]
- Moo, L. d. R. C.; González, A. K.; Rodríguez-Zapata, L. C.; Suarez, V.; Castaño, E. Expression of RNA polymerase IV and V in Oryza sativa. Electronic Journal of Biotechnology 2012, 15, 9-9. [Google Scholar] [CrossRef]
- Huang, Y.; Kendall, T.; Forsythe, E. S.; Dorantes-Acosta, A.; Li, S.; Caballero-Perez, J.; Chen, X.; Arteaga-Vazquez, M.; Beilstein, M. A.; Mosher, R. A. Ancient Origin and Recent Innovations of RNA Polymerase IV and V. Mol Biol Evol 2015, 32, 1788–1799. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, X. Q.; Xie, H. T.; Zhao, S. S.; Wu, J. G. Multifaceted roles of RNA polymerase IV in plant growth and development. J Exp Bot 2020, 71, 5725–5732. [Google Scholar] [CrossRef]
- Barba-Aliaga, M.; Alepuz, P.; Perez-Ortin, J. E. Eukaryotic RNA Polymerases: The Many Ways to Transcribe a Gene. Front Mol Biosci 2021, 8, 663209. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.; Thuriaux, P.; Soutourina, J. Structure-function analysis of RNA polymerases I and III. Curr Opin Struct Biol 2009, 19, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Morcillo, M.; Taylor, N. M.; Gruene, T.; Legrand, P.; Rashid, U. J.; Ruiz, F. M.; Steuerwald, U.; Muller, C. W.; Fernandez-Tornero, C. Solving the RNA polymerase I structural puzzle. Acta Crystallogr D Biol Crystallogr 2014, 70 Pt 10, 2570–2582. [Google Scholar] [CrossRef]
- Moir, R. D.; Willis, I. M. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim Biophys Acta 2013, 1829, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Tornero, C.; Bottcher, B.; Rashid, U. J.; Muller, C. W. Analyzing RNA polymerase III by electron cryomicroscopy. RNA Biol 2011, 8, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Turowski, T. W.; Boguta, M. Specific Features of RNA Polymerases I and III: Structure and Assembly. Front Mol Biosci 2021, 8, 680090. [Google Scholar] [CrossRef] [PubMed]
- Armache, K. J.; Mitterweger, S.; Meinhart, A.; Cramer, P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem 2005, 280, 7131–7134. [Google Scholar] [CrossRef] [PubMed]
- Perez-Ortin, J. E.; Mena, A.; Barba-Aliaga, M.; Singh, A.; Chavez, S.; Garcia-Martinez, J. Cell volume homeostatically controls the rDNA repeat copy number and rRNA synthesis rate in yeast. PLoS Genet 2021, 17, e1009520. [Google Scholar] [CrossRef] [PubMed]
- Tan, E. H.; Blevins, T.; Ream, T. S.; Pikaard, C. S. Functional consequences of subunit diversity in RNA polymerases II and V. Cell Rep 2012, 1, 208–214. [Google Scholar] [CrossRef]
- Ream, T. S.; Haag, J. R.; Wierzbicki, A. T.; Nicora, C. D.; Norbeck, A. D.; Zhu, J. K.; Hagen, G.; Guilfoyle, T. J.; Pasa-Tolic, L.; Pikaard, C. S. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 2009, 33, 192–203. [Google Scholar] [CrossRef]
- Pikaard, C. S.; Tucker, S. RNA-silencing enzymes Pol IV and Pol V in maize: more than one flavor? PLoS Genet 2009, 5, e1000736. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhou, W.; Zhang, X.; Li, Y. Roles of long non-coding RNAs in plant immunity. PLoS Pathog 2023, 19, e1011340. [Google Scholar] [CrossRef] [PubMed]
- Haag, J. R.; Brower-Toland, B.; Krieger, E. K.; Sidorenko, L.; Nicora, C. D.; Norbeck, A. D.; Irsigler, A.; LaRue, H.; Brzeski, J.; McGinnis, K.; Ivashuta, S.; Pasa-Tolic, L.; Chandler, V. L.; Pikaard, C. S. Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Rep 2014, 9, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Marcussen, T.; Oxelman, B.; Skog, A.; Jakobsen, K. S. Evolution of plant RNA polymerase IV/V genes: evidence of subneofunctionalization of duplicated NRPD2/NRPE2-like paralogs in Viola (Violaceae). BMC Evol Biol 2010, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- He, X. J.; Hsu, Y. F.; Pontes, O.; Zhu, J.; Lu, J.; Bressan, R. A.; Pikaard, C.; Wang, C. S.; Zhu, J. K. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev 2009, 23, 318–330. [Google Scholar] [CrossRef]
- Fernandez-Parras, I.; Ramirez-Tejero, J. A.; Luque, F.; Navarro, F. Several Isoforms for Each Subunit Shared by RNA Polymerases are Differentially Expressed in the Cultivated Olive Tree (Olea europaea L.). Front Mol Biosci 2021, 8, 679292. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, J. T.; Seetharam, A. S.; Hufford, M. B.; Beilstein, M. A.; Mosher, R. A. Evidence for a Unique DNA-Dependent RNA Polymerase in Cereal Crops. Mol Biol Evol 2018, 35, 2454–2462. [Google Scholar] [CrossRef]
- Chakraborty, T.; Trujillo, J. T.; Kendall, T.; Mosher, R. A. A null allele of the pol IV second subunit impacts stature and reproductive development in Oryza sativa. Plant J 2022, 111, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Bohmdorfer, G.; Rowley, M. J.; Kucinski, J.; Zhu, Y.; Amies, I.; Wierzbicki, A. T. RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J 2014, 79, 181–191. [Google Scholar] [CrossRef]
- Wierzbicki, A. T.; Haag, J. R.; Pikaard, C. S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 2008, 135, 635–648. [Google Scholar] [CrossRef]
- Liu, X.; Hao, L.; Li, D.; Zhu, L.; Hu, S. Long non-coding RNAs and their biological roles in plants. Genomics Proteomics Bioinformatics 2015, 13, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Vandivier, L. E.; Tu, B.; Gao, L.; Won, S. Y.; Zheng, B.; Gregory, B. D.; Chen, X. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res 2015, 25, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. V.; Chekanova, J. A. Long Noncoding RNAs in Plants. Adv Exp Med Biol 2017, 1008, 133–154. [Google Scholar]
- Chen, L.; Zhu, Q. H.; Kaufmann, K. Long non-coding RNAs in plants: emerging modulators of gene activity in development and stress responses. Planta 2020, 252, 92. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Kaya, S. B.; Cagirici, H. B. Long Non-coding RNA in Plants in the Era of Reference Sequences. Front Plant Sci 2020, 11, 276. [Google Scholar] [CrossRef]
- Wierzbicki, A. T.; Blevins, T.; Swiezewski, S. Long Noncoding RNAs in Plants. Annu Rev Plant Biol 2021, 72, 245–271. [Google Scholar] [CrossRef] [PubMed]
- Rai, M. I.; Alam, M.; Lightfoot, D. A.; Gurha, P.; Afzal, A. J. Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Heo, J. B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef]
- Shin, J. H.; Chekanova, J. A. Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis. PLoS Genet 2014, 10, e1004612. [Google Scholar] [CrossRef]
- Kim, D. H.; Sung, S. Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev Cell 2017, 40, 302–312 e4. [Google Scholar]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Zhang, T.; Qi, Y.; Gerstein, M. B.; Guo, Y.; Lu, Z. J. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J 2014, 80, 848–861. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, Y.; Dong, J.; Sun, Y.; Lim, B. L.; Liu, D.; Lu, Z. J. Systematic characterization of novel lncRNAs responding to phosphate starvation in Arabidopsis thaliana. BMC Genomics 2016, 17, 655. [Google Scholar] [CrossRef]
- Li, S.; Yamada, M.; Han, X.; Ohler, U.; Benfey, P. N. High-Resolution Expression Map of the Arabidopsis Root Reveals Alternative Splicing and lincRNA Regulation. Dev Cell 2016, 39, 508–522. [Google Scholar] [CrossRef]
- Gutiérrez, F.; Arnaud, T.; Garrido, A. Contribution of polyphenols to the oxidative stability of virgin olive oil. Journal of the Science of Food and Agriculture 2001, 81, 1463–1470. [Google Scholar] [CrossRef]
- Donaire, L.; Pedrola, L.; Rosa Rde, L.; Llave, C. High-throughput sequencing of RNA silencing-associated small RNAs in olive (Olea europaea L.). PLoS One 2011, 6, e27916. [Google Scholar]
- Conde, C.; Delrot, S.; Geros, H. Physiological, biochemical and molecular changes occurring during olive development and ripening. J Plant Physiol 2008, 165, 1545–1562. [Google Scholar] [CrossRef] [PubMed]
- Moret, M.; Ramírez-Tejero, J. A.; Serrano, A.; Ramírez-Yera, E.; Cueva-López, M. D.; Belaj, A.; León, L.; de la Rosa, R.; Bombarely, A.; Luque, F. Identification of Genetic Markers and Genes Putatively Involved in Determining Olive Fruit Weight. Plants 2023, 12. [Google Scholar]
- Ramírez-Tejero, J. A.; Jiménez-Ruiz, J.; Leyva-Pérez, M. O.; Barroso, J. B.; Luque, F. Gene Expression Pattern in Olive Tree Organs (Olea europaea L.). Genes 2020, 11. [Google Scholar]
- Leyva-Pérez, M. O.; Valverde-Corredor, A.; Valderrama, R.; Jiménez-Ruiz, J.; Muñoz-Mérida, A.; Trelles, O.; Barroso, J. B.; Mercado-Blanco, J.; Luque, F. Early and delayed long-term transcriptional changes and short-term transient responses during cold acclimation in olive leaves. DNA Res 2015, 22, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Ruiz, J.; Ramírez Tejero, J.; Fernández Pozo, N.; Leyva-Pérez, M. D. L. O.; Yan, H.; de la Rosa, R.; Belaj, A.; Montes, E.; Rodríguez-Ariza, M.; Navarro, F.; Barroso, J.; Beuzón, C.; Valpuesta, V.; Bombarely, A.; Luque, F. Transposon activation is a major driver in the genome evolution of cultivated olive trees ( Olea europaea L.). The Plant Genome 2020, e20010. [Google Scholar]
- Kim, D.; Paggi, J. M.; Park, C.; Bennett, C.; Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G. M.; Leek, J. T.; Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Res 2020, 9. [Google Scholar]
- Chan, P. P.; Lin, B. Y.; Mak, A. J.; Lowe, T. M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Kang, Y. J.; Yang, D. C.; Kong, L.; Hou, M.; Meng, Y. Q.; Wei, L.; Gao, G. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F. J.; Llorens, C.; Roig, F. J.; Parmaksiz, I.; Dundar, E.; Xie, F.; Zhang, B.; Ipek, A.; Uranbey, S.; Erayman, M.; Ilhan, E.; Badad, O.; Ghazal, H.; Lightfoot, D. A.; Kasarla, P.; Colantonio, V.; Tombuloglu, H.; Hernandez, P.; Mete, N.; Cetin, O.; Van Montagu, M.; Yang, H.; Gao, Q.; Dorado, G.; Van de Peer, Y. Genome of wild olive and the evolution of oil biosynthesis. Proc Natl Acad Sci U S A 2017, 114, E9413–E9422. [Google Scholar] [CrossRef] [PubMed]
- Popova, O. V.; Dinh, H. Q.; Aufsatz, W.; Jonak, C. The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol Plant 2013, 6, 396–410. [Google Scholar] [CrossRef]
- Jiménez-Ruiz, J.; Leyva-Pérez, M. d. l. O.; Schilirò, E.; Barroso, J. B.; Bombarely, A.; Mueller, L.; Mercado-Blanco, J.; Luque, F. Transcriptomic Analysis of Olea europaea L. Roots during the Verticillium dahliae Early Infection Process. The Plant Genome 2017, 10, 1–15. [Google Scholar] [CrossRef]
- Luo, J.; Hall, B. D. A multistep process gave rise to RNA polymerase IV of land plants. J Mol Evol 2007, 64, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Jones, A. M.; Searle, I.; Patel, K.; Vogler, H.; Hubner, N. C.; Baulcombe, D. C. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol 2009, 16, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, H. Step-wise and lineage-specific diversification of plant RNA polymerase genes and origin of the largest plant-specific subunits. New Phytol 2015, 207, 1198–1212. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Pérez, M. O.; Jiménez-Ruiz, J.; Gómez-Lama Cabanas, C.; Valverde-Corredor, A.; Barroso, J. B.; Luque, F.; Mercado-Blanco, J. Tolerance of olive (Olea europaea) cv Frantoio to Verticillium dahliae relies on both basal and pathogen-induced differential transcriptomic responses. New Phytol 2018, 217, 671–686. [Google Scholar] [CrossRef]
- Chekanova, J. A.; Shaw, R. J.; Belostotsky, D. A. Analysis of an essential requirement for the poly(A) binding protein function using cross-species complementation. Curr Biol 2001, 11, 1207–1214. [Google Scholar] [CrossRef]
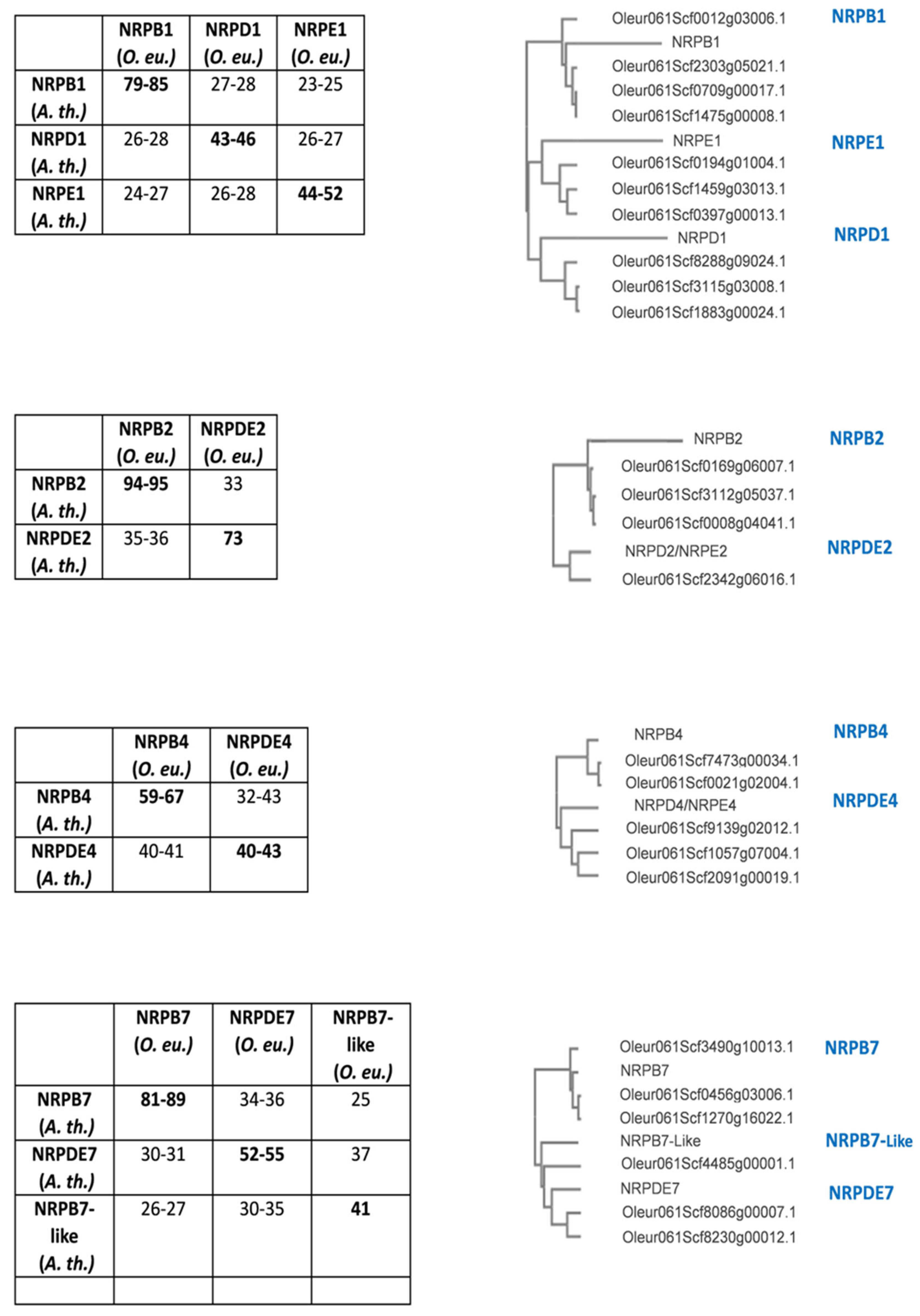
| RNA pol II | RNA pol IV | RNA pol V | |||
|---|---|---|---|---|---|
| NRPB1 | Oleur061Scf2303g05021.1 | NRPD1 | Oleur061Scf8288g09024.1 Oleur061Scf3115g03008.1 Oleur061Scf1883g00024.1 |
NRPE1 | Oleur061Scf1459g03013.1 Oleur061Scf0397g00013.1 Oleur061Scf0194g01004.1 |
| Oleur061Scf0709g00017.1 | |||||
| Oleur061Scf1475g00008.1 | |||||
| Oleur061Scf0012g03006.1 | |||||
| NRPB2 | Oleur061Scf0169g06007.1 Oleur061Scf0008g04041.1 Oleur061Scf3112g05037.1 |
NRPDE2 | Oleur061Scf2342g06016.1 | ||
| NRPB4 | Oleur061Scf7473g00034.1 Oleur061Scf0021g02004.1 |
NRPDE4 | Oleur061Scf9139g02012.1 | ||
| Oleur061Scf1057g07004.1 | |||||
| Oleur061Scf2091g00019.1 | |||||
| NRPB7 | Oleur061Scf0456g03006.1 | NRPDE7 | Oleur061Scf8086g00007.1 Oleur061Scf8230g00012.1 |
||
| Oleur061Scf1270g16022.1 | |||||
| Oleur061Scf3490g10013.1 | |||||
| Oleur061Scf0186g07027.1 | |||||
| Oleur061Scf0397g02002.1 | |||||
| NRPB7-like | Oleur061Scf4485g00001.1 | ||||
| Oleur061Scf7934g03011.1 | |||||
| LncRNA type | lncRNAs | Average RPKMs | |||
|---|---|---|---|---|---|
| up regulated | down regulated | flower | 15 days AFB | p-value | |
| Intergenic | 106 | 237 | 525.43 | 594.10 | 0.0399 |
| Intronic | 13 | 22 | 234.96 | 336.53 | 0.0023 |
| Antisense | 24 | 14 | 2484.33 | 2046.71 | 0.4484 |
| Total | 143 | 273 | 718.68 | 733.22 | 0.8365 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





