Submitted:
17 October 2023
Posted:
17 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Main Content
Context
Methods
Sample collection and sequencing
Genome assembly, annotation and assessment
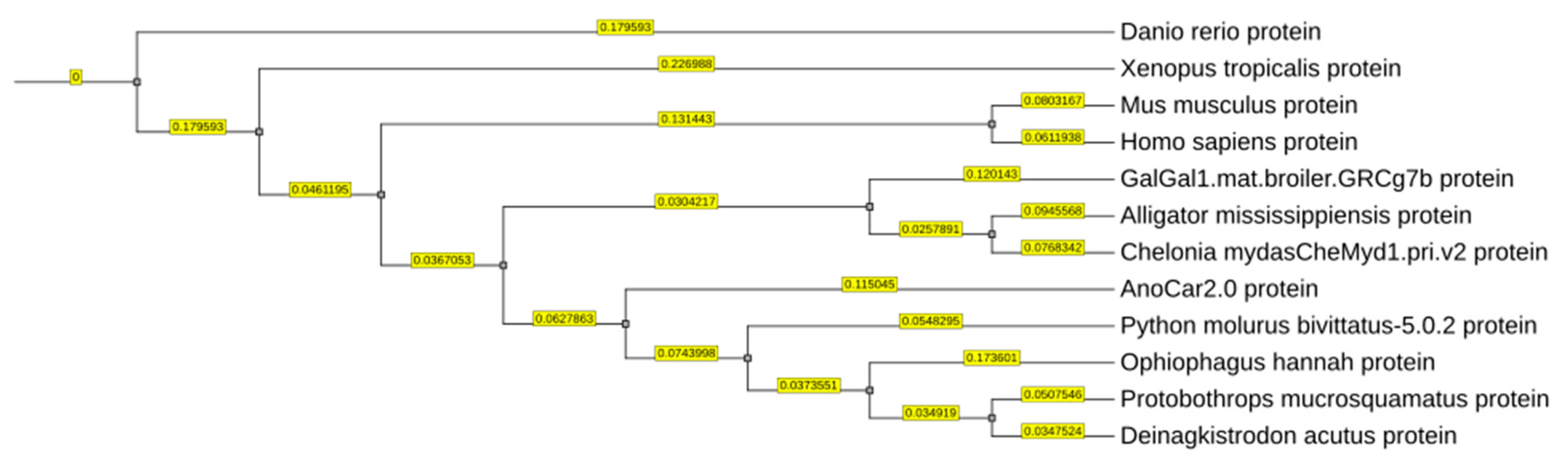
Results
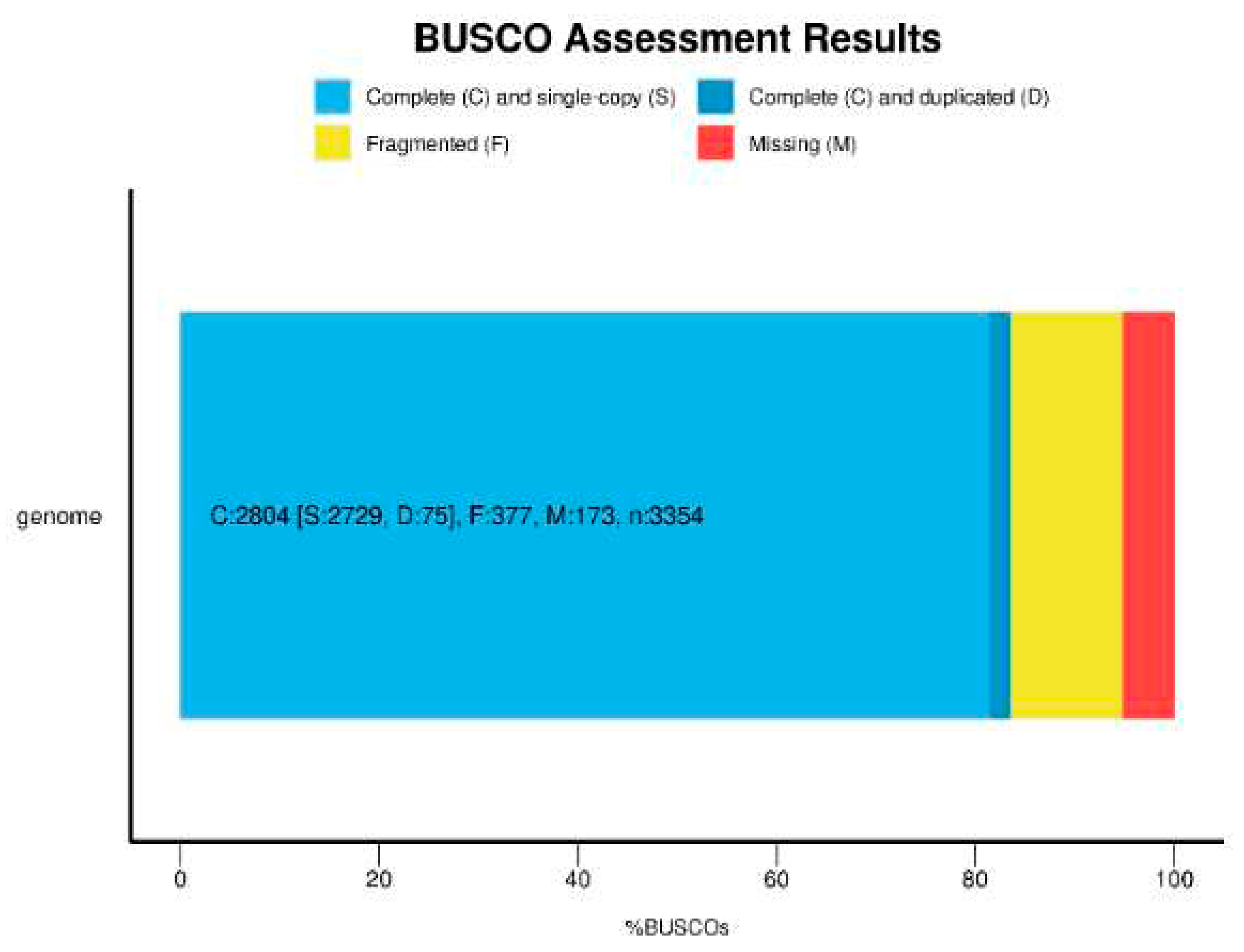
Data validation and quality control
Author Contributions
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Reuse Potential
References
- Liu C-C, Wu C-J, Hsiao Y-C, Yang Y-H, Liu K-L, Huang G-J, et al.. Snake venom proteome of Protobothrops mucrosquamatus in Taiwan: Delaying venom-induced lethality in a rodent model by inhibition of phospholipase A2 activity with varespladib. Journal of Proteomics. 2021. [CrossRef]
- Valenta J, Stach Z, Otahal M. Protobothrops mangshanensis bite: First clinical report of envenoming and its treatment. Biomedical papers of the Medical Faculty of the University Palacký, Olomouc, Czechoslovakia. 2012. [CrossRef]
- He Y, Sun W-Q, Li J-L, Zhang S, Li J, You H-J, et al.. Yuanmaotoufu Shedu Duxing Ji Huifu Guilv[Toxicity and Recovery of Protobothrops Mucrosquamatus Venoms]. Xiandai Shengwu Yixue Jinzhan. 2016; 0:1623–62016;
- Mao Y-C, Liu P-Y, Chiang L-C, Lee C-H, Lai C-S, Lai K-L, et al.. Clinical manifestations and treatments of Protobothrops mucrosquamatus bite and associated factors for wound necrosis and subsequent debridement and finger or toe amputation surgery. Clinical Toxicology. Taylor & Francis; 2021. [CrossRef]
- Zeng F, Chen C, Chen X, Zhang L, Liu M. Small Incisions Combined with Negative-Pressure Wound Therapy for Treatment of Protobothrops Mucrosquamatus Bite Envenomation: A New Treatment Strategy. Med Sci Monit. 2019. [CrossRef]
- Lin C-C, Chen Y-C, Goh ZNL, Seak C-K, Seak JC-Y, Shi-Ying G, et al.. Wound Infections of Snakebites from the Venomous Protobothrops mucrosquamatus and Viridovipera stejnegeri in Taiwan: Bacteriology, Antibiotic Susceptibility, and Predicting the Need for Antibiotics—A BITE Study. Toxins. Multidisciplinary Digital Publishing Institute; 2020. [CrossRef]
- Offor BC, Muller B, Piater LA. A Review of the Proteomic Profiling of African Viperidae and Elapidae Snake Venoms and Their Antivenom Neutralisation. Toxins. Multidisciplinary Digital Publishing Institute; 2022. [CrossRef]
- Whitaker R, Whitaker S. Venom, antivenom production and the medically important snakes of India. Current Science. Temporary Publisher; 103:635–432012;
- Chippaux J-P, Goyffon M. Venoms, antivenoms and immunotherapy. Toxicon. 1998. [CrossRef]
- Vogel G, Pan H, Chettri B, Yang D, Jiang K, Wang K, et al.. A New Species of the Genus Protobothrops (Squamata: Viperidae) from Southern Tibet, China and Sikkim, India. Asian Herpetological Research. 2013. [CrossRef]
- Xin H, Tao P, Demin H, Liang Z, Yinxu H, Lei Y, et al.. A New Species of the Genus Protobothrops (Squamata: Viperidae: Crotalinae) from the Dabie Mountains, Anhui, China: A New Species of the Genus Protobothrops (Squamata: Viperidae: Crotalinae) from the Dabie Mountains, Anhui, China. Asian Herpetological Research. 2012. [CrossRef]
- Tan CH. Snake Venomics: Fundamentals, Recent Updates, and a Look to the Next Decade. Toxins. Multidisciplinary Digital Publishing Institute; 2022. [CrossRef]
- Aird SD, Aggarwal S, Villar-Briones A, Tin MM-Y, Terada K, Mikheyev AS. Snake venoms are integrated systems, but abundant venom proteins evolve more rapidly. BMC Genomics. 2015. [CrossRef]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: the evolutionary novelty of venoms. Trends in Ecology & Evolution. 2013. [CrossRef]
- Aird SD, Arora J, Barua A, Qiu L, Terada K, Mikheyev AS. Population Genomic Analysis of a Pitviper Reveals Microevolutionary Forces Underlying Venom Chemistry. Genome Biology and Evolution. 2017. [CrossRef]
- Rao W, Kalogeropoulos K, Allentoft ME, Gopalakrishnan S, Zhao W, Workman CT, et al.. The rise of genomics in snake venom research: recent advances and future perspectives. GigaScience. 2022. [CrossRef]
- Wang O, Chin R, Cheng X, Wu MKY, Mao Q, Tang J, et al.. Efficient and unique cobarcoding of second-generation sequencing reads from long DNA molecules enabling cost-effective and accurate sequencing, haplotyping, and de novo assembly. Genome Res. 2019. [CrossRef]
- Liu B, Cui L, Deng Z, Ma Y, Yang D, Gong Y, et al.. The annotation pipeline for the genome of a snake. 2023. https://www.protocols.io/view/the-annotation-pipeline-for-the-genome-of-a-snake-cnv3ve8n.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, et al.. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012. [CrossRef]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 1999. [CrossRef]
- Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Research. 2007. [CrossRef]
- Smit AFA, Hubley R, Green P. RepeatModeler Open-1.0. 2008–2015. Seattle, USA: Institute for Systems Biology Available from: httpwww repeatmasker org, Last Accessed May. 1:20182015;
- arailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences Curr Protoc Bioinformatics. 2009. Chapter.
- Tempel S. Using and understanding RepeatMasker. Mobile genetic elements: protocols and genomic applications. Springer; :29–51 2012.
- Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Research. 2004. [CrossRef]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014. [CrossRef]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al.. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. Nature Publishing Group; 2013. [CrossRef]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, et al.. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008. [CrossRef]
- Mount DW. Using the Basic Local Alignment Search Tool (BLAST). Cold Spring Harb Protoc. Cold Spring Harbor Laboratory Press; 2007. [CrossRef]
- Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004. [CrossRef]
- Campbell MS, Holt C, Moore B, Yandell M. Genome Annotation and Curation Using MAKER and MAKER-P. Current Protocols in Bioinformatics. 2014. [CrossRef]
- Wick RR, Holt KE. Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Research. F1000 Research Limited London, UK; 8:21382021.
- Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology. 2015. [CrossRef]
- Margres MJ, Rautsaw RM, Strickland JL, Mason AJ, Schramer TD, Hofmann EP, et al.. The Tiger Rattlesnake genome reveals a complex genotype underlying a simple venom phenotype. Proceedings of the National Academy of Sciences. Proceedings of the National Academy of Sciences; 2021. [CrossRef]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, et al.. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014. [CrossRef]
- Kanehisa M. The KEGG Database. ‘In Silico’ Simulation of Biological Processes. John Wiley & Sons, Ltd.
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Research. 2000. [CrossRef]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015. [CrossRef]
- Guo X, Chen F, Gao F, Li L, Liu K, You L, et al.. CNSA: a data repository for archiving omics data. Database. 2020. [CrossRef]
- Chen FZ, You LJ, Yang F, Wang LN, Guo XQ, Gao F, et al.. CNGBdb: China National GeneBank DataBase. Yi Chuan. 2020. [CrossRef]

| Base Number | GC content(%) | Q20(%) | Q30(%) | ||
| WGS | fq1 | 52036970400 | 40.30 | 97.58 | 92.48 |
| fq2 | 52036970400 | 40.23 | 97.98 | 92.71 | |
| stLFR | fq1 | 104698910600 | 38.89 | 96.9 | 90.75 |
| fq2 | 136108583780 | 41.72 | 97.79 | 91.85 | |
| Statistical level | Original | Scaffold >(500)bp | ||||
| scaffold | contig | contig>(500) | scaffold | contig | ||
| Total number (>) | 203555 | 287462 | 192124 | 149173 | 232200 | |
| Total length of (bp) | 1530648812 | 1481196605 | 1457896424 | 1512499815 | 1463075630 | |
| Average length (bp) | 7519.58 | 5152.67 | 7588.31 | 10139.23 | 6300.93 | |
| N50 Length (bp) | 380005 | 36547 | 37585 | 390274 | 37334 | |
| N90 Length (bp) | 2960 | 2304 | 2773 | 3453 | 2667 | |
| Maximum length (bp) | 5566463 | 488153 | 488153 | 5566463 | 488153 | |
| GC content (%) | 39.86 | 39.86 | 39.79 | 39.8 | 39.8 | |
| Type | Repeat Size | % of genome |
| Trf | 48630912 | 3.177144 |
| Repeatmasker | 248960159 | 16.265008 |
| Proteinmask | 178699911 | 11.674782 |
| De novo | 591205406 | 38.624497 |
| Total | 630311866 | 41.179391 |
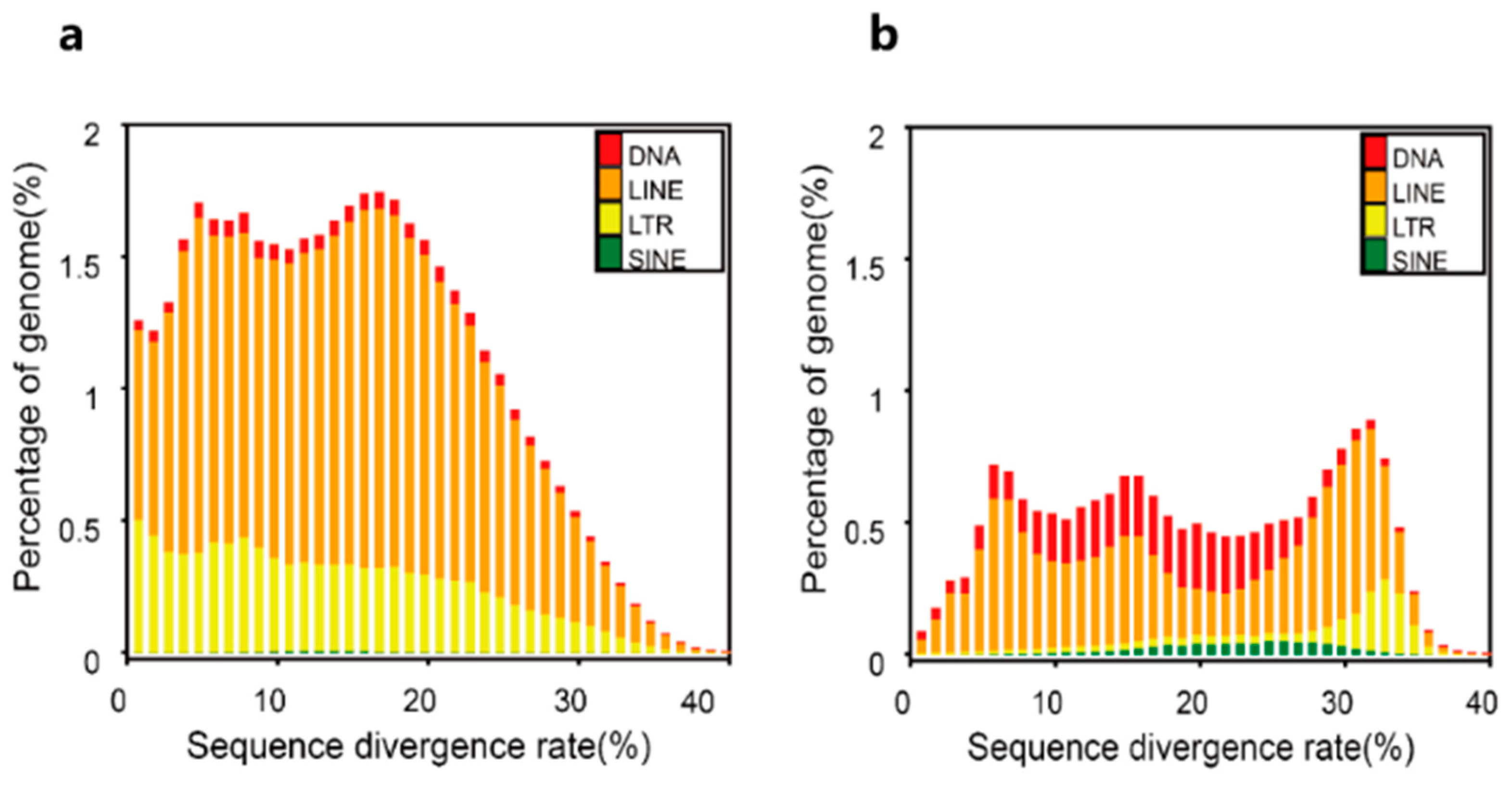
| Type | Repbase TEs | TE protiens | De novo | Combined TEs | ||||
| Length (Bp) | % in genome | Length (Bp) | % in genome | Length (Bp) | % in genome | Length (Bp) | % in genome | |
| DNA | 54802686 | 3.580357 | 2721607 | 0.177807 | 23812202 | 1.555693 | 75566775 | 4.936911 |
| LINE | 173499745 | 11.335046 | 145892994 | 9.531448 | 446008208 | 29.138507 | 494919112 | 32.333943 |
| SINE | 11128833 | 0.727066 | 0 | 0 | 1414004 | 0.092379 | 12299674 | 0.80356 |
| LTR | 27382417 | 1.788942 | 30199813 | 1.973007 | 165177572 | 10.791344 | 175979322 | 11.497041 |
| Other | 95860 | 0.006263 | 0 | 0 | 0 | 0 | 95860 | 0.006263 |
| Total | 248960159 | 16.265008 | 178699911 | 11.674782 | 588493585 | 38.447329 | 618611286 | 40.414972 |
| Type | Copy(w) | Average length(bp) | Total length(bp) | % of genome | |
| miRNA | 387 | 115.3540052 | 44642 | 0.002917 | |
| tRNA | 319 | 76.38244514 | 24366 | 0.001592 | |
| rRNA | rRNA | 75 | 111.8266667 | 8387 | 0.000548 |
| 18S | 18 | 141.5555556 | 2548 | 0.000166 | |
| 28S | 52 | 104.3269231 | 5425 | 0.000354 | |
| snRNA | snRNA | 289 | 115.6955017 | 33436 | 0.002184 |
| CD-box | 110 | 90.2 | 9922 | 0.000648 | |
| HACA-box | 66 | 144.7575758 | 9554 | 0.000624 | |
| splicing | 98 | 112.1734694 | 10993 | 0.000718 |
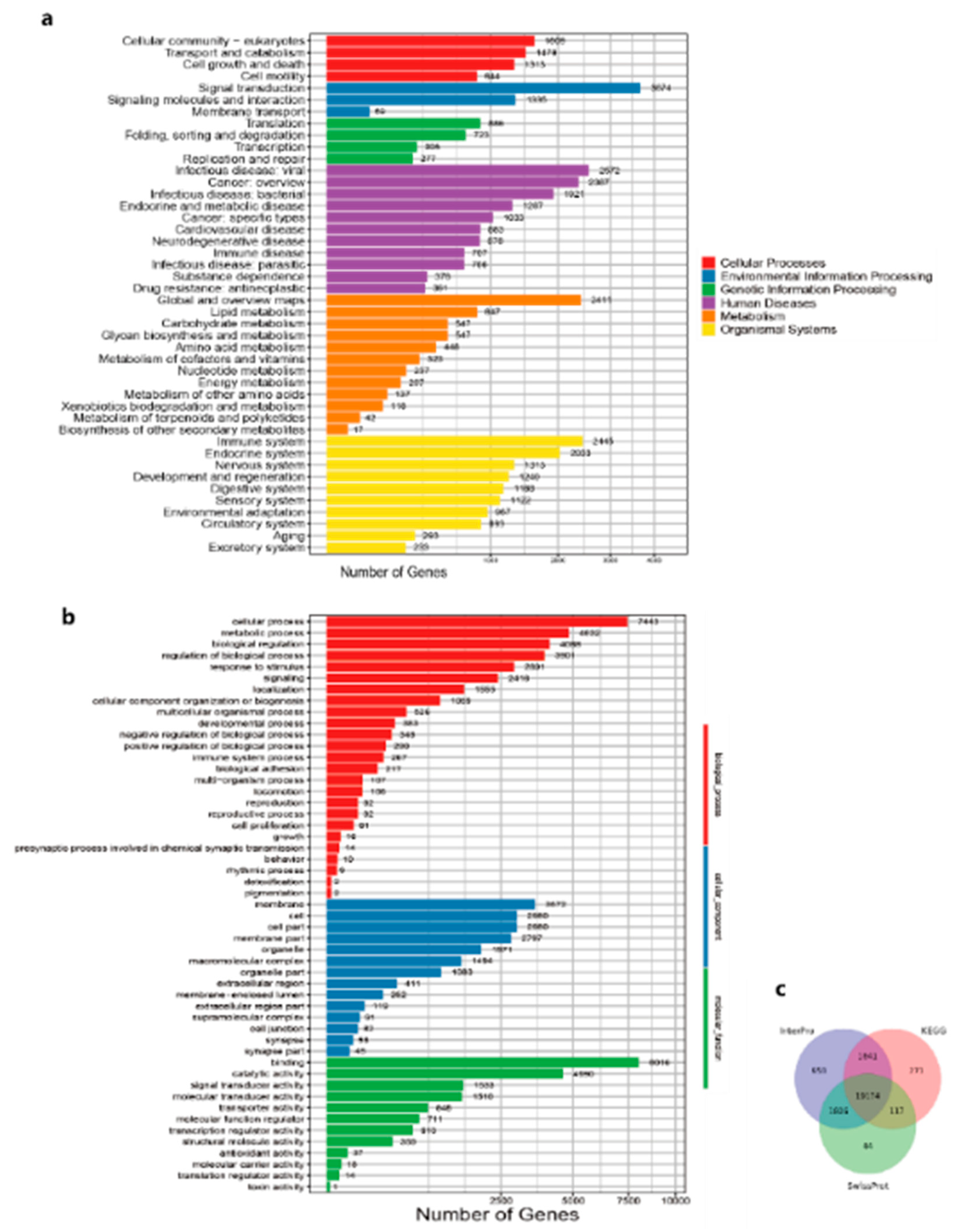
| Values | Total | Swissprot-Annotated | KEGG-Annotated | TrEMBL-Annotated | Interpro-Annotated | GO-Annotated | Overall |
| Number | 24,799 | 21,141 | 21,203 | 23,741 | 23,579 | 15,322 | 24,296 |
| Percentage | 100% | 85.25% | 85.50% | 95.73% | 95.08% | 61.78% | 97.97% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





