Submitted:
17 October 2023
Posted:
18 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction.
2. Materials.
3. Methods.
3.1. Input Feature vectors.
3.1.1. Complementarity descriptors.
3.1.2. Accessibility descriptors.
3.1.3. Interfacial contact network descriptors.
3.1.4. Size descriptors.
3.2. Training and Performance.
3.3. Output Features.
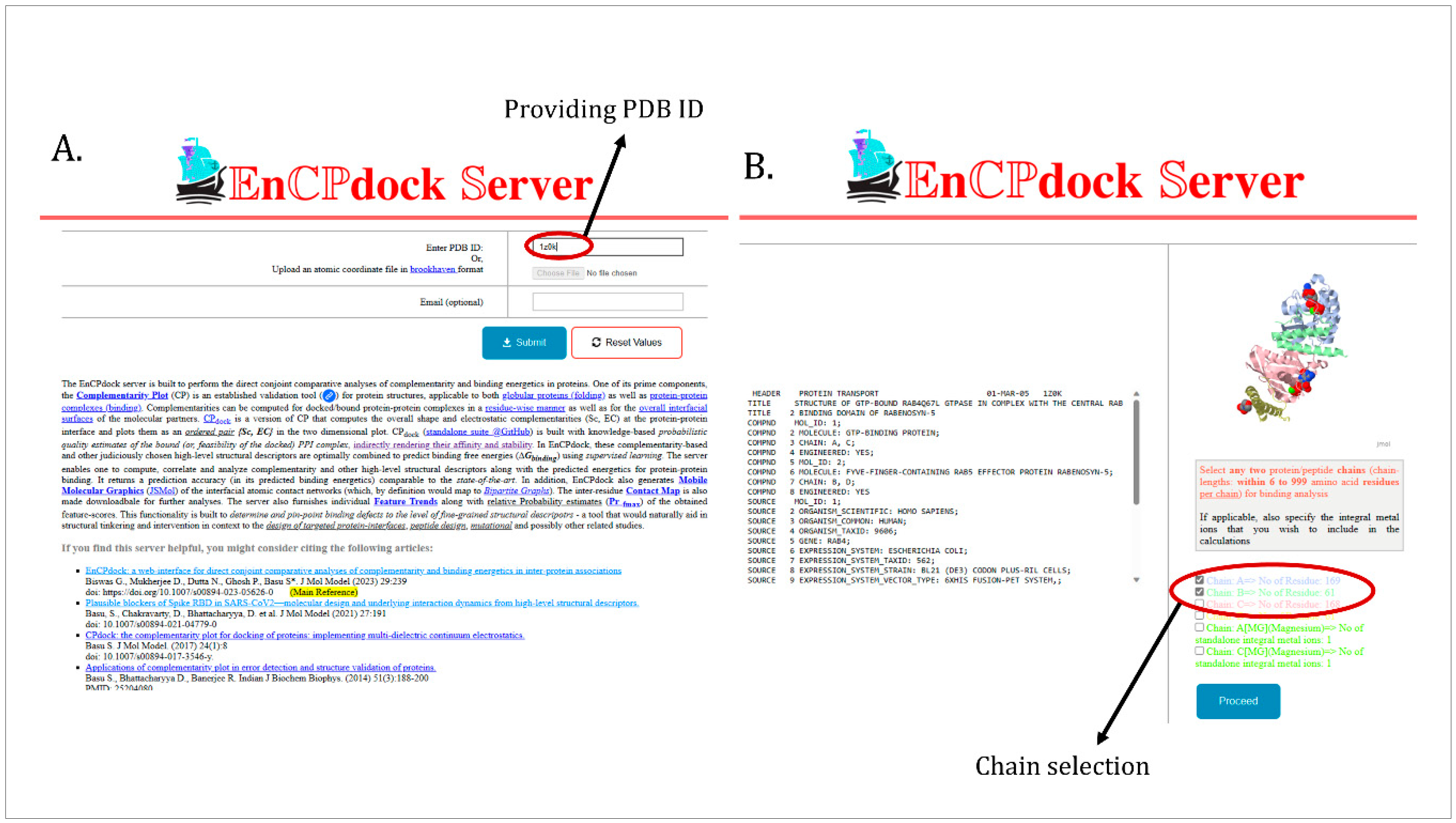
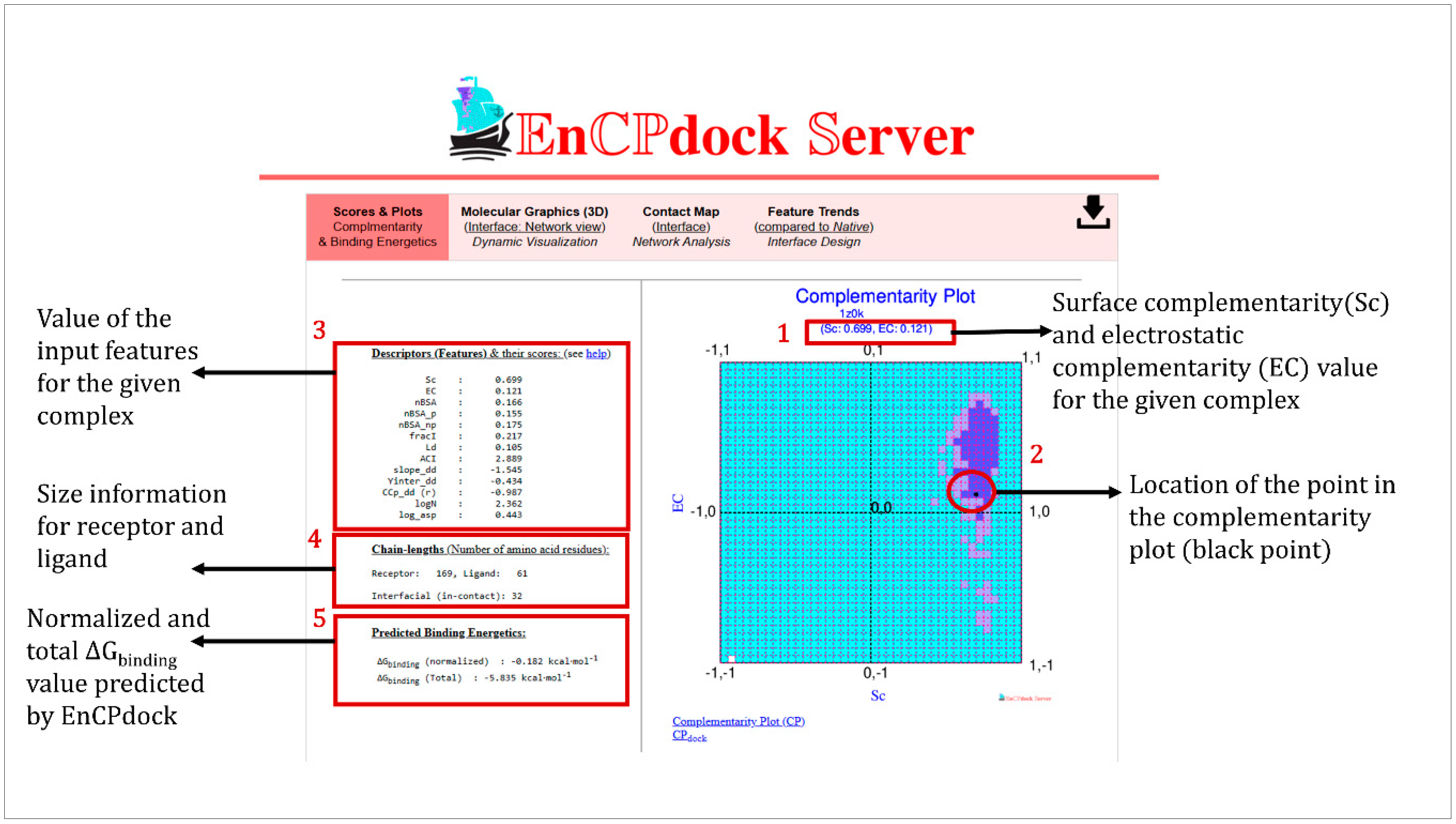
3.3.1. Scores and Plots.
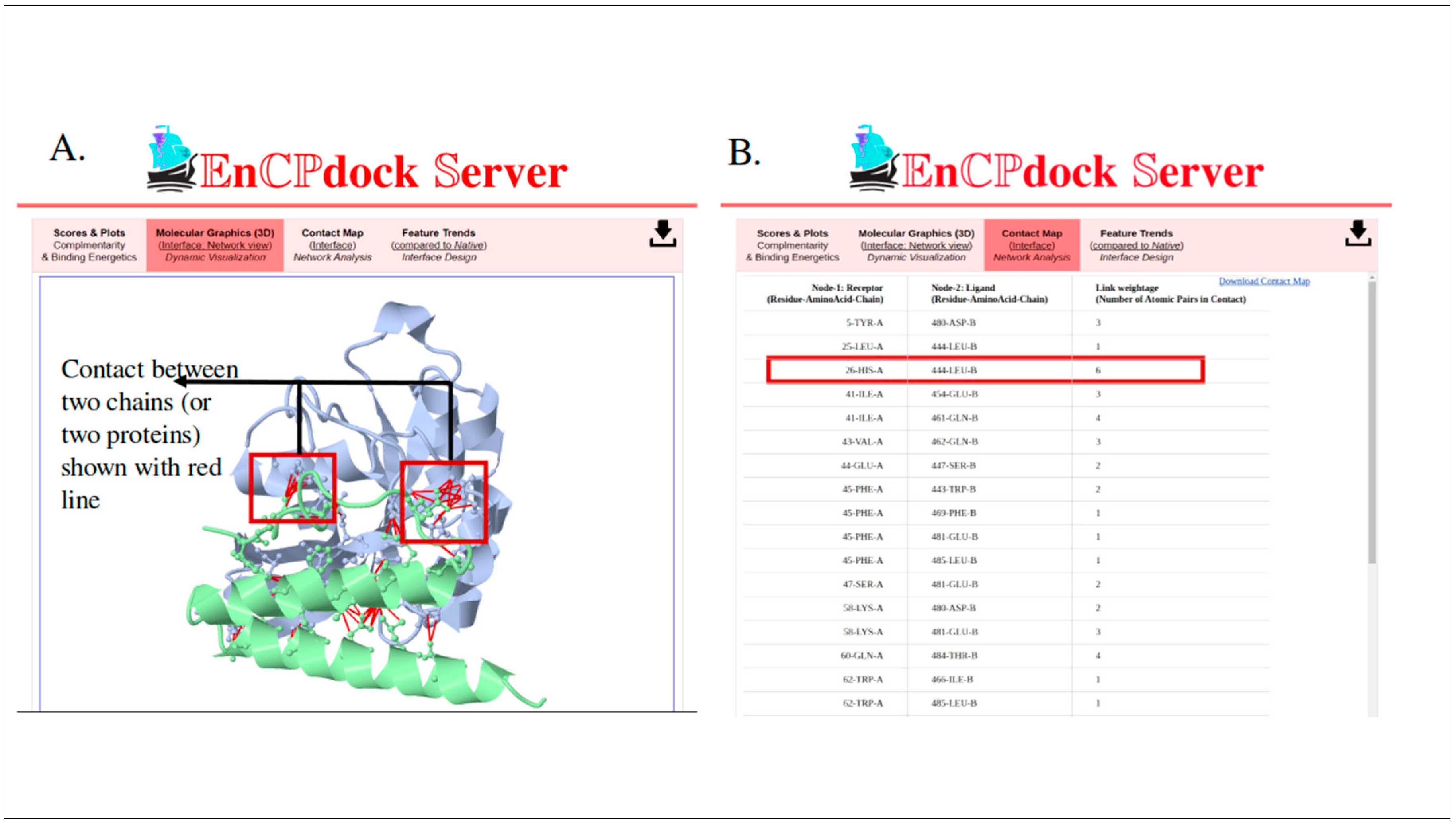
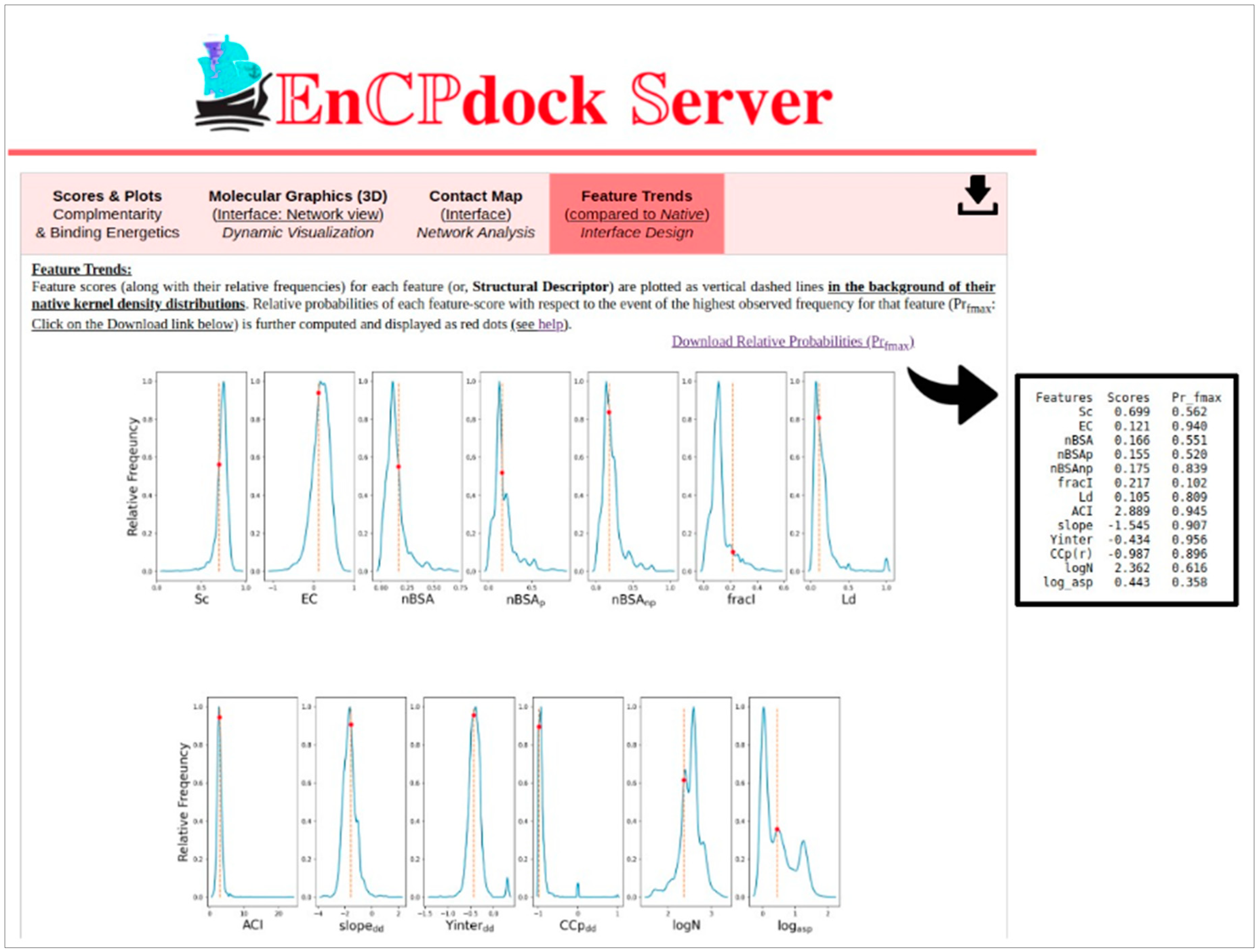
3.1.2. Molecular graphics, contact maps, and feature trends:
4. Conclusion.
Top 9 sets hitting the same highest correlation of r=0.745 × 10 models for the ten-fold cross validation (for each set) |
References
- Feng Y, Wang Q, Wang T (2017) Drug Target Protein-Protein Interaction Networks: A Systematic Perspective. BioMed Res Int 2017:1–13. https://doi.org/10.1155/2017/1289259. [CrossRef]
- Sable R, Jois S (2015) Surfing the Protein-Protein Interaction Surface Using Docking Methods: Application to the Design of PPI Inhibitors. Molecules 20:11569–11603. https://doi.org/10.3390/molecules200611569. [CrossRef]
- Keskin O, Gursoy A, Ma B, Nussinov R (2008) Principles of Protein−Protein Interactions: What are the Preferred Ways For Proteins To Interact? Chem Rev 108:1225–1244. https://doi.org/10.1021/cr040409x. [CrossRef]
- Zhang QC, Petrey D, Deng L, et al (2012) Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature 490:556–560. https://doi.org/10.1038/nature11503. [CrossRef]
- Bryant P, Pozzati G, Elofsson A (2022) Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun 13:1265. https://doi.org/10.1038/s41467-022-28865-w. [CrossRef]
- Lazaridis T (2000) Effective energy functions for protein structure prediction. Curr Opin Struct Biol 10:139–145. https://doi.org/10.1016/S0959-440X(00)00063-4. [CrossRef]
- Lazaridis T, Karplus M (1997) “New view” of protein folding reconciled with the old through multiple unfolding simulations. Science 278:1928–1931. https://doi.org/10.1126/science.278.5345.1928. [CrossRef]
- MacKerell ADJr, Bashford D, Bellott M, et al (1998) All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B 102:3586–3616. https://doi.org/10.1021/jp973084f. [CrossRef]
- Brooks BR, Bruccoleri RE, Olafson BD, et al (1983) CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217. https://doi.org/10.1002/jcc.540040211. [CrossRef]
- Lazaridis T, Karplus M (1999) Discrimination of the native from misfolded protein models with an energy function including implicit solvation. J Mol Biol 288:477–487. https://doi.org/10.1006/jmbi.1999.2685. [CrossRef]
- Sippl MJ (1995) Knowledge-based potentials for proteins. Curr Opin Struct Biol 5:229–235. https://doi.org/10.1016/0959-440x(95)80081-6. [CrossRef]
- Simons KT, Ruczinski I, Kooperberg C, et al (1999) Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins 34:82–95. https://doi.org/10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [CrossRef]
- Thomas PD, Dill KA (1996) Statistical potentials extracted from protein structures: how accurate are they? J Mol Biol 257:457–469. https://doi.org/10.1006/jmbi.1996.0175. [CrossRef]
- Simons KT, Kooperberg C, Huang E, Baker D (1997) Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol 268:209–225. https://doi.org/10.1006/jmbi.1997.0959. [CrossRef]
- Chen F, Liu H, Sun H, et al (2016) Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein-protein binding free energies and re-rank binding poses generated by protein-protein docking. Phys Chem Chem Phys 18:22129–22139. https://doi.org/10.1039/c6cp03670h. [CrossRef]
- Duarte Ramos Matos G, Kyu DY, Loeffler HH, et al (2017) Approaches for Calculating Solvation Free Energies and Enthalpies Demonstrated with an Update of the FreeSolv Database. J Chem Eng Data 62:1559–1569. https://doi.org/10.1021/acs.jced.7b00104. [CrossRef]
- Gohlke H, Case DA (2004) Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex Ras-Raf. J Comput Chem 25:238–250. https://doi.org/10.1002/jcc.10379. [CrossRef]
- Brown SP, Muchmore SW (2006) High-throughput calculation of protein-ligand binding affinities: modification and adaptation of the MM-PBSA protocol to enterprise grid computing. J Chem Inf Model 46:999–1005. https://doi.org/10.1021/ci050488t. [CrossRef]
- Rodrigues CHM, Pires DEV, Ascher DB (2021) mmCSM-PPI: predicting the effects of multiple point mutations on protein–protein interactions. Nucleic Acids Res 49:W417–W424. https://doi.org/10.1093/nar/gkab273. [CrossRef]
- Liu X, Luo Y, Li P, et al (2021) Deep geometric representations for modeling effects of mutations on protein-protein binding affinity. PLOS Comput Biol 17:e1009284. https://doi.org/10.1371/journal.pcbi.1009284. [CrossRef]
- Wang M, Cang Z, Wei G-W (2020) A topology-based network tree for the prediction of protein–protein binding affinity changes following mutation. Nat Mach Intell 2:116–123. https://doi.org/10.1038/s42256-020-0149-6. [CrossRef]
- Romero-Molina S, Ruiz-Blanco YB, Mieres-Perez J, et al (2022) PPI-Affinity: A Web Tool for the Prediction and Optimization of Protein–Peptide and Protein–Protein Binding Affinity. J Proteome Res 21:1829–1841. https://doi.org/10.1021/acs.jproteome.2c00020. [CrossRef]
- Jankauskaite J, Jiménez-García B, Dapkunas J, et al (2019) SKEMPI 2.0: an updated benchmark of changes in protein-protein binding energy, kinetics and thermodynamics upon mutation. Bioinforma Oxf Engl 35:462–469. https://doi.org/10.1093/bioinformatics/bty635. [CrossRef]
- Xue LC, Rodrigues JP, Kastritis PL, et al (2016) PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinforma Oxf Engl 32:3676–3678. https://doi.org/10.1093/bioinformatics/btw514. [CrossRef]
- Liu S, Zhang C, Zhou H, Zhou Y (2004) A physical reference state unifies the structure-derived potential of mean force for protein folding and binding. Proteins Struct Funct Bioinforma 56:93–101. https://doi.org/10.1002/prot.20019. [CrossRef]
- Ravikant DVS, Elber R (2010) PIE-efficient filters and coarse grained potentials for unbound protein-protein docking. Proteins 78:400–419. https://doi.org/10.1002/prot.22550. [CrossRef]
- Abbasi WA, Yaseen A, Hassan FU, et al (2020) ISLAND: in-silico proteins binding affinity prediction using sequence information. BioData Min 13:20. https://doi.org/10.1186/s13040-020-00231-w. [CrossRef]
- Biswas G, Mukherjee D, Dutta N, et al (2023) EnCPdock: a web-interface for direct conjoint comparative analyses of complementarity and binding energetics in inter-protein associations. J Mol Model 29:239. https://doi.org/10.1007/s00894-023-05626-0. [CrossRef]
- Blanco JD, Radusky L, Climente-González H, Serrano L (2018) FoldX accurate structural protein-DNA binding prediction using PADA1 (Protein Assisted DNA Assembly 1). Nucleic Acids Res 46:3852–3863. https://doi.org/10.1093/nar/gky228. [CrossRef]
- Basu S, Bhattacharyya D, Banerjee R (2012) Self-complementarity within proteins: Bridging the gap between binding and folding. Biophys J 102:2605–2614. https://doi.org/10.1016/j.bpj.2012.04.029. [CrossRef]
- Winn MD, Ballard CC, Cowtan KD, et al (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:. https://doi.org/10.1107/S0907444910045749. [CrossRef]
- Lawrence MC, Colman PM (1993) Shape complementarity at protein/protein interfaces. J Mol Biol 234:946–950. https://doi.org/10.1002/bip.360340711. [CrossRef]
- Connolly ML (1983) Analytical molecular surface calculation. J Appl Crystallogr 16:548–558. https://doi.org/10.1107/S0021889883010985. [CrossRef]
- Xu D, Zhang Y (2009) Generating triangulated macromolecular surfaces by euclidean distance transform. PLoS ONE 4:. https://doi.org/10.1371/ journal.pone.0008140. [CrossRef]
- Li L, Li C, Sarkar S, et al (2012) DelPhi: a comprehensive suite for DelPhi software and associated resources. BMC Biophys 5:. https://doi.org/10.1186/2046-1682-5-9. [CrossRef]
- Li L, Li C, Zhang Z, Alexov E (2013) On the Dielectric “Constant” of Proteins: Smooth Dielectric Function for Macromolecular Modeling and Its Implementation in DelPhi. J Chem Theory Comput 9:2126–2136. https://doi.org/10.1021/ct400065j. [CrossRef]
- McCoy AJ, Chandana Epa V, Colman PM (1997) Electrostatic complementarity at protein/protein interfaces. J Mol Biol 268:570–584. https://doi.org/10.1006/jmbi.1997.0987. [CrossRef]
- Basu S (2017) CPdock: the complementarity plot for docking of proteins: implementing multi-dielectric continuum electrostatics. J Mol Model 24:8. https://doi.org/10.1007/s00894-017-3546-y. [CrossRef]
- Basu S, Bhattacharyya D, Banerjee R (2014) Applications of complementarity plot in error detection and structure validation of proteins. Indian J Biochem Biophys 51:188–200.
- Basu S, Chakravarty D, Bhattacharyya D, et al (2021) Plausible blockers of Spike RBD in SARS-CoV2—molecular design and underlying interaction dynamics from high-level structural descriptors. J Mol Model 27:191. https://doi.org/10.1007/s00894-021-04779-0. [CrossRef]
- Joachims T (2002) Learning to Classify Text Using Support Vector Machines.
- Schymkowitz J, Borg J, Stricher F, et al (2005) The FoldX web server: An online force field. Nucleic Acids Res 33:382–388. https://doi.org/10.1093/nar/gki387. [CrossRef]
- Kandathil SM, Garza-Fabre M, Handl J, Lovell SC (2018) Improved fragment-based protein structure prediction by redesign of search heuristics. Sci Rep 8:13694. https://doi.org/10.1038/s41598-018-31891-8. [CrossRef]
- Hubbard SSJ, Thornton JJM (1993) “NACCESS”, computer program. Dep. Biochem. Mol. Biol. Univ. Coll. Lond.
- Banerjee R, Sen M, Bhattacharya D, Saha P (2003) The jigsaw puzzle model: search for conformational specificity in protein interiors. J Mol Biol 333:211–226. [CrossRef]
- Berman HM, Westbrook J, Feng Z, et al (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235. [CrossRef]
- Kastritis PL, Moal IH, Hwang H, et al (2011) A structure-based benchmark for protein–protein binding affinity. Protein Sci 20:482–491. https://doi.org/10.1002/pro.580. [CrossRef]
- Vreven T, Moal IH, Vangone A, et al (2015) Updates to the Integrated Protein–Protein Interaction Benchmarks: Docking Benchmark Version 5 and Affinity Benchmark Version 2. J Mol Biol 427:3031–3041. https://doi.org/10.1016/j.jmb.2015.07.016. [CrossRef]
- Jemimah S, Yugandhar K, Michael Gromiha M (2017) PROXiMATE: a database of mutant protein–protein complex thermodynamics and kinetics. Bioinformatics 33:2787–2788. https://doi.org/10.1093/bioinformatics/btx312. [CrossRef]
- Choromański K, Matuszak M, Miȩkisz J (2013) Scale-Free Graph with Preferential Attachment and Evolving Internal Vertex Structure. J Stat Phys 151:1175–1183. https://doi.org/10.1007/s10955-013-0749-1. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




