1. Introduction
The analysis of phylogenetic data presents several challenges for epidemiologists and microbiologists, as well as for computer scientists and engineers. One of the major challenges is taking into account data from several sources, such as data from surveillance databases to rapidly detect microbial outbreaks, and data generated by different sequence based typing methods. The appearance of NGS technologies further increased this challenge by the substantial growth on the amount of genomic data that can be used to characterize a population.
Phylogenetic analyses usually consist of four main steps: obtaining relevant typing data and ancillary data, inferring phylogenetic trees and descendent patterns, integrating typing and ancillary data, and providing suitable information visualizations of trees and integrated data. There are more than one alternative tool to achieve these main steps, which choice usually depends on the type of data and results of interest. Several tools to support phylogenetic analyses have been proposed, varying from standalone applications to integrative web applications that include tools and algorithms used in these tasks. Some of them are standalone tools that are focused on inferring phylogenetic trees, such Phylip [
1], START [
2], eBURST [
3] and goeBURST [
4]; others are focused on visualization and integration of data and exploration of results, such as Dendroscope [
5], TreeView [
6] and Evolview [
7]. Online web services dedicated to one phylogenetic tool have been also proposed, such as PhyML [
8], FastME [
9] and BOOSTER [
10], and that target in general just one step of phylogenetic analysis. These approaches imply often to transform and reformat their output to serve as input data for other tools, in addition to manage their storage. Thus, users have to build these procedures manually as well as to wait for and collect results in order to proceed to the next steps. This is time-consuming, inefficient and more prone to human error, as no automation is involved. To overcome these problems, standalone integrative applications, i.e., which can execute all steps previously described for phylogenetic analyses, have been proposed, such as SplitsTree4 [
11] and PHYLOViZ [
12]. Also, web integrative applications have emerged, such as PHYLOViZ Online[
13], GrapeTree [
14] and Phylogeny.fr [
15] providing the possibility of remote cloud-based execution and storage management. But cloud-based execution mechanisms provided by these solutions are often only partial and do not scale. Moreover, new tools are not easily integrated. Recently, Phylogeny.fr as evolved to NGPhylogeny.fr, a web integrative application that relies on the Galaxy workflow system [
16] for executing tools and algorithms over data. It is however a specific implementation for NGPhylogeny.fr [
17], and the integration between NGPhylogeny.fr and the worflow system is not generic and modular to adapt to other web integrative applications.
In this work we introduce FLOWViZ, an Apache Airflow based workflow middleware for computational phylogenetics, that abstracts contracts and a core API for defining and specifying data-centric workflows based on containerized tools. It has been designed to be: (i) interoperable by allowing seamless integration with different web integrative applications, that want to provide workflow building, through contracts and loosely-coupled relationships among components; (ii) scalable by supporting large-scale analyses over on cloud and HPC premises; (iii) flexible by allowing to add and use external tools, along with the bundled ones inside the web application itself, which is achieved by assuring that tools comply with defined contracts, that specify rules and operation guidelines for a correct tool execution and configuration; (iv) user-friendly by giving a web API and interface through which users can manage, build and execute workflows along with the access of complete and detailed results and logs. To accomplish these features, FLOWViZ is composed by two components: (i) a web client which supplies the user with both an API and a graphical user interface allowing it to add its own tools, build workflows and get results; (ii) a server which includes, among other features, the contracts that should be complied by tools and mechanisms to interact with Apache Airflow instances for scheduling and monitoring workflows execution.
We describe in this paper the key aspects and choices that guided the design and implementation process of FLOWViZ. It is organized as follows.
Section 2 presents the design principles and the architecture, namely the main components and interactions among them.
Section 3 details the implementation choices. An use case is provided in
Section 4, namely the integration of FLOWViZ middleware in the PHYLOViZ web application, which illustrates how data-centric workflows can be built and used in this context. Finally, in
Section 5 we discuss our approach and results, as well as possible future work, and we summarize the main contributions.
2. FLOWViZ
FLOWViZ middleware aims to provide a bridge between workflow clients, such as web applications, and data-centric workflow systems. Although we chose to rely on Apache Airflow, FLOWViZ design principles and architecture are independent of the underlying workflow engine. The choice of Apache Airflow relied on the comparison of its features among other task-based data-centric workflow systems [
18,
19]. Apache AirFlow [
20] is higly scalable and it can be deployed on cloud and HPC environments, it supports several workflow models, and it provides both a graphical user interface (GUI) and a application programming interface (API). Others such as Galaxy [
21], Kepler [
22] and Taverna [
23] only support a GUI. Pegasus [
24], Nextflow [
25], Swift [
26] and Snakemake [
27] provide only a command line interface. An application programming interface is particularly relevant when developing a middleware framework, to be able to programmatically define the tasks to be executed. Unlike most of the other workflow managers, it supports all the following workflow models [
28]:
procedural, since the model explicitly specifies the sequence of steps and tasks known as control-flow;
script-based, because the composition of nodes is described using scripting languages;
event-based, since workflows are characterized by a discrete set of states; and
adaptive, having the possibility to react dynamically to runtime situations and exceptions, such as a failure in pre-processing an input file.
We describe next the design principles and the architecture, detailing the different modules. The domain model will be also presented and discussed.
2.1. Architecture
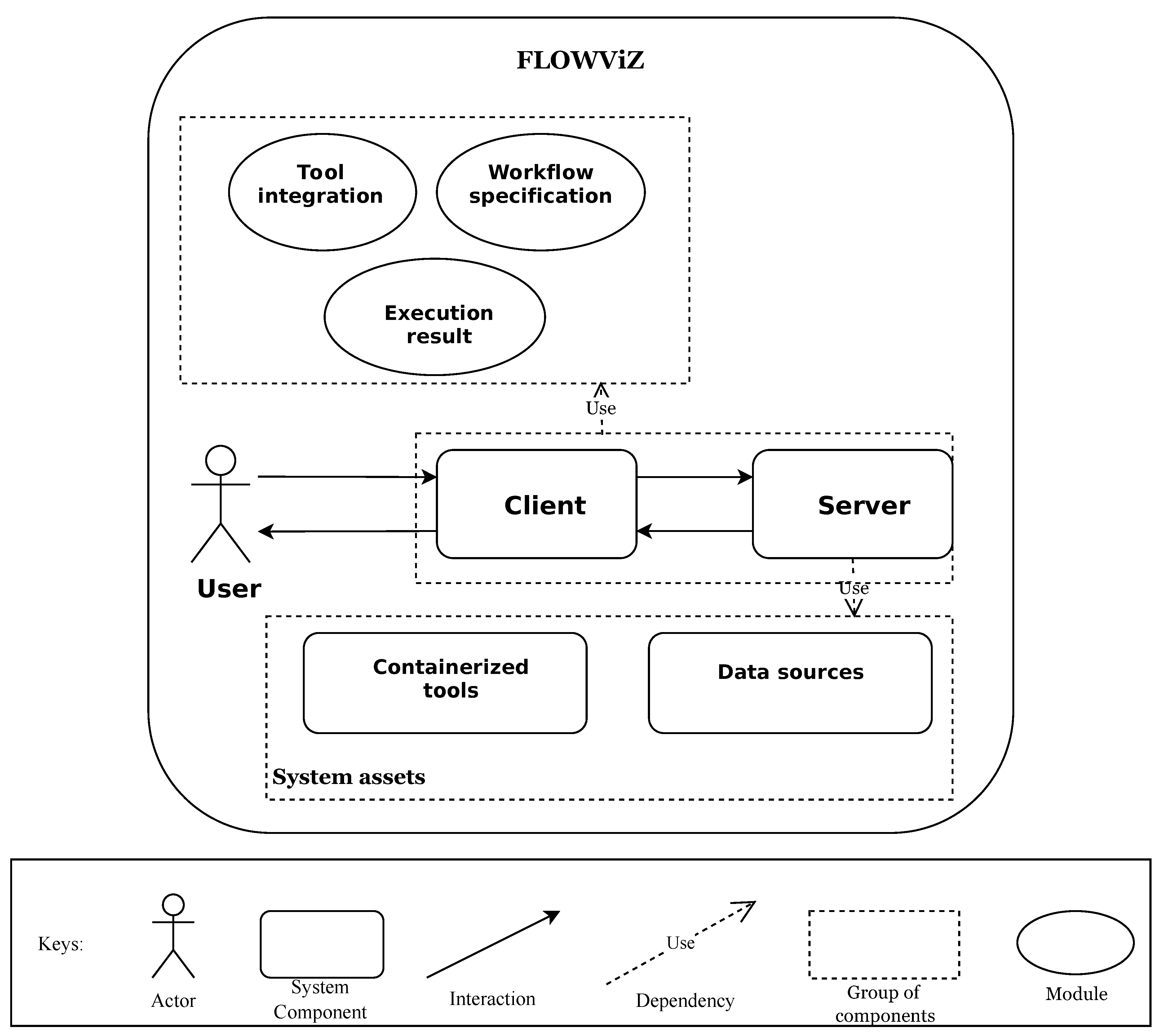
FLOWViZ middleware supports the following functionalities: (i)
tools integration; (ii)
workflow design and specification; (iii) and
workflow execution and monitoring. It main usage scenario is to be integrated as a component. In order to support required functionalities, FLOWViZ has two main components, the
client and the
server, as depicted in
Figure 1. The web client supplies the user with a GUI that exposes the three functionalities previously referred. The server component serves as a middleware between the web client and both the workflow system and a database, which is responsible for storing the information about users, tools and workflows. Moreover, the server provides the necessary API endpoints that make the execution of the three main functionalities possible, while providing user authentication and account management.
The workflow system interacts both with the database and the server. It retrieves information from the database, in order to dynamically create and execute workflows submitted by the user, and provides the server with information regarding execution of each workflow, which latterly the server will deliver to the client.
Web applications that rely on FLOWViZ can use the server API directly, submitting tools and workflows specifications, as well as monitoring their execution. To achieve a seamless tool integration with main web applications, tools to be used in workflows must be containerized and their usage must be specified in compliance with FLOWViZ contracts. Moreover, the common core dependencies between FLOWViZ and the web application, for instance if they are deployed on the same server, must be also by a dependency contract. But if there are no dependencies, the FLOWViZ will have to be deployed standalone, meaning that it cannot be integrated as an internal module and share object instances. Instead, it will be an external component that communicates with the web application or phylogenetic framework and its tools over the network.
The database is a requirement, since FLOWViZ need to have access to the storage of tool contracts, which specifies for instance how to invoke the tool, to the submitted workflows and to the user credentials. Tool contracts and workflows contain information which can point out to external containerized tools and data sources, i.e. the system assets.
2.2. Modules
The functionalities supported by FLOWViZ rely on three functional modules: (i) tool integration module; (ii) workflow specification module; (iii) execution result module. The tool integration module (i) is responsible for integrating new phylogenetic tools within the framework via contracts; the workflow building module (ii) supports designing workflows and scheduling their execution, either through the GUI or directly through the server API; the result production (iii) retrieves workflow execution logs and builds a final report related to the workflow execution.
2.2.1. Tool Integration Module
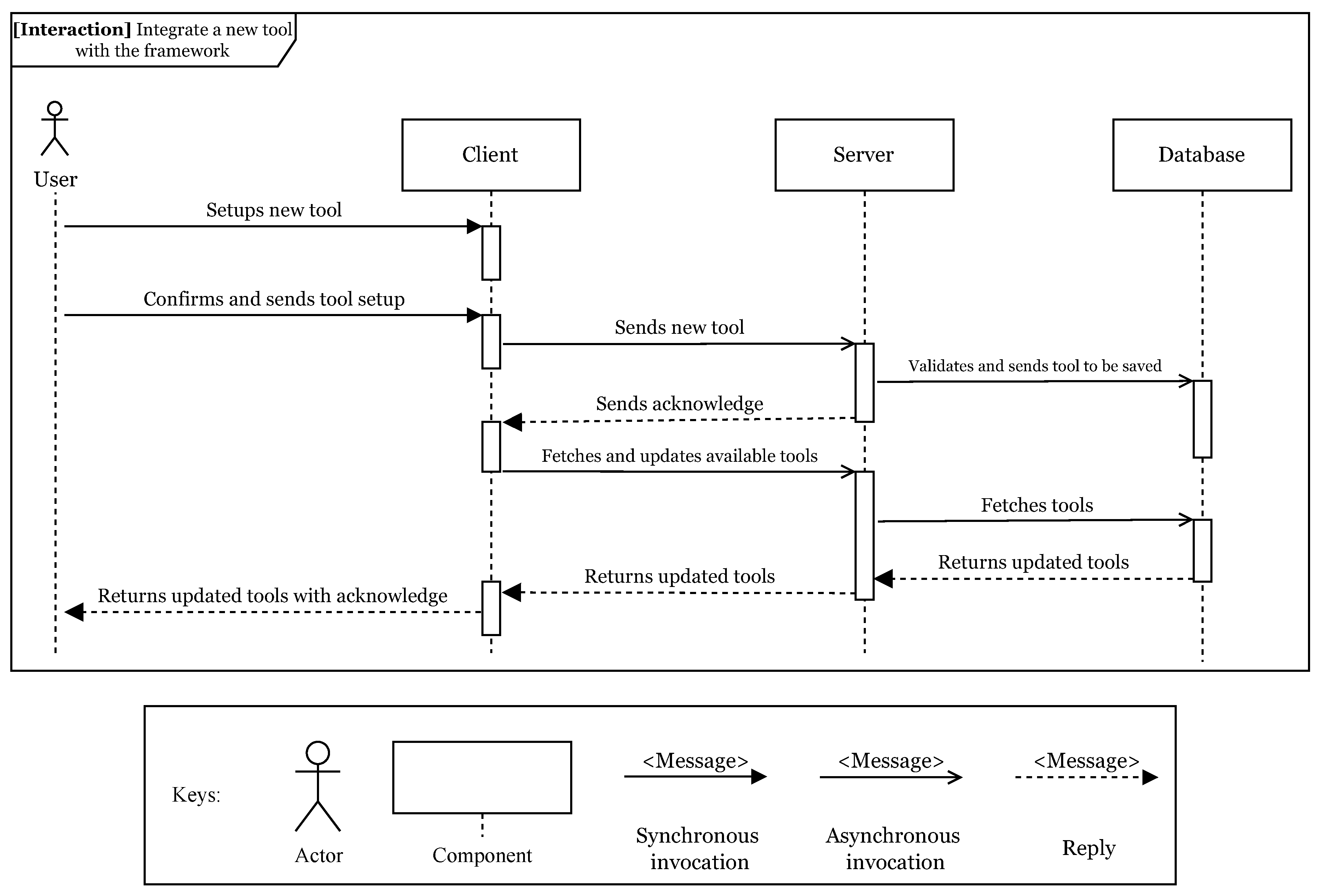
The
tool integration module (i) supports the setup and addition of new tools to FLOWViZ. This is done by specifying the usage contract for the tool.
Figure 2 shows the interaction diagram of this module.
By using the web client, the user can configure the tool contract. When concluded, it is sent to the server, validated and if successful, saved into the database. A web application can also submit tool contracts directly through the server API. After the contract being stored, the tool is integrated with the middleware and its documentation can be accessed in the web client and through the server API. Notice that the tool must be containerized, accessible to the workflow system. The user can then build workflows, if all the necessary tools are already integrated.
2.2.2. Workflow Specification Module
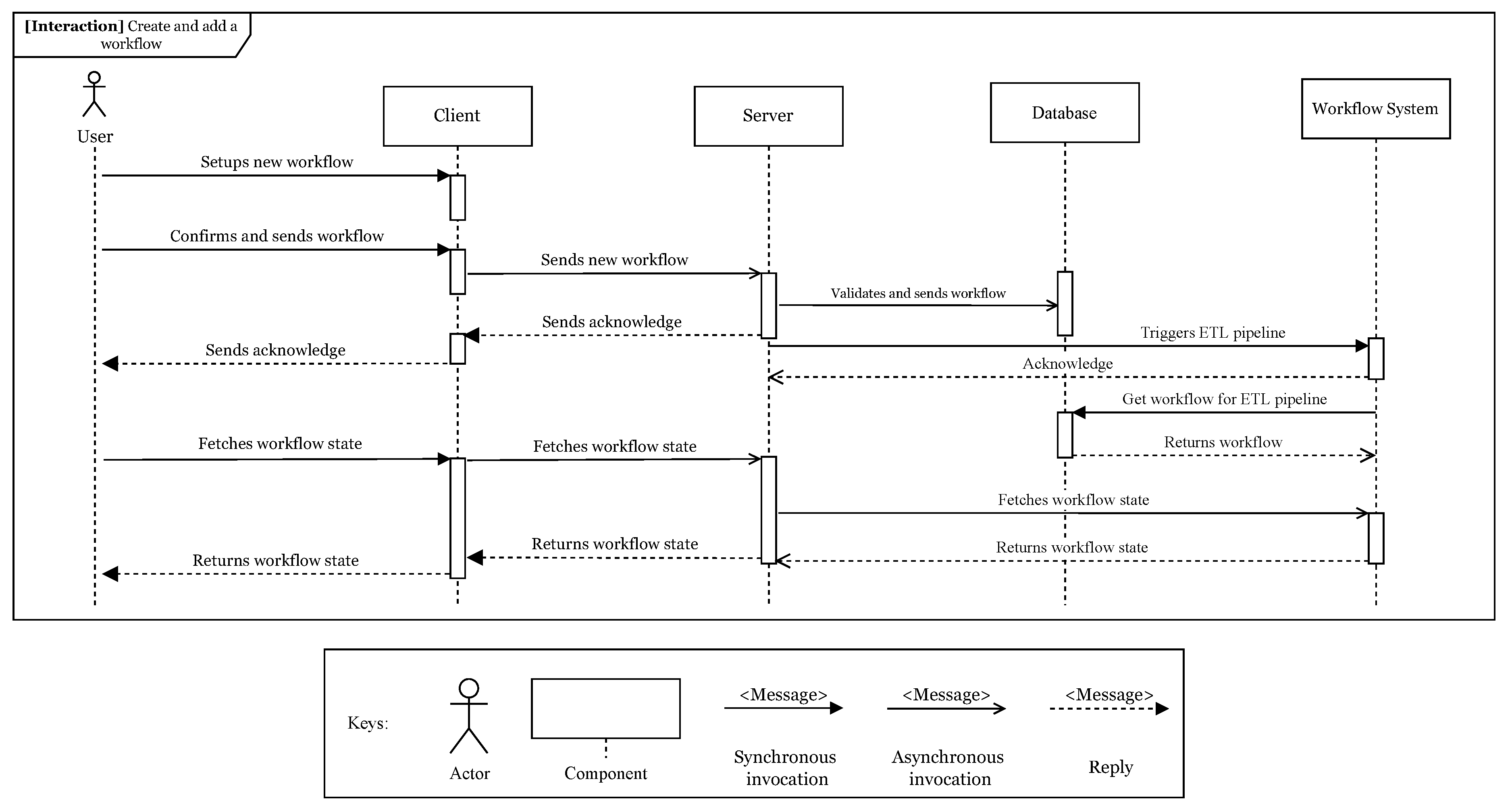
As depicted in
Figure 3, the workflow specification module allows designing workflows and it is responsible for managing workflows.
During a workflow specification, the user specifies the workflow using the graphical editor provided in the web client. Each tool will be represented as a node inside the editor and can be configured as a task or step of the workflow. The user must also connect the nodes, in order to let the framework infer the data flow between each involved tool. When the user finishes the setup, the web client will send the workflow to the server. A web application can also send a specified workflow directly through the server API. When it reaches the server, the workflow will go through a validation and, if successful, it will be saved into the database. If the database save procedure has executed successfully, the server will then trigger the Airflow ETL pipeline, by sending an HTTP request to its REST API, which notifies that a new user workflow was created. The ETL pipeline, as it will be detailed ahead, will transform the workflow contract into the Airflow directed acyclic graph (DAG) and, then, starts its execution. Its execution will occur at the date and time configured by the user and the execution result status can be retrieved later.
2.2.3. Execution Result Module
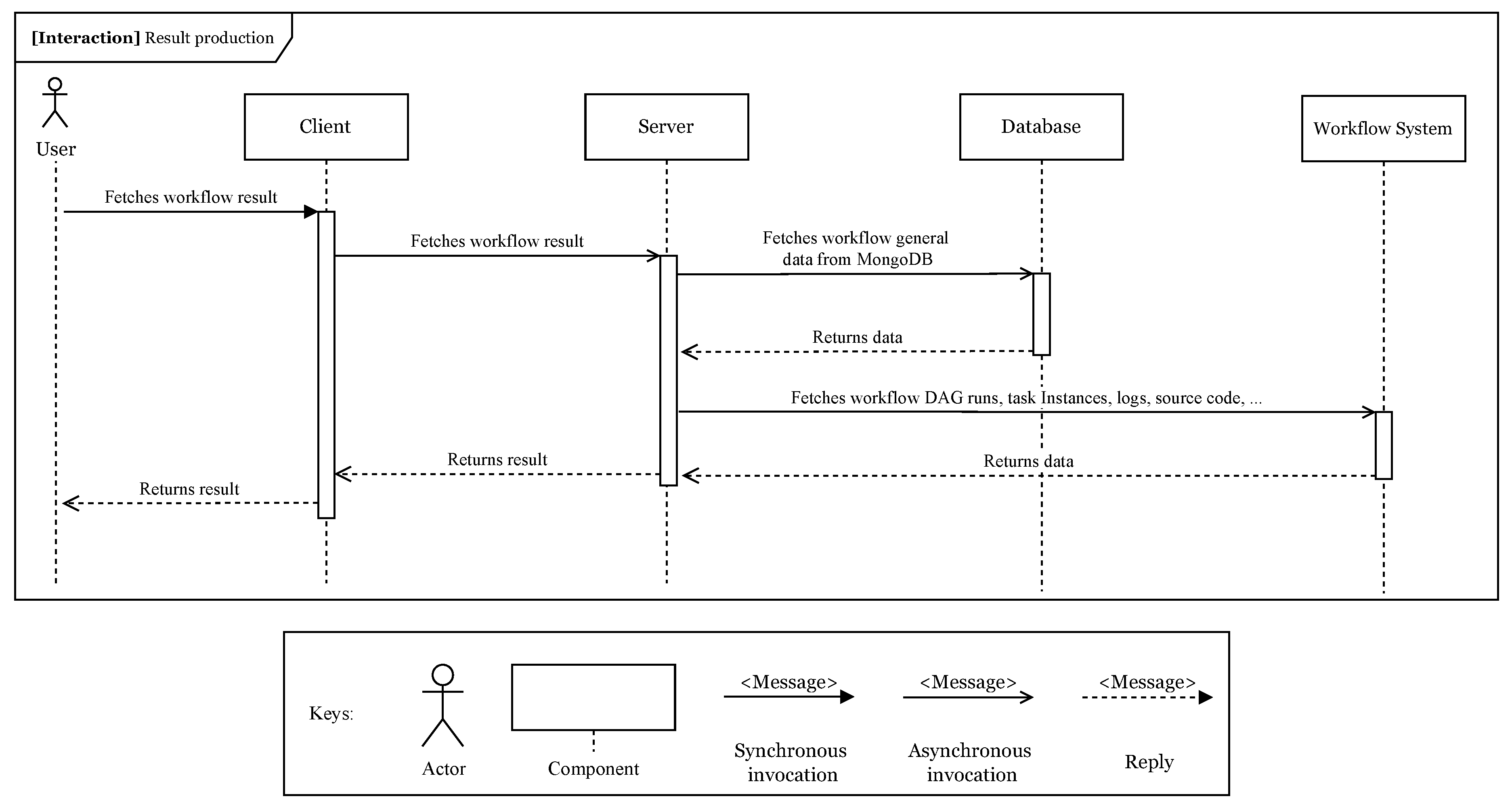
The
execution result module (iii) delivers the workflow log back to the client, so the user can check it. When the workflow execution finishes, the server provides the client with endpoints, which fetch data from the Airflow REST API, in order to retrieve results for a specific workflow.
Figure 4 displays the general use case implementation of this module. When the workflow execution finishes, the user can check its execution logs and the dynamically generated DAG source code, created by the ETL pipeline. It can also access output files or data generated in each step.
2.3. Domain Model
To support the functionalities described before, FLOWViZ stores tool contracts, submitted workflows and user credentials. Therefore, the domain model includes three main entities, namely Tool, Workflow and User.
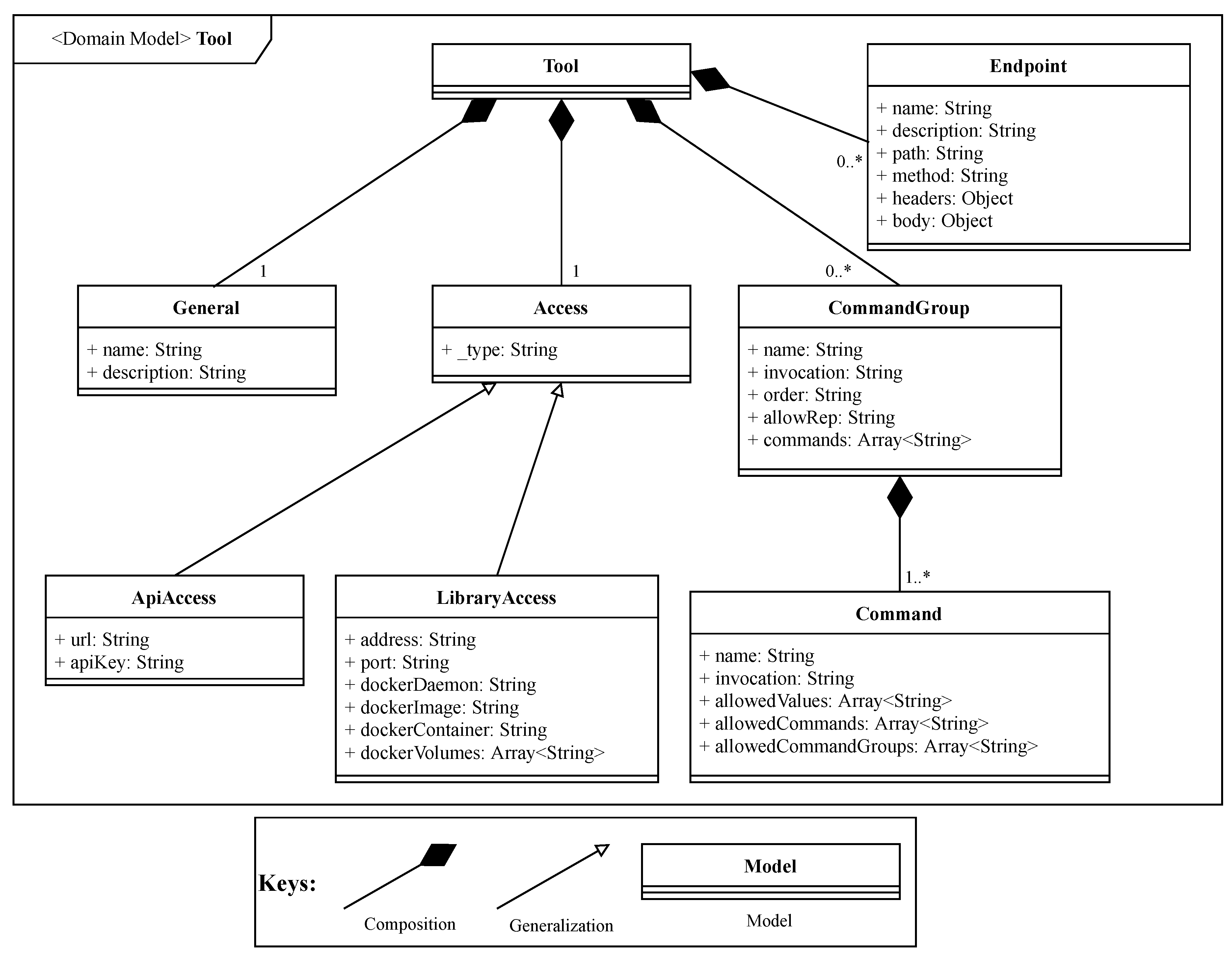
The
Tool, as depicted in
Figure 5, represents the contract of an integrated tool. The
Access of a tool can be through a docker container (
LibraryAccess) or an API (
ApiAccess). Although we have selected docker containers, more container engines can be used, with little effort.
In the case of tool libraries which include also standalone tools, it is necessary to define the tool command tree and establish the command hierarchy. This is supported by specifying a CommandGroup, which may contain more than one Command.
The CommandGroup includes properties such as: invocation property, which allows the user to specify an alias to invoke a specific command group or command; order property, that defines the invocation priority of a specific command or command group; allowRep property, that defines if a command or command group can be invoked again, after a previous invocation. Each CommandGroup is composed by one or more commands included in the tool library. The sub-properties which compose a Command and their functions are the following: name that represents the designation of the command; invocation, which specify how the command can be invoked; allowedValues, which are the values that the specific command accepts; allowedCommands, that gives the list of commands which can be invoked after the current command invocation; allowedCommandGroups which gives the list of command groups that can be invoked after the current command invocation.
In the case of an tool accessed through an API, it is necessary to define the Endpoint, which includes: the method, which specifies the HTTP method accepted by the endpoint; the headers, for specifying the HTTP headers that are accepted by the endpoint; the body, for specifying the body that is accepted by the endpoint.
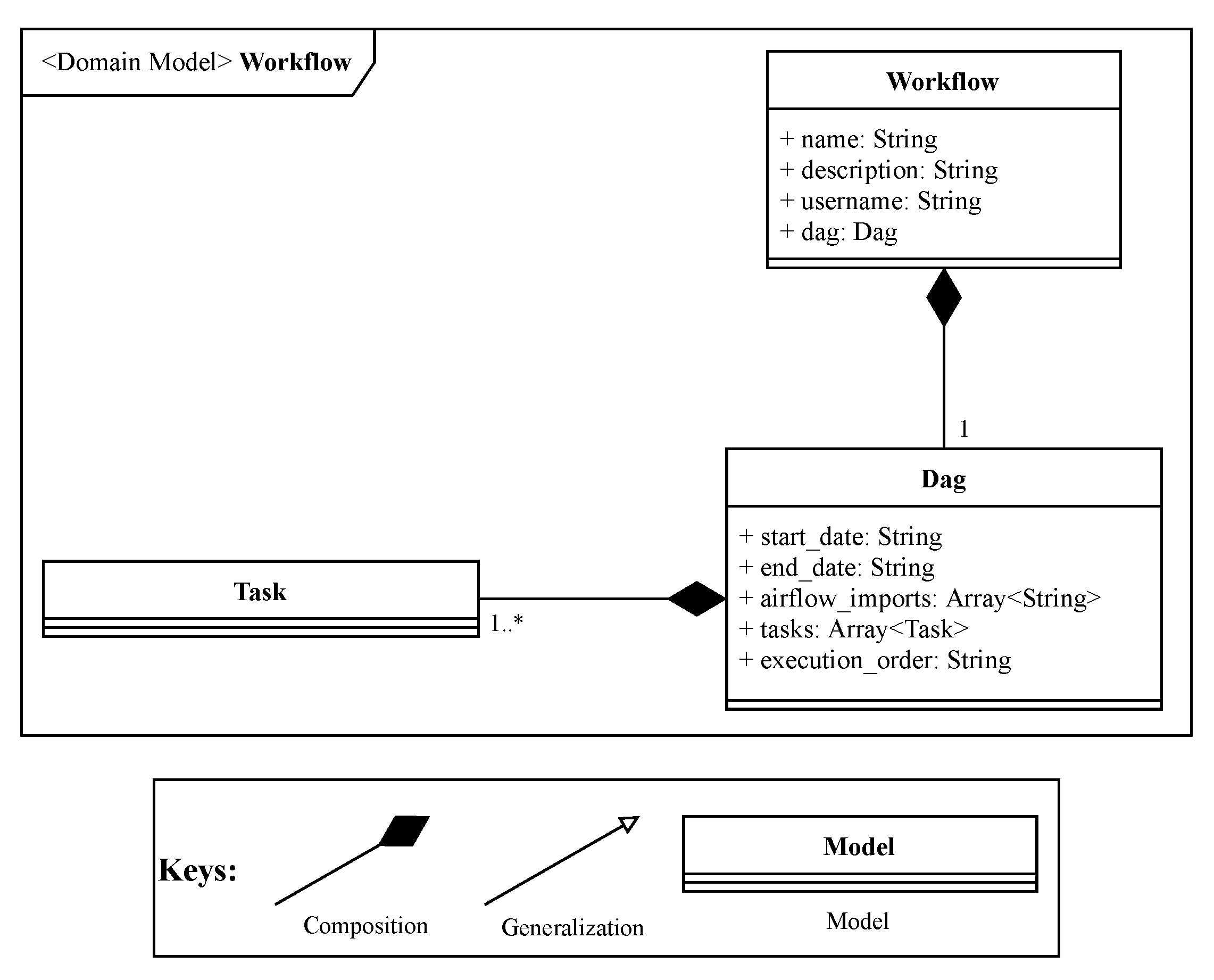
The workflow entity, depicted in
Figure 6 represents a workflow to be executed by Apache Airflow. This entity is responsible for binding a workflow to the user who submitted it, as Apache Airflow does not have a way to separate workflows for each user. Later, in
Section 3, it will be detailed how Apache Airflow will process the user created workflows.
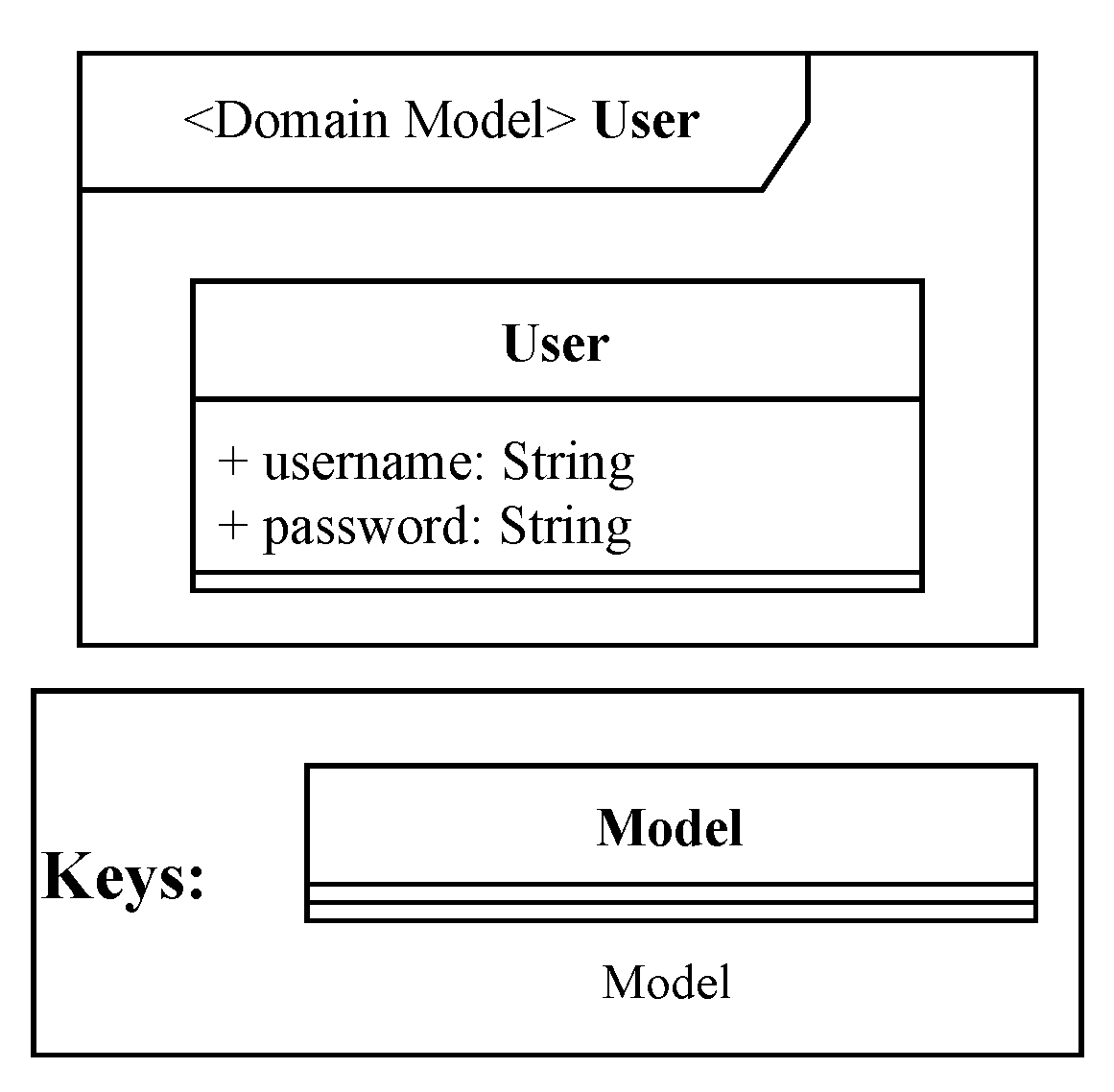
The user model, depicted by
Figure 7, represents a FLOWViZ user, and it is mainly used during the registration and login procedures. The
password property is always stored in the hashed form, in order to be unusable in case of a data leak.
3. FLOWViZ Implementation
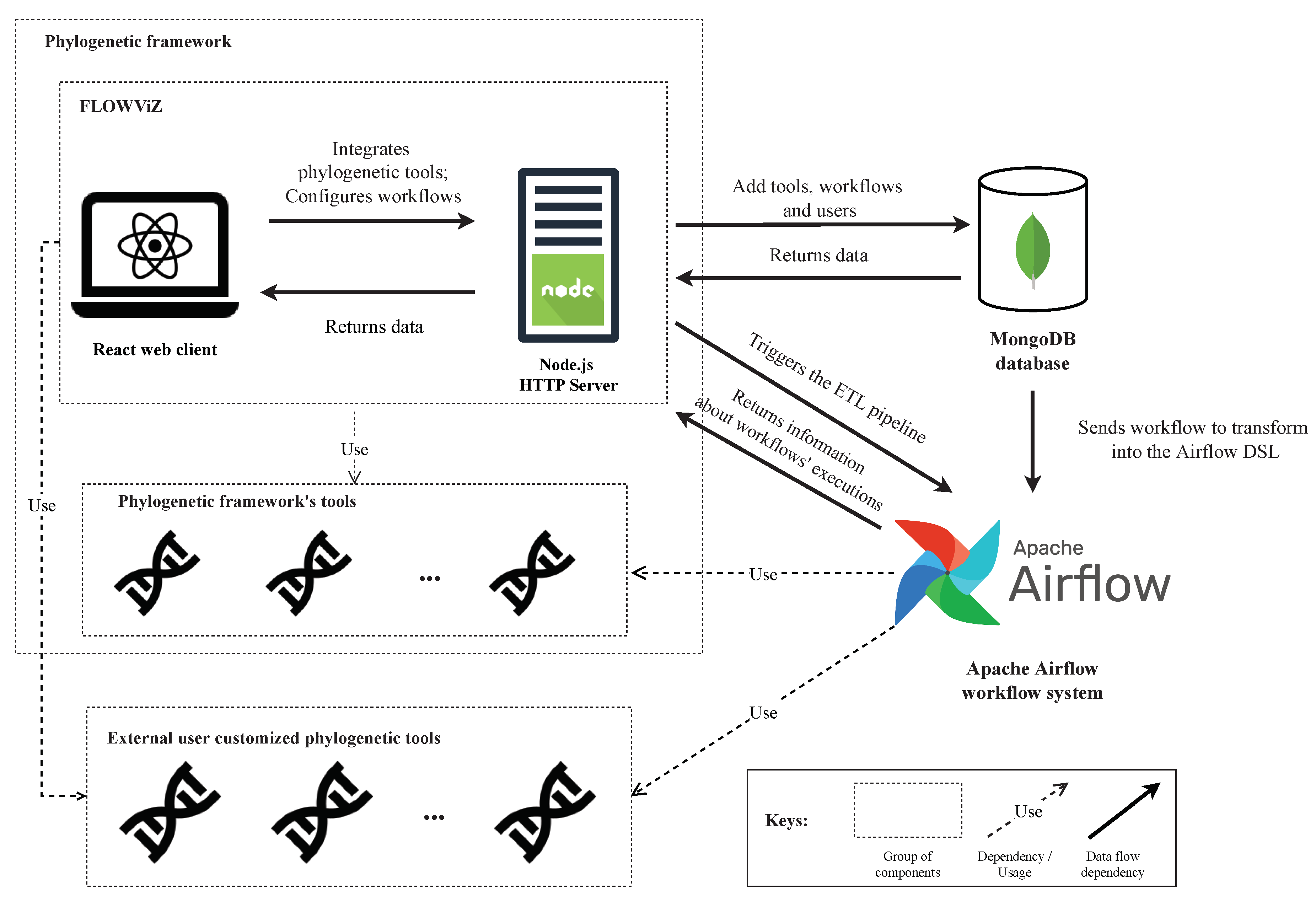
As depicted in
Figure 8, FLOWViZ interacts with a database (e.g., MongoDB) and the Apache Airflow workflow system. FLOWViZ includes a React web client and an HTTP Express server, built with Node.js, and hence both written in JavaScript. The reference implementation is publicly available at
https://github.com/DIVA-IPL-Project/FLOWViZ.
3.1. Server
The server is mainly responsible to: (i) receive, validate and save tool contracts into the database, namely in the tool model collection; (ii) receive, validate and save user workflows into the database, namely in the workflow model collection; (iii) manage users access; (iv) supply the client with integrated tools, created workflows and workflow execution logs. To perform these operations it was designed a REST API, described in
Table 1.
The server was also built to be extensible, since it can be integrated as a dependency along with another Node.js applications that meet the minimum requirements. Therefore, FLOWViZ can be integrated with an web application via an established contract. If the application has common core dependencies, such as the Express server dependency, the Express instance can be shared with the FLOWViZ server, if specified in the contract. This way, FLOWViZ will behave as an extension or a plugin of the framework. FLOWViZ is also able to be integrated with frameworks that do not contain common dependencies or technologies, however, the deployment of FLOWViZ will have to be standalone, meaning that it can not be integrated as an internal module and share object instances, as depicted in
Figure 8, but it will be an external module that communicates with the framework and its tools over the network. Self-hosting FLOWViZ does not require exposing tools using remote instances; in the
localhost environment, the user or developer only needs to specify the tools contracts.
3.1.1. Tools
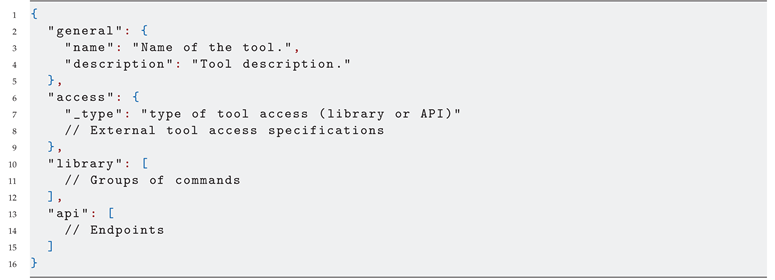
Listing 1 shows the generic tool contract. As mentioned before, there exists two alternatives for configuring a tool: if it has CLI support, the user can configure it as a library and configure each command invocation and settings; or if the tool exposes an API, the user can set up each exposed endpoint, providing the allowed structure for the HTTP body and headers. Although the listing has both api and library fields, the access will bound the kind of configuration that will be used for a given tool.
|
Listing 1: Generic tool contract |
 |
3.1.2. Worflows
The main goal of Apache Airflow is to schedule and execute users workflows, that are sent from the server and saved into MongoDB. To this end, an
ETL pipeline was created, in order to retrieve users workflows from the database and transform them into Airflow
DAGs, to be executable by the workflow system. An
ETL pipeline is a set of processes that
extracts information from a data source, namely, from the MongoDB
workflow collection,
transforms the extracted information into another format and, finally,
loads the transformed information into a new
DAG (Directed Acyclic Graph) to be executed by the workflow system.
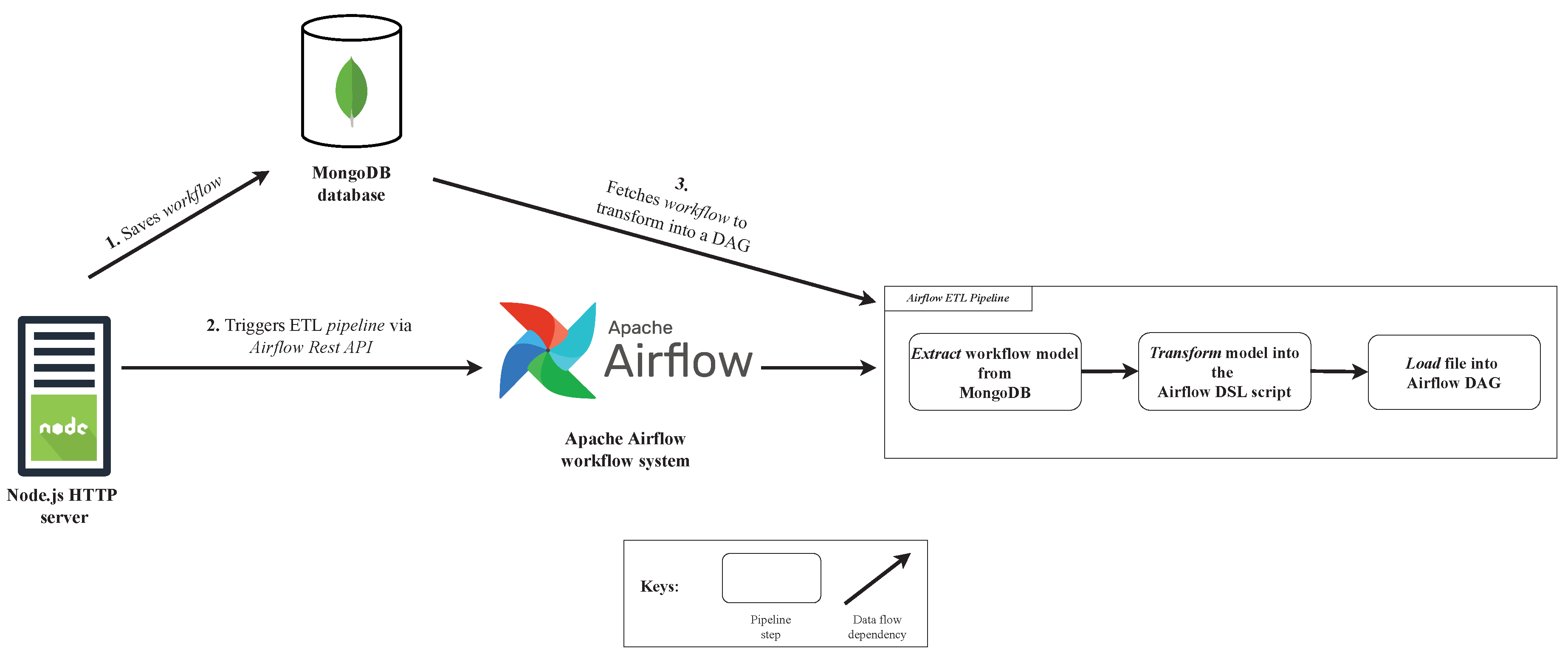
Figure 9 shows the use case of the developed
ETL pipeline inside the system architecture.
Observing
Figure 9, when a client sends a workflow to the server, it will first go through a validation and, if it succeeds, it will be saved into database (1). If the workflow was successfully saved, the server will then notify the workflow system that there is a new user workflow to be parsed and executed at a certain date and time, that was previously configured by the user (2). This notification will trigger the Airflow ETL pipeline via the workflow system REST API, which will retrieve the workflow from the database and parse it to an Airflow DAG, in order to be executable by the workflow system (3).
The workflow system starts by fetching the workflow specification from the database, using the name of the workflow and its user, i.e., the metadata that is included in the HTTP request that performs the trigger of the ETL pipeline. After the retrieval of the workflow, the extract function will extract the dag property from the workflow model in the database and send it to the next function. The interaction between the workflow system and the database is made by a specific Airflow provider, that had to be explicitly installed. The second function will transform the workflow fetched from the database using the dag property and the generate_dag function will generate a new Airflow DAG – an Airflow DSL script. Finally, Airflow will load the script into the Airflow DAG collection, which will be recognized by the workflow system and executed at the configured date and time.
3.1.3. User Access Management
When the user is authenticated, workflows are associated with its username, so only the user can manage its own workflows. The adopted authentication strategy relies on passport-jwt, which requires the user to authenticate using a signed and valid JSON Web Token (JWT), previously generated by the server and sent within the HTTP header. Sensitive information are safely stored into the database, using the hashing library Argon2 [
29].
3.2. Client
The web client is built with React [
30] and written in JavaScript. It uses the CSS framework Material-UI [
31], which provides React with out-of-the-box components that follow the Material Design of Google. It allows to seamlessly build the application graphical user interface. Moreover, for building workflows, it was used the component React Flow. With this customizable component, the web client includes an editor when it is possible to create an interactive flow, where nodes represent the integrated tools and edges represent data dependencies and the execution flow.
4. Use Case
Our use case consists of using FLOWViZ middleware within PHYLOViZ [
13], namely in its new version deployed at
https://web.phyloviz.net/. We note data all data analysis tasks in this new version of PHYLOViZ are defined as data-centric workflows. We illustrate next how tools are configured and how workflows are defined and sumitted for asynchronous execution.
The workflow described next relies on phylolib [
32], a set of tools for inferring phylognetic trees from typing data, and on another two helper tools to deal with datasets. These tools are containerized and cached locally in our environment. But they can be stored elsewhere, as is the case of phylolib container available at
https://hub.docker.com/r/luanab/phylolib.
Listings 2 and 3 show the JSON objects that describe the workflow and underlying tools, respectively. In this use case, tools are integrated through direct submission to FLOWViZ server API, with Docker volumes being customized accordingly to a given PHYLOViZ project. The described tools could however be also integrated through the web client, which would then submit them to the FLOWViZ server. In both situations, the JSON objects are validated by FLOWViZ server component. If the contract is valid, it will be saved into the database. At this point, tools become successfully integrated, and the user can build customized workflows using such tools.
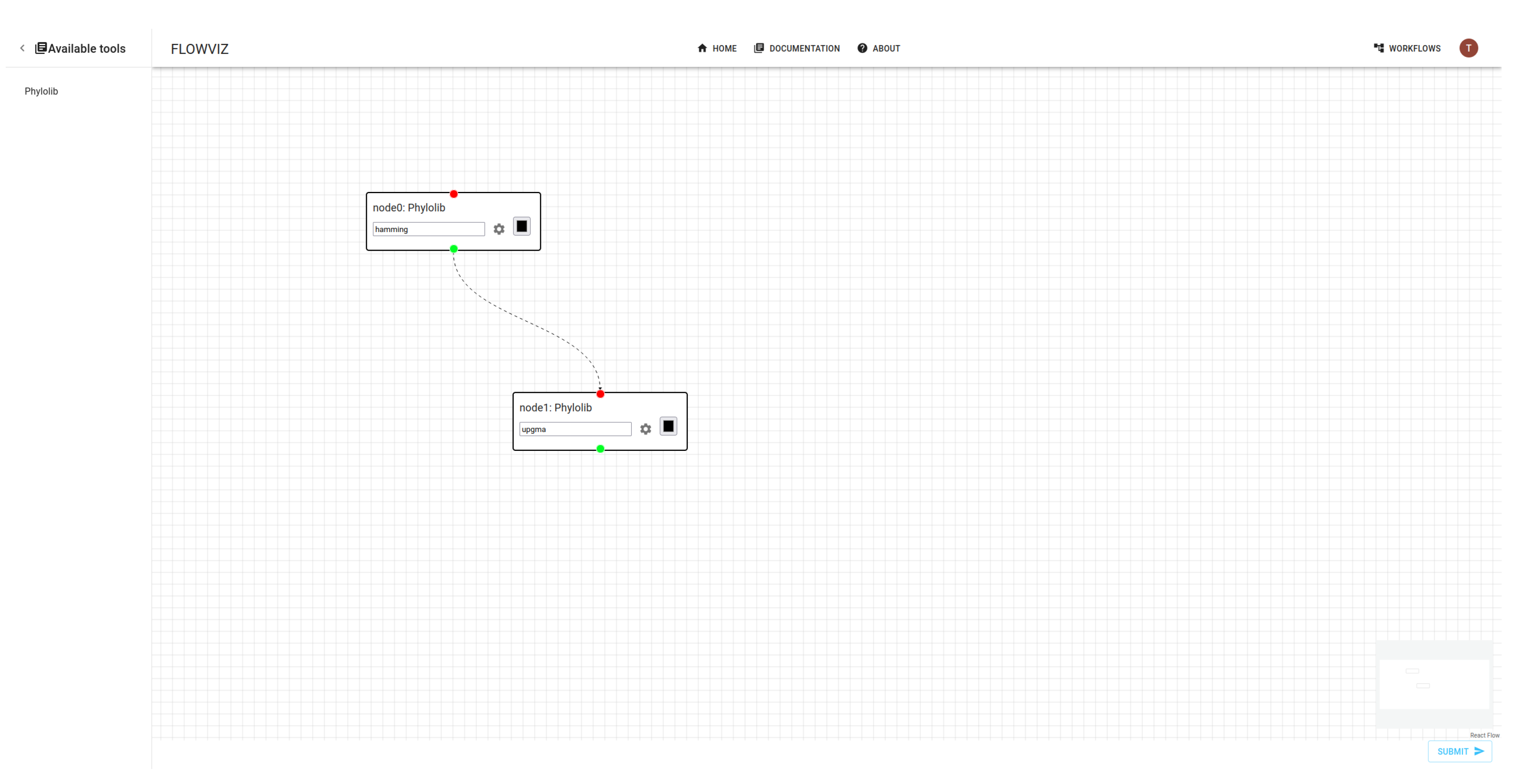
Workflows can be specified using the web client provided editor – the
whiteboard, where the user can graphically draw and configure the workflow tasks (see
Figure 10). The user can also submit a workflow specification directly to the FLOWViZ server API as mentioned before, which is the approach followed in our use case.
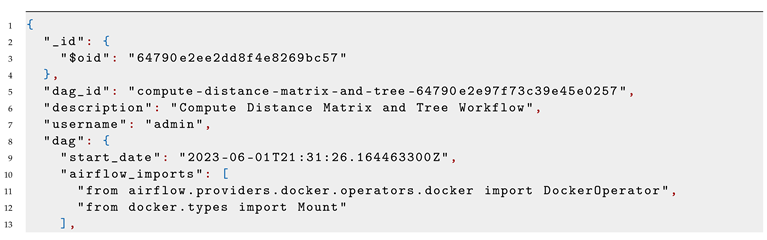
When a workflow is submitted to the FLOWViZ server, it is also validated. If it succeeds, the workflow data will be saved into the database (see Listing 4) and an HTTP request, containing the name of the workflow and its correspondent user, will be sent to the Airflow REST API, that will then fetch from the database the correspondent user workflow and parse it to an Airflow DSL script or DAG. The DAG will then be executed at the pre-configured date and time, which will produce results, that can be easily consulted via the web client. In our use case, PHYLOViZ relies on FLOWViZ server API to check executions status. An exemple of an execution log is shown in Listing 5, namely for the fourth task in the workflow shown in Listing 2.
|
Listing 2: Workflow JSON sumitted to FLOWViZ server API. |
 |
 |
|
Listing 3: Tools JSON sumitted to FLOWViZ server API. |
 |
 |
 |
 |
|
Listing 4: FLOWViZ internal workflow instance representation. |
 |
 |
 |
|
Listing 5: Tool execution log. |
 |
5. Conclusion
This paper introduces FLOWViZ, a tool integration and workflow management middleware for phylogenetic analysis frameworks. It provides both a client and a server components, allowing the user to build workflows, through a user-friendly web client interface, enabling it to add its own phylogenetic tools and use them in its own built workflows. It aims to allow and facilitate the integration of workflows and their execution offloading within data-centric phylogenetic analysis frameworks.
The advantages are a greater scalability and interoperability, as tools can be continuously integrated, while the workflow system workers can be also scaled-out to handle larger workloads. Tool interoperability is also supported by using contracts and loosely coupled relationship between components, which allows seamless integration within phylogenetic frameworks, requiring the developer to only add the phylogenetic framework bundled tools to the FLOWViZ configuration.
The proposed architecture was tested and materialized into an application prototype, composed by two main components: (i) a React web client and (ii) an HTTP server, both written in JavaScript. With these two components it is possible to: (i) integrate external phylogenetic tools, (ii) build workflows with the previously integrated tools and, finally, (iii) retrieve results and logs from the underlying workflow system.
We plan to extend FLOWViZ to support integration with other data-centric workflow systems, since the architecture was designed independently of the underlying workflow engine. This will allow data-centric web platforms to configure different execution environments according to their needs, when using FLOWViZ as middleware, without effort.
Author Contributions
CV, ML and APF designed the solution and proposed the software architecture. ML developed the software. APF and ML tested the software. All authors wrote, read and approved the final manuscript.
Funding
This research was funded by European Union’s Horizon 2020 research and innovation program under Grant Agreement No. 951970 (OLISSIPO project), by the Fundação para a Ciência e a Tecnologia (FCT) through projects NGPHYLO PTDC/CCI-BIO/29676/2017 and UIDB/50021/2020, and by the Polytechnic Institute of Lisbon trough project IPL/2021/DIVA.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Felsenstein, J. PHYLIP (phylogeny inference package) version 3.695. Distributed by the 2013. [Google Scholar]
- Jolley, K.A.; Feil, E.; Chan, M.S.; Maiden, M.C.J. Sequence type analysis and recombinational tests (START). Bioinformatics 2001, 17, 1230–1231. [Google Scholar] [CrossRef]
- Feil, E.J.; Li, B.C.; Aanensen, D.M.; Hanage, W.P.; Spratt, B.G. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. Journal of bacteriology 2004, 186, 1518–1530. [Google Scholar] [CrossRef]
- Francisco, A.P.; Bugalho, M.; Ramirez, M.; Carriço, J.A. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC bioinformatics 2009, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Systematic biology 2012, 61, 1061–1067. [Google Scholar] [CrossRef]
- Page, R.D. Tree View: An application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic acids research 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Molecular biology and evolution 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Lemoine, F.; Domelevo Entfellner, J.B.; Wilkinson, E.; Correia, D.; Dávila Felipe, M.; De Oliveira, T.; Gascuel, O. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 2018, 556, 452–456. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Molecular biology and evolution 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.P.; Carriço, J.A.; Vaz, C. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2016, 33, 128–129. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Gonçalves, B.; Francisco, A.P.; Vaz, C.; Ramirez, M.; Carriço, J.A. PHYLOViZ Online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic acids research 2016, 44, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome research 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Goecks, J.; Nekrutenko, A.; Taylor, J.; team@ galaxyproject. org, G.T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome biology 2010, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny. fr: new generation phylogenetic services for non-specialists. Nucleic acids research 2019, 47, W260–W265. [Google Scholar] [CrossRef] [PubMed]
- Ramon-Cortes, C.; Alvarez, P.; Lordan, F.; Alvarez, J.; Ejarque, J.; Badia, R.M. A survey on the Distributed Computing stack. Computer Science Review 2021, 42, 100422. [Google Scholar] [CrossRef]
- Liu, J.; Pacitti, E.; Valduriez, P.; Mattoso, M. A survey of data-intensive scientific workflow management. Journal of Grid Computing 2015, 13, 457–493. [Google Scholar] [CrossRef]
- Finnigan, L.; Toner, E. Building and Maintaining Metadata Aggregation Workflows Using Apache Airflow. Temple University Libraries 2021. [Google Scholar]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research 2018, 46, W537–W544. Available online: https://www.lanl.gov/errors/system-notification.php. [CrossRef] [PubMed]
- Altintas, I.; Berkley, C.; Jaeger, E.; Jones, M.; Ludascher, B.; Mock, S. Kepler: an extensible system for design and execution of scientific workflows. In Proceedings of the Proceedings. 16th International Conference on Scientific and Statistical Database Management, 2004, 2004. IEEE; pp. 423–424. [Google Scholar]
- Hull, D.; Wolstencroft, K.; Stevens, R.; Goble, C.; Pocock, M.R.; Li, P.; Oinn, T. Taverna: a tool for building and running workflows of services. Nucleic acids research 2006, 34, W729–W732. [Google Scholar] [CrossRef] [PubMed]
- Deelman, E.; Vahi, K.; Juve, G.; Rynge, M.; Callaghan, S.; Maechling, P.J.; Mayani, R.; Chen, W.; Da Silva, R.F.; Livny, M.; et al. Pegasus, a workflow management system for science automation. Future Generation Computer Systems 2015, 46, 17–35. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Floden, E.W.; Magis, C.; Palumbo, E.; Notredame, C. Nextflow: un outil efficace pour l’amélioration de la stabilité numérique des calculs en analyse génomique. Biologie Aujourd’hui 2017, 211, 233–237. [Google Scholar] [CrossRef]
- Wilde, M.; Hategan, M.; Wozniak, J.M.; Clifford, B.; Katz, D.S.; Foster, I. Swift: A language for distributed parallel scripting. Parallel Computing 2011, 37, 633–652. [Google Scholar] [CrossRef]
- Köster, J.; Rahmann, S. Snakemake — a scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. Available online: https://academic.oup.com/bioinformatics/article/28/19/2520/290322. [CrossRef]
- Matskin, M.; Tahmasebi, S.; Layegh, A.; Payberah, A.H.; Thomas, A.; Nikolov, N.; Roman, D. A survey of big data pipeline orchestration tools from the perspective of the datacloud project. In Proceedings of the Proc. 23rd Int. Conf. Data Analytics Management Data Intensive Domains (DAMDID/RCDL 2021). CEUR-WS; 2021; pp. 63–78. [Google Scholar]
- Biryukov, A.; Dinu, D.; Khovratovich, D. Argon2: new generation of memory-hard functions for password hashing and other applications. In Proceedings of the 2016 IEEE European Symposium on Security and Privacy (EuroS&, 2016, P). IEEE; pp. 292–302.
- Accomazzo, A.; Murray, N.; Lerner, A. Fullstack React: The Complete Guide to ReactJS and Friends; Fullstack.io, 2017.
- Boduch, A. React Material-UI Cookbook: Build Captivating User Experiences Using React and Material-UI; Packt Publishing, 2019.
- Silva, L. Library of efficient algorithms for phylogenetic analysis. CoRR, 2012; abs/2012.12697. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).