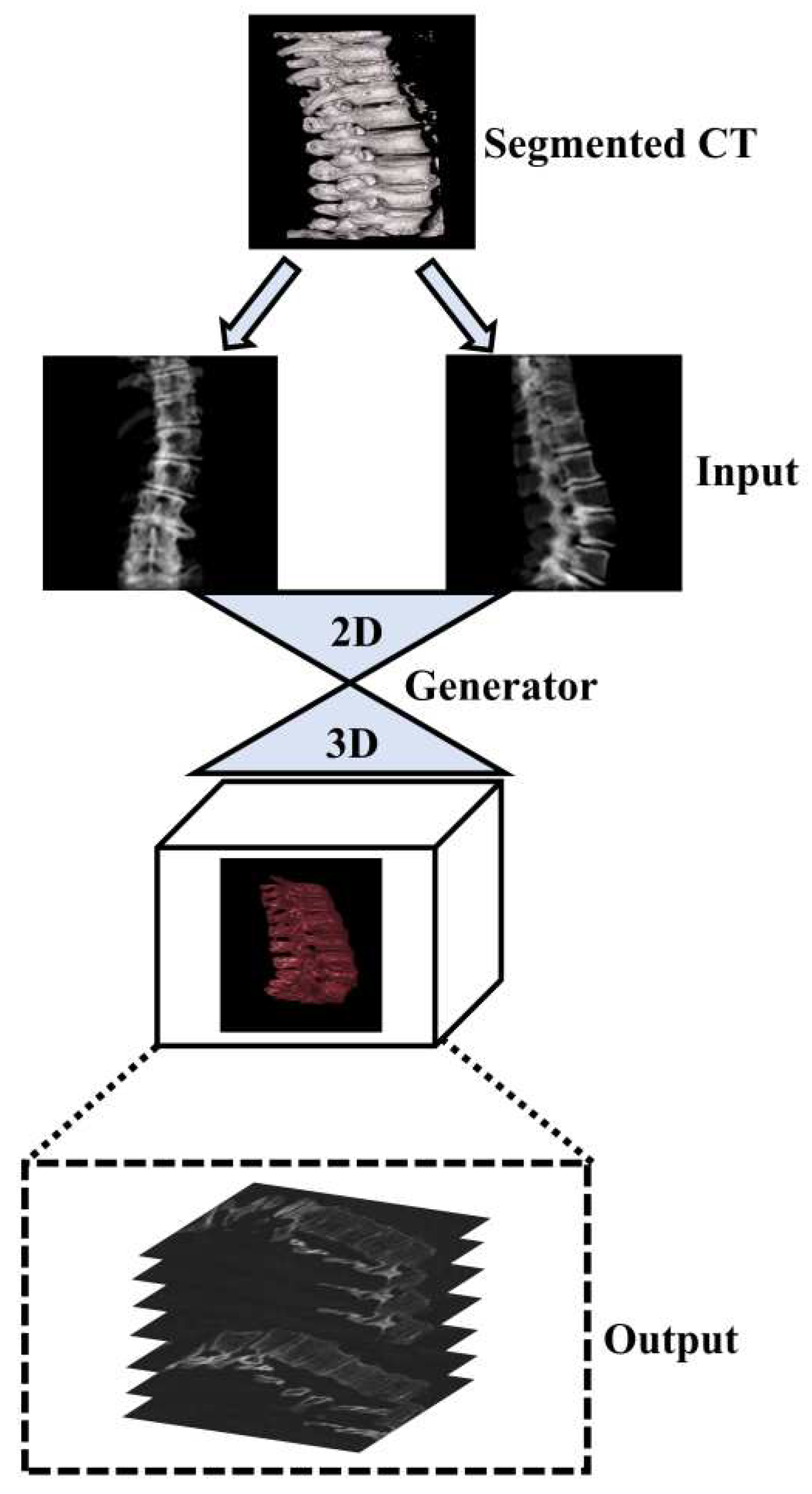
1. Introduction
Computed Tomography (CT) scans are widely used in medical imaging due to their high-resolution and detailed insights into the internal anatomy of patients [
1]. In the field of spinal imaging, CT scans play a critical role in the diagnosis and management of various spinal disorders, providing accurate information on bone structure, alignment, and pathological changes [
2]. However, cumulative radiation exposure for patients, particularly in the context of repeated diagnostic procedures, is a concern. Additionally, the cost of CT scans can be prohibitive, limiting their accessibility for healthcare providers and patients in resource-constrained settings [
3].
Conventional X-ray imaging is associated with lower radiation exposure and cost compared to CT scans, making it a more accessible imaging modality. However, X-ray images are inherently two-dimensional (2D) and may not provide the same level of anatomical detail as 3D CT scans, particularly when it comes to evaluating spinal vertebrae structures [
4]. Thus, there is a growing interest in developing novel techniques that can leverage the advantages of X-ray imaging while providing 3D information akin to CT scans [
1].
In recent years, 3D reconstruction has gained significant attention in the field of medical imaging, particularly for its potential to improve diagnosis, treatment planning, and patient outcomes [
5]. The ability to generate 3D representations of anatomical structures from 2D images offers several benefits, some of which are outlined below [
6,
7,
8]:
1)Enhanced Visualization and Interpretation: 3D reconstruction provides a more comprehensive view of complex anatomical structures compared to traditional 2D imaging. This enhanced visualization enables healthcare professionals to better understand the spatial relationships between different structures, leading to more accurate diagnoses and improved decision-making.
2)Improved Surgical Planning and Navigation: In the context of spinal surgery, 3D reconstructions can facilitate preoperative planning by allowing surgeons to assess the extent of spinal deformities or pathological conditions, as well as the ideal surgical approach. Additionally, 3D reconstructions can be used intraoperatively to guide surgical navigation, thereby increasing the precision and safety of the procedure.
3)Patient-Specific Modeling and Simulation: 3D reconstructions can be employed to create patient-specific models of spinal structures, which can be used for biomechanical analyses, finite element simulations, or personalized implant design. These patient-specific models may contribute to the development of more effective and personalized treatment strategies, ultimately improving patient outcomes.
4)Enhanced Patient Communication and Education: 3D reconstructions can facilitate communication between healthcare providers and patients by providing a more intuitive understanding of the patient’s condition, the proposed treatment plan, and the expected outcomes. This improved communication can lead to better patient engagement, satisfaction, and adherence to treatment recommendations.
5)Reduced Radiation Exposure: By generating 3D reconstructions from a limited number of 2D X-ray images, the proposed technique has the potential to reduce the cumulative radiation dose associated with traditional CT scans. This reduction in radiation exposure is particularly important for patients who require repeated imaging over time, such as those with progressive spinal disorders or those undergoing long-term follow-up after surgical intervention.
6)Cost-efficiency and availability: CT scanners frequently have a higher price tag compared to X-ray machines, which can make them less accessible in resource-limited settings or developing countries.
In summary, 3D reconstruction offers numerous benefits in the field of spinal imaging, with the potential to improve diagnostic accuracy, facilitate surgical planning, enable patient-specific modeling and simulation, enhance patient communication, reduce radiation exposure, and improve cost-effectiveness and availability in resource-limited settings.
Generative Adversarial Networks (GANs) have emerged as powerful deep learning tools capable of synthesizing realistic images from different modalities. Recent studies have demonstrated the potential of GANs for generating 3D images from chest 2D X-ray projections, thus bridging the gap between conventional X-ray imaging and CT scans [
8]. In this study, we applied the GAN-based framework for reconstructing 3D spinal vertebrae structures from synthetic biplanar X-ray images as presented by Ying et al. (X2CT-GAN), focusing specifically on anterior and lateral views of spinal vertebrae[
8]. In contrast to previous work [
8,
9] which applied X2CT-GAN for 3D reconstruction in medical settings, we focussed on segmented vertebrae to specifically focus on the region of interest (spinal vertebrae) while reducing unnecessary information and, thus computational cost. This approach leverages a novel feature fusion technique based on X2CT-GAN to combine information from both views and employs a combination of Mean Squared Error (MSE) loss and adversarial loss to train the generator, resulting in high-quality synthetic 3D spinal vertebrae CTs. By incorporating a focused view of the spinal vertebrae through segmentation, this approach reduces unnecessary information which could affect the synthetic generation of CTs. The general concept is illustrated in
Figure 1.
2. Materials and Methods
2.1. Dataset and segmentation
In this study, we employed a COLONOG subset [
10] of the CTSpine1K dataset [
11], which is a large and comprehensive collection of spine CT data with segmentation masks. The data can be obtained upon request from the authors of the CTSpine 1Kdataset (
https://github.com/MIRACLE-Center/CTSpine1K). The prospective study to obtain the COLONOG subset was reviewed and approved by each participating institution’s institutional review board, and subjects gave their informed consent to participate and to have their private health information accessed for the purposes of the study. All data were anonymized when accessed through the database. A total of n=440 CT data were processed. The data were acquired using multidetector-row CT scanners with a minimum of 16 rows while patients were in supine and prone positions. The CT images were obtained using the following specifications: collimation of 0.5 to 1.0 mm, pitch ranging from 0.98 to 1.5, a 512 by 512 matrix, and a field of view adjusted to fit the patient. The effective mAs was 50, and the peak voltage was 120 kV. A standard reconstruction algorithm was utilized, and images of patients in both prone and supine positions were reconstructed with slice thicknesses between 1.0 and 1.25 mm and a reconstruction interval of 0.8 mm [
10].
As the benchmark, a fully supervised method was employed to train a deep network for spinal vertebrae segmentation using the nnUnet model [
11]. The nnUnet model has outperformed other methods in various medical image segmentation tasks in recent years, making it the acknowledged baseline for medical image segmentation [
12]. Essentially a U-Net, nnUnet features a specific network architecture and design parameters that self-adapt to the dataset’s characteristics, along with robust data augmentation [
11,
12]. The 3D full-resolution U-net architecture was used to accommodate the large volume of high-resolution 3D images in the dataset [
11]. Further details about the nnUnet model can be found in the original publication [
11].
To facilitate the annotation process, the segmentation network was trained using the public datasets from the VerSe’19 [
13]and VerSe’20 Challenges [
14], employing the nnUnet algorithm. As most of the samples from the VerSe’ Challenge were cropped, cases that had complete CT images and consistent image spacing between images and their corresponding ground truth were selected. Next, the trained segmentation model was assigned to junior annotators to predict segmentation masks and refine the labels based on the predictions. These refined labels were subsequently reviewed by two senior annotators for further improvement. In cases where the senior annotator encountered difficulties in determining the annotations, input was sought from experienced spine surgeons [
11]. To ensure the final quality of the annotations, coordinators conducted a random double-check, and any incorrect cases were corrected by the annotators. The human-corrected annotations and corresponding images were added to the training data to develop a more powerful model. To expedite the annotation process, the database and retrained the deep learning model were updated after every 100 cases. This iterative process continued until the annotation task was completed. The entire annotation process was performed using ITK-SNAP software, and segmentation masks were saved in NIfTI format [
11].
2.2. Data preprocessing and synthetic x-ray generation
In the initial stage of data preprocessing, we cropped the original CT data based on their respective segmentation masks (see section 2.1). We implemented this procedure in Python by first defining a function that took two input arguments: the CT image and its corresponding segmentation mask. This function identified the non-zero elements in the segmentation mask and subsequently determined the minimum and maximum indices along the z, y, and x-axes. The function then returned the cropped CT data with dimensions ranging from the minimum to the maximum indices along each axis. We saved the cropped CT data as a new NIfTI file in the output directory with the same affine transformation as the original CT data.
Acquiring a sufficient number of original synchronized X-ray and CT images for machine learning purposes poses significant practical and ethical challenges. One major obstacle in training the X2CT-GAN model is the scarcity of paired X-ray and CT data from patients. Obtaining such paired data from patients is not only costly but also raises ethical concerns due to the additional radiation exposure involved. To address this issue, we have opted to train our network using synthesized X-rays generated from the ground truth CT dataset, as proposed before [
8]. In this study, we employ a CT volume to simulate two X-rays corresponding to the posterior-anterior and lateral views. This process is achieved through the utilization of digitally reconstructed radiographs (DRR) technology [
15]. By leveraging this approach, we can effectively generate the required paired X-ray and CT data for training the X2CT-GAN model without subjecting patients to unnecessary radiation risks. Consequently, the method strikes a balance between the need for accurate and diverse training data and the ethical concerns associated with obtaining such data from human subjects. To generate synthetic X-ray images from the cropped CT data, we used a custom-made Python script in the 3D Slicer software (Python extension of 3D Slicer) [
16]. For each subfolder containing the cropped CT data, the script modified the 3D viewer to display a black-white gradient and removed the bounding box and orientation axes labels. It then loaded the CT volume in 3D Slicer and switched to the one-up 3D view layout. The script defined a function which centered the 3D view and slice views to fill the background. This function was applied before setting up the volume rendering display. The volume rendering was set up by creating a default volume rendering display node and making it visible. The script then applied a ‘CT-X-ray’ volume rendering preset from the DRRGenerator module [
17] to the display node. To adjust the scalar opacity mapping, a six-point transfer function was defined and set as the scalar opacity for the volume property node. The function was called again to ensure that the view was centered. Next, the script rotated the 3D view and captured screenshots of the synthetic X-ray images in different orientations, such as the anterior and lateral views. The captured images were saved to disk in the respective subfolders as PNG files with appropriate filenames indicating the orientation. After processing each CT volume, the script cleared the 3D Slicer scene to prepare for the next iteration. By following this procedure, we generated synthetic X-ray images from the corresponding segmented CT data, which were then used for further analysis and experimentation.
The generated images were then preprocessed using the following method. A function was used to preprocess the synthetic x-ray images. This function takes an image as input and applies a series of image processing operations to it, including converting the image to grayscale, thresholding to keep only the brighter parts of the image, performing morphological operations to remove small noise and fill small gaps, finding the largest contour in the binary image, cropping the original image using the bounding box of the largest contour, normalizing the cropped image, padding the image to make it square, and resizing the padded image to the desired output size. The function was then applied to each synthetic x-ray image to produce a preprocessed image for the anterior and lateral view. Another function was implemented to load the CT images from NIfTI files. For each folder in the input folder, the code read in the preprocessed anterior and lateral x-ray images, as well as the CT image in NIfTI format. The preprocessed x-ray images and the CT image were then combined into a single HDF5 file for the current folder. The HDF5 file contained three datasets: “ct”, “xray1”, and “xray2”, corresponding to the CT image, synthetic anterior x-ray image, and synthetic lateral x-ray image, respectively. The data were then randomly split into a train set (80%) and a test set (20%) for further analyses.
2.3. Model training and evaluation
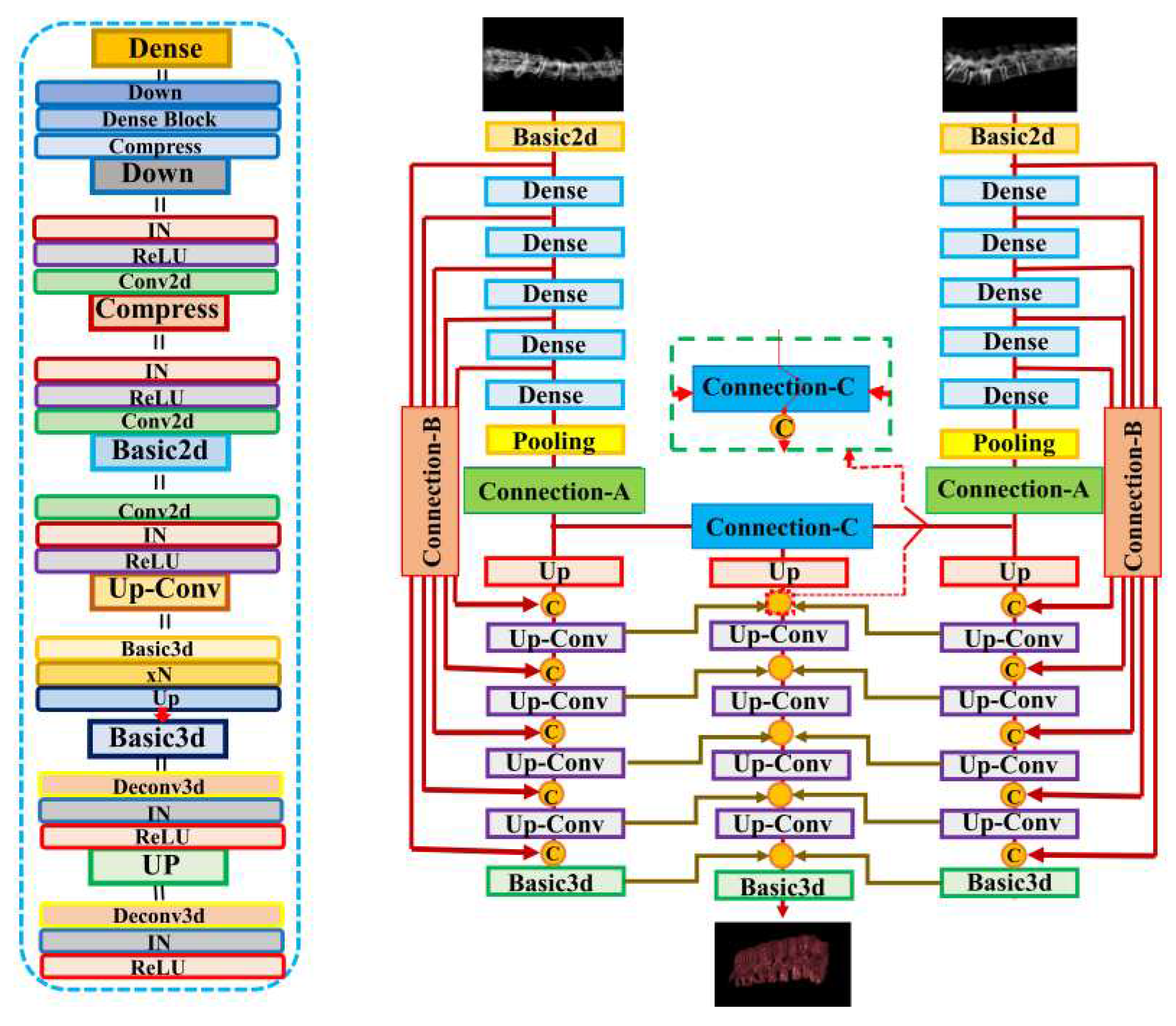
In this study, we applied the X2CT-GAN model to fuse X-ray and CT images using a deep learning approach. The model was trained using a multiview network architecture consisting of a dense UNet fused with transposed convolutions as the generator and a basic 3D discriminator with instance normalization (
Figure 1). The GAN loss was computed using least squares. The generator network employed ReLU activation functions and a conditional discriminator with no dropout layers. The model’s training parameters included a learning rate of 0.0002 (lr: 0.0002), Adam optimizer with beta1 set to 0.5, and beta2 set to 0.99. The training process employed a batch size of 1 due to limited computational resources (NVIDIA GTX 3090; 24GB) and did not utilize weight decay. Data augmentation was applied to the images, with a fine size of 128x128, and images were resized to 150x150. The CT and X-ray images were normalized using the provided mean and standard deviation values. Various loss functions and weighting parameters were used to optimize the model, such as identity loss, feature matching loss, map projection, and GAN. A detailed list of parameters, configurations, and functions used can be found in the code provided in the data availability section. During the training process, the discriminator and generator were optimized using the specified configuration parameters. The loss and metrics were logged, and the model was saved at specified intervals. The learning rate was updated at the end of each epoch.
Figure 2.
Illustration of the X2CT-GAN as proposed by Ying et al. [
8]. In contrast to Ying et al. [
8] who used unsegmented chest x-rays, we used segmented biplanar x-ray inputs and constructed 3D models focusing specifically on the spinal vertebrae.
Figure 2.
Illustration of the X2CT-GAN as proposed by Ying et al. [
8]. In contrast to Ying et al. [
8] who used unsegmented chest x-rays, we used segmented biplanar x-ray inputs and constructed 3D models focusing specifically on the spinal vertebrae.
For validation, the model was evaluated using a separate dataset, and the performance metrics were computed and logged. We used multiple metrics for evaluation to provide a sophisticated evaluation of the approach:1)Mean Absolute Error (MAE), 2)Cosine Similarity, 3)Peak Signal-to-Noise Ratio (PSNR), 4)Structural Similarity Index Measure (SSIM), 4)3D Peak Signal-to-Noise Ratio (PSNR-3D).
First, the model and datasets were initialized, setting them to the evaluation mode in the X2CT-GAN model. We then iterated through the dataset, using the model to generate CT images. For each generated CT image, the metrics were calculated by comparing the generated CT image with the corresponding ground truth CT image. The metrics are as follows:
- -
Mean Absolute Error (MAE): MAE calculates the average absolute difference between the predicted and the ground truth CT images. It gives an idea of the magnitude of the errors without considering their direction. MAE was calculated for each slice (MAE0) and for the entire 3D volume (MAE).
- -
Cosine Similarity: cosine similarity measures the cosine of the angle between the predicted and the ground truth CT images, providing a similarity score between -1 and 1.
- -
Peak Signal-to-Noise Ratio (PSNR): PSNR measures the ratio between the maximum possible power of a signal and the power of the noise.
- -
Structural Similarity Index Measure (SSIM): SSIM measures the structural similarity between the predicted and the ground truth CT images.
- -
3D Peak Signal-to-Noise Ratio (PSNR-3D): PSNR-3D is an extension of the PSNR metric for 3D images and measures the 3D image quality.
After calculating the metrics for each image, we computed the average value for each metric across the entire dataset. These average values represent the overall performance of the model using the specified evaluation metrics. Visualization was performed by importing the ground truth and synthetic CT data in 3D Slicer and using the CT-bone preset for visualization. All analyses were performed using Python. The code for the custom Python scripts for 3D Slicer, the preprocessing scripts, and the configuration file is available from the data availability section. The original code of the X2CT-GAN is available from the repository provided by Ying et al. [
8] and the modified code used for the present study is available from the data availability section.
3. Results
3.1. Quantitative results
The evaluation of the X2CT-GAN model for generating synthetic CT scans based on biplanar X-ray inputs yielded promising results across various performance metrics. MAE and MSE were employed to assess the model’s accuracy. For the MAE per slice (MAE0), an average of 0.034 was recorded, while the MSE per slice (MSE0) obtained an average value of 0.005. The overall MAE and MSE were 85.359 and 31439.027, respectively. Cosine similarity, which measures the angular distance between the generated synthetic CT scans and the ground truth, resulted in an average score of 0.484, indicating a reasonably good level of agreement between the two sets of data. PSNR is a metric that helps quantify the quality of the reconstructed image by comparing it to the original image. The average PSNR for the 3D reconstructed images (PSNR-3D) was 27.432. Furthermore, the PSNR values for individual channels, which can be interpreted as comparisons between the real and generated CT images in three different orientations or planes, were 28.817 (PSNR-1), 28.764 (PSNR-2), and 27.599 (PSNR-3), resulting in an average PSNR (PSNR-avg) of 28.394. Notably, PSNR-avg is the average of the PSNR calculated for each of the three separate views or planes (PSNR-1, PSNR-2, and PSNR-3). This means that the PSNR is calculated independently for each view and then averaged to obtain a single value that represents the overall PSNR across all the views. PSNR-3D, in contrast, computes the PSNR for the entire 3D volumetric data as a whole. It directly compares the entire 3D real CT data to the generated 3D CT data and calculates the PSNR in one go without separating the data into individual views or planes. SSIM was also employed to evaluate the model, which quantifies the perceived visual quality of the generated synthetic CT scans. The SSIM was 0.468. The results are presented in
Table 1.
3.2. Qualitative results
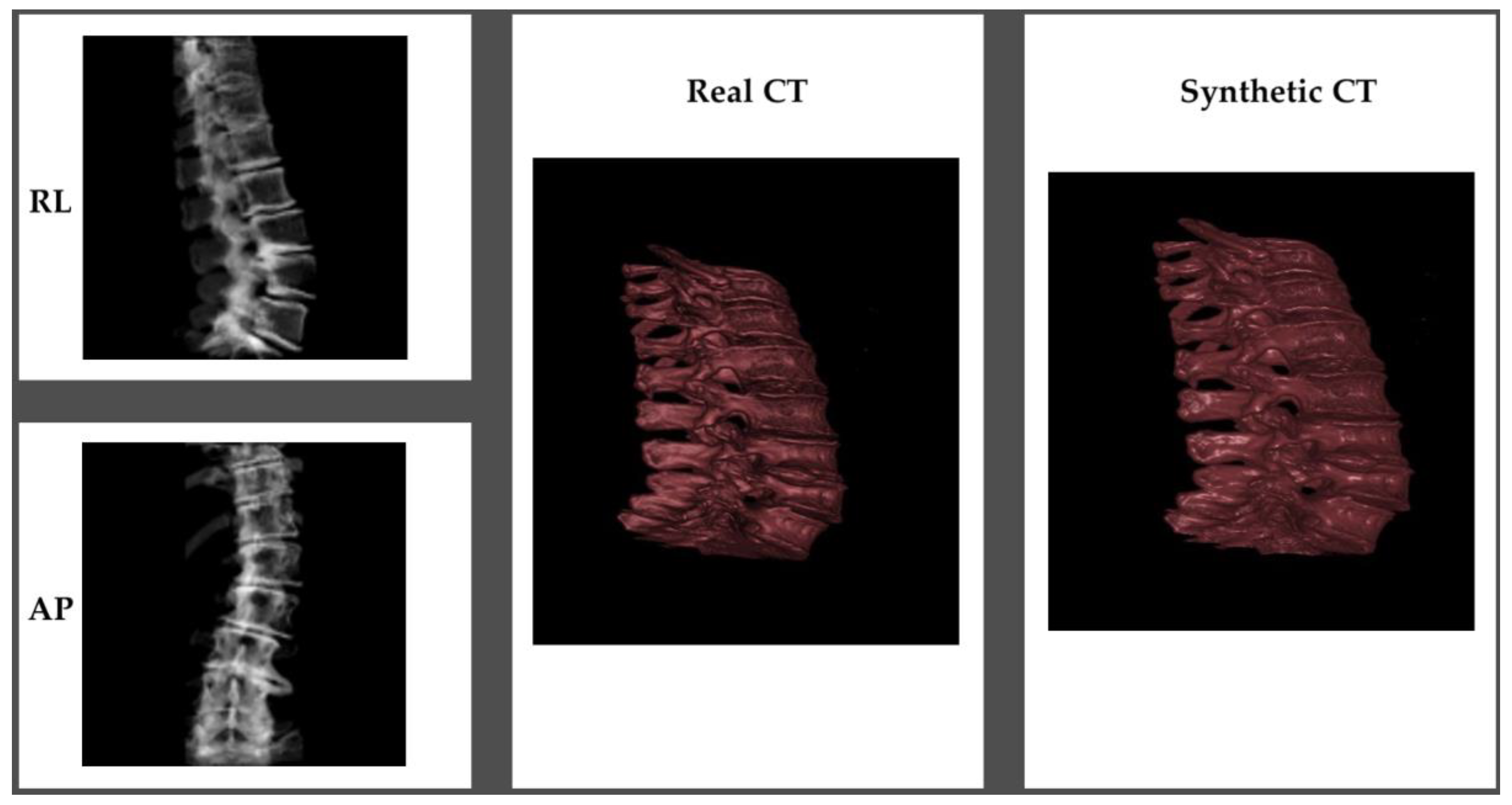
An example of the synthetic CT generated from a biplanar x-ray input is shown in
Figure 3. The results were rated qualitatively by importing the real CT and synthetic CT into 3D Slicer. Although the synthetic CT provided an impressive 3D model of the spinal vertebrae based on only 2D inputs, there were structural differences in the form of the vertebrae and artificial-looking surfaces of the bone. Especially, fine structures of the bone were not appropriately reflected.
The quantitative evaluation metrics also provide useful insights into the qualitative differences between the ground truth and synthetic CT volumes. The MAE between the synthetic and ground truth CTs was found to be 85.359, indicating that there are noticeable deviations in the intensities of the voxels. The MSE value of 31439.027 further supports this observation, suggesting that the errors are not only localized but also significant in some areas. The PSNR values obtained for the individual channels (PSNR-1, PSNR-2, and PSNR-3) were 28.817, 28.764, and 27.599, respectively, with an average PSNR (PSNR-avg) of 28.394. These values signify that the overall contrast and dynamic range of the synthetic CT are relatively close to the ground truth. However, the lower PSNR value for the third channel indicates that some parts of the synthetic CT may have less accurate contrast and intensity representation compared to the original CT. This could be attributed to the insufficient capture of fine bone structures, leading to the artificial appearance of bone surfaces. The SSIM was 0.468. This value implies that the structural similarity between the synthetic and ground truth CTs is only moderate, reflecting the differences in the form of the vertebrae and the artificial-looking surfaces of the bone. The moderate Cosine Similarity value of 0.484 also suggests that the overall orientation and shape of the fine structures within the synthetic CT might not be accurately represented.
In summary, while the synthetic CT generated by the X2CT-GAN demonstrates a remarkable capability to create 3D spinal vertebrae models from synthetic 2D biplanar x-ray inputs, there are still some limitations in accurately capturing the fine bone structures and maintaining the precise morphology of the vertebrae. The artificial appearance of the bone surfaces and the differences in contrast and intensity representation in some areas indicate that further improvements to the X2CT-GAN model are necessary to achieve a more accurate and consistent performance in generating synthetic CTs from biplanar x-ray inputs.
4. Discussion
In this study, we applied a GAN framework to reconstruct 3D spinal vertebrae structures from synthetic biplanar X-ray images, specifically focusing on anterior and lateral views. The results demonstrated the effectiveness of the approach in reconstructing 3D spinal vertebrae structures from biplanar X-rays, although some limitations in accurately capturing the fine bone structures and maintaining the precise morphology of the vertebrae were present.
GANs, first introduced by Goodfellow et al. in 2014 [
18], have revolutionized the field of deep learning by offering a novel approach to unsupervised learning. GANs consist of two neural networks, a generator and a discriminator, which compete against each other in a game-theoretic framework. The generator learns to create realistic synthetic data samples, while the discriminator learns to distinguish between real and generated samples. Through this adversarial process, the generator progressively improves its ability to generate more convincing data [
19]. This study applied the X2CT-GAN architecture as introduced by Ying et al. [
8] with a novel approach of focussing on segmented regions of interest for synthetic 3D reconstruction. In comparison to previous GAN-based methods, the X2CT-GAN offers significant improvements. One key enhancement is the incorporation of a feature fusion technique that effectively combines information from multiple X-ray views, enabling a more accurate 3D reconstruction. Additionally, the architecture optimizes the generator network using a combination of MSE loss and adversarial loss, resulting in higher-quality synthetic 3D images with better structural consistency and finer anatomical details. In the medical imaging domain, GANs have shown promise in a variety of applications, including data augmentation, image synthesis, and image-to-image translation [
20]. The ability of GANs to generate high-quality synthetic images has been particularly valuable in addressing challenges related to the limited availability of labeled data, data privacy concerns, and the need for multi-modal image synthesis [
21]. Recent advancements in GAN architectures, such as conditional GANs [
22], have further expanded the scope of applications in medical imaging. By incorporating additional information as input, conditional GANs can be trained to generate images with specific desired characteristics, making them well-suited for tasks such as 3D image reconstruction from 2D projections. Studies exploring the use of GANs for 3D image generation from chest 2D X-ray projections have demonstrated their potential in bridging the gap between conventional X-ray imaging and CT scans [
8]. In this context, the application of GANs for 3D spinal vertebrae reconstruction from biplanar X-rays represents a promising direction in leveraging the power of deep learning to enhance diagnostic capabilities while reducing the cost and radiation exposure associated with traditional CT scans.
Notably, the objective of this work is not to supplant CT scans with biplanar x-rays. Although our proposed method can reconstruct the overall structure of spinal vertebrae, finer anatomical details may still exhibit some artifacts, as seen in our results. Nevertheless, the developed approach has the potential to find specialized applications in clinical practice, especially when trained with larger datasets and more advanced techniques such as cross-modality transfer learning. For instance, the approach could be employed to measure the dimensions of spinal vertebrae, assess vertebral alignment, or detect anatomical abnormalities in the reconstructed 3D volume on a macroscopical scale. Moreover, the method may be utilized for treatment planning in radiation therapy, preoperative planning, and intra-operative guidance during minimally invasive spinal procedures. Further, this technique can be used for educational purposes for students and residents when only biplanar x-rays of patients are available. As a valuable addition to low-cost x-ray machines, this technique can provide healthcare professionals with an artificial CT-like 3D volume of spinal vertebrae, offering clinical insights with reduced cost and radiation exposure.
5. Conclusions
In summary, while the synthetic CT generated by the X2CT-GAN demonstrates a remarkable capability to create 3D spinal vertebrae models from synthetic 2D biplanar x-ray inputs for use in many areas, there are still some limitations in accurately capturing the fine bone structures and maintaining the precise morphology of the vertebrae. The artificial appearance of the bone surfaces and the differences in contrast and intensity representation in some areas indicate that further improvements to the model are necessary to achieve a more accurate and consistent performance in generating synthetic CTs from biplanar x-ray inputs. Future work could focus on refining the model architecture, applying cross-modality learning and utilizing novel loss functions to enhance the overall quality of the synthetic CT volumes.
Author Contributions
Conceptualization, Babak Saravi, Gernot Lang and Frank Hassel; Formal analysis, Babak Saravi, Hamza Eren Guzel, Alisia Zink, Sara Ülkümen, Jakob Wollborn, Gernot Lang and Frank Hassel; Investigation, Babak Saravi, Alisia Zink, Sara Ülkümen, Sebastien Couillard-Despres and Jakob Wollborn; Methodology, Babak Saravi, Sara Ülkümen, Sebastien Couillard-Despres, Gernot Lang and Frank Hassel; Project administration, Sebastien Couillard-Despres, Gernot Lang and Frank Hassel; Resources, Sebastien Couillard-Despres, Jakob Wollborn, Gernot Lang and Frank Hassel; Software, Babak Saravi; Supervision, Sebastien Couillard-Despres, Jakob Wollborn, Gernot Lang and Frank Hassel; Validation, Hamza Eren Guzel, Sebastien Couillard-Despres, Jakob Wollborn and Frank Hassel; Visualization, Babak Saravi; Writing – original draft, Babak Saravi; Writing – review & editing, Hamza Eren Guzel, Alisia Zink, Sara Ülkümen, Jakob Wollborn, Gernot Lang and Frank Hassel.
Funding
This research received no external funding.
Institutional Review Board Statement
In this study, we employed a [
10] of the CTSpine1K dataset [
11], which is a large and comprehensive collection of spine CT data with segmentation masks publicly available. A total of 15 clinical sites participated in the COLONOG study, and approval was obtained from the Institutional Review Board at each site prior to the commencement of the study.
Data Availability Statement
Acknowledgments
The article processing charge was funded by the Baden-Wuerttemberg Ministry of Science, Research and Art, and the University of Freiburg in the funding program Open Access Publishing. Joimax GmbH provided fellowship support for B.S.
Conflicts of Interest
The authors of the present manuscript declare no conflict of interest.
References
- Withers, P.J.; Bouman, C.; Carmignato, S.; Cnudde, V.; Grimaldi, D.; Hagen, C.K.; Maire, E.; Manley, M.; Du Plessis, A.; Stock, S.R. X-Ray Computed Tomography. Nat Rev Methods Primers 2021, 1, 18. [Google Scholar] [CrossRef]
- Ahmad, Z.; Mobasheri, R.; Das, T.; Vaidya, S.; Mallik, S.; El-Hussainy, M.; Casey, A. How to Interpret Computed Tomography of the Lumbar Spine. Ann R Coll Surg Engl 2014, 96, 502–507. [Google Scholar] [CrossRef]
- Kim, J.S.M.; Dong, J.Z.; Brener, S.; Coyte, P.C.; Rampersaud, Y.R. Cost-Effectiveness Analysis of a Reduction in Diagnostic Imaging in Degenerative Spinal Disorders. Healthc Policy 2011, 7, e105–121. [Google Scholar] [CrossRef]
- Alshamari, M.; Geijer, M.; Norrman, E.; Lidén, M.; Krauss, W.; Wilamowski, F.; Geijer, H. Low Dose CT of the Lumbar Spine Compared with Radiography: A Study on Image Quality with Implications for Clinical Practice. Acta Radiol 2016, 57, 602–611. [Google Scholar] [CrossRef]
- Khan, U.; Yasin, A.; Abid, M.; Shafi, I.; Khan, S.A. A Methodological Review of 3D Reconstruction Techniques in Tomographic Imaging. J Med Syst 2018, 42, 190. [Google Scholar] [CrossRef]
- Yuniarti, A.; Suciati, N. A Review of Deep Learning Techniques for 3D Reconstruction of 2D Images. In Proceedings of the 2019 12th International Conference on Information & Communication Technology and System (ICTS); IEEE: Surabaya, Indonesia, July, 2019; pp. 327–331. [Google Scholar]
- Han, X.-F.; Laga, H.; Bennamoun, M. Image-Based 3D Object Reconstruction: State-of-the-Art and Trends in the Deep Learning Era. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 1578–1604. [Google Scholar] [CrossRef]
- Ying, X.; Guo, H.; Ma, K.; Wu, J.; Weng, Z.; Zheng, Y. X2CT-GAN: Reconstructing CT from Biplanar X-Rays with Generative Adversarial Networks. 2019. [Google Scholar] [CrossRef]
- Yang, C.-J.; Lin, C.-L.; Wang, C.-K.; Wang, J.-Y.; Chen, C.-C.; Su, F.-C.; Lee, Y.-J.; Lui, C.-C.; Yeh, L.-R.; Fang, Y.-H.D. Generative Adversarial Network (GAN) for Automatic Reconstruction of the 3D Spine Structure by Using Simulated Bi-Planar X-Ray Images. Diagnostics (Basel) 2022, 12, 1121. [Google Scholar] [CrossRef]
- Johnson, C.D.; Chen, M.-H.; Toledano, A.Y.; Heiken, J.P.; Dachman, A.; Kuo, M.D.; Menias, C.O.; Siewert, B.; Cheema, J.I.; Obregon, R.G.; et al. Accuracy of CT Colonography for Detection of Large Adenomas and Cancers. N Engl J Med 2008, 359, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, C.; Hui, Y.; Li, Q.; Li, J.; Luo, S.; Sun, M.; Quan, Q.; Yang, S.; Hao, Y.; et al. CTSpine1K: A Large-Scale Dataset for Spinal Vertebrae Segmentation in Computed Tomography. 2021. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Sekuboyina, A.; Husseini, M.E.; Bayat, A.; Löffler, M.; Liebl, H.; Li, H.; Tetteh, G.; Kukačka, J.; Payer, C.; Štern, D.; et al. VerSe: A Vertebrae Labelling and Segmentation Benchmark for Multi-Detector CT Images. 2020. [Google Scholar] [CrossRef]
- Löffler, M.T.; Sekuboyina, A.; Jacob, A.; Grau, A.-L.; Scharr, A.; El Husseini, M.; Kallweit, M.; Zimmer, C.; Baum, T.; Kirschke, J.S. A Vertebral Segmentation Dataset with Fracture Grading. Radiology: Artificial Intelligence 2020, 2, e190138. [Google Scholar] [CrossRef]
- Milickovic, N.; Baltast, D.; Giannouli, S.; Lahanas, M.; Zamboglou, N. CT Imaging Based Digitally Reconstructed Radiographs and Their Application in Brachytherapy. Phys Med Biol 2000, 45, 2787–2800. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magn Reson Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Levine, L.; Levine, M. DRRGenerator: A Three-Dimensional Slicer Extension for the Rapid and Easy Development of Digitally Reconstructed Radiographs. JCIS 2020, 10, 69. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. Communications of the ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Jin, L.; Tan, F.; Jiang, S. Generative Adversarial Network Technologies and Applications in Computer Vision. Comput Intell Neurosci 2020, 2020, 1459107. [Google Scholar] [CrossRef]
- Gong, M.; Chen, S.; Chen, Q.; Zeng, Y.; Zhang, Y. Generative Adversarial Networks in Medical Image Processing. Curr Pharm Des 2021, 27, 1856–1868. [Google Scholar] [CrossRef]
- Figueira, A.; Vaz, B. Survey on Synthetic Data Generation, Evaluation Methods and GANs. Mathematics 2022, 10, 2733. [Google Scholar] [CrossRef]
- Ding, X.; Wang, Y.; Xu, Z.; Welch, W.J.; Wang, Z.J. Continuous Conditional Generative Adversarial Networks: Novel Empirical Losses and Label Input Mechanisms. 2020. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).