Submitted:
17 October 2023
Posted:
19 October 2023
You are already at the latest version
Abstract
Keywords:
Introduction
Host signaling and virus life cycles
Epigenetic modifications
DNA methylation
Histone modifications.
RNA modifications
miRNA
Viral infections and Epigenetic modifications
Virus infection promotes epigenetic modifications in the immune system and host genes
Viral infection affects Host miRNA expression
Summary
Conclusions and Future directions
Author Contributions
Funding
Conflicts of Interest
References
- Forterre, P. Defining life: the virus viewpoint. Orig Life Evol Biosph 2010, 40, 151–160. [Google Scholar] [CrossRef]
- Caspar, D.L.; Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol 1962, 27, 1–24. [Google Scholar] [CrossRef]
- Wilen, C.B.; Tilton, J.C.; Doms, R.W. HIV: cell binding and entry. Cold Spring Harb Perspect Med 2012, 2. [Google Scholar] [CrossRef]
- Zlotnick, A. Theoretical aspects of virus capsid assembly. J Mol Recognit 2005, 18, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.J.; Tichelaar, W.; Myers, N.; Burns, N.R.; Butcher, S.J.; Kingsman, A.J.; Fuller, S.D.; Saibil, H.R. Cryo-electron microscopy structure of yeast Ty retrotransposon virus-like particles. J Virol 1997, 71, 6863–6868. [Google Scholar] [CrossRef]
- Wilson, D.P. Protruding Features of Viral Capsids Are Clustered on Icosahedral Great Circles. PLoS One 2016, 11, e0152319. [Google Scholar] [CrossRef]
- Scheiffele, P.; Rietveld, A.; Wilk, T.; Simons, K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem 1999, 274, 2038–2044. [Google Scholar] [CrossRef] [PubMed]
- Louis, J.M.; Weber, I.T.; Tozser, J.; Clore, G.M.; Gronenborn, A.M. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv Pharmacol 2000, 49, 111–146. [Google Scholar] [CrossRef] [PubMed]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A 2014, 111, E3900–3909. [Google Scholar] [CrossRef]
- Garcia-Sastre, A.; Egorov, A.; Matassov, D.; Brandt, S.; Levy, D.E.; Durbin, J.E.; Palese, P.; Muster, T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 1998, 252, 324–330. [Google Scholar] [CrossRef]
- Neuman, B.W.; Angelini, M.M.; Buchmeier, M.J. Does form meet function in the coronavirus replicative organelle? Trends Microbiol 2014, 22, 642–647. [Google Scholar] [CrossRef]
- Schubert, K.; Karousis, E.D.; Jomaa, A.; Scaiola, A.; Echeverria, B.; Gurzeler, L.A.; Leibundgut, M.; Thiel, V.; Muhlemann, O.; Ban, N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 2020, 27, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Drosten, C.; Gunther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.; et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003, 348, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, H.; Geisbert, T.W. Ebola haemorrhagic fever. Lancet 2011, 377, 849–862. [Google Scholar] [CrossRef]
- Taylor, L.H.; Latham, S.M.; Woolhouse, M.E. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 2001, 356, 983–989. [Google Scholar] [CrossRef]
- Tellier, R.; Li, Y.; Cowling, B.J.; Tang, J.W. Recognition of aerosol transmission of infectious agents: a commentary. BMC Infect Dis 2019, 19, 101. [Google Scholar] [CrossRef]
- Morawska, L.; Cao, J. Airborne transmission of SARS-CoV-2: The world should face the reality. Environ Int 2020, 139, 105730. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D. ; World Health Organization-Coordinated Global Rotavirus Surveillance, N. Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000-2013. Clin Infect Dis, 2. [CrossRef]
- Cohen, M.S.; Chen, Y.Q.; McCauley, M.; Gamble, T.; Hosseinipour, M.C.; Kumarasamy, N.; Hakim, J.G.; Kumwenda, J.; Grinsztejn, B.; Pilotto, J.H.; et al. Antiretroviral Therapy for the Prevention of HIV-1 Transmission. N Engl J Med 2016, 375, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Schillie, S.; Vellozzi, C.; Reingold, A.; Harris, A.; Haber, P.; Ward, J.W.; Nelson, N.P. Prevention of Hepatitis B Virus Infection in the United States: Recommendations of the Advisory Committee on Immunization Practices. MMWR Recomm Rep 2018, 67, 1–31. [Google Scholar] [CrossRef]
- Jackson, A.C. Current and future approaches to the therapy of human rabies. Antiviral Res 2013, 99, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Harris, E. Dengue. Lancet 2015, 385, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Saiz, J.C.; Vazquez-Calvo, A.; Blazquez, A.B.; Merino-Ramos, T.; Escribano-Romero, E.; Martin-Acebes, M.A. Zika Virus: the Latest Newcomer. Front Microbiol 2016, 7, 496. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Gambaryan, A.S.; Tuzikov, A.B.; Piskarev, V.E.; Yamnikova, S.S.; Lvov, D.K.; Robertson, J.S.; Bovin, N.V.; Matrosovich, M.N. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6'-sialyl(N-acetyllactosamine). Virology 1997, 232, 345–350. [Google Scholar] [CrossRef]
- Matrosovich, M.; Tuzikov, A.; Bovin, N.; Gambaryan, A.; Klimov, A.; Castrucci, M.R.; Donatelli, I.; Kawaoka, Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 2000, 74, 8502–8512. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Kartenbeck, J.; Helenius, A. Membrane fusion activity of influenza virus. EMBO J 1982, 1, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Klenk, H.D.; Garten, W. Host cell proteases controlling virus pathogenicity. Trends Microbiol 1994, 2, 39–43. [Google Scholar] [CrossRef]
- Hogle, J.M. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu Rev Microbiol 2002, 56, 677–702. [Google Scholar] [CrossRef]
- Lakadamyali, M.; Rust, M.J.; Zhuang, X. Endocytosis of influenza viruses. Microbes and Infection 2004, 6, 929–936. [Google Scholar] [CrossRef]
- Hirose, T.; Fujita, K.; Kusumoto, S.; Oki, Y.; Murata, Y.; Sugiyama, T.; Ishida, H.; Shirai, T.; Nakashima, M.; Yamaoka, T.; et al. Association of pharmacokinetics and pharmacogenomics with safety and efficacy of gefitinib in patients with EGFR mutation positive advanced non-small cell lung cancer. Lung Cancer 2016, 93, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Hojjat-Farsangi, M. Small-molecule inhibitors of the receptor tyrosine kinases: promising tools for targeted cancer therapies. Int J Mol Sci 2014, 15, 13768–13801. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Qian, W.; Sun, X.; Jiang, S. Small-molecule inhibitors, immune checkpoint inhibitors, and more: FDA-approved novel therapeutic drugs for solid tumors from 1991 to 2021. J Hematol Oncol 2022, 15, 143. [Google Scholar] [CrossRef] [PubMed]
- Knight, Z.A. Small molecule inhibitors of the PI3-kinase family. Curr Top Microbiol Immunol 2010, 347, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Bekerman, E.; Neveu, G.; Shulla, A.; Brannan, J.; Pu, S.Y.; Wang, S.; Xiao, F.; Barouch-Bentov, R.; Bakken, R.R.; Mateo, R.; et al. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J Clin Invest 2017, 127, 1338–1352. [Google Scholar] [CrossRef] [PubMed]
- Eaaswarkhanth, M.; Al Madhoun, A.; Al-Mulla, F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int J Infect Dis 2020, 96, 459–460. [Google Scholar] [CrossRef] [PubMed]
- Keck, F.; Ataey, P.; Amaya, M.; Bailey, C.; Narayanan, A. Phosphorylation of Single Stranded RNA Virus Proteins and Potential for Novel Therapeutic Strategies. Viruses 2015, 7, 5257–5273. [Google Scholar] [CrossRef]
- Kumar, R.; Barua, S.; Tripathi, B.N.; Kumar, N. Role of ROCK signaling in virus replication. Virus Res 2023, 329, 199105. [Google Scholar] [CrossRef] [PubMed]
- Chander, Y.; Kumar, R.; Khandelwal, N.; Singh, N.; Shringi, B.N.; Barua, S.; Kumar, N. Role of p38 mitogen-activated protein kinase signalling in virus replication and potential for developing broad spectrum antiviral drugs. Rev Med Virol 2021, 31, 1–16. [Google Scholar] [CrossRef]
- Adamson, A.L.; Darr, D.; Holley-Guthrie, E.; Johnson, R.A.; Mauser, A.; Swenson, J.; Kenney, S. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J Virol 2000, 74, 1224–1233. [Google Scholar] [CrossRef]
- Sreekanth, G.P.; Yenchitsomanus, P.T.; Limjindaporn, T. Role of mitogen-activated protein kinase signaling in the pathogenesis of dengue virus infection. Cell Signal 2018, 48, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. Activation of the c-Jun NH(2)-terminal kinase pathway by coronavirus infectious bronchitis virus promotes apoptosis independently of c-Jun. Cell Death Dis 2017, 8, 3215. [Google Scholar] [CrossRef] [PubMed]
- Voss, K.; Amaya, M.; Mueller, C.; Roberts, B.; Kehn-Hall, K.; Bailey, C.; Petricoin, E., 3rd; Narayanan, A. Inhibition of host extracellular signal-regulated kinase (ERK) activation decreases new world alphavirus multiplication in infected cells. Virology 2014, 468-470, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Hou, X.; Peng, H.; Zhang, L.; Li, Y.; Gu, Z.; Jiang, Q.; Shi, M.; Ji, Y.; Jiang, J. MEK/ERK signaling pathway is required for enterovirus 71 replication in immature dendritic cells. Virol J 2014, 11, 227. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Sun, F.; Wang, L.; Gao, M.; Xie, Y.; Sun, Y.; Liu, H.; Yuan, Y.; Yi, W.; Huang, Z.; et al. Virus-induced p38 MAPK activation facilitates viral infection. Theranostics 2020, 10, 12223–12240. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Sharma, S.; Kumar, R.; Tripathi, B.N.; Barua, S.; Ly, H.; Rouse, B.T. Host-Directed Antiviral Therapy. Clin Microbiol Rev 2020, 33. [Google Scholar] [CrossRef] [PubMed]
- Keating, J.A.; Striker, R. Phosphorylation events during viral infections provide potential therapeutic targets. Rev Med Virol 2012, 22, 166–181. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ye, C.; Wan, C.; Li, G.; Peng, L.; Peng, Y.; Fang, R. The Roles of c-Jun N-Terminal Kinase (JNK) in Infectious Diseases. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Shimojima, M.; Ikeda, Y.; Kawaoka, Y. The mechanism of Axl-mediated Ebola virus infection. J Infect Dis 2007, 196 Suppl 2, S259–263. [Google Scholar] [CrossRef]
- Zheng, K.; Kitazato, K.; Wang, Y. Viruses exploit the function of epidermal growth factor receptor. Rev Med Virol 2014, 24, 274–286. [Google Scholar] [CrossRef]
- Wang, X.; Huong, S.M.; Chiu, M.L.; Raab-Traub, N.; Huang, E.S. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature 2003, 424, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.L.; Amornphimoltham, P.; Schmidt, M.; Wilson, P.A.; Gutkind, J.S.; Chiorini, J.A. Epidermal growth factor receptor is a co-receptor for adeno-associated virus serotype 6. Nat Med 2010, 16, 662–664. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Liang, Y.; Parslow, T.G.; Liang, Y. Receptor tyrosine kinase inhibitors block multiple steps of influenza a virus replication. J Virol 2011, 85, 2818–2827. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.; Saul, S.; Ghita, L.; Sahoo, M.K.; Ye, C.; Bhalla, N.; Lo, C.-W.; Jin, J.; Park, J.-G.; Martinez-Gualda, B.; et al. Numb-associated kinases are required for SARS-CoV-2 infection and are cellular targets for antiviral strategies. Antiviral Research 2022, 204, 105367. [Google Scholar] [CrossRef] [PubMed]
- Neveu, G.; Ziv-Av, A.; Barouch-Bentov, R.; Berkerman, E.; Mulholland, J.; Einav, S. AP-2-Associated Protein Kinase 1 and Cyclin G-Associated Kinase Regulate Hepatitis C Virus Entry and Are Potential Drug Targets. Journal of Virology 2015, 89, 4387–4404. [Google Scholar] [CrossRef] [PubMed]
- Urits, I.; Israel, J.; Hakobyan, H.; Yusin, G.; Lassiter, G.; Fackler, N.; Berger, A.A.; Kassem, H.; Kaye, A.; Viswanath, O. Baricitinib for the treatment of rheumatoid arthritis. Reumatologia 2020, 58, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Richardson, P.; Griffin, I.; Tucker, C.; Smith, D.; Oechsle, O.; Phelan, A.; Stebbing, J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 2020, 395, e30–e31. [Google Scholar] [CrossRef] [PubMed]
- Richardson, P.J.; Stebbing, J. Baricitinib as the treatment of choice for hospitalised individuals with COVID-19. EClinicalMedicine 2022, 49, 101493. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J Nat Prod 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Lu, A.T.; Fei, Z.; Haghani, A.; Robeck, T.R.; Zoller, J.A.; Li, C.Z.; Lowe, R.; Yan, Q.; Zhang, J.; Vu, H.; et al. Universal DNA methylation age across mammalian tissues. Nature Aging, 1038; -6. [Google Scholar] [CrossRef]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Musselman, C.A.; Lalonde, M.E.; Côté, J.; Kutateladze, T.G. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 2012, 19, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet 2009, 43, 143–166. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Van Emburgh, B.O.; Robertson, K.D. DNA methylation in development and human disease. Mutat Res 2008, 647, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Tao, Q.; Peng, J.; Soo, H.M.; Wu, W.; Ying, J.; Fields, C.R.; Delmas, A.L.; Liu, X.; Qiu, J.; et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet 2008, 17, 690–709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Wang, C.; Wang, X. TET (Ten-eleven translocation) family proteins: structure, biological functions and applications. Signal Transduction and Targeted Therapy 2023, 8, 297. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nature Reviews Genetics 2018, 19, 371–384. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Ohlsson, R.; Henikoff, S. The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006, 7, 21–33. [Google Scholar] [CrossRef]
- Kuo, M.H.; Allis, C.D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 1998, 20, 615–626. [Google Scholar] [CrossRef]
- Dokmanovic, M.; Clarke, C.; Marks, P.A. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res 2007, 5, 981–989. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Klose, R.J. The molecular principles of gene regulation by Polycomb repressive complexes. Nat Rev Mol Cell Biol 2021, 22, 815–833. [Google Scholar] [CrossRef] [PubMed]
- Healey, M.A.; Hu, R.; Beck, A.H.; Collins, L.C.; Schnitt, S.J.; Tamimi, R.M.; Hazra, A. Association of H3K9me3 and H3K27me3 repressive histone marks with breast cancer subtypes in the Nurses' Health Study. Breast Cancer Res Treat 2014, 147, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Mason, C.E. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet 2014, 15, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Kwa, F.A.A.; Jackson, D.E. Manipulating the epigenome for the treatment of disorders with thrombotic complications. Drug Discovery Today 2018, 23, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Tavares, V.; Afonso, A.; Garcia, J.; Cerqueira, F.; Medeiros, R. Plasmatic MicroRNAs and Treatment Outcomes of Patients with Metastatic Castration-Resistant Prostate Cancer: A Hospital-Based Cohort Study and In Silico Analysis. International Journal of Molecular Sciences 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, Q.; Wen, X.; Hong, X.; Yang, X.; Tang, X.; Zhang, P.; Lei, Y.; Sun, Y.; Zhang, J.; et al. EZH2-DNMT1-mediated epigenetic silencing of miR-142-3p promotes metastasis through targeting ZEB2 in nasopharyngeal carcinoma. Cell Death & Differentiation 2019, 26, 1089–1106. [Google Scholar] [CrossRef]
- Chhabra, R. miRNA and methylation: a multifaceted liaison. Chembiochem 2015, 16, 195–203. [Google Scholar] [CrossRef]
- Duursma, A.M.; Kedde, M.; Schrier, M.; le Sage, C.; Agami, R. miR-148 targets human DNMT3b protein coding region. Rna 2008, 14, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Zhou, Z.; Zhang, X. Effects of miR-195-5p on cell proliferation and apoptosis in gestational diabetes mellitus via targeting EZH2. Mol Med Rep 2020, 22, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat Biotechnol 2010, 28, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Obata, Y.; Furusawa, Y.; Hase, K. Epigenetic modifications of the immune system in health and disease. Immunol Cell Biol 2015, 93, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Smale, S.T.; Tarakhovsky, A.; Natoli, G. Chromatin contributions to the regulation of innate immunity. Annu Rev Immunol 2014, 32, 489–511. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.; Cullen, B.R. Epigenetic and epitranscriptomic regulation of viral replication. Nat Rev Microbiol 2020, 18, 559–570. [Google Scholar] [CrossRef]
- Tsai, K.; Cullen, B.R. Epigenetic and epitranscriptomic regulation of viral replication. Nature Reviews Microbiology 2020, 18, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Knipe, D.M. Nuclear sensing of viral DNA, epigenetic regulation of herpes simplex virus infection, and innate immunity. Virology 2015, 479-480, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Knipe, D.M.; Raja, P.; Lee, J. Viral gene products actively promote latent infection by epigenetic silencing mechanisms. Curr Opin Virol 2017, 23, 68–74. [Google Scholar] [CrossRef]
- Tsai, K.; Jaguva Vasudevan, A.A.; Martinez Campos, C.; Emery, A.; Swanstrom, R.; Cullen, B.R. Acetylation of Cytidine Residues Boosts HIV-1 Gene Expression by Increasing Viral RNA Stability. Cell Host Microbe 2020, 28, 306–312. [Google Scholar] [CrossRef]
- Kee, J.; Thudium, S.; Renner, D.M.; Glastad, K.; Palozola, K.; Zhang, Z.; Li, Y.; Lan, Y.; Cesare, J.; Poleshko, A.; et al. SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature 2022, 610, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.K.; Arora, P.; Pagano, J.S.; Jang, K.L. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res 2007, 67, 5771–5778. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Vogel, J.L.; Narayanan, A.; Peng, H.; Kristie, T.M. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med 2009, 15, 1312–1317. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- García-Sastre, A.; Biron, C.A. Type 1 interferons and the virus-host relationship: a lesson in détente. Science 2006, 312, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Eisfeld, A.J.; Schafer, A.; Josset, L.; Sims, A.C.; Proll, S.; Fan, S.; Li, C.; Neumann, G.; Tilton, S.C.; et al. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 2014, 5, e01174–01114. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol 2020, 92, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.; Morris, P.; Dickman, M.J.; Dockrell, D.H. The therapeutic potential of epigenetic manipulation during infectious diseases. Pharmacol Ther 2016, 167, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Vivekanandan, P.; Daniel, H.D.; Kannangai, R.; Martinez-Murillo, F.; Torbenson, M. Hepatitis B virus replication induces methylation of both host and viral DNA. J Virol 2010, 84, 4321–4329. [Google Scholar] [CrossRef]
- Zheng, D.L.; Zhang, L.; Cheng, N.; Xu, X.; Deng, Q.; Teng, X.M.; Wang, K.S.; Zhang, X.; Huang, J.; Han, Z.G. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol 2009, 50, 377–387. [Google Scholar] [CrossRef]
- Laurson, J.; Khan, S.; Chung, R.; Cross, K.; Raj, K. Epigenetic repression of E-cadherin by human papillomavirus 16 E7 protein. Carcinogenesis 2010, 31, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Burgers, W.A.; Blanchon, L.; Pradhan, S.; de Launoit, Y.; Kouzarides, T.; Fuks, F. Viral oncoproteins target the DNA methyltransferases. Oncogene 2007, 26, 1650–1655. [Google Scholar] [CrossRef] [PubMed]
- Holland, D.; Hoppe-Seyler, K.; Schuller, B.; Lohrey, C.; Maroldt, J.; Dürst, M.; Hoppe-Seyler, F. Activation of the enhancer of zeste homologue 2 gene by the human papillomavirus E7 oncoprotein. Cancer Res 2008, 68, 9964–9972. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Peters, G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 2006, 7, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Toyooka, S.; Pass, H.I.; Shivapurkar, N.; Fukuyama, Y.; Maruyama, R.; Toyooka, K.O.; Gilcrease, M.; Farinas, A.; Minna, J.D.; Gazdar, A.F. Aberrant methylation and simian virus 40 tag sequences in malignant mesothelioma. Cancer Res 2001, 61, 5727–5730. [Google Scholar] [PubMed]
- Slack, A.; Cervoni, N.; Pinard, M.; Szyf, M. DNA methyltransferase is a downstream effector of cellular transformation triggered by simian virus 40 large T antigen. J Biol Chem 1999, 274, 10105–10112. [Google Scholar] [CrossRef]
- Ferrari, R.; Pellegrini, M.; Horwitz, G.A.; Xie, W.; Berk, A.J.; Kurdistani, S.K. Epigenetic reprogramming by adenovirus e1a. Science 2008, 321, 1086–1088. [Google Scholar] [CrossRef]
- Li, J.; Hao, D.; Wang, L.; Wang, H.; Wang, Y.; Zhao, Z.; Li, P.; Deng, C.; Di, L.J. Epigenetic targeting drugs potentiate chemotherapeutic effects in solid tumor therapy. Sci Rep 2017, 7, 4035. [Google Scholar] [CrossRef] [PubMed]
- El Baba, R.; Herbein, G. Management of epigenomic networks entailed in coronavirus infections and COVID-19. Clin Epigenetics 2020, 12, 118. [Google Scholar] [CrossRef]
- Zheng, B.; Zhou, J.; Wang, H. Host microRNAs and exosomes that modulate influenza virus infection. Virus Res 2020, 279, 197885. [Google Scholar] [CrossRef]
- Moghoofei, M.; Najafipour, S.; Mostafaei, S.; Tavakoli, A.; Bokharaei-Salim, F.; Ghorbani, S.; Javanmard, D.; Ghaffari, H.; Monavari, S.H. MicroRNAs Profiling in HIV, HCV, and HIV/HCV Co-Infected Patients. Curr HIV Res 2021, 19, 27–34. [Google Scholar] [CrossRef]
- Kunden, R.D.; Khan, J.Q.; Ghezelbash, S.; Wilson, J.A. The Role of the Liver-Specific microRNA, miRNA-122 in the HCV Replication Cycle. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Panigrahi, M.; Thibault, P.A.; Wilson, J.A. MicroRNA 122 Affects both the Initiation and the Maintenance of Hepatitis C Virus Infections. J Virol 2022, 96, e0190321. [Google Scholar] [CrossRef]
- Yousefpouran, S.; Mostafaei, S.; Manesh, P.V.; Iranifar, E.; Bokharaei-Salim, F.; Nahand, J.S.; Mirzaei, H.; Taran, M.; Babaei, F.; Sayad, B.; et al. The assessment of selected MiRNAs profile in HIV, HBV, HCV, HIV/HCV, HIV/HBV Co-infection and elite controllers for determination of biomarker. Microb Pathog 2020, 147, 104355. [Google Scholar] [CrossRef]
- Haneklaus, M.; Gerlic, M.; O'Neill, L.A.; Masters, S.L. miR-223: infection, inflammation and cancer. J Intern Med 2013, 274, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Morales, L.; Oliveros, J.C.; Enjuanes, L.; Sola, I. Contribution of Host miRNA-223-3p to SARS-CoV-Induced Lung Inflammatory Pathology. mBio 2022, 13, e0313521. [Google Scholar] [CrossRef] [PubMed]
- Tycowski, K.T.; Guo, Y.E.; Lee, N.; Moss, W.N.; Vallery, T.K.; Xie, M.; Steitz, J.A. Viral noncoding RNAs: more surprises. Genes Dev 2015, 29, 567–584. [Google Scholar] [CrossRef]
- Cazalla, D.; Yario, T.; Steitz, J.A. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 2010, 328, 1563–1566. [Google Scholar] [CrossRef]
- Guo, Y.E.; Riley, K.J.; Iwasaki, A.; Steitz, J.A. Alternative capture of noncoding RNAs or protein-coding genes by herpesviruses to alter host T cell function. Mol Cell 2014, 54, 67–79. [Google Scholar] [CrossRef]
- Abedini, M.; Zhang, C. Performance assessment of concrete and steel material models in ls-dyna for enhanced numerical simulation, a state of the art review. Archives of Computational Methods in Engineering 2021, 28, 2921–2942. [Google Scholar] [CrossRef]
- Kitazato, K.; Wang, Y.; Kobayashi, N. Viral infectious disease and natural products with antiviral activity. Drug Discov Ther 2007, 1, 14–22. [Google Scholar] [PubMed]
- Abookleesh, F.L.; Al-Anzi, B.S.; Ullah, A. Potential Antiviral Action of Alkaloids. Molecules 2022, 27, 903. [Google Scholar] [CrossRef] [PubMed]
- Denaro, M.; Smeriglio, A.; Barreca, D.; De Francesco, C.; Occhiuto, C.; Milano, G.; Trombetta, D. Antiviral activity of plants and their isolated bioactive compounds: An update. Phytother Res 2020, 34, 742–768. [Google Scholar] [CrossRef] [PubMed]
- Behl, T.; Rocchetti, G.; Chadha, S.; Zengin, G.; Bungau, S.; Kumar, A.; Mehta, V.; Uddin, M.S.; Khullar, G.; Setia, D.; et al. Phytochemicals from Plant Foods as Potential Source of Antiviral Agents: An Overview. Pharmaceuticals (Basel) 2021, 14. [Google Scholar] [CrossRef] [PubMed]
- Jassim, S.A.; Naji, M.A. Novel antiviral agents: a medicinal plant perspective. J Appl Microbiol 2003, 95, 412–427. [Google Scholar] [CrossRef] [PubMed]
- Astani, A.; Reichling, J.; Schnitzler, P. Screening for antiviral activities of isolated compounds from essential oils. Evid Based Complement Alternat Med 2011, 2011, 253643. [Google Scholar] [CrossRef] [PubMed]
- Gavanji, S.; Sayedipour, S.S.; Larki, B.; Bakhtari, A. Antiviral activity of some plant oils against herpes simplex virus type 1 in Vero cell culture. Journal of Acute Medicine 2015, 5, 62–68. [Google Scholar] [CrossRef]
- Feriotto, G.; Marchetti, N.; Costa, V.; Beninati, S.; Tagliati, F.; Mischiati, C. Chemical Composition of Essential Oils from Thymus vulgaris, Cymbopogon citratus, and Rosmarinus officinalis, and Their Effects on the HIV-1 Tat Protein Function. Chem Biodivers 2018, 15. [Google Scholar] [CrossRef]
- Mori, K.; Obossou, E.K.; Suwa, S.; Miura, S.; Oh, S.-H.; Jinbo, N.; Ishibashi, Y.; Shikamoto, Y.; Hosono, T.; Toda, T. Human Immunodeficiency Virus Type 1 (HIV-1) Reverse Transcriptase Inhibitory Effect of Cymbopogon Nardus Essential Oil.
- Ma, L.; Yao, L. Antiviral Effects of Plant-Derived Essential Oils and Their Components: An Updated Review. Molecules 2020, 25. [Google Scholar] [CrossRef]
- Naithani, R.; Huma, L.C.; Holland, L.E.; Shukla, D.; McCormick, D.L.; Mehta, R.G.; Moriarty, R.M. Antiviral activity of phytochemicals: a comprehensive review. Mini Rev Med Chem 2008, 8, 1106–1133. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Lim, C.H.; Song, J.H.; Baek, S.H.; Kwon, D.H. Antiviral activity of raoulic acid from Raoulia australis against Picornaviruses. Phytomedicine 2009, 16, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.T.; Chen, T.Y.; Lin, S.C.; Chung, C.Y.; Lin, T.C.; Wang, G.H.; Anderson, R.; Lin, C.C.; Richardson, C.D. Broad-spectrum antiviral activity of chebulagic acid and punicalagin against viruses that use glycosaminoglycans for entry. BMC Microbiol 2013, 13, 187. [Google Scholar] [CrossRef] [PubMed]
- Cotin, S.; Calliste, C.A.; Mazeron, M.C.; Hantz, S.; Duroux, J.L.; Rawlinson, W.D.; Ploy, M.C.; Alain, S. Eight flavonoids and their potential as inhibitors of human cytomegalovirus replication. Antiviral Res 2012, 96, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Lyu, S.Y.; Rhim, J.Y.; Park, W.B. Antiherpetic activities of flavonoids against herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2) in vitro. Arch Pharm Res 2005, 28, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.E.; Min, J.S.; Jang, M.S.; Lee, J.Y.; Shin, Y.S.; Song, J.H.; Kim, H.R.; Kim, S.; Jin, Y.H.; Kwon, S. Natural Bis-Benzylisoquinoline Alkaloids-Tetrandrine, Fangchinoline, and Cepharanthine, Inhibit Human Coronavirus OC43 Infection of MRC-5 Human Lung Cells. Biomolecules 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Luganini, A.; Mercorelli, B.; Messa, L.; Palù, G.; Gribaudo, G.; Loregian, A. The isoquinoline alkaloid berberine inhibits human cytomegalovirus replication by interfering with the viral Immediate Early-2 (IE2) protein transactivating activity. Antiviral Res 2019, 164, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Qiu, M.; Chu, Y.; Chen, D.; Wang, X.; Su, A.; Wu, Z. Downregulation of cellular c-Jun N-terminal protein kinase and NF-κB activation by berberine may result in inhibition of herpes simplex virus replication. Antimicrob Agents Chemother 2014, 58, 5068–5078. [Google Scholar] [CrossRef] [PubMed]
- Gómez, L.A.; Stashenko, E.; Ocazionez, R.E. Comparative study on in vitro activities of citral, limonene and essential oils from Lippia citriodora and L. alba on yellow fever virus. Nat Prod Commun 2013, 8, 249–252. [Google Scholar] [CrossRef]
- Haddad, J.G.; Picard, M.; Bénard, S.; Desvignes, C.; Desprès, P.; Diotel, N.; El Kalamouni, C. Ayapana triplinervis Essential Oil and Its Main Component Thymohydroquinone Dimethyl Ether Inhibit Zika Virus at Doses Devoid of Toxicity in Zebrafish. Molecules 2019, 24, 3447. [Google Scholar] [CrossRef]
- Vimalanathan, S.; Hudson, J.B. Anti-influenza virus activity of essential oils and vapors. American Journal of Essential Oils and Natural Products 2014, 2, 47–53. [Google Scholar]
- Mokni, R.E.; Youssef, F.S.; Jmii, H.; Khmiri, A.; Bouazzi, S.; Jlassi, I.; Jaidane, H.; Dhaouadi, H.; Ashour, M.L.; Hammami, S. The Essential Oil of Tunisian Dysphania ambrosioides and its Antimicrobial and Antiviral Properties. Journal of Essential Oil Bearing Plants 2019, 22, 282–294. [Google Scholar] [CrossRef]
- Elaissi, A.; Rouis, Z.; Salem, N.A.; Mabrouk, S.; ben Salem, Y.; Salah, K.B.; Aouni, M.; Farhat, F.; Chemli, R.; Harzallah-Skhiri, F.; et al. Chemical composition of 8 eucalyptus species' essential oils and the evaluation of their antibacterial, antifungal and antiviral activities. BMC Complement Altern Med 2012, 12, 81. [Google Scholar] [CrossRef] [PubMed]
- Nabaweya, A. Ibrahim, S.S.E.-H., Magdy M. D. Mohammed, Mohamed A. Farid, Nayera A. M. AbdelWahed, Mohamed A. Ali, Eman A. W. El-Abd. Chemical Composition, Antiviral against avian Influenza (H5N1) Virus and Antimicrobial activities of the Essential Oils of the Leaves and Fruits of Fortunella margarita, Lour. Swingle, Growing in Egypt, 5: 1: 2015; Vol. Volume, 2015; 5. [Google Scholar]
- Sharifi-Rad, J.; Salehi, B.; Schnitzler, P.; Ayatollahi, S.A.; Kobarfard, F.; Fathi, M.; Eisazadeh, M.; Sharifi-Rad, M. Susceptibility of herpes simplex virus type 1 to monoterpenes thymol, carvacrol, p-cymene and essential oils of Sinapis arvensis L., Lallemantia royleana Benth. and Pulicaria vulgaris Gaertn. Cell Mol Biol (Noisy-le-grand) 2017, 63, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Pilau, M.R.; Alves, S.H.; Weiblen, R.; Arenhart, S.; Cueto, A.P.; Lovato, L.T. Antiviral activity of the Lippia graveolens (Mexican oregano) essential oil and its main compound carvacrol against human and animal viruses. Braz J Microbiol 2011, 42, 1616–1624. [Google Scholar] [CrossRef] [PubMed]
- Garozzo, A.; Timpanaro, R.; Stivala, A.; Bisignano, G.; Castro, A. Activity of Melaleuca alternifolia (tea tree) oil on Influenza virus A/PR/8: study on the mechanism of action. Antiviral Res 2011, 89, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Civitelli, L.; Panella, S.; Marcocci, M.E.; De Petris, A.; Garzoli, S.; Pepi, F.; Vavala, E.; Ragno, R.; Nencioni, L.; Palamara, A.T.; et al. In vitro inhibition of herpes simplex virus type 1 replication by Mentha suaveolens essential oil and its main component piperitenone oxide. Phytomedicine 2014, 21, 857–865. [Google Scholar] [CrossRef] [PubMed]
- da Silva, J.K.R.; Figueiredo, P.L.B.; Byler, K.G.; Setzer, W.N. Essential Oils as Antiviral Agents, Potential of Essential Oils to Treat SARS-CoV-2 Infection: An In-Silico Investigation. International Journal of Molecular Sciences 2020, 21, 3426. [Google Scholar] [CrossRef] [PubMed]
- Cagno, V.; Sgorbini, B.; Sanna, C.; Cagliero, C.; Ballero, M.; Civra, A.; Donalisio, M.; Bicchi, C.; Lembo, D.; Rubiolo, P. In vitro anti-herpes simplex virus-2 activity of Salvia desoleana Atzei & V. Picci essential oil. PLoS One 2017, 12, e0172322. [Google Scholar] [CrossRef]
- Toujani, M.M.; Rittà, M.; Civra, A.; Genovese, S.; Epifano, F.; Ghram, A.; Lembo, D.; Donalisio, M. Inhibition of HSV-2 infection by pure compounds from Thymus capitatus extract in vitro. Phytother Res 2018, 32, 1555–1563. [Google Scholar] [CrossRef]
- Shin, W.J.; Lee, K.H.; Park, M.H.; Seong, B.L. Broad-spectrum antiviral effect of Agrimonia pilosa extract on influenza viruses. Microbiol Immunol 2010, 54, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Rezazadeh, F.; Moshaverinia, M.; Motamedifar, M.; Alyaseri, M. Assessment of Anti HSV-1 Activity of Aloe Vera Gel Extract: an In Vitro Study. J Dent (Shiraz) 2016, 17, 49–54. [Google Scholar] [PubMed]
- Makau, J.N.; Watanabe, K.; Mohammed, M.M.D.; Nishida, N. Antiviral Activity of Peanut (Arachis hypogaea L.) Skin Extract Against Human Influenza Viruses. J Med Food 2018, 21, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Namazi, R.; Zabihollahi, R.; Behbahani, M.; Rezaei, A. Inhibitory Activity of Avicennia marina, a Medicinal Plant in Persian Folk Medicine, against HIV and HSV. Iran J Pharm Res 2013, 12, 435–443. [Google Scholar] [PubMed]
- Prinsloo, G.; Marokane, C.K.; Street, R.A. Anti-HIV activity of southern African plants: Current developments, phytochemistry and future research. Journal of ethnopharmacology 2018, 210, 133–155. [Google Scholar] [CrossRef] [PubMed]
- Mushi, N.F.; Mbwambo, Z.H.; Innocent, E.; Tewtrakul, S. Antibacterial, anti-HIV-1 protease and cytotoxic activities of aqueous ethanolic extracts from Combretum adenogonium Steud. Ex A. Rich (Combretaceae). BMC Complementary and Alternative Medicine 2012, 12, 163. [Google Scholar] [CrossRef] [PubMed]
- Churqui, M.P.; Lind, L.; Thörn, K.; Svensson, A.; Savolainen, O.; Aranda, K.T.; Eriksson, K. Extracts of Equisetum giganteum L and Copaifera reticulate Ducke show strong antiviral activity against the sexually transmitted pathogen herpes simplex virus type 2. Journal of ethnopharmacology 2018, 210, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, S.; Côté, I.; Pichette, A.; Gauthier, C.; Ouellet, M.; Nagau-Lavoie, F.; Mshvildadze, V.; Legault, J. Chemical composition and anti-herpes simplex virus type 1 (HSV-1) activity of extracts from Cornus canadensis. BMC Complement Altern Med 2017, 17, 123. [Google Scholar] [CrossRef]
- Hossan, M.S.; Fatima, A.; Rahmatullah, M.; Khoo, T.J.; Nissapatorn, V.; Galochkina, A.V.; Slita, A.V.; Shtro, A.A.; Nikolaeva, Y.; Zarubaev, V.V.; et al. Antiviral activity of Embelia ribes Burm. f. against influenza virus in vitro. Arch Virol 2018, 163, 2121–2131. [Google Scholar] [CrossRef]
- Cho, W.K.; Weeratunga, P.; Lee, B.H.; Park, J.S.; Kim, C.J.; Ma, J.Y.; Lee, J.S. Epimedium koreanum Nakai displays broad spectrum of antiviral activity in vitro and in vivo by inducing cellular antiviral state. Viruses 2015, 7, 352–377. [Google Scholar] [CrossRef]
- Derksen, A.; Kühn, J.; Hafezi, W.; Sendker, J.; Ehrhardt, C.; Ludwig, S.; Hensel, A. Antiviral activity of hydroalcoholic extract from Eupatorium perfoliatum L. against the attachment of influenza A virus. Journal of ethnopharmacology 2016, 188, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Gyuris, A.; Szlávik, L.; Minárovits, J.; Vasas, A.; Molnár, J.; Hohmann, J. Antiviral activities of extracts of Euphorbia hirta L. against HIV-1, HIV-2 and SIVmac251. In Vivo 2009, 23, 429–432. [Google Scholar] [PubMed]
- Ghosh, M.; Civra, A.; Rittà, M.; Cagno, V.; Mavuduru, S.G.; Awasthi, P.; Lembo, D.; Donalisio, M. Ficus religiosa L. bark extracts inhibit infection by herpes simplex virus type 2 in vitro. Arch Virol 2016, 161, 3509–3514. [Google Scholar] [CrossRef] [PubMed]
- Bonvicini, F.; Lianza, M.; Mandrone, M.; Poli, F.; Gentilomi, G.A.; Antognoni, F. Hemidesmus indicus (L.) R. Br. extract inhibits the early step of herpes simplex type 1 and type 2 replication. New Microbiol 2018, 41, 187–194. [Google Scholar] [PubMed]
- Shoji, M.; Woo, S.Y.; Masuda, A.; Win, N.N.; Ngwe, H.; Takahashi, E.; Kido, H.; Morita, H.; Ito, T.; Kuzuhara, T. Anti-influenza virus activity of extracts from the stems of Jatropha multifida Linn. collected in Myanmar. BMC Complement Altern Med 2017, 17, 96. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.Y.; Chang, H.W.; Lin, C.F.; Liu, C.J.; Hsieh, C.F.; Horng, J.T. Characterization of the anti-influenza activity of the Chinese herbal plant Paeonia lactiflora. Viruses 2014, 6, 1861–1875. [Google Scholar] [CrossRef]
- Ojha, D.; Das, R.; Sobia, P.; Dwivedi, V.; Ghosh, S.; Samanta, A.; Chattopadhyay, D. Pedilanthus tithymaloides Inhibits HSV Infection by Modulating NF-κB Signaling. PLoS One 2015, 10, e0139338. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.; Price, J.; Brindley, M.A.; Widrlechner, M.P.; Qu, L.; McCoy, J.A.; Murphy, P.; Hauck, C.; Maury, W. Inhibition of HIV-1 infection by aqueous extracts of Prunella vulgaris L. Virol J 2011, 8, 188. [Google Scholar] [CrossRef] [PubMed]
- Bisignano, C.; Mandalari, G.; Smeriglio, A.; Trombetta, D.; Pizzo, M.M.; Pennisi, R.; Sciortino, M.T. Almond Skin Extracts Abrogate HSV-1 Replication by Blocking Virus Binding to the Cell. Viruses 2017, 9. [Google Scholar] [CrossRef]
- Karimi, A.; Rafieian-Kopaei, M.; Moradi, M.T.; Alidadi, S. Anti-Herpes Simplex Virus Type-1 Activity and Phenolic Content of Crude Ethanol Extract and Four Corresponding Fractions of Quercus brantii L Acorn. J Evid Based Complementary Altern Med 2017, 22, 455–461. [Google Scholar] [CrossRef]
- Reichling, J.; Neuner, A.; Sharaf, M.; Harkenthal, M.; Schnitzler, P. Antiviral activity of Rhus aromatica (fragrant sumac) extract against two types of herpes simplex viruses in cell culture. Pharmazie 2009, 64, 538–541. [Google Scholar] [PubMed]
- Nocchi, S.R.; Companhoni, M.V.; de Mello, J.C.; Dias Filho, B.P.; Nakamura, C.V.; Carollo, C.A.; Silva, D.B.; Ueda-Nakamura, T. Antiviral Activity of Crude Hydroethanolic Extract from Schinus terebinthifolia against Herpes simplex Virus Type 1. Planta Med 2017, 83, 509–518. [Google Scholar] [CrossRef]
- Di Sotto, A.; Di Giacomo, S.; Amatore, D.; Locatelli, M.; Vitalone, A.; Toniolo, C.; Rotino, G.L.; Lo Scalzo, R.; Palamara, A.T.; Marcocci, M.E.; et al. A Polyphenol Rich Extract from Solanum melongena L. DR2 Peel Exhibits Antioxidant Properties and Anti-Herpes Simplex Virus Type 1 Activity In Vitro. Molecules 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- Boff, L.; Silva, I.T.; Argenta, D.F.; Farias, L.M.; Alvarenga, L.F.; Pádua, R.M.; Braga, F.C.; Leite, J.P.; Kratz, J.M.; Simões, C.M. Strychnos pseudoquina A. St. Hil.: a Brazilian medicinal plant with promising in vitro antiherpes activity. J Appl Microbiol 2016, 121, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Benassi-Zanqueta, É.; Marques, C.F.; Valone, L.M.; Pellegrini, B.L.; Bauermeister, A.; Ferreira, I.C.P.; Lopes, N.P.; Nakamura, C.V.; Dias Filho, B.P.; Natali, M.R.M.; et al. Evaluation of anti-HSV-1 activity and toxicity of hydroethanolic extract of Tanacetum parthenium (L.) Sch.Bip. (Asteraceae). Phytomedicine 2019, 55, 249–254. [Google Scholar] [CrossRef]
- Hsieh, C.F.; Chen, Y.L.; Lin, C.F.; Ho, J.Y.; Huang, C.H.; Chiu, C.H.; Hsieh, P.W.; Horng, J.T. An extract from Taxodium distichum targets hemagglutinin- and neuraminidase-related activities of influenza virus in vitro. Sci Rep 2016, 6, 36015. [Google Scholar] [CrossRef]
- Donalisio, M.; Cagno, V.; Civra, A.; Gibellini, D.; Musumeci, G.; Rittà, M.; Ghosh, M.; Lembo, D. The traditional use of Vachellia nilotica for sexually transmitted diseases is substantiated by the antiviral activity of its bark extract against sexually transmitted viruses. Journal of ethnopharmacology 2018, 213, 403–408. [Google Scholar] [CrossRef]
- Hafidh, R.R.; Abdulamir, A.S.; Abu Bakar, F.; Sekawi, Z.; Jahansheri, F.; Jalilian, F.A. Novel antiviral activity of mung bean sprouts against respiratory syncytial virus and herpes simplex virus -1: an in vitro study on virally infected Vero and MRC-5 cell lines. BMC Complement Altern Med 2015, 15, 179. [Google Scholar] [CrossRef]
- Khole, S.; Mittal, S.; Jagadish, N.; Ghosh, D.; Gadgil, V.; Sinkar, V.; Ghaskadbi, S. Andrographolide enhances redox status of liver cells by regulating microRNA expression. Free Radical Biology and Medicine 2019, 130, 397–407. [Google Scholar] [CrossRef]
- Li, F.; Khanom, W.; Sun, X.; Paemanee, A.; Roytrakul, S.; Wang, D.; Smith, D.R.; Zhou, G.C. Andrographolide and Its 14-Aryloxy Analogues Inhibit Zika and Dengue Virus Infection. Molecules 2020, 25. [Google Scholar] [CrossRef]
- Latif, R.; Wang, C.Y. Andrographolide as a potent and promising antiviral agent. Chin J Nat Med 2020, 18, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Mishra, K.P.; Ganju, L. Broad-spectrum antiviral properties of andrographolide. Arch Virol 2017, 162, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Malat, P.; Ekalaksananan, T.; Heawchaiyaphum, C.; Suebsasana, S.; Roytrakul, S.; Yingchutrakul, Y.; Pientong, C. Andrographolide Inhibits Epstein-Barr Virus Lytic Reactivation in EBV-Positive Cancer Cell Lines through the Modulation of Epigenetic-Related Proteins. Molecules 2022, 27. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Chen, L.; Wu, W.; Yang, J.; Yang, Z.; Liu, S. Andrographolide inhibits influenza A virus-induced inflammation in a murine model through NF-κB and JAK-STAT signaling pathway. Microbes Infect 2017, 19, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.; Kaur, P.; Shukla, S.; Abbas, A.; Fu, P.; Gupta, S. Plant flavone apigenin inhibits HDAC and remodels chromatin to induce growth arrest and apoptosis in human prostate cancer cells: in vitro and in vivo study. Mol Carcinog 2012, 51, 952–962. [Google Scholar] [CrossRef] [PubMed]
- Zakaryan, H.; Arabyan, E.; Oo, A.; Zandi, K. Flavonoids: promising natural compounds against viral infections. Arch Virol 2017, 162, 2539–2551. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Khan, M.A.; Mashwani, Z.U.; Ullah, N.; Nadhman, A. Therapeutic potential of medicinal plants against COVID-19: The role of antiviral medicinal metabolites. Biocatal Agric Biotechnol 2021, 31, 101890. [Google Scholar] [CrossRef]
- Sharma, R.; Bhattu, M.; Tripathi, A.; Verma, M.; Acevedo, R.; Kumar, P.; Rajput, V.D.; Singh, J. Potential medicinal plants to combat viral infections: A way forward to environmental biotechnology. Environmental Research 2023, 227, 115725. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Miao, J.; Shao, Q.; Gao, Y.; Hong, L. Apigenin suppresses influenza A virus-induced RIG-I activation and viral replication. J Med Virol, 1002. [Google Scholar] [CrossRef]
- Chiang, L.C.; Ng, L.T.; Cheng, P.W.; Chiang, W.; Lin, C.C. Antiviral activities of extracts and selected pure constituents of Ocimum basilicum. Clin Exp Pharmacol Physiol 2005, 32, 811–816. [Google Scholar] [CrossRef]
- Ożarowski, M.; Karpiński, T.M. The Effects of Propolis on Viral Respiratory Diseases. Molecules 2023, 28. [Google Scholar] [CrossRef]
- Ohno, M.; Shibata, C.; Kishikawa, T.; Yoshikawa, T.; Takata, A.; Kojima, K.; Akanuma, M.; Kang, Y.J.; Yoshida, H.; Otsuka, M.; et al. The flavonoid apigenin improves glucose tolerance through inhibition of microRNA maturation in miRNA103 transgenic mice. Sci Rep 2013, 3, 2553. [Google Scholar] [CrossRef] [PubMed]
- Meerson, A.; Khatib, S.; Mahajna, J. Natural Products Targeting Cancer Stem Cells for Augmenting Cancer Therapeutics. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhou, W.; Zhu, H.; Zhou, P.; Shi, X. Baicalin benefits the anti-HBV therapy via inhibiting HBV viral RNAs. Toxicol Appl Pharmacol 2017, 323, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Li, B.Q.; Fu, T.; Yan, Y.D.; Baylor, N.W.; Ruscetti, F.W.; Kung, H.F. Inhibition of HIV infection by baicalin--a flavonoid compound purified from Chinese herbal medicine. Cell Mol Biol Res 1993, 39, 119–124. [Google Scholar] [PubMed]
- Zandi, K.; Teoh, B.-T.; Sam, S.-S.; Wong, P.-F.; Mustafa, M.R.; AbuBakar, S. Novel antiviral activity of baicalein against dengue virus. BMC Complementary and Alternative Medicine 2012, 12, 214. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Kuang, X.-P.; Zhou, Q.-Q.; Yan, C.-Y.; Li, W.; Gong, H.-B.; Kurihara, H.; Li, W.-X.; Li, Y.-F.; He, R.-R. Inhibitory effects of baicalein against herpes simplex virus type 1. Acta Pharmaceutica Sinica B 2020, 10, 2323–2338. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, H.; Hu, P.; Qing, Y.; Wang, X.; Zhu, M.; Wang, H.; Wang, Z.; Xu, J.; Guo, Q.; et al. Natural HDAC-1/8 inhibitor baicalein exerts therapeutic effect in CBF-AML. Clin Transl Med 2020, 10, e154. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Yao, Q.; An, Y.; Fan, L.; Wang, J.; Li, H. Baicalin suppresses the progression of Type 2 diabetes-induced liver tumor through regulating METTL3/m(6)A/HKDC1 axis and downstream p-JAK2/STAT1/clevaged Capase3 pathway. Phytomedicine 2022, 94, 153823. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Li, J.; Bie, B.; Shi, M.; Zhu, M.; Tian, J.; Zhu, K.; Sun, J.; Mu, Y.; Li, Z.; et al. miR-3,178 contributes to the therapeutic action of baicalein against hepatocellular carcinoma cells via modulating HDAC10. Phytother Res 2023, 37, 295–309. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, S.; Ma, Q.; Xiao, D.; Chen, L. Berberine enhances the AMPK activation and autophagy and mitigates high glucose-induced apoptosis of mouse podocytes. Eur J Pharmacol 2017, 794, 106–114. [Google Scholar] [CrossRef]
- Lou, G.; Liu, Y.; Wu, S.; Xue, J.; Yang, F.; Fu, H.; Zheng, M.; Chen, Z. The p53/miR-34a/SIRT1 Positive Feedback Loop in Quercetin-Induced Apoptosis. Cell Physiol Biochem 2015, 35, 2192–2202. [Google Scholar] [CrossRef] [PubMed]
- Lü, Y.; Han, B.; Yu, H.; Cui, Z.; Li, Z.; Wang, J. Berberine regulates the microRNA-21-ITGΒ4-PDCD4 axis and inhibits colon cancer viability. Oncol Lett 2018, 15, 5971–5976. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, L.; Hu, D.; Yang, Y. Berberine Inhibits Herpes Simplex Virus 1 Replication in HEK293T Cells. Comput Math Methods Med 2022, 2022, 7137401. [Google Scholar] [CrossRef]
- Hayashi, K.; Minoda, K.; Nagaoka, Y.; Hayashi, T.; Uesato, S. Antiviral activity of berberine and related compounds against human cytomegalovirus. Bioorg Med Chem Lett 2007, 17, 1562–1564. [Google Scholar] [CrossRef] [PubMed]
- Mahata, S.; Bharti, A.C.; Shukla, S.; Tyagi, A.; Husain, S.A.; Das, B.C. Berberine modulates AP-1 activity to suppress HPV transcription and downstream signaling to induce growth arrest and apoptosis in cervical cancer cells. Mol Cancer 2011, 10, 39. [Google Scholar] [CrossRef]
- Ratanakomol, T.; Roytrakul, S.; Wikan, N.; Smith, D.R. Berberine Inhibits Dengue Virus through Dual Mechanisms. Molecules 2021, 26. [Google Scholar] [CrossRef]
- Zha, W.; Liang, G.; Xiao, J.; Studer, E.J.; Hylemon, P.B.; Pandak, W.M., Jr.; Wang, G.; Li, X.; Zhou, H. Berberine inhibits HIV protease inhibitor-induced inflammatory response by modulating ER stress signaling pathways in murine macrophages. PLoS One 2010, 5, e9069. [Google Scholar] [CrossRef]
- Hung, T.C.; Jassey, A.; Liu, C.H.; Lin, C.J.; Lin, C.C.; Wong, S.H.; Wang, J.Y.; Yen, M.H.; Lin, L.T. Berberine inhibits hepatitis C virus entry by targeting the viral E2 glycoprotein. Phytomedicine 2019, 53, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, J.Q.; Kim, Y.J.; Wu, J.; Wang, Q.; Hao, Y. In vivo and in vitro antiviral effects of berberine on influenza virus. Chin J Integr Med 2011, 17, 444–452. [Google Scholar] [CrossRef]
- Pizzorno, A.; Padey, B.; Dubois, J.; Julien, T.; Traversier, A.; Dulière, V.; Brun, P.; Lina, B.; Rosa-Calatrava, M.; Terrier, O. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. Antiviral Res 2020, 181, 104878. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, H.; Kim, S.W.; Lee, N.R.; Yi, C.M.; Heo, J.; Kim, B.J.; Kim, N.J.; Inn, K.S. An Effective Antiviral Approach Targeting Hepatitis B Virus with NJK14047, a Novel and Selective Biphenyl Amide p38 Mitogen-Activated Protein Kinase Inhibitor. Antimicrob Agents Chemother 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Adewole, K.E.; Ishola, A.A. A Computational Approach to Investigate the HDAC6 and HDAC10 Binding Propensity of Psidium guajava-derived Compounds as Potential Anticancer Agents. Curr Drug Discov Technol 2021, 18, 423–436. [Google Scholar] [CrossRef]
- Peyrat, L.A.; Eparvier, V.; Eydoux, C.; Guillemot, J.C.; Litaudon, M.; Stien, D. Betulinic Acid, The First Lupane-Type Triterpenoid Isolated from Both a Phomopsis sp. and Its Host Plant Diospyros carbonaria Benoist. Chem Biodivers 2017, 14. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Li, H.; Gou, Y.; Zhang, H.; Vlessidis, A.G.; Zhou, H.; Evmiridis, N.P.; Liu, Z. Betulinic acid-mediated inhibitory effect on hepatitis B virus by suppression of manganese superoxide dismutase expression. Febs j 2009, 276, 2599–2614. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Gupta, K.P. Modulation of miR-203 and its regulators as a function of time during the development of 7, 12 dimethylbenz [a] anthracene induced mouse skin tumors in presence or absence of the antitumor agents. Toxicol Appl Pharmacol 2014, 278, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Pant, K.; Mishra, A.K.; Pradhan, S.M.; Nayak, B.; Das, P.; Shalimar, D.; Saraya, A.; Venugopal, S.K. Butyrate inhibits HBV replication and HBV-induced hepatoma cell proliferation via modulating SIRT-1/Ac-p53 regulatory axis. Mol Carcinog 2019, 58, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.H.; Min, J.S.; Kwon, S. Cardamonin as a p38 MAPK Signaling Pathway Activator Inhibits Human Coronavirus OC43 Infection in Human Lung Cells. Nutrients 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Ryu, E.; Son, M.; Lee, M.; Lee, K.; Cho, J.Y.; Cho, S.; Lee, S.K.; Lee, Y.M.; Cho, H.; Sung, G.H.; et al. Cordycepin is a novel chemical suppressor of Epstein-Barr virus replication. Oncoscience 2014, 1, 866–881. [Google Scholar] [CrossRef] [PubMed]
- Rabie, A.M. Potent Inhibitory Activities of the Adenosine Analogue Cordycepin on SARS-CoV-2 Replication. ACS Omega 2022, 7, 2960–2969. [Google Scholar] [CrossRef]
- Rose, K.M.; Bell, L.E.; Jacob, S.T. Specific inhibition of chromatin-associated poly(A) synthesis in vitro by cordycepin 5'-triphosphate. Nature 1977, 267, 178–180. [Google Scholar] [CrossRef]
- Mahy, B.W.; Cox, N.J.; Armstrong, S.J.; Barry, R.D. Multiplication of influenza virus in the presence of cordycepin, an inhibitor of cellular RNA synthesis. Nat New Biol 1973, 243, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.E.; Weiler, B.E.; Charubala, R.; Pfleiderer, W.; Leserman, L.; Sobol, R.W.; Suhadolnik, R.J.; Schröder, H.C. Cordycepin analogues of 2',5'-oligoadenylate inhibit human immunodeficiency virus infection via inhibition of reverse transcriptase. Biochemistry 1991, 30, 2027–2033. [Google Scholar] [CrossRef] [PubMed]
- Lonai, P.; Declève, A.; Kaplan, H.S. Spontaneous induction of endogenous murine leukemia virus-related antigen expression during short-term in vitro incubation of mouse lymphocytes. Proc Natl Acad Sci U S A 1974, 71, 2008–2012. [Google Scholar] [CrossRef] [PubMed]
- Doetsch, P.W.; Suhadolnik, R.J.; Sawada, Y.; Mosca, J.D.; Flick, M.B.; Reichenbach, N.L.; Dang, A.Q.; Wu, J.M.; Charubala, R.; Pfleiderer, W.; et al. Core (2'-5')oligoadenylate and the cordycepin analog: inhibitors of Epstein--Barr virus-induced transformation of human lymphocytes in the absence of interferon. Proc Natl Acad Sci U S A 1981, 78, 6699–6703. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, N.; Yang, L.; Song, X.Q. Bioactive natural products in COVID-19 therapy. Front Pharmacol 2022, 13, 926507. [Google Scholar] [CrossRef] [PubMed]
- Hudlikar, R.R.; Sargsyan, D.; Wu, R.; Su, S.; Zheng, M.; Kong, A.N. Triterpenoid corosolic acid modulates global CpG methylation and transcriptome of tumor promotor TPA induced mouse epidermal JB6 P+ cells. Chem Biol Interact 2020, 321, 109025. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wu, R.; Li, W.; Gao, L.; Yang, Y.; Li, P.; Kong, A.N. The triterpenoid corosolic acid blocks transformation and epigenetically reactivates Nrf2 in TRAMP-C1 prostate cells. Mol Carcinog 2018, 57, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhou, H.; An, Y.; Shen, K.; Yu, L. Biological effects of corosolic acid as an anti-inflammatory, anti-metabolic syndrome and anti-neoplasic natural compound. Oncol Lett 2021, 21, 84. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Chen, Y.; Cui, G.H.; Zhou, J.F. Curcumin, a potent anti-tumor reagent, is a novel histone deacetylase inhibitor regulating B-NHL cell line Raji proliferation. Acta Pharmacol Sin 2005, 26, 603–609. [Google Scholar] [CrossRef]
- Shu, L.; Khor, T.O.; Lee, J.H.; Boyanapalli, S.S.; Huang, Y.; Wu, T.Y.; Saw, C.L.; Cheung, K.L.; Kong, A.N. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. Aaps j 2011, 13, 606–614. [Google Scholar] [CrossRef]
- Wei, Z.Q.; Zhang, Y.H.; Ke, C.Z.; Chen, H.X.; Ren, P.; He, Y.L.; Hu, P.; Ma, D.Q.; Luo, J.; Meng, Z.J. Curcumin inhibits hepatitis B virus infection by down-regulating cccDNA-bound histone acetylation. World J Gastroenterol 2017, 23, 6252–6260. [Google Scholar] [CrossRef] [PubMed]
- Sohn, E.J.; Bak, K.M.; Nam, Y.K.; Park, H.T. Upregulation of microRNA 344a-3p is involved in curcumin induced apoptosis in RT4 schwannoma cells. Cancer Cell Int 2018, 18, 199. [Google Scholar] [CrossRef] [PubMed]
- Butnariu, M.; Quispe, C.; Koirala, N.; Khadka, S.; Salgado-Castillo, C.M.; Akram, M.; Anum, R.; Yeskaliyeva, B.; Cruz-Martins, N.; Martorell, M.; et al. Bioactive Effects of Curcumin in Human Immunodeficiency Virus Infection Along with the Most Effective Isolation Techniques and Type of Nanoformulations. Int J Nanomedicine 2022, 17, 3619–3632. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-Y.; Shien, J.-H.; Tiley, L.; Chiou, S.-S.; Wang, S.-Y.; Chang, T.-J.; Lee, Y.-J.; Chan, K.-W.; Hsu, W.-L. Curcumin inhibits influenza virus infection and haemagglutination activity. Food Chemistry 2010, 119, 1346–1351. [Google Scholar] [CrossRef]
- Prusty, B.K.; Das, B.C. Constitutive activation of transcription factor AP-1 in cervical cancer and suppression of human papillomavirus (HPV) transcription and AP-1 activity in HeLa cells by curcumin. Int J Cancer 2005, 113, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Jennings, M.R.; Parks, R.J. Curcumin as an Antiviral Agent. Viruses 2020, 12, 1242. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.; Okla, M.; Chung, S. Ellagic acid inhibits adipocyte differentiation through coactivator-associated arginine methyltransferase 1-mediated chromatin modification. The Journal of Nutritional Biochemistry 2014, 25, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Dinata, R.; Nisa, N.; Arati, C.; Rasmita, B.; Uditraj, C.; Siddhartha, R.; Bhanushree, B.; Saeed-Ahmed, L.; Manikandan, B.; Bidanchi, R.M.; et al. Repurposing immune boosting and anti-viral efficacy of Parkia bioactive entities as multi-target directed therapeutic approach for SARS-CoV-2: exploration of lead drugs by drug likeness, molecular docking and molecular dynamics simulation methods. J Biomol Struct Dyn, 1080; -39. [Google Scholar] [CrossRef]
- Li, F.; Qasim, S.; Li, D.; Dou, Q.P. Updated review on green tea polyphenol epigallocatechin-3-gallate as a cancer epigenetic regulator. Semin Cancer Biol 2022, 83, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Henning, S.M.; Wang, P.; Carpenter, C.L.; Heber, D. Epigenetic effects of green tea polyphenols in cancer. Epigenomics 2013, 5, 729–741. [Google Scholar] [CrossRef]
- Moseley, V.R.; Morris, J.; Knackstedt, R.W.; Wargovich, M.J. Green tea polyphenol epigallocatechin 3-gallate, contributes to the degradation of DNMT3A and HDAC3 in HCT 116 human colon cancer cells. Anticancer Res 2013, 33, 5325–5333. [Google Scholar]
- Meeran, S.M.; Patel, S.N.; Chan, T.H.; Tollefsbol, T.O. A novel prodrug of epigallocatechin-3-gallate: differential epigenetic hTERT repression in human breast cancer cells. Cancer Prev Res (Phila) 2011, 4, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, Z.; Rasheed, N.; Al-Shaya, O. Epigallocatechin-3-O-gallate modulates global microRNA expression in interleukin-1β-stimulated human osteoarthritis chondrocytes: potential role of EGCG on negative co-regulation of microRNA-140-3p and ADAMTS5. Eur J Nutr 2018, 57, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Kim, K.; Lee, M.J.; Lee, J.; Choi, S.; Kim, K.S.; Ko, J.M.; Han, H.; Kim, S.Y.; Youn, H.J.; et al. Epigallocatechin Gallate-Mediated Alteration of the MicroRNA Expression Profile in 5α-Dihydrotestosterone-Treated Human Dermal Papilla Cells. Ann Dermatol 2016, 28, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chang, L.; Qu, Y.; Liang, J.; Jin, W.; Xia, X. Tea polyphenols inhibit the proliferation, migration, and invasion of melanoma cells through the down-regulation of TLR4. Int J Immunopathol Pharmacol 2018, 32, 394632017739531. [Google Scholar] [CrossRef] [PubMed]
- Arffa, M.L.; Zapf, M.A.; Kothari, A.N.; Chang, V.; Gupta, G.N.; Ding, X.; Al-Gayyar, M.M.; Syn, W.; Elsherbiny, N.M.; Kuo, P.C.; et al. Epigallocatechin-3-Gallate Upregulates miR-221 to Inhibit Osteopontin-Dependent Hepatic Fibrosis. PLoS One 2016, 11, e0167435. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, V.; Varizhuk, A.; Kozlovskaya, L.; Shtro, A.; Lebedeva, O.; Komissarov, A.; Vedekhina, T.; Manuvera, V.; Zubkova, O.; Eremeev, A.; et al. EGCG as an anti-SARS-CoV-2 agent: Preventive versus therapeutic potential against original and mutant virus. Biochimie 2021, 191, 27–32. [Google Scholar] [CrossRef] [PubMed]
- da Silva-Júnior, E.F.; Silva, L.R. Multi-target Approaches of Epigallocatechin-3-O-gallate (EGCG) and its Derivatives against Influenza Viruses. Curr Top Med Chem 2022, 22, 1485–1500. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Tanabe, H.; Suzuki, T.; Saeki, K.; Hara, Y. Applications of a Standardized Green Tea Catechin Preparation for Viral Warts and Human Papilloma Virus-Related and Unrelated Cancers. Molecules 2020, 25. [Google Scholar] [CrossRef]
- Mekky, R.Y.; El-Ekiaby, N.; El Sobky, S.A.; Elemam, N.M.; Youness, R.A.; El-Sayed, M.; Hamza, M.T.; Esmat, G.; Abdelaziz, A.I. Epigallocatechin gallate (EGCG) and miR-548m reduce HCV entry through repression of CD81 receptor in HCV cell models. Arch Virol 2019, 164, 1587–1595. [Google Scholar] [CrossRef]
- Date, A.A.; Destache, C.J. Natural polyphenols: potential in the prevention of sexually transmitted viral infections. Drug Discovery Today 2016, 21, 333–341. [Google Scholar] [CrossRef]
- Shorobi, F.M.; Nisa, F.Y.; Saha, S.; Chowdhury, M.A.H.; Srisuphanunt, M.; Hossain, K.H.; Rahman, M.A. Quercetin: A Functional Food-Flavonoid Incredibly Attenuates Emerging and Re-Emerging Viral Infections through Immunomodulatory Actions. Molecules 2023, 28. [Google Scholar] [CrossRef]
- Carneiro, B.M.; Batista, M.N.; Braga, A.C.S.; Nogueira, M.L.; Rahal, P. The green tea molecule EGCG inhibits Zika virus entry. Virology 2016, 496, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Huang, P.; Wang, X.; Wu, J.; Wu, M.; Huang, J. Galangin-induced down-regulation of BACE1 by epigenetic mechanisms in SH-SY5Y cells. Neuroscience 2015, 294, 172–181. [Google Scholar] [CrossRef]
- Deng, X.; Zuo, M.; Pei, Z.; Xie, Y.; Yang, Z.; Zhang, Z.; Jiang, M.; Kuang, D. MicroRNA-455-5p Contributes to Cholangiocarcinoma Growth and Mediates Galangin's Anti-Tumor Effects. J Cancer 2021, 12, 4710–4721. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.J.; Afolayan, A.J.; Taylor, M.B.; Erasmus, D. Antiviral activity of galangin isolated from the aerial parts of Helichrysum aureonitens. Journal of ethnopharmacology 1997, 56, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Kopytko, P.; Piotrowska, K.; Janisiak, J.; Tarnowski, M. Garcinol-A Natural Histone Acetyltransferase Inhibitor and New Anti-Cancer Epigenetic Drug. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Parasramka, M.A.; Ali, S.; Banerjee, S.; Deryavoush, T.; Sarkar, F.H.; Gupta, S. Garcinol sensitizes human pancreatic adenocarcinoma cells to gemcitabine in association with microRNA signatures. Mol Nutr Food Res 2013, 57, 235–248. [Google Scholar] [CrossRef]
- Ahmad, A.; Sarkar, S.H.; Bitar, B.; Ali, S.; Aboukameel, A.; Sethi, S.; Li, Y.; Bao, B.; Kong, D.; Banerjee, S.; et al. Garcinol regulates EMT and Wnt signaling pathways in vitro and in vivo, leading to anticancer activity against breast cancer cells. Mol Cancer Ther 2012, 11, 2193–2201. [Google Scholar] [CrossRef]
- Corona, A.; Seibt, S.; Schaller, D.; Schobert, R.; Volkamer, A.; Biersack, B.; Tramontano, E. Garcinol from Garcinia indica inhibits HIV-1 reverse transcriptase-associated ribonuclease H. Arch Pharm (Weinheim) 2021, 354, e2100123. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Quispe, C.; Imran, M.; Rauf, A.; Nadeem, M.; Gondal, T.A.; Ahmad, B.; Atif, M.; Mubarak, M.S.; Sytar, O.; et al. Genistein: An Integrative Overview of Its Mode of Action, Pharmacological Properties, and Health Benefits. Oxid Med Cell Longev 2021, 2021, 3268136. [Google Scholar] [CrossRef]
- Ma, J.; Cheng, L.; Liu, H.; Zhang, J.; Shi, Y.; Zeng, F.; Miele, L.; Sarkar, F.H.; Xia, J.; Wang, Z. Genistein down-regulates miR-223 expression in pancreatic cancer cells. Curr Drug Targets 2013, 14, 1150–1156. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, I.; Ito, A.; Hirai, G.; Nishimura, S.; Kawasaki, H.; Saitoh, H.; Kimura, K.; Sodeoka, M.; Yoshida, M. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol 2009, 16, 133–140. [Google Scholar] [CrossRef]
- Ibrahim, M.A.; Ramadan, H.H.; Mohammed, R.N. Evidence that Ginkgo Biloba could use in the influenza and coronavirus COVID-19 infections. J Basic Clin Physiol Pharmacol 2021, 32, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.L.D.; Lam, W.C.; Yang, W.; Chan, K.W.; Sze, S.C.W.; Miao, J.; Yung, K.K.L.; Bian, Z.; Wong, V.T. Potential Targets for Treatment of Coronavirus Disease 2019 (COVID-19): A Review of Qing-Fei-Pai-Du-Tang and Its Major Herbs. Am J Chin Med 2020, 48, 1051–1071. [Google Scholar] [CrossRef] [PubMed]
- Omer, M.O.; Almalki, W.H.; Shahid, I.; Khuram, S.; Altaf, I.; Imran, S. Comparative study to evaluate the anti-viral efficacy of Glycyrrhiza glabra extract and ribavirin against the Newcastle disease virus. Pharmacognosy Res 2014, 6, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Huan, C.; Xu, Y.; Zhang, W.; Guo, T.; Pan, H.; Gao, S. Research Progress on the Antiviral Activity of Glycyrrhizin and its Derivatives in Liquorice. Front Pharmacol 2021, 12, 680674. [Google Scholar] [CrossRef] [PubMed]
- Wolkerstorfer, A.; Kurz, H.; Bachhofner, N.; Szolar, O.H. Glycyrrhizin inhibits influenza A virus uptake into the cell. Antiviral Res 2009, 83, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Meng, T.; Wang, Y.; Tang, W. A Review of the Antiviral Activities of Glycyrrhizic Acid, Glycyrrhetinic Acid and Glycyrrhetinic Acid Monoglucuronide. Pharmaceuticals 2023, 16, 641. [Google Scholar] [CrossRef] [PubMed]
- Fiore, C.; Eisenhut, M.; Krausse, R.; Ragazzi, E.; Pellati, D.; Armanini, D.; Bielenberg, J. Antiviral effects of Glycyrrhiza species. Phytother Res 2008, 22, 141–148. [Google Scholar] [CrossRef]
- Chen, F.; Chan, K.H.; Jiang, Y.; Kao, R.Y.; Lu, H.T.; Fan, K.W.; Cheng, V.C.; Tsui, W.H.; Hung, I.F.; Lee, T.S.; et al. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J Clin Virol 2004, 31, 69–75. [Google Scholar] [CrossRef]
- Yang, R.; Liu, H.; Bai, C.; Wang, Y.; Zhang, X.; Guo, R.; Wu, S.; Wang, J.; Leung, E.; Chang, H.; et al. Chemical composition and pharmacological mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction against Coronavirus Disease 2019 (COVID-19): In silico and experimental study. Pharmacol Res 2020, 157, 104820. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Huang, S.; Li, Y.; Li, Y.; Li, D.; Wu, P.; Wang, Q.; Zheng, X.; Zhang, K. Glycyrrhizic acid from licorice down-regulates inflammatory responses via blocking MAPK and PI3K/Akt-dependent NF-κB signalling pathways in TPA-induced skin inflammation. Medchemcomm 2018, 9, 1502–1510. [Google Scholar] [CrossRef] [PubMed]
- El Omari, N.; Bakrim, S.; Bakha, M.; Lorenzo, J.M.; Rebezov, M.; Shariati, M.A.; Aboulaghras, S.; Balahbib, A.; Khayrullin, M.; Bouyahya, A. Natural Bioactive Compounds Targeting Epigenetic Pathways in Cancer: A Review on Alkaloids, Terpenoids, Quinones, and Isothiocyanates. Nutrients 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Yang, L.; Xiao, L.; Xia, X.; Dong, X.; Zhong, J.; Liu, Y.; Li, N.; Chen, L.; Li, H.; et al. Grifolin directly targets ERK1/2 to epigenetically suppress cancer cell metastasis. Oncotarget 2015, 6, 42704–42716. [Google Scholar] [CrossRef] [PubMed]
- Fabiani, R.; Vella, N.; Rosignoli, P. Epigenetic Modifications Induced by Olive Oil and Its Phenolic Compounds: A Systematic Review. Molecules 2021, 26, 273. [Google Scholar] [CrossRef] [PubMed]
- Juli, G.; Oliverio, M.; Bellizzi, D.; Gallo Cantafio, M.E.; Grillone, K.; Passarino, G.; Colica, C.; Nardi, M.; Rossi, M.; Procopio, A.; et al. Anti-tumor Activity and Epigenetic Impact of the Polyphenol Oleacein in Multiple Myeloma. Cancers (Basel) 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Lee-Huang, S.; Huang, P.L.; Zhang, D.; Lee, J.W.; Bao, J.; Sun, Y.; Chang, Y.T.; Zhang, J.; Huang, P.L. Discovery of small-molecule HIV-1 fusion and integrase inhibitors oleuropein and hydroxytyrosol: Part I. fusion [corrected] inhibition. Biochemical and biophysical research communications 2007, 354, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Gomes, N.G.M.; Valentão, P.; Andrade, P.B.; Pereira, R.B. Plitidepsin to treat multiple myeloma. Drugs Today (Barc) 2020, 56, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Li, D.; Marcial, D.; Khan, M.; Lin, M.H.; Snape, N.; Ghildyal, R.; Harrich, D.; Spann, K. The eukaryotic elongation factor 1A is critical for genome replication of the paramyxovirus respiratory syncytial virus. PLoS One 2014, 9, e114447. [Google Scholar] [CrossRef]
- White, K.M.; Rosales, R.; Yildiz, S.; Kehrer, T.; Miorin, L.; Moreno, E.; Jangra, S.; Uccellini, M.B.; Rathnasinghe, R.; Coughlan, L.; et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 2021, 371, 926–931. [Google Scholar] [CrossRef]
- Reuschl, A.-K.; Thorne, L.G.; Zuliani-Alvarez, L.; Bouhaddou, M.; Obernier, K.; Hiatt, J.; Soucheray, M.; Turner, J.; Fabius, J.M.; Nguyen, G.T. Host-directed therapies against early-lineage SARS-CoV-2 retain efficacy against B. 1.1. 7 variant. BioRxiv 2021. [Google Scholar]
- Kala, R.; Shah, H.N.; Martin, S.L.; Tollefsbol, T.O. Epigenetic-based combinatorial resveratrol and pterostilbene alters DNA damage response by affecting SIRT1 and DNMT enzyme expression, including SIRT1-dependent γ-H2AX and telomerase regulation in triple-negative breast cancer. BMC cancer 2015, 15, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, P.Y.; Marshall, G.M. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer research 2009, 69, 1702–1705. [Google Scholar] [CrossRef] [PubMed]
- Ter Ellen, B.M.; Dinesh Kumar, N.; Bouma, E.M.; Troost, B.; van de Pol, D.P.I.; van der Ende-Metselaar, H.H.; Apperloo, L.; van Gosliga, D.; van den Berge, M.; Nawijn, M.C.; et al. Resveratrol and Pterostilbene Inhibit SARS-CoV-2 Replication in Air-Liquid Interface Cultured Human Primary Bronchial Epithelial Cells. Viruses 2021, 13. [Google Scholar] [CrossRef]
- Lee, W.J.; Chen, Y.R.; Tseng, T.H. Quercetin induces FasL-related apoptosis, in part, through promotion of histone H3 acetylation in human leukemia HL-60 cells. Oncology reports 2011, 25, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Kedhari Sundaram, M.; Hussain, A.; Haque, S.; Raina, R.; Afroze, N. Quercetin modifies 5'CpG promoter methylation and reactivates various tumor suppressor genes by modulating epigenetic marks in human cervical cancer cells. J Cell Biochem 2019, 120, 18357–18369. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.; Ijaz, B.; Ahmad, Z.; Farooq, N.; Sarwar, M.B.; Husnain, T. Modification of miRNA Expression through plant extracts and compounds against breast cancer: Mechanism and translational significance. Phytomedicine 2020, 68, 153168. [Google Scholar] [CrossRef]
- Huang, Q.; Chen, L.; Luo, M.; Lv, H.; Luo, D.; Li, T.; Huang, S.; Xie, L.; Teng, Y.; Liu, Z.; et al. HIV-1-Induced miR-146a Attenuates Monocyte Migration by Targeting CCL5 in Human Primary Macrophages. AIDS Res Hum Retroviruses 2018, 34, 580–589. [Google Scholar] [CrossRef]
- Sonoki, H.; Sato, T.; Endo, S.; Matsunaga, T.; Yamaguchi, M.; Yamazaki, Y.; Sugatani, J.; Ikari, A. Quercetin Decreases Claudin-2 Expression Mediated by Up-Regulation of microRNA miR-16 in Lung Adenocarcinoma A549 Cells. Nutrients 2015, 7, 4578–4592. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, Q.; Chen, J.; Chen, Z. Quercetin Enhances Cisplatin Sensitivity of Human Osteosarcoma Cells by Modulating microRNA-217-KRAS Axis. Mol Cells 2015, 38, 638–642. [Google Scholar] [CrossRef]
- Zhou, J.; Gong, J.; Ding, C.; Chen, G. Quercetin induces the apoptosis of human ovarian carcinoma cells by upregulating the expression of microRNA-145. Mol Med Rep 2015, 12, 3127–3131. [Google Scholar] [CrossRef] [PubMed]
- Di Petrillo, A.; Orrù, G.; Fais, A.; Fantini, M.C. Quercetin and its derivates as antiviral potentials: A comprehensive review. Phytother Res 2022, 36, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Zarenezhad, E.; Abdulabbas, H.T.; Kareem, A.S.; Kouhpayeh, S.A.; Barbaresi, S.; Najafipour, S.; Mazarzaei, A.; Sotoudeh, M.; Ghasemian, A. Protective role of flavonoids quercetin and silymarin in the viral-associated inflammatory bowel disease: an updated review. Arch Microbiol 2023, 205, 252. [Google Scholar] [CrossRef] [PubMed]
- Brito, J.C.M.; Lima, W.G.; Cordeiro, L.P.B.; da Cruz Nizer, W.S. Effectiveness of supplementation with quercetin-type flavonols for treatment of viral lower respiratory tract infections: Systematic review and meta-analysis of preclinical studies. Phytother Res 2021, 35, 4930–4942. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, O.; Fontanes, V.; Raychaudhuri, S.; Loo, R.; Loo, J.; Arumugaswami, V.; Sun, R.; Dasgupta, A.; French, S.W. The heat shock protein inhibitor Quercetin attenuates hepatitis C virus production. Hepatology 2009, 50, 1756–1764. [Google Scholar] [CrossRef] [PubMed]
- Parashar, G.; Capalash, N. Promoter methylation-independent reactivation of PAX1 by curcumin and resveratrol is mediated by UHRF1. Clinical and experimental medicine 2016, 16, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Torres, E.; Hernández-Oliveras, A.; Meneses-Morales, I.; Rodríguez, G.; Fuentes-García, G.; Zarain-Herzberg, Á. Resveratrol up-regulates ATP2A3 gene expression in breast cancer cell lines through epigenetic mechanisms. The International Journal of Biochemistry & Cell Biology 2019, 113, 37–47. [Google Scholar]
- Borra, M.T.; Smith, B.C.; Denu, J.M. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem 2005, 280, 17187–17195. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Michaille, J.J. Resveratrol, MicroRNAs, Inflammation, and Cancer. J Nucleic Acids 2011, 2011, 102431. [Google Scholar] [CrossRef]
- Bai, T.; Dong, D.S.; Pei, L. Synergistic antitumor activity of resveratrol and miR-200c in human lung cancer. Oncology reports 2014, 31, 2293–2297. [Google Scholar] [CrossRef]
- Smoliga, J.M.; Blanchard, O.L. Recent data do not provide evidence that resveratrol causes 'mainly negative' or 'adverse' effects on exercise training in humans. J Physiol 2013, 591, 5251–5252. [Google Scholar] [CrossRef] [PubMed]
- Docherty, J.J.; Smith, J.S.; Fu, M.M.; Stoner, T.; Booth, T. Effect of topically applied resveratrol on cutaneous herpes simplex virus infections in hairless mice. Antiviral Res 2004, 61, 19–26. [Google Scholar] [CrossRef]
- Ayipo, Y.O.; Yahaya, S.N.; Alananzeh, W.A.; Babamale, H.F.; Mordi, M.N. Pathomechanisms, therapeutic targets and potent inhibitors of some beta-coronaviruses from bench-to-bedside. Infect Genet Evol 2021, 93, 104944. [Google Scholar] [CrossRef] [PubMed]
- Docherty, J.J.; Sweet, T.J.; Bailey, E.; Faith, S.A.; Booth, T. Resveratrol inhibition of varicella-zoster virus replication in vitro. Antiviral Res 2006, 72, 171–177. [Google Scholar] [CrossRef]
- Sasivimolphan, P.; Lipipun, V.; Likhitwitayawuid, K.; Takemoto, M.; Pramyothin, P.; Hattori, M.; Shiraki, K. Inhibitory activity of oxyresveratrol on wild-type and drug-resistant varicella-zoster virus replication in vitro. Antiviral Res 2009, 84, 95–97. [Google Scholar] [CrossRef]
- Lehman, C.W.; Kehn-Hall, K.; Aggarwal, M.; Bracci, N.R.; Pan, H.C.; Panny, L.; Lamb, R.A.; Lin, S.C. Resveratrol Inhibits Venezuelan Equine Encephalitis Virus Infection by Interfering with the AKT/GSK Pathway. Plants (Basel) 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Eladwy, R.A.; Vu, H.T.; Shah, R.; Li, C.G.; Chang, D.; Bhuyan, D.J. The Fight against the Carcinogenic Epstein-Barr Virus: Gut Microbiota, Natural Medicines, and Beyond. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Zhou, L.; Wang, Y.; Yu, C.; Wang, Z.; Liu, H.; Wei, H.; Han, L.; Cheng, J.; Wang, F.; et al. Mechanisms and Therapeutic Strategies of Viral Myocarditis Targeting Autophagy. Front Pharmacol 2022, 13, 843103. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Li, Q.; Zhang, F.; Li, X.; You, H.; Pan, X.; Zheng, K.; Tang, R. Sirtuins as Potential Therapeutic Targets for Hepatitis B Virus Infection. Front Med (Lausanne) 2021, 8, 751516. [Google Scholar] [CrossRef]
- Alway, S.E.; McCrory, J.L.; Kearcher, K.; Vickers, A.; Frear, B.; Gilleland, D.L.; Bonner, D.E.; Thomas, J.M.; Donley, D.A.; Lively, M.W.; et al. Resveratrol Enhances Exercise-Induced Cellular and Functional Adaptations of Skeletal Muscle in Older Men and Women. J Gerontol A Biol Sci Med Sci 2017, 72, 1595–1606. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Q.; Chen, H. DNA methylation and histone modifications of Wnt genes by genistein during colon cancer development. Carcinogenesis 2013, 34, 1756–1763. [Google Scholar] [CrossRef] [PubMed]
- Barros, T.M.B.; Lima, A.P.B.; Almeida, T.C.; da Silva, G.N. Inhibition of urinary bladder cancer cell proliferation by silibinin. Environ Mol Mutagen 2020, 61, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Kauntz, H.; Bousserouel, S.; Gossé, F.; Raul, F. Epigenetic effects of the natural flavonolignan silibinin on colon adenocarcinoma cells and their derived metastatic cells. Oncol Lett 2013, 5, 1273–1277. [Google Scholar] [CrossRef] [PubMed]
- Bouyahya, A.; Mechchate, H.; Oumeslakht, L.; Zeouk, I.; Aboulaghras, S.; Balahbib, A.; Zengin, G.; Kamal, M.A.; Gallo, M.; Montesano, D.; et al. The Role of Epigenetic Modifications in Human Cancers and the Use of Natural Compounds as Epidrugs: Mechanistic Pathways and Pharmacodynamic Actions. Biomolecules 2022, 12, 367. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, M.M.; Motamed, N.; Ranji, N.; Majidi, M.; Falahi, F. Silibinin-Induced Apoptosis and Downregulation of MicroRNA-21 and MicroRNA-155 in MCF-7 Human Breast Cancer Cells. jbc 2016, 19, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.H.; Jassey, A.; Hsu, H.Y.; Lin, L.T. Antiviral Activities of Silymarin and Derivatives. Molecules 2019, 24. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Galicia-Vázquez, G.; Cencic, R.; Mills, J.R.; Katigbak, A.; Porco, J.A., Jr.; Pelletier, J. CRISPR-Mediated Drug-Target Validation Reveals Selective Pharmacological Inhibition of the RNA Helicase, eIF4A. Cell Rep 2016, 15, 2340–2347. [Google Scholar] [CrossRef] [PubMed]
- Biedenkopf, N.; Lange-Grünweller, K.; Schulte, F.W.; Weißer, A.; Müller, C.; Becker, D.; Becker, S.; Hartmann, R.K.; Grünweller, A. The natural compound silvestrol is a potent inhibitor of Ebola virus replication. Antiviral Res 2017, 137, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Elgner, F.; Sabino, C.; Basic, M.; Ploen, D.; Grünweller, A.; Hildt, E. Inhibition of Zika Virus Replication by Silvestrol. Viruses 2018, 10. [Google Scholar] [CrossRef]
- Henss, L.; Scholz, T.; Grünweller, A.; Schnierle, B.S. Silvestrol Inhibits Chikungunya Virus Replication. Viruses 2018, 10. [Google Scholar] [CrossRef]
- Müller, C.; Schulte, F.W.; Lange-Grünweller, K.; Obermann, W.; Madhugiri, R.; Pleschka, S.; Ziebuhr, J.; Hartmann, R.K.; Grünweller, A. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antiviral Res 2018, 150, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.; Obermann, W.; Schulte, F.W.; Lange-Grünweller, K.; Oestereich, L.; Elgner, F.; Glitscher, M.; Hildt, E.; Singh, K.; Wendel, H.G.; et al. Comparison of broad-spectrum antiviral activities of the synthetic rocaglate CR-31-B (-) and the eIF4A-inhibitor Silvestrol. Antiviral Res 2020, 175, 104706. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.; Obermann, W.; Karl, N.; Wendel, H.G.; Taroncher-Oldenburg, G.; Pleschka, S.; Hartmann, R.K.; Grünweller, A.; Ziebuhr, J. The rocaglate CR-31-B (-) inhibits SARS-CoV-2 replication at non-cytotoxic, low nanomolar concentrations in vitro and ex vivo. Antiviral Res 2021, 186, 105012. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.-L.; Zhou, S.-J.; Zhang, X.-M.; Chen, H.-Q.; Liu, W. Sulforaphane suppresses in vitro and in vivo lung tumorigenesis through downregulation of HDAC activity. Biomedicine & Pharmacotherapy 2016, 78, 74–80. [Google Scholar]
- Ordonez, A.A.; Bullen, C.K.; Villabona-Rueda, A.F.; Thompson, E.A.; Turner, M.L.; Merino, V.F.; Yan, Y.; Kim, J.; Davis, S.L.; Komm, O.; et al. Sulforaphane exhibits antiviral activity against pandemic SARS-CoV-2 and seasonal HCoV-OC43 coronaviruses in vitro and in mice. Commun Biol 2022, 5, 242. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-S.; Chen, W.-C.; Tseng, C.-K.; Lin, C.-K.; Hsu, Y.-C.; Chen, Y.-H.; Lee, J.-C. Sulforaphane suppresses hepatitis C virus replication by up-regulating heme oxygenase-1 expression through PI3K/Nrf2 pathway. PloS one 2016, 11, e0152236. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, C.; Guo, Y.; Su, Z.-Y.; Yang, Y.; Shu, L.; Kong, A.-N.T. Blocking of JB6 cell transformation by tanshinone IIA: epigenetic reactivation of Nrf2 antioxidative stress pathway. The AAPS journal 2014, 16, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Valipour, M. Therapeutic prospects of naturally occurring p38 MAPK inhibitors tanshinone IIA and pinocembrin for the treatment of SARS-CoV-2-induced CNS complications. Phytother Res, 1002. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, C.; Li, W.; Wu, R.; Guo, Y.; Cheng, D.; Yang, Y.; Androulakis, I.P.; Kong, A.N. Pharmacokinetics and Pharmacodynamics of the Triterpenoid Ursolic Acid in Regulating the Antioxidant, Anti-inflammatory, and Epigenetic Gene Responses in Rat Leukocytes. Mol Pharm 2017, 14, 3709–3717. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, J.; Liu, T.; Fang, J.; Wan, J.; Zhao, J.; Li, W.; Liu, J.; Zhao, X.; Chen, S. Anti-viral effects of urosolic acid on guinea pig cytomegalovirus in vitro. J Huazhong Univ Sci Technolog Med Sci 2012, 32, 883–887. [Google Scholar] [CrossRef]
- Bungau, S.; Vesa, C.M.; Abid, A.; Behl, T.; Tit, D.M.; Purza, A.L.; Pasca, B.; Todan, L.M.; Endres, L. Withaferin A-A Promising Phytochemical Compound with Multiple Results in Dermatological Diseases. Molecules 2021, 26. [Google Scholar] [CrossRef]
- Royston, K.J.; Udayakumar, N.; Lewis, K.; Tollefsbol, T.O. A novel combination of withaferin A and sulforaphane inhibits epigenetic machinery, cellular viability and induces apoptosis of breast cancer cells. International journal of molecular sciences 2017, 18, 1092. [Google Scholar] [CrossRef]
- Choe, J.; Har Yong, P.; Xiang Ng, Z. The Efficacy of Traditional Medicinal Plants in Modulating the Main Protease of SARS-CoV-2 and Cytokine Storm. Chem Biodivers 2022, 19, e202200655. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Zhang, G.; Tang, B.; Liu, Y.; Fu, X.; Zhang, X. Promising Anti-influenza Properties of Active Constituent of Withania somnifera Ayurvedic Herb in Targeting Neuraminidase of H1N1 Influenza: Computational Study. Cell Biochem Biophys 2015, 72, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Shaik, H.A.; Sinha, R.K.; Shah, B.R. Andrographolide: A Herbal-Chemosynthetic Approach for Enhancing Immunity, Combating Viral Infections, and Its Implication on Human Health. Molecules 2021, 26. [Google Scholar] [CrossRef]
- Theerawatanasirikul, S.; Lueangaramkul, V.; Thangthamniyom, N.; Chankeeree, P.; Semkum, P.; Lekcharoensuk, P. Andrographolide and Deoxyandrographolide Inhibit Protease and IFN-Antagonist Activities of Foot-and-Mouth Disease Virus 3C(pro). Animals (Basel) 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lee, E.M.; Sun, X.; Wang, D.; Tang, H.; Zhou, G.C. Design, synthesis and discovery of andrographolide derivatives against Zika virus infection. Eur J Med Chem 2020, 187, 111925. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.X.; Xue, H.J.; Ye, W.C.; Fang, B.H.; Liu, Y.H.; Yuan, S.H.; Yu, P.; Wang, Y.Q. Activity of andrographolide and its derivatives against influenza virus in vivo and in vitro. Biol Pharm Bull 2009, 32, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Gupta, S. Apigenin: a promising molecule for cancer prevention. Pharm Res 2010, 27, 962–978. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.H.; Chien, M.H.; Lin, W.L.; Wen, Y.C.; Chow, J.M.; Chen, C.K.; Kuo, T.C.; Lee, W.J. Inhibition of MDA-MB-231 breast cancer cell proliferation and tumor growth by apigenin through induction of G2/M arrest and histone H3 acetylation-mediated p21(WAF1/CIP1) expression. Environ Toxicol 2017, 32, 434–444. [Google Scholar] [CrossRef]
- Husain, K.; Villalobos-Ayala, K.; Laverde, V.; Vazquez, O.A.; Miller, B.; Kazim, S.; Blanck, G.; Hibbs, M.L.; Krystal, G.; Elhussin, I.; et al. Apigenin Targets MicroRNA-155, Enhances SHIP-1 Expression, and Augments Anti-Tumor Responses in Pancreatic Cancer. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Tao, Y.; Zhan, S.; Wang, Y.; Zhou, G.; Liang, H.; Chen, X.; Shen, H. Baicalin, the major component of traditional Chinese medicine Scutellaria baicalensis induces colon cancer cell apoptosis through inhibition of oncomiRNAs. Sci Rep 2018, 8, 14477. [Google Scholar] [CrossRef] [PubMed]
- Cicero, A.F.; Baggioni, A. Berberine and Its Role in Chronic Disease. Adv Exp Med Biol 2016, 928, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Warowicka, A.; Nawrot, R.; Goździcka-Józefiak, A. Antiviral activity of berberine. Arch Virol 2020, 165, 1935–1945. [Google Scholar] [CrossRef] [PubMed]
- Varghese, F.S.; Thaa, B.; Amrun, S.N.; Simarmata, D.; Rausalu, K.; Nyman, T.A.; Merits, A.; McInerney, G.M.; Ng, L.F.P.; Ahola, T. The Antiviral Alkaloid Berberine Reduces Chikungunya Virus-Induced Mitogen-Activated Protein Kinase Signaling. J Virol 2016, 90, 9743–9757. [Google Scholar] [CrossRef] [PubMed]
- Zuco, V.; Supino, R.; Righetti, S.C.; Cleris, L.; Marchesi, E.; Gambacorti-Passerini, C.; Formelli, F. Selective cytotoxicity of betulinic acid on tumor cell lines, but not on normal cells. Cancer Letters 2002, 175, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S. Betulinic Acid for cancer treatment and prevention. Int J Mol Sci 2008, 9, 1096–1107. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jutooru, I.; Lei, P.; Kim, K.; Lee, S.O.; Brents, L.K.; Prather, P.L.; Safe, S. Betulinic acid targets YY1 and ErbB2 through cannabinoid receptor-dependent disruption of microRNA-27a:ZBTB10 in breast cancer. Mol Cancer Ther 2012, 11, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Voon, F.L.; Sulaiman, M.R.; Akhtar, M.N.; Idris, M.F.; Akira, A.; Perimal, E.K.; Israf, D.A.; Ming-Tatt, L. Cardamonin (2',4'-dihydroxy-6'-methoxychalcone) isolated from Boesenbergia rotunda (L.) Mansf. inhibits CFA-induced rheumatoid arthritis in rats. Eur J Pharmacol 2017, 794, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Masuda, M.; Sakurai, A.; Sakakibara, M. Medicinal uses of the mushroom Cordyceps militaris: current state and prospects. Fitoterapia 2010, 81, 961–968. [Google Scholar] [CrossRef]
- Ling, J.Y.; Sun, Y.J.; Zhang, H.; Lv, P.; Zhang, C.K. Measurement of cordycepin and adenosine in stroma of Cordyceps sp. by capillary zone electrophoresis (CZE). J Biosci Bioeng 2002, 94, 371–374. [Google Scholar] [CrossRef]
- Hou, W.; Li, Y.; Zhang, Q.; Wei, X.; Peng, A.; Chen, L.; Wei, Y. Triterpene acids isolated from Lagerstroemia speciosa leaves as alpha-glucosidase inhibitors. Phytother Res 2009, 23, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Ratovitski, E.A. Anticancer Natural Compounds as Epigenetic Modulators of Gene Expression. Curr Genomics 2017, 18, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Hewlings, S.J.; Kalman, D.S. Curcumin: A Review of Its Effects on Human Health. Foods 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Hassan, F.U.; Rehman, M.S.; Khan, M.S.; Ali, M.A.; Javed, A.; Nawaz, A.; Yang, C. Curcumin as an Alternative Epigenetic Modulator: Mechanism of Action and Potential Effects. Front Genet 2019, 10, 514. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Yoo, H.S.; Kim, J.C.; Park, C.S.; Choi, M.S.; Kim, M.; Choi, H.; Min, J.S.; Kim, Y.S.; Yoon, S.W.; et al. Antiviral effect of Curcuma longa Linn extract against hepatitis B virus replication. Journal of ethnopharmacology 2009, 124, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Zahedipour, F.; Hosseini, S.A.; Sathyapalan, T.; Majeed, M.; Jamialahmadi, T.; Al-Rasadi, K.; Banach, M.; Sahebkar, A. Potential effects of curcumin in the treatment of COVID-19 infection. Phytother Res 2020, 34, 2911–2920. [Google Scholar] [CrossRef] [PubMed]
- Sordillo, P.P.; Helson, L. Curcumin suppression of cytokine release and cytokine storm. A potential therapy for patients with Ebola and other severe viral infections. In Vivo 2015, 29, 1–4. [Google Scholar] [PubMed]
- Wen, C.C.; Kuo, Y.H.; Jan, J.T.; Liang, P.H.; Wang, S.Y.; Liu, H.G.; Lee, C.K.; Chang, S.T.; Kuo, C.J.; Lee, S.S.; et al. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J Med Chem 2007, 50, 4087–4095. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, J.; Quispe, C.; Castillo, C.M.S.; Caroca, R.; Lazo-Vélez, M.A.; Antonyak, H.; Polishchuk, A.; Lysiuk, R.; Oliinyk, P.; De Masi, L.; et al. Ellagic Acid: A Review on Its Natural Sources, Chemical Stability, and Therapeutic Potential. Oxid Med Cell Longev 2022, 2022, 3848084. [Google Scholar] [CrossRef]
- Prasanth, M.I.; Sivamaruthi, B.S.; Chaiyasut, C.; Tencomnao, T. A Review of the Role of Green Tea (Camellia sinensis) in Antiphotoaging, Stress Resistance, Neuroprotection, and Autophagy. Nutrients 2019, 11. [Google Scholar] [CrossRef]
- Ngwe Tun, M.M.; Luvai, E.; Nwe, K.M.; Toume, K.; Mizukami, S.; Hirayama, K.; Komatsu, K.; Morita, K. Anti-SARS-CoV-2 activity of various PET-bottled Japanese green teas and tea compounds in vitro. Arch Virol 2022, 167, 1547–1557. [Google Scholar] [CrossRef]
- Bag, A.; Bag, N. Tea Polyphenols and Prevention of Epigenetic Aberrations in Cancer. J Nat Sci Biol Med 2018, 9, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Huang, M.; Wei, Y. Diversity of the reaction mechanisms of SAM-dependent enzymes. Acta Pharmaceutica Sinica B 2021, 11, 632–650. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, V.; Vaid, M.; Katiyar, S.K. (-)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis 2011, 32, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Ciesielski, O.; Biesiekierska, M.; Balcerczyk, A. Epigallocatechin-3-gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells. Molecules 2020, 25. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.Z.; Wang, Y.; Ai, N.; Hou, Z.; Sun, Y.; Lu, H.; Welsh, W.; Yang, C.S. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res 2003, 63, 7563–7570. [Google Scholar] [PubMed]
- Guo, C.; Huang, Q.; Wang, Y.; Yao, Y.; Li, J.; Chen, J.; Wu, M.; Zhang, Z.; E, M.; Qi, H.; et al. Therapeutic application of natural products: NAD(+) metabolism as potential target. Phytomedicine 2023, 114, 154768. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, J.; Buer, J.; Pietschmann, T.; Steinmann, E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br J Pharmacol 2013, 168, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- Ahn, W.S.; Huh, S.W.; Bae, S.M.; Lee, I.P.; Lee, J.M.; Namkoong, S.E.; Kim, C.K.; Sin, J.I. A major constituent of green tea, EGCG, inhibits the growth of a human cervical cancer cell line, CaSki cells, through apoptosis, G(1) arrest, and regulation of gene expression. DNA and cell biology 2003, 22, 217–224. [Google Scholar] [CrossRef]
- Stockfleth, E.; Meyer, T. Sinecatechins (Polyphenon E) ointment for treatment of external genital warts and possible future indications. Expert Opinion on Biological Therapy 2014, 14, 1033–1043. [Google Scholar] [CrossRef]
- Mekky, R.Y.; El-Ekiaby, N.M.; Hamza, M.T.; Elemam, N.M.; El-Sayed, M.; Esmat, G.; Abdelaziz, A.I. Mir-194 is a hepatocyte gate keeper hindering HCV entry through targeting CD81 receptor. J Infect 2015, 70, 78–87. [Google Scholar] [CrossRef]
- Kong, Y.; Feng, Z.; Chen, A.; Qi, Q.; Han, M.; Wang, S.; Zhang, Y.; Zhang, X.; Yang, N.; Wang, J.; et al. The Natural Flavonoid Galangin Elicits Apoptosis, Pyroptosis, and Autophagy in Glioblastoma. Front Oncol 2019, 9, 942. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, V.; Tuli, H.S.; Kaur, J.; Aggarwal, D.; Parashar, G.; Chaturvedi Parashar, N.; Kulkarni, S.; Kaur, G.; Sak, K.; Kumar, M.; et al. Garcinol Exhibits Anti-Neoplastic Effects by Targeting Diverse Oncogenic Factors in Tumor Cells. Biomedicines 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Padhye, S.; Ahmad, A.; Oswal, N.; Sarkar, F.H. Emerging role of Garcinol, the antioxidant chalcone from Garcinia indica Choisy and its synthetic analogs. J Hematol Oncol 2009, 2, 38. [Google Scholar] [CrossRef] [PubMed]
- Saadat, N.; Gupta, S.V. Potential role of garcinol as an anticancer agent. J Oncol 2012, 2012, 647206. [Google Scholar] [CrossRef]
- Aggarwal, V.; Tuli, H.S.; Kaur, J.; Aggarwal, D.; Parashar, G.; Chaturvedi Parashar, N.; Kulkarni, S.; Kaur, G.; Sak, K.; Kumar, M.; et al. Garcinol Exhibits Anti-Neoplastic Effects by Targeting Diverse Oncogenic Factors in Tumor Cells. Biomedicines 2020, 8, 103. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Jo, J.; Leem, J.; Park, K.-K. Inhibition of p300 by Garcinol Protects against Cisplatin-Induced Acute Kidney Injury through Suppression of Oxidative Stress, Inflammation, and Tubular Cell Death in Mice. Antioxidants 2020, 9, 1271. [Google Scholar] [CrossRef]
- Balasubramanyam, K.; Altaf, M.; Varier, R.A.; Swaminathan, V.; Ravindran, A.; Sadhale, P.P.; Kundu, T.K. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem 2004, 279, 33716–33726. [Google Scholar] [CrossRef]
- Biersack, B. Current state of phenolic and terpenoidal dietary factors and natural products as non-coding RNA/microRNA modulators for improved cancer therapy and prevention. Noncoding RNA Res 2016, 1, 12–34. [Google Scholar] [CrossRef]
- Fang, M.Z.; Chen, D.; Sun, Y.; Jin, Z.; Christman, J.K.; Yang, C.S. Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res 2005, 11, 7033–7041. [Google Scholar] [CrossRef]
- Gerstmeier, J.; Seegers, J.; Witt, F.; Waltenberger, B.; Temml, V.; Rollinger, J.M.; Stuppner, H.; Koeberle, A.; Schuster, D.; Werz, O. Ginkgolic Acid is a Multi-Target Inhibitor of Key Enzymes in Pro-Inflammatory Lipid Mediator Biosynthesis. Front Pharmacol 2019, 10, 797. [Google Scholar] [CrossRef] [PubMed]
- Flotho, A.; Melchior, F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 2013, 82, 357–385. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; Campbell, M.; Chang, P.C. SUMO modification of a heterochromatin histone demethylase JMJD2A enables viral gene transactivation and viral replication. PLoS Pathog 2017, 13, e1006216. [Google Scholar] [CrossRef] [PubMed]
- Bhutta, M.S.; Shechter, O.; Gallo, E.S.; Martin, S.D.; Jones, E.; Doncel, G.F.; Borenstein, R. Ginkgolic Acid Inhibits Herpes Simplex Virus Type 1 Skin Infection and Prevents Zosteriform Spread in Mice. Viruses 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.M.; Yan, S.; Jamaluddin, S.; Weakley, S.M.; Liang, Z.; Siwak, E.B.; Yao, Q.; Chen, C. Ginkgolic acid inhibits HIV protease activity and HIV infection in vitro. Med Sci Monit 2012, 18, Br293–298. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, G.; Cornara, L.; Soares, S.; Rodrigues, F.; Oliveira, M. Liquorice (Glycyrrhiza glabra): A phytochemical and pharmacological review. Phytother Res 2018, 32, 2323–2339. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.G.; Zhao, T.T.; Lu, N.; Yang, Y.A.; Zhu, H.L. Research Progress of Glycyrrhizic Acid on Antiviral Activity. Mini Rev Med Chem 2019, 19, 826–832. [Google Scholar] [CrossRef]
- Cao, L.; Ding, W.; Jia, R.; Du, J.; Wang, T.; Zhang, C.; Gu, Z.; Yin, G. Anti-inflammatory and hepatoprotective effects of glycyrrhetinic acid on CCl4-induced damage in precision-cut liver slices from Jian carp (Cyprinus carpio var. jian) through inhibition of the nf-kƁ pathway. Fish & Shellfish Immunology 2017, 64, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 2003, 361, 2045–2046. [Google Scholar] [CrossRef]
- Yue, G.G.; Chan, B.C.; Kwok, H.F.; To, M.H.; Hon, K.L.; Fung, K.P.; Lau, C.B.; Leung, P.C. Screening for anti-inflammatory and bronchorelaxant activities of 12 commonly used Chinese herbal medicines. Phytother Res 2012, 26, 915–925. [Google Scholar] [CrossRef]
- Kwon, H.S.; Oh, S.M.; Kim, J.K. Glabridin, a functional compound of liquorice, attenuates colonic inflammation in mice with dextran sulphate sodium-induced colitis. Clin Exp Immunol 2008, 151, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Q.; Xue, C.C.; Zhou, Z.W.; Li, C.G.; Du, Y.M.; Liang, J.; Zhou, S.F. In vitro and in vivo neuroprotective effect and mechanisms of glabridin, a major active isoflavan from Glycyrrhiza glabra (licorice). Life Sci 2008, 82, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Wu, L.Y.; Hou, M.F.; Tsai, E.M.; Lee, J.N.; Liang, H.L.; Jong, Y.J.; Hung, C.H.; Kuo, P.L. Glabridin, an isoflavan from licorice root, inhibits migration, invasion and angiogenesis of MDA-MB-231 human breast adenocarcinoma cells by inhibiting focal adhesion kinase/Rho signaling pathway. Mol Nutr Food Res 2011, 55, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Mu, J.; Wang, X.; Ye, X.; Si, L.; Ning, S.; Li, Z.; Li, Y. The repressive effect of miR-148a on TGF beta-SMADs signal pathway is involved in the glabridin-induced inhibition of the cancer stem cells-like properties in hepatocellular carcinoma cells. PLoS One 2014, 9, e96698. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Li, Y.; Mu, J.; Hu, C.; Zhou, M.; Wang, X.; Si, L.; Ning, S.; Li, Z. Glabridin inhibits cancer stem cell-like properties of human breast cancer cells: An epigenetic regulation of miR-148a/SMAd2 signaling. Mol Carcinog 2016, 55, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Ning, S.; Wang, X.; Si, L.; Jiang, F.; Li, Y.; Li, Z. The repressive effect of miR-520a on NF-κB/IL-6/STAT-3 signal involved in the glabridin-induced anti-angiogenesis in human breast cancer cells. RSC Advances 2015, 5, 34257–34264. [Google Scholar] [CrossRef]
- Zhou, C.; Li, X.; Du, W.; Feng, Y.; Kong, X.; Li, Y.; Xiao, L.; Zhang, P. Antitumor effects of ginkgolic acid in human cancer cell occur via cell cycle arrest and decrease the Bcl-2/Bax ratio to induce apoptosis. Chemotherapy 2010, 56, 393–402. [Google Scholar] [CrossRef]
- Lü, J.-M.; Yan, S.; Jamaluddin, S.; Weakley, S.M.; Liang, Z.; Siwak, E.B.; Yao, Q.; Chen, C. Ginkgolic acid inhibits HIV protease activity and HIV infection in vitro. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research 2012, 18, BR293. [Google Scholar] [CrossRef]
- Ye, M.; Liu, J.K.; Lu, Z.X.; Zhao, Y.; Liu, S.F.; Li, L.L.; Tan, M.; Weng, X.X.; Li, W.; Cao, Y. Grifolin, a potential antitumor natural product from the mushroom Albatrellus confluens, inhibits tumor cell growth by inducing apoptosis in vitro. FEBS Lett 2005, 579, 3437–3443. [Google Scholar] [CrossRef]
- Nediani, C.; Ruzzolini, J.; Romani, A.; Calorini, L. Oleuropein, a Bioactive Compound from Olea europaea L., as a Potential Preventive and Therapeutic Agent in Non-Communicable Diseases. Antioxidants (Basel) 2019, 8. [Google Scholar] [CrossRef]
- Juli, G.; Oliverio, M.; Bellizzi, D.; Gallo Cantafio, M.E.; Grillone, K.; Passarino, G.; Colica, C.; Nardi, M.; Rossi, M.; Procopio, A. Anti-tumor activity and epigenetic impact of the polyphenol oleacein in multiple myeloma. Cancers 2019, 11, 990. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Kumagai, H. Characteristics, biosynthesis, decomposition, metabolism and functions of the garlic odour precursor, S-allyl-L-cysteine sulfoxide. Exp Ther Med 2020, 19, 1528–1535. [Google Scholar] [CrossRef] [PubMed]
- Tolo, F.M.; Rukunga, G.M.; Muli, F.W.; Njagi, E.N.; Njue, W.; Kumon, K.; Mungai, G.M.; Muthaura, C.N.; Muli, J.M.; Keter, L.K.; et al. Anti-viral activity of the extracts of a Kenyan medicinal plant Carissa edulis against herpes simplex virus. Journal of ethnopharmacology 2006, 104, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.T.; Hsu, W.C.; Lin, C.C. Antiviral natural products and herbal medicines. J Tradit Complement Med 2014, 4, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.; Cole, L.L.; Davis, L.E.; Lockwood, S.J.; Simmons, V.; Wild, G.C. Antiviral properties of garlic: in vitro effects on influenza B, herpes simplex and coxsackie viruses. Planta Med 1985, 51, 460–461. [Google Scholar] [CrossRef] [PubMed]
- Abiy, E.; Berhe, A. Anti-bacterial effect of garlic (Allium sativum) against clinical isolates of Staphylococcus aureus and Escherichia coli from patients attending Hawassa Referral Hospital, Ethiopia. Journal of Infectious Diseases and Treatment 2016, 2, 1–5. [Google Scholar] [CrossRef]
- Gebreyohannes, G.; Gebreyohannes, M. Medicinal values of garlic: A review. International Journal of Medicine and Medical Sciences 2013, 5, 401–408. [Google Scholar]
- Lawson, L.D.; Bauer, R. Phytomedicines of Europe: chemistry and biological activity; ACS Publications: 1998.
- Borlinghaus, J.; Albrecht, F.; Gruhlke, M.C.; Nwachukwu, I.D.; Slusarenko, A.J. Allicin: chemistry and biological properties. Molecules 2014, 19, 12591–12618. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jiao, H.; Zhao, J.; Wang, X.; Sun, S.; Lin, H. Allicin Alleviates Reticuloendotheliosis Virus-Induced Immunosuppression via ERK/Mitogen-Activated Protein Kinase Pathway in Specific Pathogen-Free Chickens. Front Immunol 2017, 8, 1856. [Google Scholar] [CrossRef]
- Weber, N.D.; Andersen, D.O.; North, J.A.; Murray, B.K.; Lawson, L.D.; Hughes, B.G. In vitro virucidal effects of Allium sativum (garlic) extract and compounds. Planta Med 1992, 58, 417–423. [Google Scholar] [CrossRef]
- Arreola, R.; Quintero-Fabián, S.; López-Roa, R.I.; Flores-Gutiérrez, E.O.; Reyes-Grajeda, J.P.; Carrera-Quintanar, L.; Ortuño-Sahagún, D. Immunomodulation and anti-inflammatory effects of garlic compounds. J Immunol Res 2015, 2015, 401630. [Google Scholar] [CrossRef] [PubMed]
- Metwally, D.M.; Al-Olayan, E.M.; Alanazi, M.; Alzahrany, S.B.; Semlali, A. Antischistosomal and anti-inflammatory activity of garlic and allicin compared with that of praziquantel in vivo. BMC Complement Altern Med 2018, 18, 135. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.; Laxdal, V.A.; Yu, M.; Raney, B.L. Antioxidant activity of allicin, an active principle in garlic. Mol Cell Biochem 1995, 148, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Amagase, H. Clarifying the real bioactive constituents of garlic. J Nutr 2006, 136, 716s–725s. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.C.; Cottrell, S.L.; Plummer, S.; Lloyd, D. Antimicrobial properties of Allium sativum (garlic). Applied microbiology and biotechnology 2001, 57, 282–286. [Google Scholar] [CrossRef] [PubMed]
- El-Saber Batiha, G.; Magdy Beshbishy, A.; L, G.W.; Elewa, Y.H.A.; A, A.A.-S.; Abd El-Hack, M.E.; Taha, A.E.; Y, M.A.-E.; Prasad Devkota, H. Chemical Constituents and Pharmacological Activities of Garlic (Allium sativum L.): A Review. Nutrients 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Trio, P.Z.; You, S.; He, X.; He, J.; Sakao, K.; Hou, D.X. Chemopreventive functions and molecular mechanisms of garlic organosulfur compounds. Food Funct 2014, 5, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Li, H.; Cui, W.; Dong, Y. Treatment of hepatitis caused by cytomegalovirus with allitridin injection--an experimental study. J Tongji Med Univ 1999, 19, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Keyaerts, E.; Vijgen, L.; Pannecouque, C.; Van Damme, E.; Peumans, W.; Egberink, H.; Balzarini, J.; Van Ranst, M. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antiviral Res 2007, 75, 179–187. [Google Scholar] [CrossRef]
- Liu, Z.F.; Fang, F.; Dong, Y.S.; Li, G.; Zhen, H. Experimental study on the prevention and treatment of murine cytomegalovirus hepatitis by using allitridin. Antiviral Res 2004, 61, 125–128. [Google Scholar] [CrossRef]
- Terrasson, J.; Xu, B.; Li, M.; Allart, S.; Davignon, J.L.; Zhang, L.H.; Wang, K.; Davrinche, C. Activities of Z-ajoene against tumour and viral spreading in vitro. Fundam Clin Pharmacol 2007, 21, 281–289. [Google Scholar] [CrossRef]
- Walder, R.; Kalvatchev, Z.; Garzaro, D.; Barrios, M.; Apitz-Castro, R. In vitro suppression of HIV-1 replication by ajoene [(e)-(z)-4,5,9-trithiadodeca-1,6,11-triene-9 oxide]. Biomed Pharmacother 1997, 51, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.; Troupin, A.; Londono-Renteria, B.; Colpitts, T.M. Garlic Organosulfur Compounds Reduce Inflammation and Oxidative Stress during Dengue Virus Infection. Viruses 2017, 9. [Google Scholar] [CrossRef]
- Li, Y.N.; Huang, F.; Liu, X.L.; Shu, S.N.; Huang, Y.J.; Cheng, H.J.; Fang, F. Allium sativum-derived allitridin inhibits Treg amplification in cytomegalovirus infection. J Med Virol 2013, 85, 493–500. [Google Scholar] [CrossRef]
- Yi, X.; Feng, F.; Xiang, Z.; Ge, L. The effects of allitridin on the expression of transcription factors T-bet and GATA-3 in mice infected by murine cytomegalovirus. J Med Food 2005, 8, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Kaschula, C.H.; Hunter, R.; Parker, M.I. Garlic-derived anticancer agents: structure and biological activity of ajoene. Biofactors 2010, 36, 78–85. [Google Scholar] [CrossRef]
- Yi, L.; Su, Q. Molecular mechanisms for the anti-cancer effects of diallyl disulfide. Food Chem Toxicol 2013, 57, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Bae, E.Y.; Song, J.H.; Baek, S.H.; Kwon, D.H. Inhibitory effects of orobol 7-O-D-glucoside from banaba (Lagerstroemia speciosa L.) on human rhinoviruses replication. Lett Appl Microbiol 2010, 51, 1–5. [Google Scholar] [CrossRef]
- Pevear, D.C.; Tull, T.M.; Seipel, M.E.; Groarke, J.M. Activity of pleconaril against enteroviruses. Antimicrob Agents Chemother 1999, 43, 2109–2115. [Google Scholar] [CrossRef]
- Burgett, A.W.; Poulsen, T.B.; Wangkanont, K.; Anderson, D.R.; Kikuchi, C.; Shimada, K.; Okubo, S.; Fortner, K.C.; Mimaki, Y.; Kuroda, M.; et al. Natural products reveal cancer cell dependence on oxysterol-binding proteins. Nat Chem Biol 2011, 7, 639–647. [Google Scholar] [CrossRef]
- Albulescu, L.; Bigay, J.; Biswas, B.; Weber-Boyvat, M.; Dorobantu, C.M.; Delang, L.; van der Schaar, H.M.; Jung, Y.S.; Neyts, J.; Olkkonen, V.M.; et al. Uncovering oxysterol-binding protein (OSBP) as a target of the anti-enteroviral compound TTP-8307. Antiviral Res 2017, 140, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.L.; Severance, Z.C.; Bensen, R.C.; Le-McClain, A.T.; Malinky, C.A.; Mettenbrink, E.M.; Nuñez, J.I.; Reddig, W.J.; Blewett, E.L.; Burgett, A.W.G. Differing activities of oxysterol-binding protein (OSBP) targeting anti-viral compounds. Antiviral Res 2019, 170, 104548. [Google Scholar] [CrossRef] [PubMed]
- Albulescu, L.; Strating, J.R.; Thibaut, H.J.; van der Linden, L.; Shair, M.D.; Neyts, J.; van Kuppeveld, F.J. Broad-range inhibition of enterovirus replication by OSW-1, a natural compound targeting OSBP. Antiviral Res 2015, 117, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Álvarez, S.; Pardal, E.; Sánchez-Nieto, D.; Navarro, M.; Caballero, M.D.; Mateos, M.V.; Martín, A. Plitidepsin: design, development, and potential place in therapy. Drug Des Devel Ther 2017, 11, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Duke, S.O. Benefits of Resveratrol and Pterostilbene to Crops and Their Potential Nutraceutical Value to Mammals. Agriculture 2022, 12, 368. [Google Scholar] [CrossRef]
- Anand David, A.V.; Arulmoli, R.; Parasuraman, S. Overviews of Biological Importance of Quercetin: A Bioactive Flavonoid. Pharmacogn Rev 2016, 10, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Noratto, G.D.; Kim, Y.; Talcott, S.T.; Mertens-Talcott, S.U. Flavonol-rich fractions of yaupon holly leaves (Ilex vomitoria, Aquifoliaceae) induce microRNA-146a and have anti-inflammatory and chemopreventive effects in intestinal myofibroblast CCD-18Co cells. Fitoterapia 2011, 82, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Pop, S.; Enciu, A.M.; Tarcomnicu, I.; Gille, E.; Tanase, C. Phytochemicals in cancer prevention: modulating epigenetic alterations of DNA methylation. Phytochemistry Reviews 2019, 18, 1005–1024. [Google Scholar] [CrossRef]
- Choi, H.J.; Lim, C.H.; Song, J.H.; Baek, S.H.; Kwon, D.H. Antiviral activity of raoulic acid from Raoulia australis against Picornaviruses. Phytomedicine 2009, 16, 35–39. [Google Scholar] [CrossRef]
- Saiko, P.; Pemberger, M.; Horvath, Z.; Savinc, I.; Grusch, M.; Handler, N.; Erker, T.; Jaeger, W.; Fritzer-Szekeres, M.; Szekeres, T. Novel resveratrol analogs induce apoptosis and cause cell cycle arrest in HT29 human colon cancer cells: inhibition of ribonucleotide reductase activity. Oncology reports 2008, 19, 1621–1626. [Google Scholar]
- Koushki, M.; Amiri-Dashatan, N.; Ahmadi, N.; Abbaszadeh, H.A.; Rezaei-Tavirani, M. Resveratrol: A miraculous natural compound for diseases treatment. Food science & nutrition 2018, 6, 2473–2490. [Google Scholar]
- Kala, R.; Tollefsbol, T.; Li, Y. Potential of resveratrol in inhibiting cancer and slowing aging. J Nutr Food Sci S 2012, 5. [Google Scholar] [CrossRef]
- Chen, X.; Song, X.; Zhao, X.; Zhang, Y.; Wang, Y.; Jia, R.; Zou, Y.; Li, L.; Yin, Z. Insights into the Anti-inflammatory and Antiviral Mechanisms of Resveratrol. Mediators Inflamm 2022, 2022, 7138756. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Jiang, P.; Xu, X.; Lu, W.; Yang, C.; Zhou, P.; Liu, S. Resveratrol promotes HSV-2 replication by increasing histone acetylation and activating NF-κB. Biochem Pharmacol 2020, 171, 113691. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.H.; Zang, N.; Li, S.M.; Wang, L.J.; Deng, Y.; He, Y.; Yang, X.Q.; Liu, E.M. Resveratrol Inhibits respiratory syncytial virus-induced IL-6 production, decreases viral replication, and downregulates TRIF expression in airway epithelial cells. Inflammation 2012, 35, 1392–1401. [Google Scholar] [CrossRef] [PubMed]
- Bijak, M. Silybin, a Major Bioactive Component of Milk Thistle (Silybum marianum L. Gaernt.)-Chemistry, Bioavailability, and Metabolism. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Anestopoulos, I.; Sfakianos, A.P.; Franco, R.; Chlichlia, K.; Panayiotidis, M.I.; Kroll, D.J.; Pappa, A. A novel role of silibinin as a putative epigenetic modulator in human prostate carcinoma. Molecules 2016, 22, 62. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.B.; Lucas, D.M.; Lozanski, G.; Johnson, A.J.; Su, B.-N.; Lin, T.S.; Byrd, J.C.; Kinghorn, A.D.; Grever, M.R. Silvestrol, a Rocaglate Derivative from the Indonesian Plant Aglaia foveolata, Has Significant Bcl-2- and p53-Independent Anti-Tumor Activity against Chronic Lymphocytic Leukemia Cells. Blood 2006, 108, 2600. [Google Scholar] [CrossRef]
- M de Figueiredo, S.; S Binda, N.; A Nogueira-Machado, J.; A Vieira-Filho, S.; B Caligiorne, R. The antioxidant properties of organosulfur compounds (sulforaphane). Recent Patents on Endocrine, Metabolic & Immune Drug Discovery 2015, 9, 24–39. [Google Scholar]
- Dos Santos, P.W.d.S.; Machado, A.R.T.; De Grandis, R.A.; Ribeiro, D.L.; Tuttis, K.; Morselli, M.; Aissa, A.F.; Pellegrini, M.; Antunes, L.M.G. Transcriptome and DNA methylation changes modulated by sulforaphane induce cell cycle arrest, apoptosis, DNA damage, and suppression of proliferation in human liver cancer cells. Food and Chemical Toxicology 2020, 136, 111047. [Google Scholar] [CrossRef]
- Chen, W.-C.; Huang, C.-H.; Liu, W.; Lee, J.-C. Sulforaphane suppresses dengue virus replication by inhibition of dengue protease and enhancement of antiviral interferon response through Nrf2-mediated heme oxygenase-1 induction. Antiviral Research 2022, 207, 105400. [Google Scholar] [CrossRef] [PubMed]
- Ordonez, A.A.; Bullen, C.K.; Villabona-Rueda, A.F.; Thompson, E.A.; Turner, M.L.; Davis, S.L.; Komm, O.; Powell, J.D.; D'Alessio, F.R.; Yolken, R.H.; et al. Sulforaphane exhibits in vitro and in vivo antiviral activity against pandemic SARS-CoV-2 and seasonal HCoV-OC43 coronaviruses. bioRxiv, 1101. [Google Scholar] [CrossRef]
- Zeng, A.; Wang, D.; Wu, N.; Yang, R.; Nan, G.; Bian, X.; Yang, G. Extraction of Tanshinone IIA and Cryptotanshinone from the Rhizome of Salvia miltiorrhiza Bunge: Kinetics and Modeling. Separation Science and Technology 2014, 49, 2330–2337. [Google Scholar] [CrossRef]
- Machado, D.G.; Neis, V.B.; Balen, G.O.; Colla, A.; Cunha, M.P.; Dalmarco, J.B.; Pizzolatti, M.G.; Prediger, R.D.; Rodrigues, A.L. Antidepressant-like effect of ursolic acid isolated from Rosmarinus officinalis L. in mice: evidence for the involvement of the dopaminergic system. Pharmacol Biochem Behav 2012, 103, 204–211. [Google Scholar] [CrossRef] [PubMed]
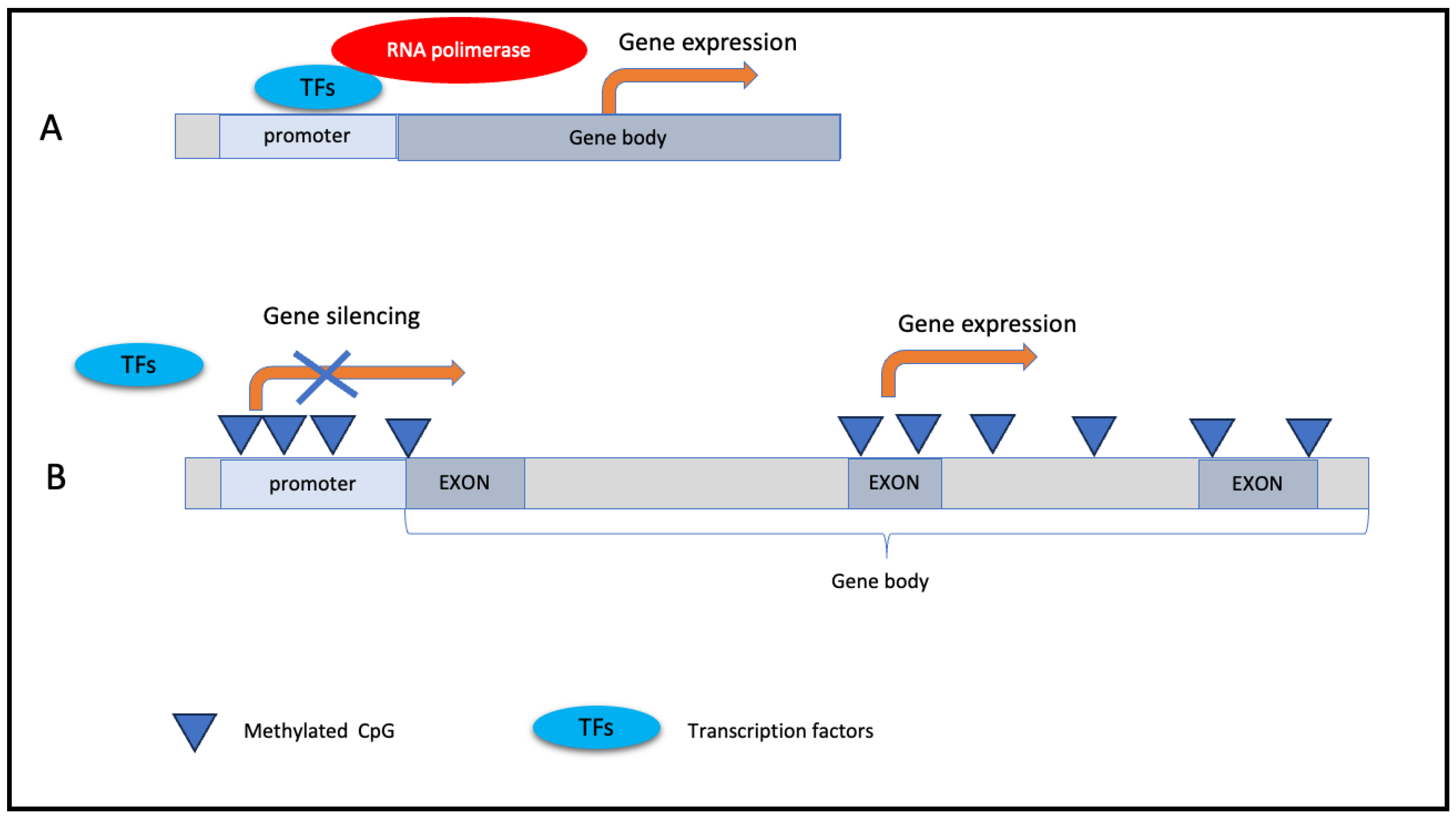
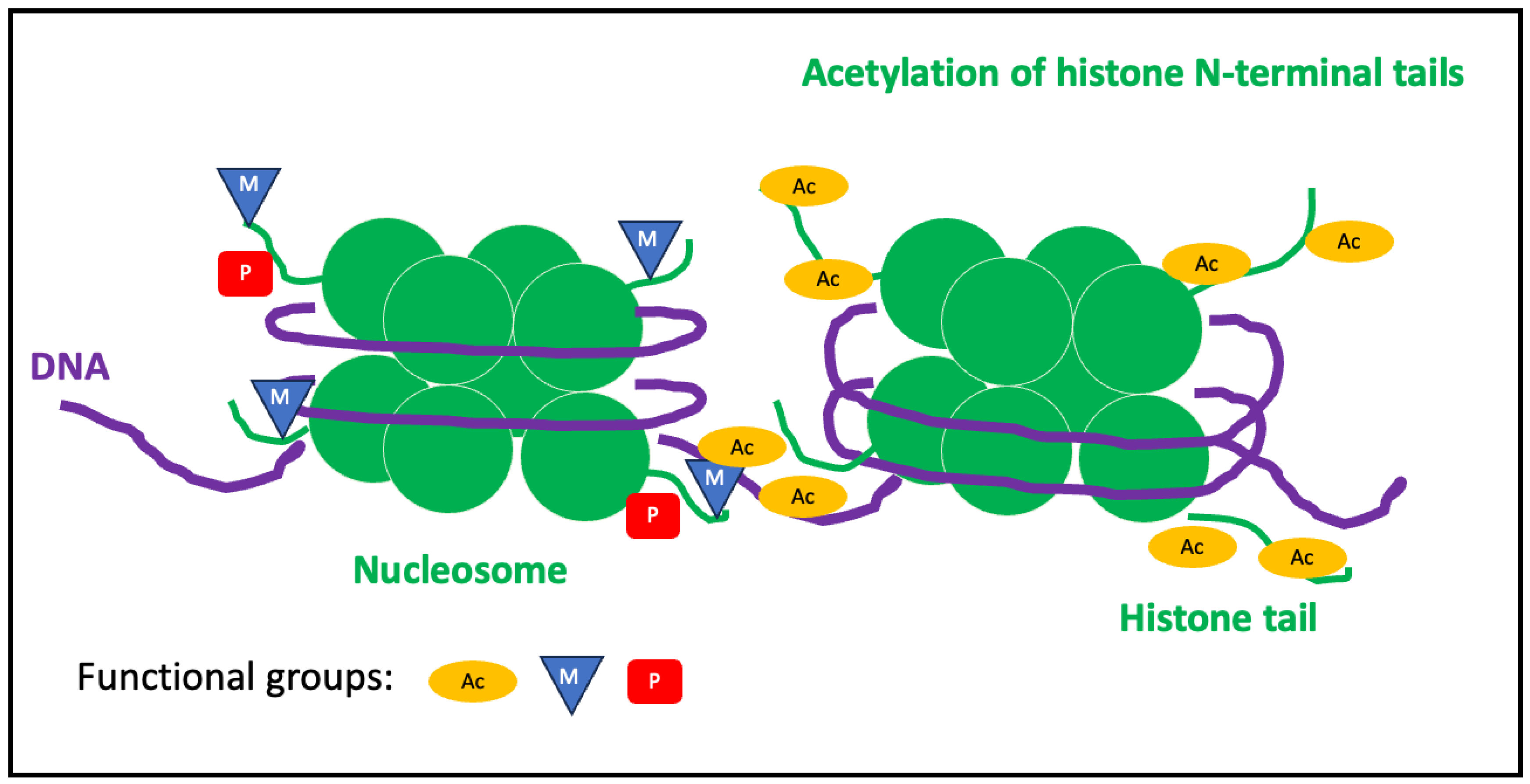
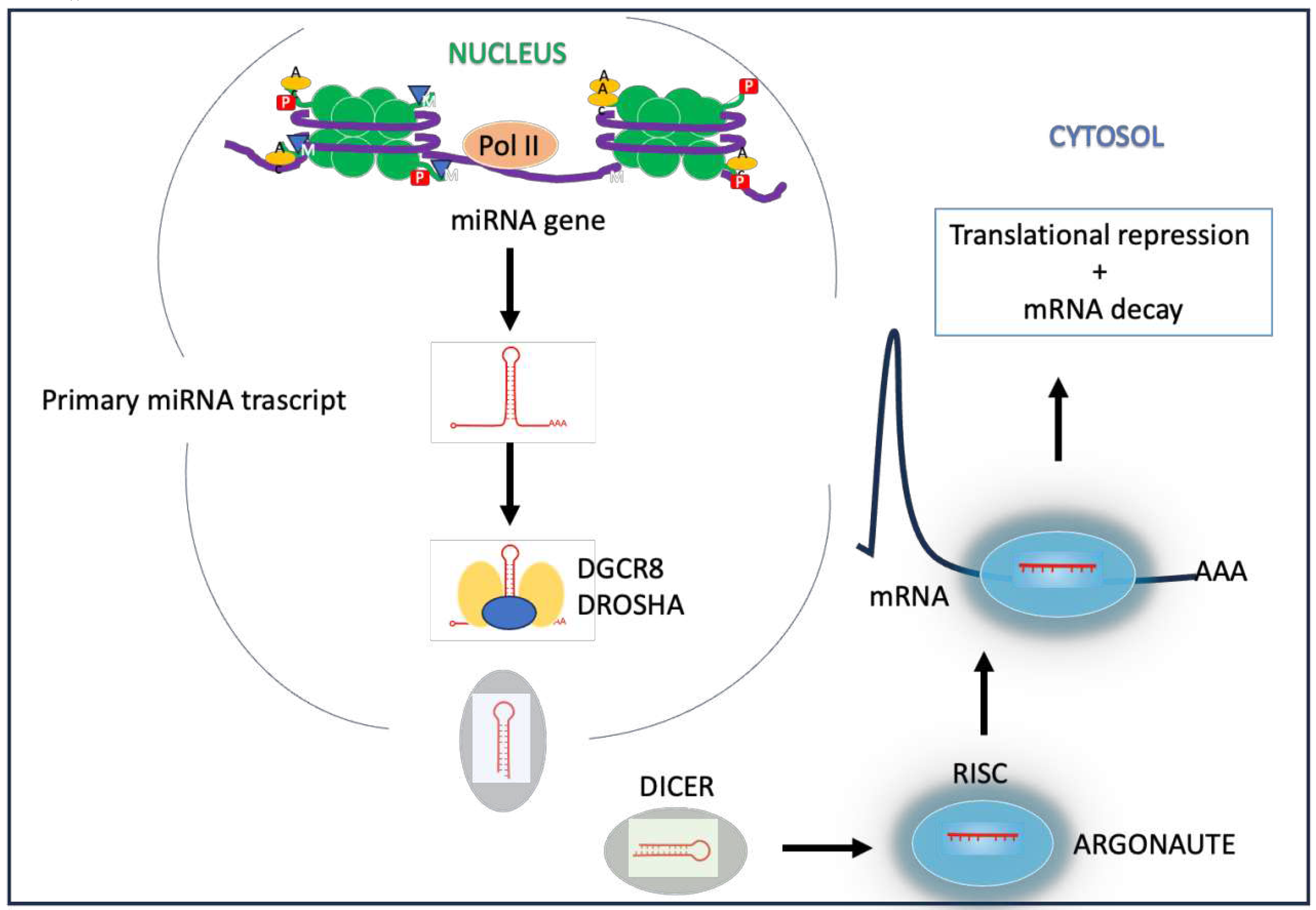
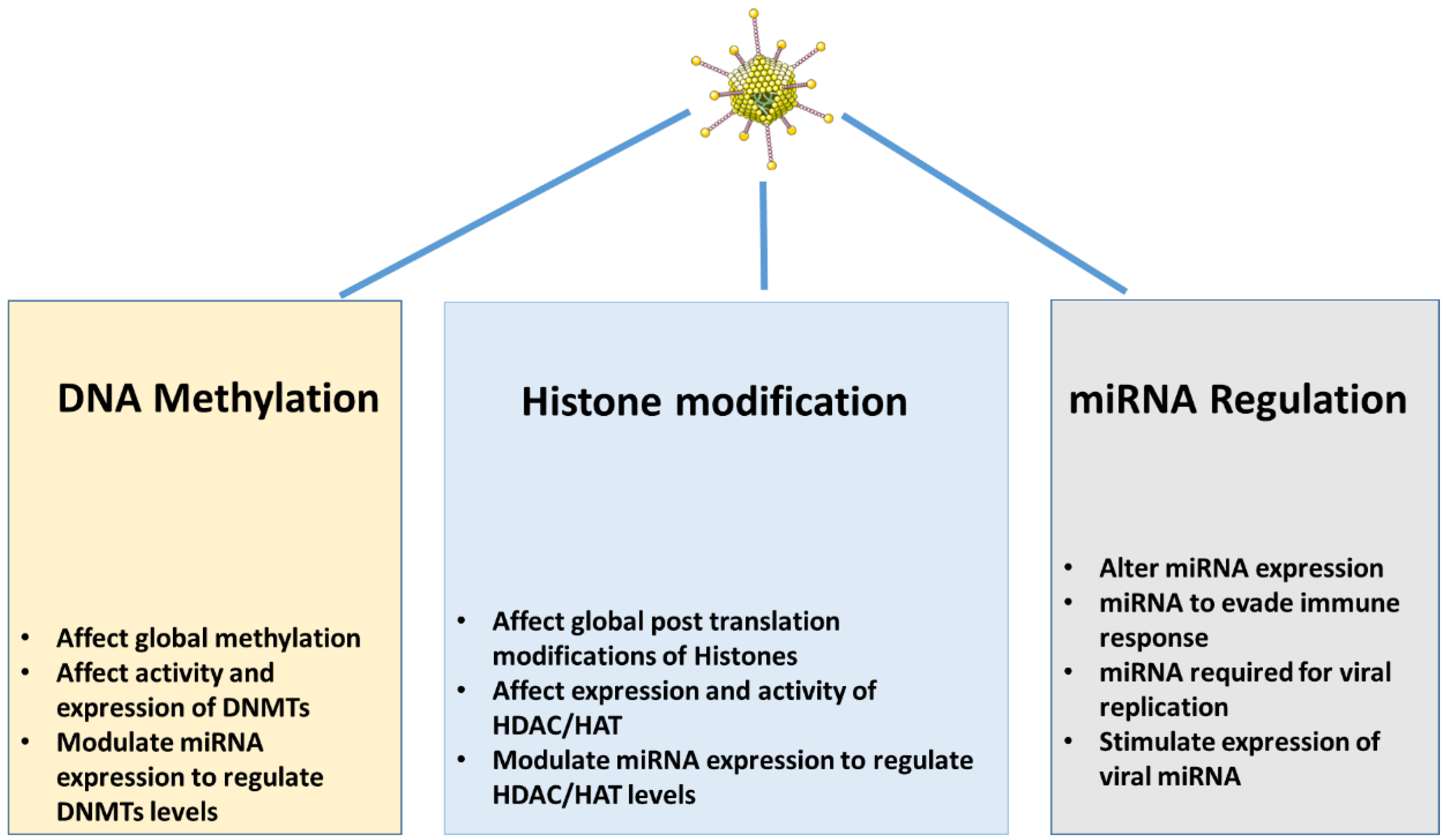
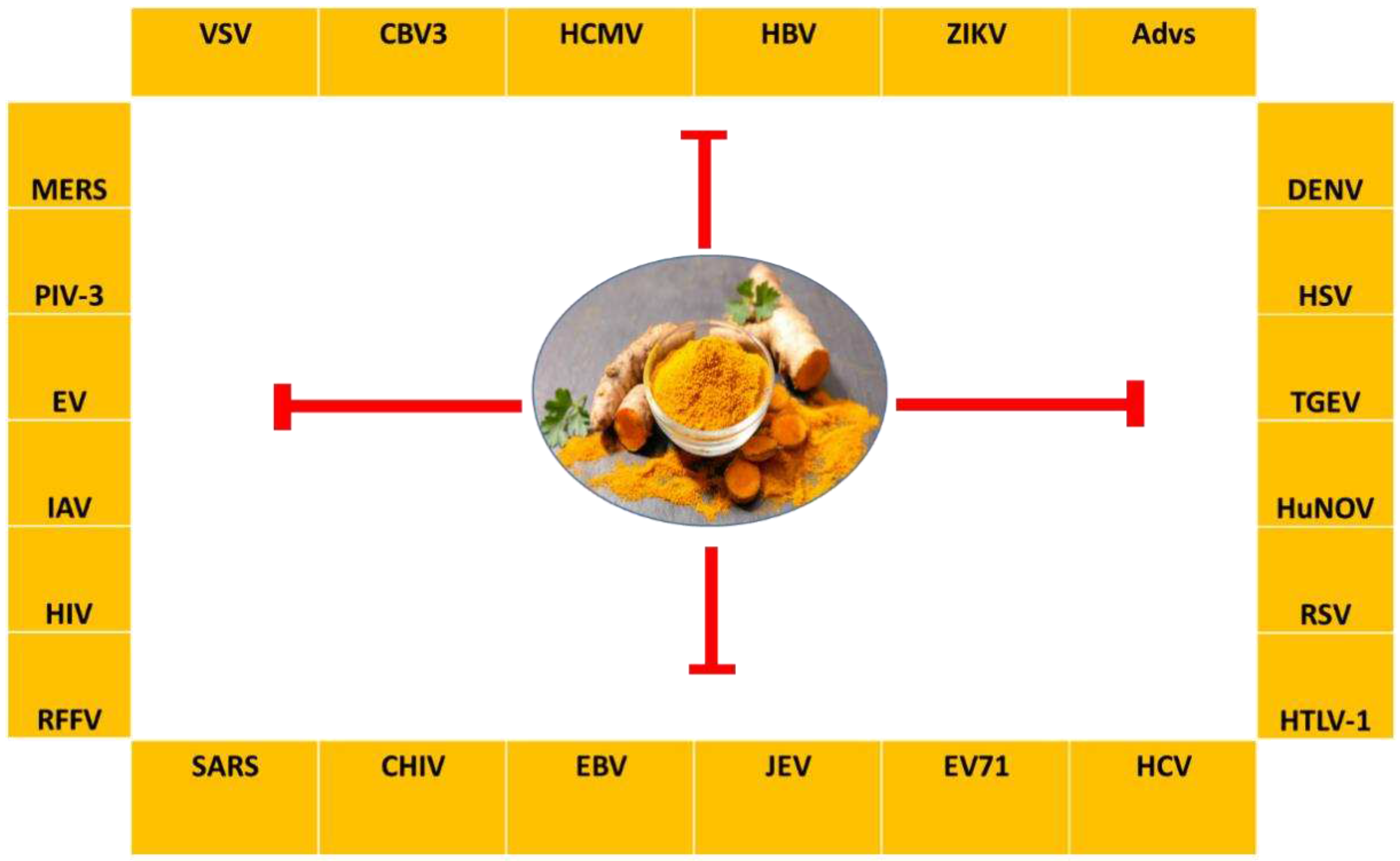
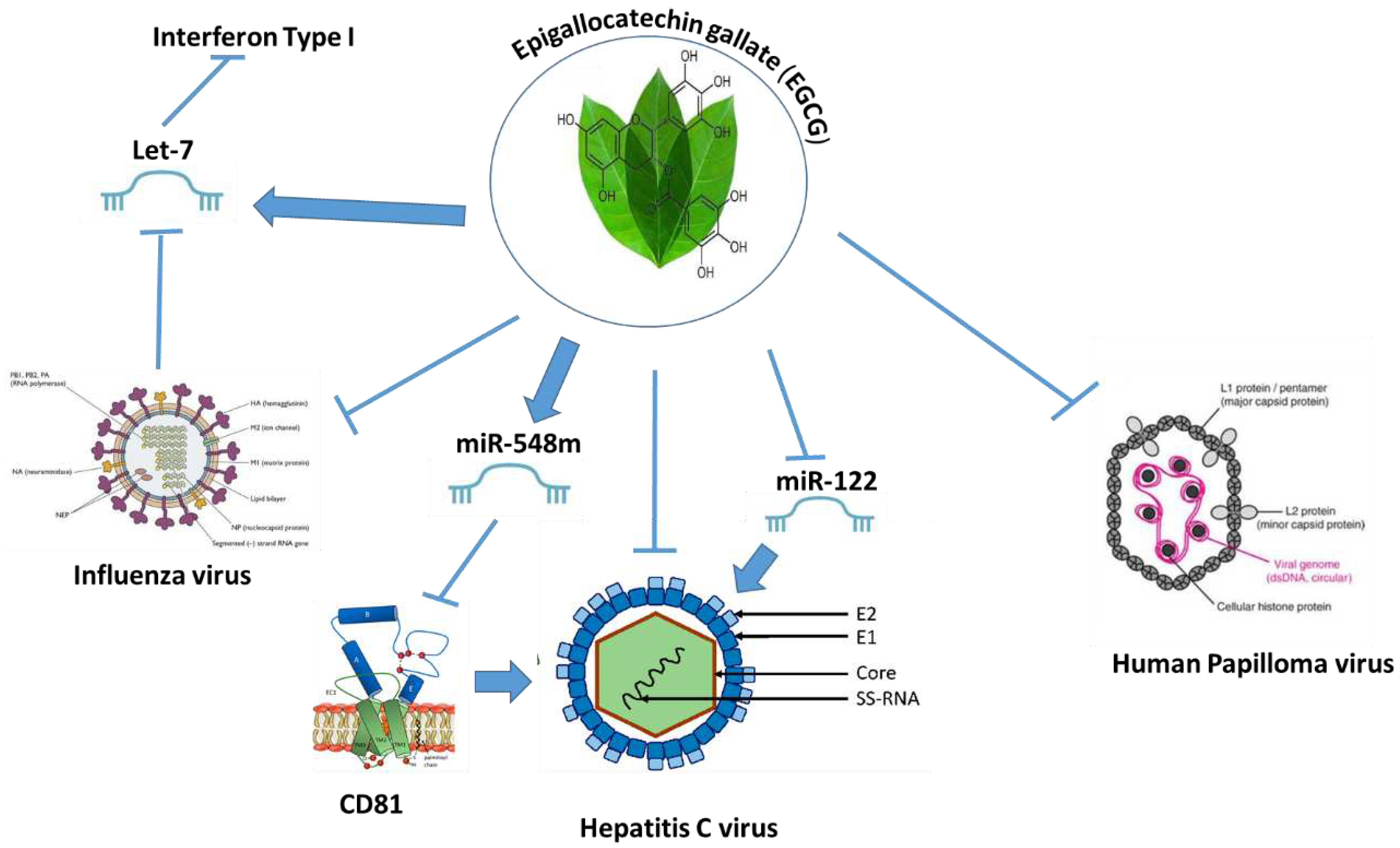
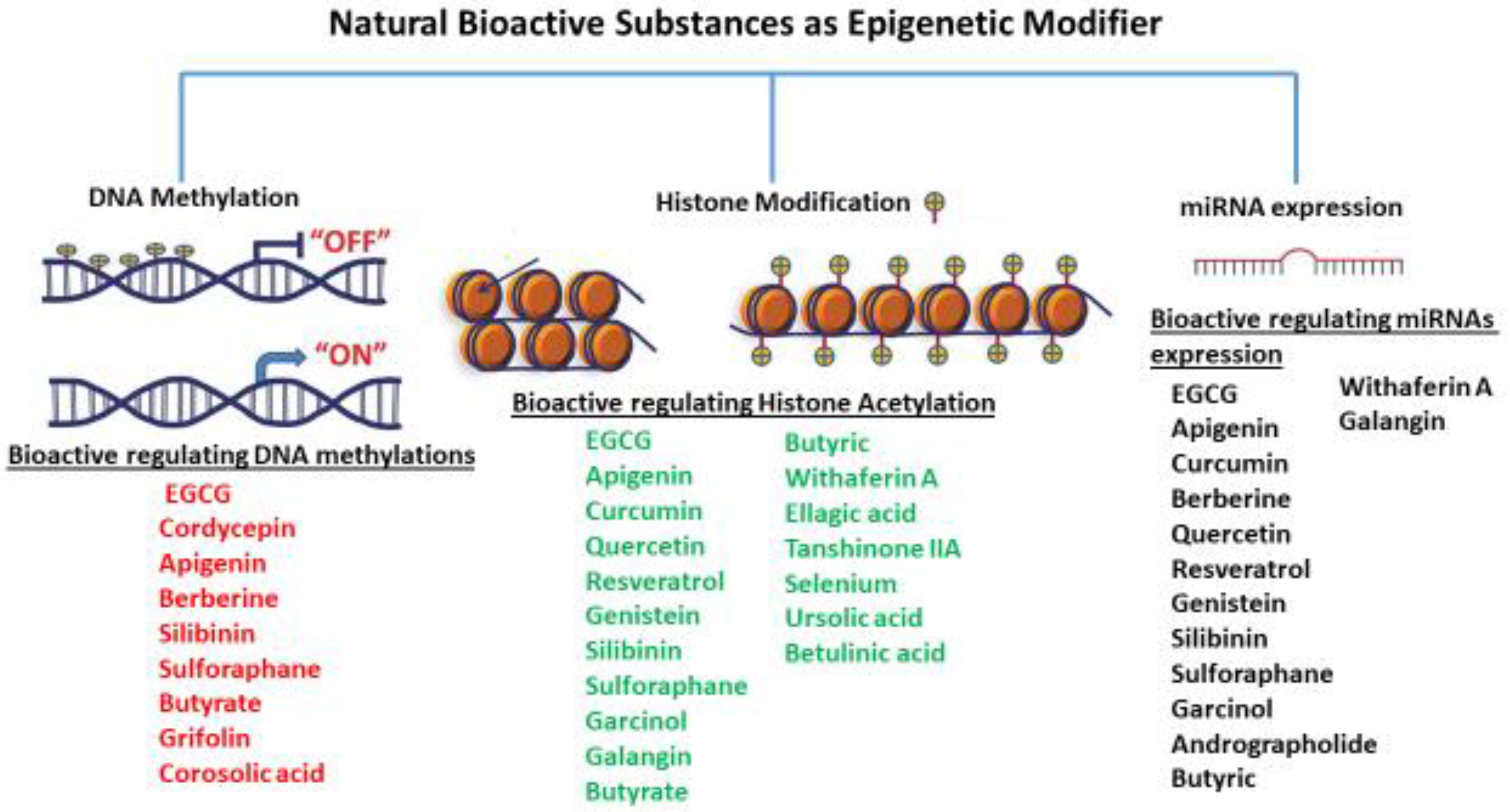
| Plant | Plant part | Main bioactive constituents | Virus | IC50 a | SIb | Mode of action | References |
|---|---|---|---|---|---|---|---|
| Aloysia citriodora Palau (Verbenaceae) | nr | geranial (18.9%), neral (15.6%), limonene (10.7%), 1,8-cineole (5.0%), spathulenol (4.7%), geraniol (2.7%) | Yellow fever virus | 19.4 μg/mL | 2.6 | nrc | [142] |
| Artemisia kermanensis Podlech (Compositae) | aerial parts | α-thujone (13.8%), camphore (10.2%), β-thujone (6.2%), p-Mentha-1,5-dien-8-ol (4.4%) | HSV-1 | 0.004% | 66.4 | nr | [130] |
| Ayapana triplinervis (Vahl) R.M.King & H.Rob. (Compositae) | aerial parts | thymohydroquinone dimethyl ether (87.1%), α-phellandrene (2.0%), β-selinene (1.9%) | Zika virus | 38 μg/mL | 12.5 | inhibitor of internalization process | [143] |
| Cananga odorata (Lam.) Hook.f. & Thomson (Annonaceae) | nr | benzyl salicylate (49.3%), benzyl benzoate (18.7%), linalool (16.6%), α-gurjunene (7.1%) | HIV-1 | 0.60 µg/mL | nr | nr | [131] |
| Cinnamomum zeylanicum Blume (Lauraceae) | leaves | eugenol (nr) | H1N1 | < 3.1 µL/mL | > 4 | intercellular | [144] |
| Citrus × bergamia Risso & Poit. (Rutaceae) | fruit peel | (–)-linalyl acetate (nr), (–)-linalool (nr), (+)-limonene (nr), γ-terpinene (nr), β-pinene (nr), α-pinene (nr), α-terpinene (nr) | H1N1 | < 3.1 µL/mL | > 5 | intercellular | [144] |
| Cymbopogon citratus (DC.) Stapf (Poaceae) | nr | cis-citral (59.2%), β-pinene (22.5%), cis-verbenol (6.1%), nerol (5.0%) | HIV-1 | 0.61 µg/mL | 1.1 | nr | [131] |
| Cymbopogon flexuosus (Nees ex Steud.) W.Watson (Poaceae) | grass | geranial (nr), neral (nr) | H1N1 | < 3.1 µL/mL | > 4 | nr | [144] |
| Cymbopogon nardus (L.) Rendle (Poaceae) | nr | nr | HIV-1 | 1.2 mg/mL | nr | nr | [132] |
| Dysphania ambrosioides (L.) Mosyakin & Clemants (Amaranthaceae) | aerial parts | cis-ascaridole (60.3%), m-cymene (22.2%), α-terpinene (1.8 %), thymol (1.1%) | Coxsackie virus B4 | 21.7 μg/mL | 74.3 | nr | [145] |
| Eucalyptus caesia Benth. (Myrtaceae) | aerial parts | 1,8-cineole (40.2%), p-cymene (14.1%), γ-terpinene (12.4%), α-pinene (7.7%), terpinen-4-ol (5.6%) | HSV-1 | 0.007% | 38.8 | nr | [130] |
| Eucalyptus globulus Labill. (Myrtaceae) | leaves | 1,8-cineole (nr), α-pinene (nr) | H1N1 | < 50µL/mL | > 0.5 | intercellular | [131] |
| 1,8-cineole (68.0%), globulol (5.4%), trans-pinocarveol (4.6%), α-pinene (3.7%) | Coxsackie virus B3 | 0.7 mg/mL | 22.8 | intercellular | [146] | ||
| Fortunella margarita Lour. Swingle (Rutaceae) | fruits | terpineol (55.5%), τ-carveol (5.5%), limonene (1.7%), muurolene (5.5%), cadinene (2.0%) | H5N1 | 6.8 µg/mL | nr | nr | [147] |
| Illicium verum Hooker f. (Schisandraceae) | fruits | (E)-anethole (80.0%) | HSV-1 | 1 µg/mL | 160 | intercellular | [129] |
| Lallemantia royleana (Benth.) Benth. (Lamiaceae) | aerial parts | (E)-pinocarvyl acetate (26.0%), pinocarvone (20.0%), verbenone (7.1%), (E)-β-ocimene (4.1%), (E)-carveol (5.3%), 3-thujen-2-one (5.1%), pulegone (4.4%), (Z)-carveol (3.5%), linalool (3.4%) | HSV-1 | 0.011% | 6.4 | intercellular | [148] |
| Lavandula officinalis Chaix (Lamiaceae) | flowers | linalyl acetate (nr), linalool (nr) | H1N1 | < 3.1 µL/mL | > 8 | intercellular | [131] |
| Lippia alba (Mill.) N.E.Br. ex Britton & P.Wilson (Verbenaceae) | nr | carvone (39.7%), limonene (30.6%), bicyclosesquiphellandrene (8.9%), piperitenone (4.5%), piperitone (2.8%), β-bourbonene (1.7%) | Yellow fever virus | 4.3 μg/mL | 30.6 | intercellular and intracellular | [142] |
| Lippia graveolens Kunth (Verbenaceae) | nr | carvacrol (56.8%), o-cymene (32.2%), γ-terpinene (3.7%) | HSV-1 | 99.6 µg/mL | 7.4 | intercellular | [149] |
| ACVR-HHV-1 | 55.9 µg/mL | 13.1 | intercellular | ||||
| Bovine viral diarrhoea virus | 78 μg/mL | 7.2 | intracellular | ||||
| Respiratory syncytial virus | 68 μg/mL | 10.8 | intercellular | ||||
| Bovine herpes virus 2 | 58.4 μg/mL | 9.7 | intercellular and intracellular | ||||
| Melaleuca alternifolia (Maiden & Betche) Cheel (Myrtaceae) | leaves | nr | HSV-1 | 13.2 µg/mL | 43 | intracellular | [150] |
| Mentha suaveolens Ehrh. (Lamiaceae) | leaves | piperitenone oxide (86.8%), α-cubebene (2.1%), pulegone (1.4%), limonene (1.4%), caryophyllene (1.3%) | HSV-1 | 5.1 µg/mL | 67 | intracellular | [151] |
| Osmunda regalis L. (Osmundaceae) | aerial parts | hexahydrofarnesyl acetone (11.8%), 2,4-di-t-butylphenol (6.8%), phytol (6.5%), neophytadiene (4.6%), 1-octadecene (4.4%), 1- eicosene (4.4%), 1-hexadecene (4.1%) | Coxsackie virus B4 | 2.2 μg/mL | 789.8 | nr | [152] |
| Pelargonium graveolens L'Hér. (Geraniaceae) | flowering aerial parts | citronellol (nr), geraniol (nr) | H1N1 | < 3.1 µL/mL | > 21 | intercellular | [131] |
| Pulicaria vulgaris Gaertn. (Compositae) | aerial parts | thymol (50.2%), p-menth-6-en-2-one (carvotanacetone, 20.2%), thymol isobutyrate (16.9%), menthan-2-one (4.3%) | HSV-1 | 0.001% | 1 | intercellular | [148] |
| Rosmarinus officinalis L. (Lamiaceae) | aerial parts | α-pinene (23.9%), verbenon (15.4%), camphor (11.0%), p-cymene (7.5%), 3-octanone (5.6%) | HSV-1 | 0.006% | 46.1 | nr | [130] |
| eucaliptol (50.6%), camphor (13.3%), α-pinene (10.1%), β-pinene (7.7%), camphene (4.6%) | HIV-1 | 0.18 µg/mL | 3.6 | nr | [131] | ||
| Salvia desoleana Atzei & V.Picci (Lamiaceae) | aerial parts | linalyl acetate (30.2%), germacrene D (18.7%), α-terpinyl acetate (16.8%), 1,8-cineole (10.2%), linalool (5.1%) | Acyclovir-resistant HSV-2 | 28.6 µg/mL | 55.2 | intercellular and intracellular | [153] |
| Satureja hortensis L. (Lamiaceae) | aerial parts | carvacrol (32.4%), γ-terpinene (32.0%), thymol (10.0%), p-cymene (6.6%), α-pinene (4.3%) | HSV-1 | 0.008% | 32.2 | nr | [130] |
| Sinapis arvensis L. (Brassicaceae) | flowers | cubenol (14.3%), 2-phyenyl isothiocyanate (7.5), dimethyl trisulfide (5.2%), thymol (4.6%), δ-cadinene (3.4%) | HSV-1 | 0.035% | 1.5 | intercellular | [148] |
| Thymbra capitata (L.) Cav. (Lamiaceae) | aerial parts | cinnamaldehyde (nr), carvacrol (nr) | HSV-1 | 17.6 µg/mL | 6.0 | intercellular | [154] |
| HSV-2 | 18.6 µg/mL | 6.9 | intercellular | ||||
| Thymus vulgaris L. (Lamiaceae) | aerial parts | 1,8-cineole (nr), terpenyl acetate (nr), borneol (nr) | H1N1 | < 3.1 µL/mL | > 4 | intercellular | [144] |
| thymol (37.8%), iso-thymol (23.2%), γ-terpinene (13.2%), β-caryophyllene (4.2%), linalool (3.3%) | HIV-1 | 1.30 | 1.6 | nr | [131] |
||
| Zataria multiflora Boiss. (Lamiaceae) | aerial parts | thymol (33.1%), carvacrol (25.9%), p-cymene (11.3%), α-pinene (3.9%) | HSV-1 | 0.003% | 55.4 | nr | [130] |
| Plant | Plant part | Extract | Main bioactive constituents | Virus | IC50 a | SIb | Mode of action | References |
| Agrimonia pilosa Ledeb. (Rosaceae) | whole plant | ethanol extract | nr | H1N1, HV A-B |
0.5-1 μg/mL | nr | block uncoating process | [155] |
| Aloe vera (L.) Burm.f. (Xanthorrhoeaceae) | leaf | Gel | nr | HSV-1 | 5% | nr | replication inhibitor | [156] |
| Arachis hypogaea L. (Leguminosae) | peanut skin | ethanol extract | nr | H1N1 | 1.3 μg/mL | 5.2 | early stages of infection inhib. | [157] |
| Avicennia marina (Forssk.) Vierh. (Acanthaceae) | leaf | methanol extract | nr | HSV-1 | 9 μg/mL | 9.1 | viral replication inhib. | [158] |
| HIV | 15 μg/mL | 5.2 | interference with replication cycle | |||||
| Centella asiatica (L.) Urb. (Apiaceae) | leaf | water extract | nr | HIV | 36 μg/mL | nr | immunomodulatory effect | [159] |
| alcoholic extract | HIV | 8 μg/mL | ||||||
| Combretum adenogonium Steud. ex A.Rich. (Combretaceae) | root | water/ethanol extract | nr | HIV-1 | 24.7 μg/mL | nr | protease inhibitor | [160] |
| Copaifera reticulate Ducke (Leguminosae) | stem, barkand leaf | water/ethanol extract | phenolics, alcohols, organic acids | HSV-2 | 50 μg/mL | nr | block virus attachment | [161] |
| Cornus canadensis L. (Cornaceae) | leaf | water/ethanol extract | tellimagrandin I and other hydrolysable tannins | HSV-1 | 9 μg/mL | nr | virus absorption inhibitor | [162] |
| Embelia ribes Burm.f. (Primulaceae) | fruit | ethylacetate extract | embelin | H1N1 | 0.2 μg/mL | 32 | block virus entry | [163] |
| Epimedium koreanum Nakai (Berberidaceae) | bark | water extract | nr | HSV | 0.20 μg/mL | 23.5 | immunomodulatory effect | [164] |
| Equisetum giganteum L. (Equisetaceae) | root and stem | water/ethanol extract | phenolics, alcohols, organic acids | HSV-2 | 18 μg/mL | nr | block virus attachment | [161] |
| Eupatorium perfoliatum L. (Compositae) | aerial part | hydroalcoholic extract | nr | H1N1 | 7 μg/mL | 52 | viral attachment inhibitor | [165] |
| Euphorbia hirta L. (Euphorbiaceae) | aerial part | methanol extract | nr | HIV-1 | 38 μg/mL | nr | RT inhib. | [166] |
| HIV-2 | 22 μg/mL | |||||||
| Ficus religiosa L. (Moraceae) | bark | methanol extract | nr | HSV-2 | 5.2 μg/mL | 31.1 | nr | [167] |
| Hemidesmus indicus L. Br. ex Schult. (Apocynaceae) | root | water/methanol extract | 2-hydroxy-4-methoxybenzaldehyde (0.41 mg/g), 3-hydroxy-4- methoxybenzaldehyde (0.16 mg/g) | HSV-1 | 66.8 μg/mL | nr | anti-ER α-glucosidase inhib. | [168] |
| 2-hydroxy-4-methoxybenzaldehyde (0.41 mg/g), 3-hydroxy-4- methoxybenzaldehyde (0.16 mg/g) | HSV-2 | 70.6 μg/mL | ||||||
| Jatropha multifida L. (Euphorbiaceae) | stem | water extract | nr | H1N1 | 25 μg/mL | nr | block virus entry | [169] |
| Paeonia lactiflora Pall. (Paeoniaceae) | root | ethanol extract | nr | H1N1 | 0.016 mg/mL | 13.5 | block several stages of infection | [170] |
| Pedilanthus tithymaloides (L.) Poit. (Euphorbiaceae) | leaf | methanol extract | 2-(3,4-dihydroxy-phenyl)-5,7-dihydroxy-chromen-4-one or luteolin | HSV-2 | 48.5 μg/mL | 9.0 | NF-B signalling pathway modulation | [171] |
| Prunella vulgaris L. (Lamiaceae) | flowers | water extract | nr | HIV | 0.8 μg/mL | nr | early post-virus binding interference | [172] |
| Prunus dulcis (Mill.) DA Webb (Rosaceae) | almond skin | methanol/HCl extract | nr | HSV-1 | 0.04 mg/mL | nr | block virus entry | [173] |
| Quercus brantii Lindl. (Fagaceae) | fruit | chloroform extract | nr | HSV-1 | 2.9 μg/mL | nr | block virus entry | [174] |
| Quercus persica Jaub. & Spach (Fagaceae) | fruit | water/ethanol extract | nr | HSV-1 | 0.26 μg/ml | nr | attachment inhib. | [164] |
| Rhus aromatica Aiton (Anacardiaceae) | root/stem bark | water extract | gallic acid | HSV-1 | 0.0005 % | nr | nr | [175] |
| Rhus aromatica Aiton (Anacardiaceae) | root/stem bark | water extract | gallic acid | HSV-2 | 0.0043% | nr | nr | [175] |
| Schinus terebinthifolia Raddi (Anacardiaceae) | bark | water/ethanol extract | condensed tannins (catechin, 5.4 mg/L) | HSV-1 | 0.21 μg/mL | < 49.0 | virucidal effect | [176] |
| Solanum melongena L. (Solenaceae) | peel | ethanol/HCl extract | delphinidin-3-rutinoside (90.3-115.0 μg/mg), chlorogenic acid (24.5-60.7 μg/mg) | HSV-1 | 83.4 μg/mL | nr | reduction of viral protein expression | [177] |
| Strychnos pseudoquina A. St.-Hil. (Loganiaceae) | stem bark | ethyl acetate extract | nr | HSV-1 | 5.29 μg/mL | nr | interference with various step of virus cycle | [178] |
| Strychnos pseudoquina A. St.-Hil. (Loganiaceae) | stem bark | ethyl acetate extract | nr | HSV-2 | 6.55 μg/mL | nr | interference with various step of virus cycle | [178] |
| Tanacetum parthenium (L.) Sch.Bip. (Compositae) | aerial parts | water/ethanol extract | chlorogenic acid, flavonoids (aglycones and glycosylated flavonoids), parthenolide | HSV-1 | 3.1 μg/mL | nr | viral replication inhib. | [179] |
| Taxodium distichum (L.) Rich. (Cupressaceae) | cone, leaf, stem | water extract | nr | H1N1 | 0.05 mg/mL | 5.6 | block virus entry | [180] |
| Vachellia nilotica (L.) P.J.H. Hurter & Mabb. (Leguminosae) | bark | methanol extract | nr | HSV-2 | 4.71 μg/mL | 30.6 | block virus attachment | [181] |
| Vachellia nilotica (L.) P.J.H. Hurter & Mabb. (Leguminosae) | bark | methanol extract | nr | HPV-16 | 1.80 μg/mL | 32.6 | block virus attachment | [181] |
| Vachellia nilotica (L.) P.J.H. Hurter & Mabb. (Leguminosae) | bark | methanol extract | nr | HSV-2 acyclovir resistant | 6.71 µg/mL | 21.5 | block virus attachment | [181] |
| Vigna radiata (L.) R.Wilczek (Leguminosae) | sprout | methanol/HCl extract | nr | HSV-1 | 7.62 μg/mL | nr | virucidal effect | [182] |
| Name | Structure | Anti-viral activity and mode of action |
| Andrographolide | 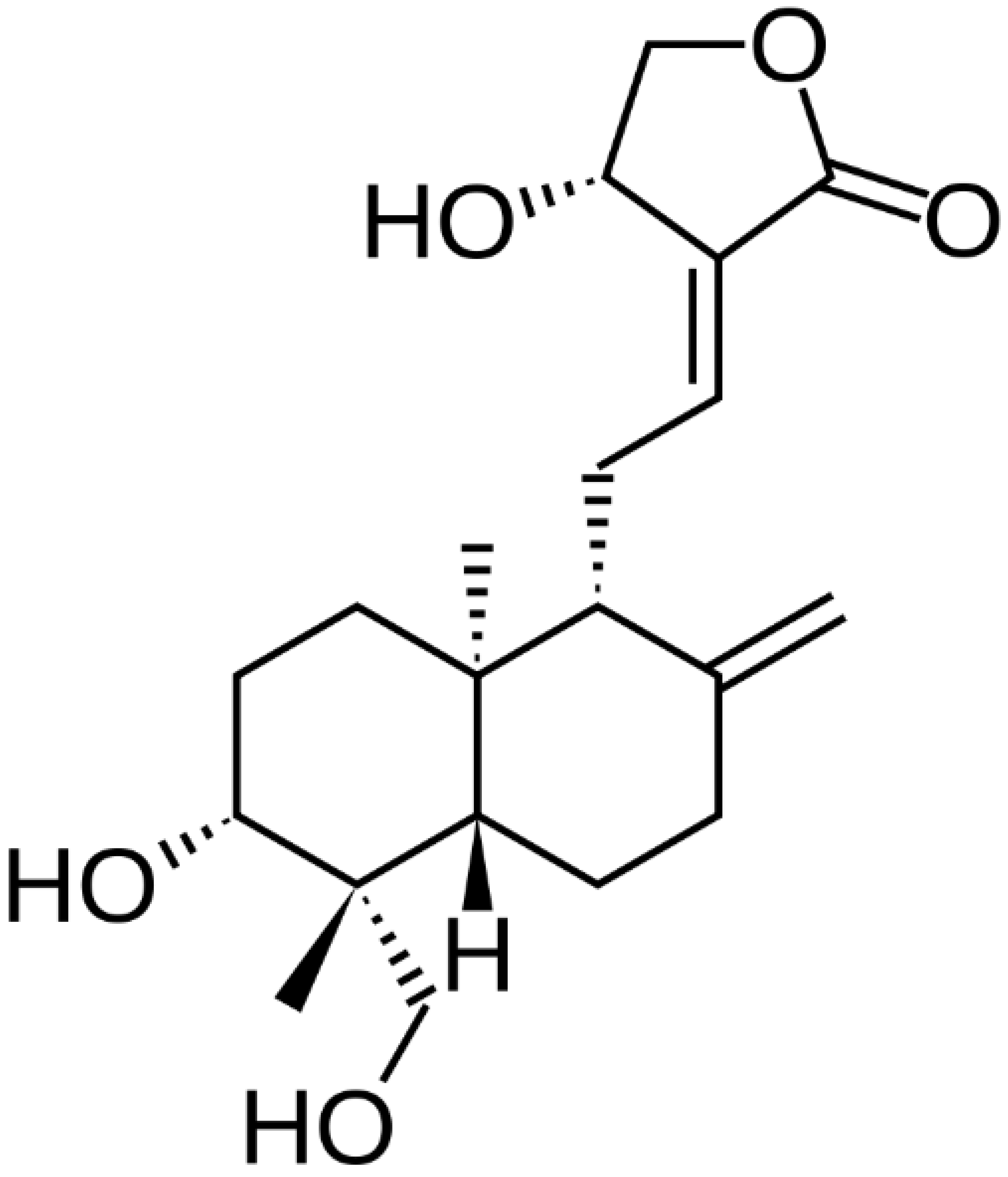 |
Modulates miR-377 to regulate HO-1. Downregulates miR-433 to regulate glutathione cysteine ligase. Upregulates miR-17, miR-224 and miR-181a [183]. Inhibits virus replications such HIV, HSV-1, HBV, HCV,ZIKV, DENV, CHIKV, FMDV, and IAV [184] [185,186]. Prevents EBV reactivation by suppressing EBV lytic genes via histone modifications [187]. Also prevents IVA-induced inflammation through modulation of NF-κB and JAK-STAT signaling pathways [188]. |
| Apigenin | A potent inhibitor of HDAC1, 3 [189]. Exhibits antiviral properties against IAV, HRV, HSV, enterovirus, HBV, HCV, EV71, CVB1 and SARS-COV2 [190,191,192,193] [194]. Anti-viral activity is attributed by inhibiting HDAC activity and chromatin remodeling [189,195]. Downregulates miR-103 [196], miR-155 [113]and miR-122 [197] to affect HCV and HIV replication [115,116,117]. |
|
| Baicalein | 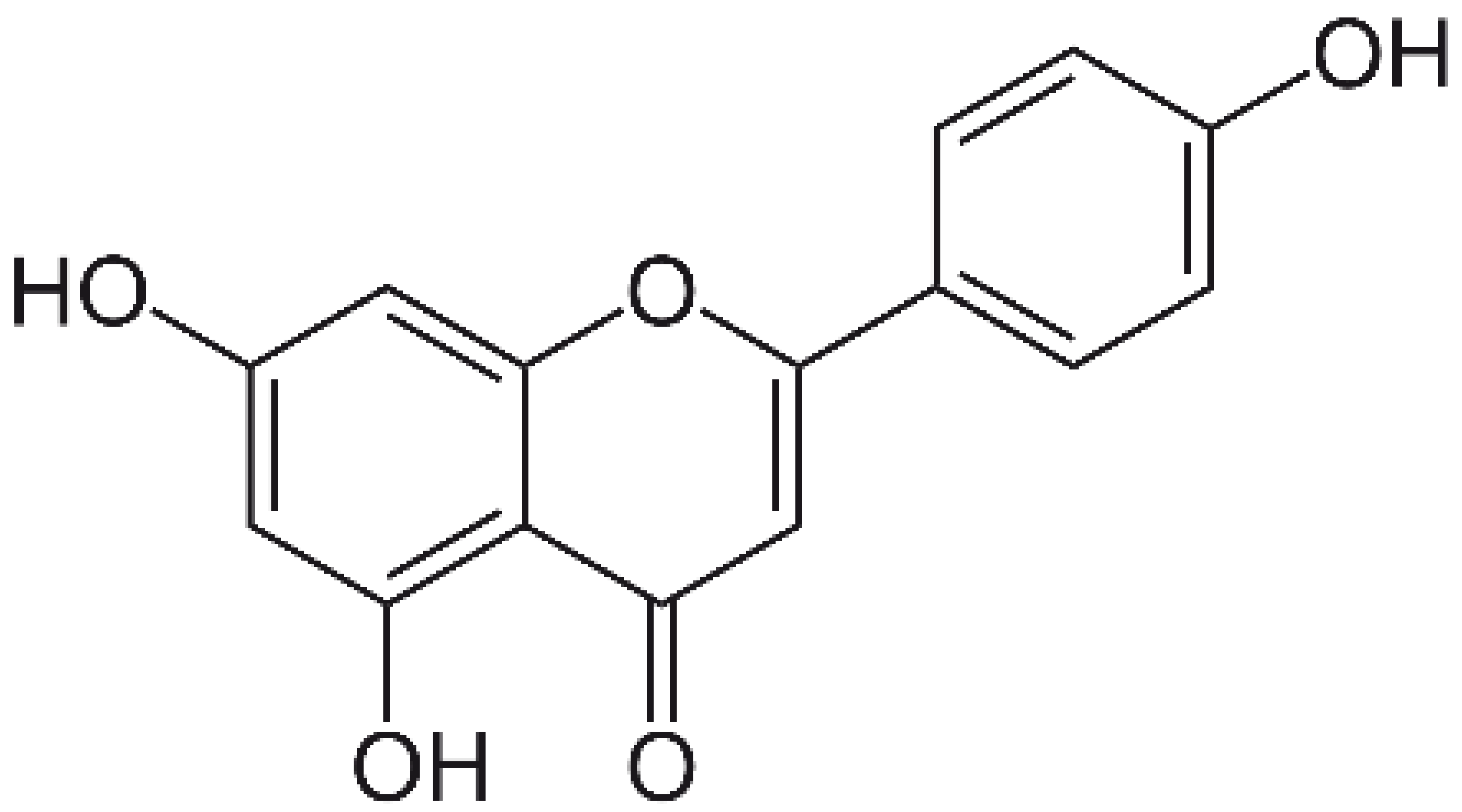 |
Inhibits HBV [198], HIV [199] , DENV [200], HSV-1 [201]. Inhibits DNMT and HDAC and affect influence epigenetic modifications [202,203]. Upregulates miR-3,178 [204] which target HDAC10 [203]. Exhibits potent antiviral activity against DENV [200] |
| Berberine | Regulates AMPK activity [205] and suppresses of SIRT1 deacetylases [206]. inhibits miR-21 expression [207]. inhibits replication of HSV [208], HCMV [209], HPV [210], DENV [211], HIV [212], HCV [213], IAV [214] and SARS-COV2 [215]. Also inhibits p38 MAPK activity [38] which might attributed for anti-viral activity [41], [41], [42], [43,44] [45], [216]. |
|
| Betulinic acid | 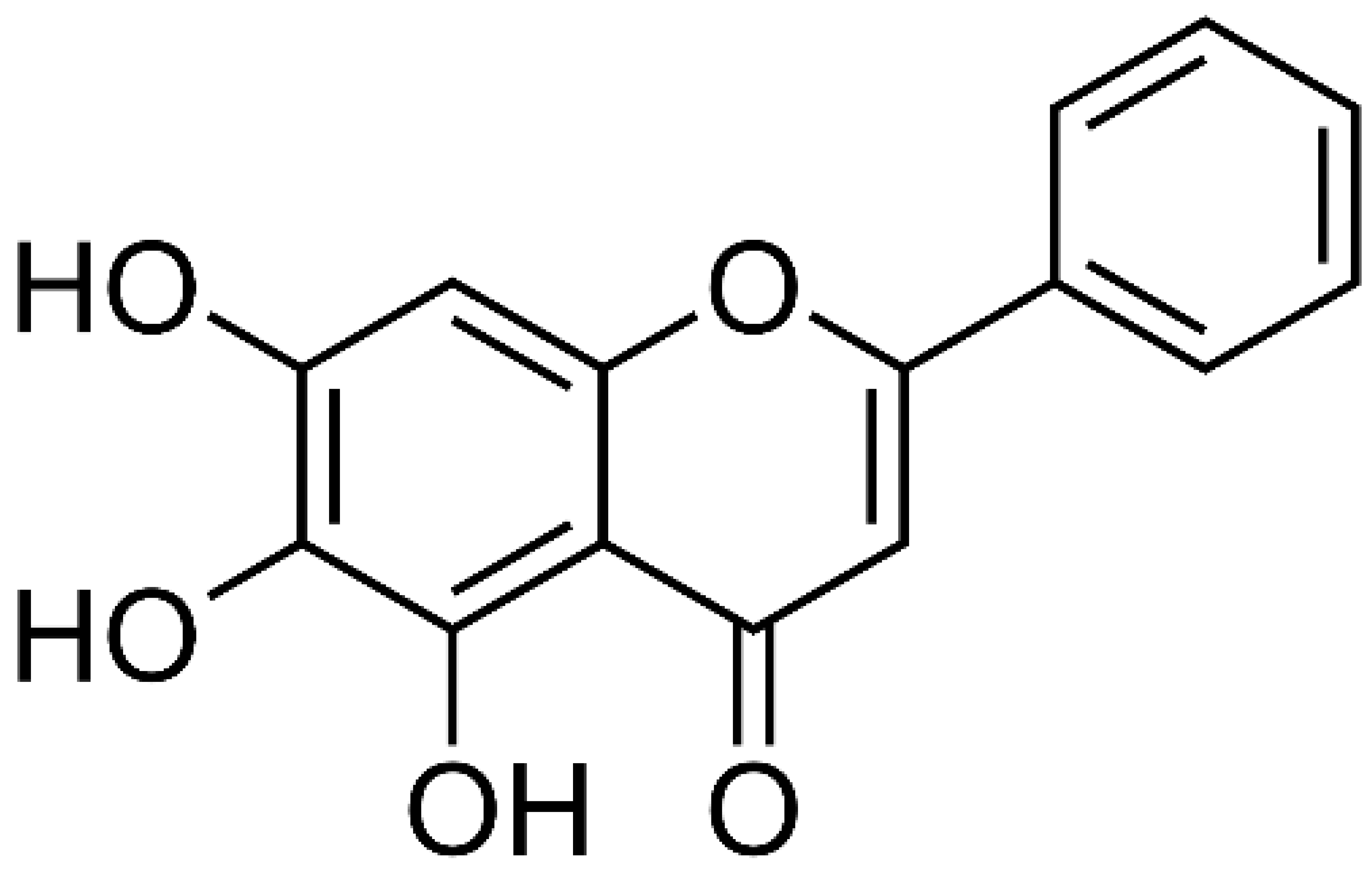 |
A computational approach predicted the capacity to alter HDAC6 and HDAC10 activity [217]. Inhibits DENV-2 NS5 polymerase activity [218] and HBV replication [219]. C-3 esterification of lead resulted in the discovery of Bevirimat, an HIV-1 maturation inhibitor. |
| Butyric acid |
Induces HDAC expression and activity by upregulation of miR-203 promoter methylation [220]. Inhibits HBV replication by reducing HBx protein expression, HBV-DNA and hepatitis B surface antigen (HBsAg) [221]. |
|
| Cardamonin | 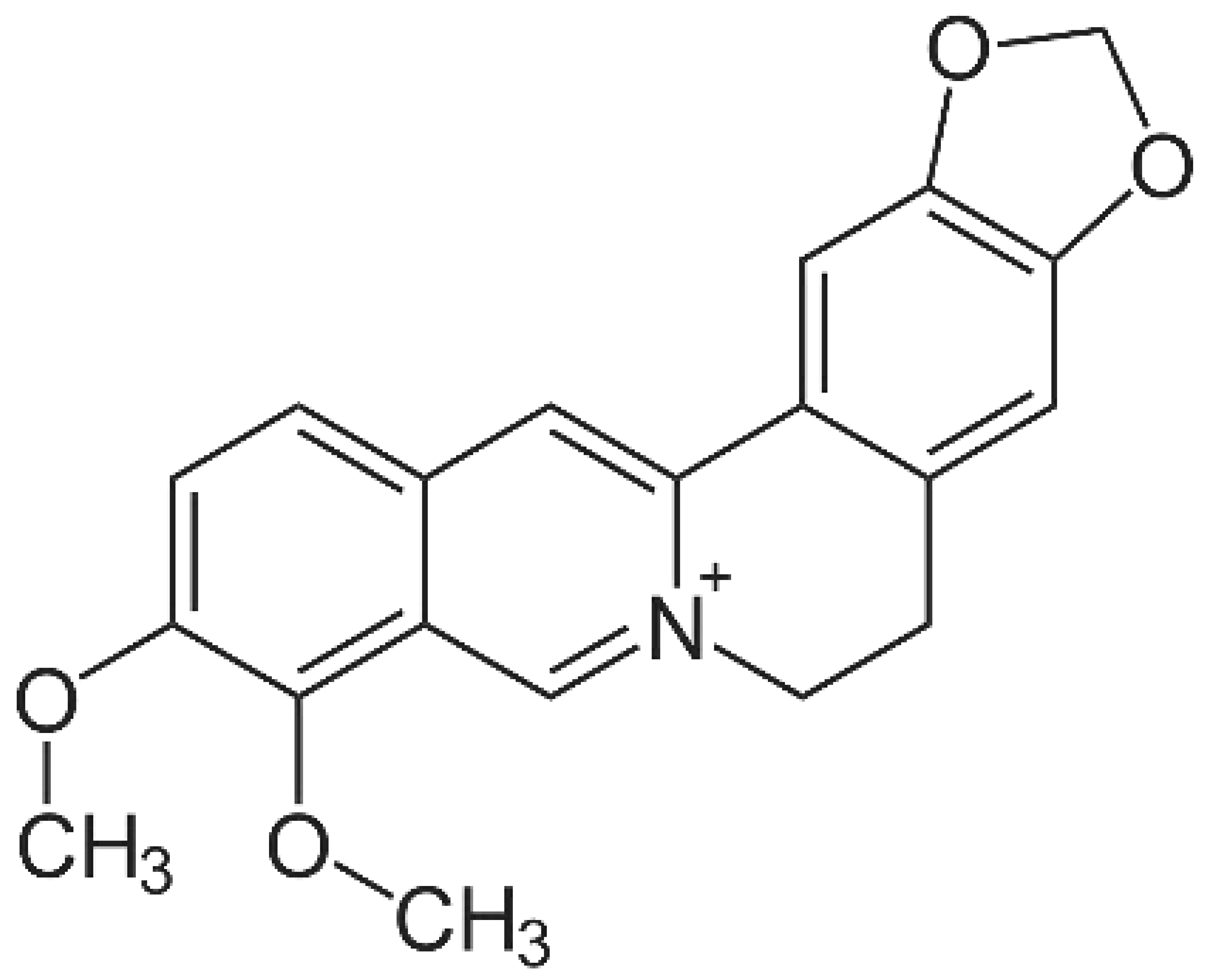 |
Exhibits antiviral action against the human coronavirus HCoV-OC43 which was mediated by induction of p38 MAPK signaling pathway [222]. |
| Cordycepin | Promotes methylation of EBV genomic sites near Fp/Qp promoters. Increases DNMT3 [223] and reducing EBV replication [224]. As adenosine derivative exhibits antiviral activity against several viruses including IAV, plant viruses, HIV, murine leukemia virus, EBV [225,226,227,228,229] and COVID-19 [230]. |
|
| Corosolic acid | Alters CpG methylation sites, resulting in altered gene expression [231]. Increases the expression of acetylated histone H3 lysine 27 (H3K27ac), while decreasing histone H3 lysine 27 trimethylation (H3K27Me3) [232]. Exhibits anti-viral activity against a number of viruses were reported [233] . |
|
| Curcumin | 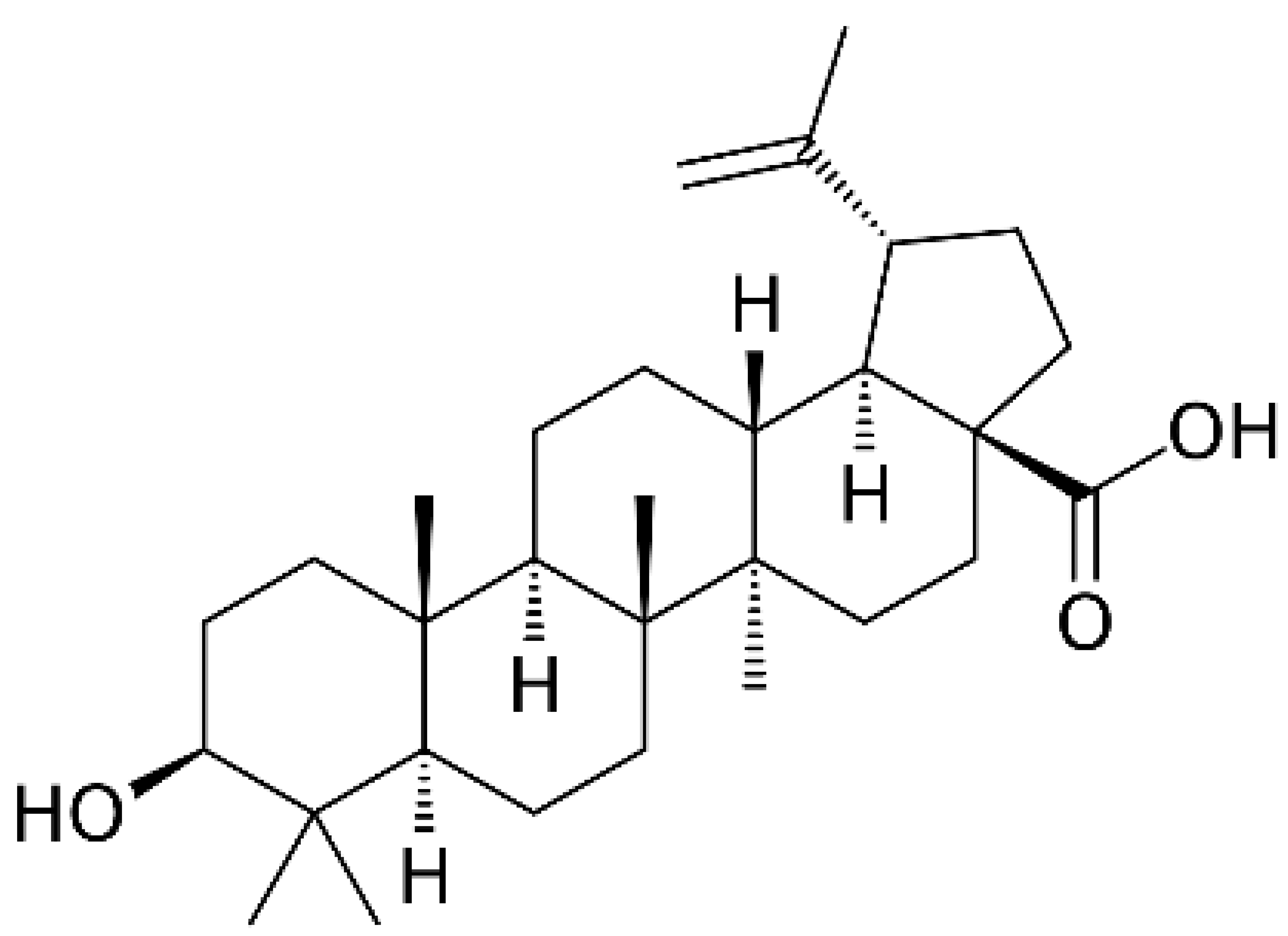 |
Decreases the expression of HDAC1, HDAC3, HDAC8 and histone acetyltransferase p300 while enhancing the expression of Ac-histone H4 protein [234]. Reduces HAT activity and inhibits DNMT [235]. Inhibition of HBV replication was attributed to a decrease in the acetylation level of cccDNA-bound histone H3 and H4 [236]. Downregulates miR-350, miR-17-2-3p, let 7e-3p, miR-1224, miR-466b-1-3p, miR-18a-5p, and miR-322-5p Upregulates miR-122-5p, miR-3473, miR-182, and miR-344a-3p [237]. inhibits the replication of various viruses, including HBV [236], HIV [238], IAV [239], HPV-18 [240], ZIKV, CHIKV, VSV, CVB3, EV71, RSV, HSV-2, KSHV and HAdV [241]. |
| Ellagic acid | Increases HDACs gene expression and histone arginine methylation [242], Decreases H3K9 acetylation and HDAC9 dissociation [242]. Inhibits SARS-CoV-2 viral entry, and replication [243]. |
|
| Epigallocatechin gallate (EGCG) | 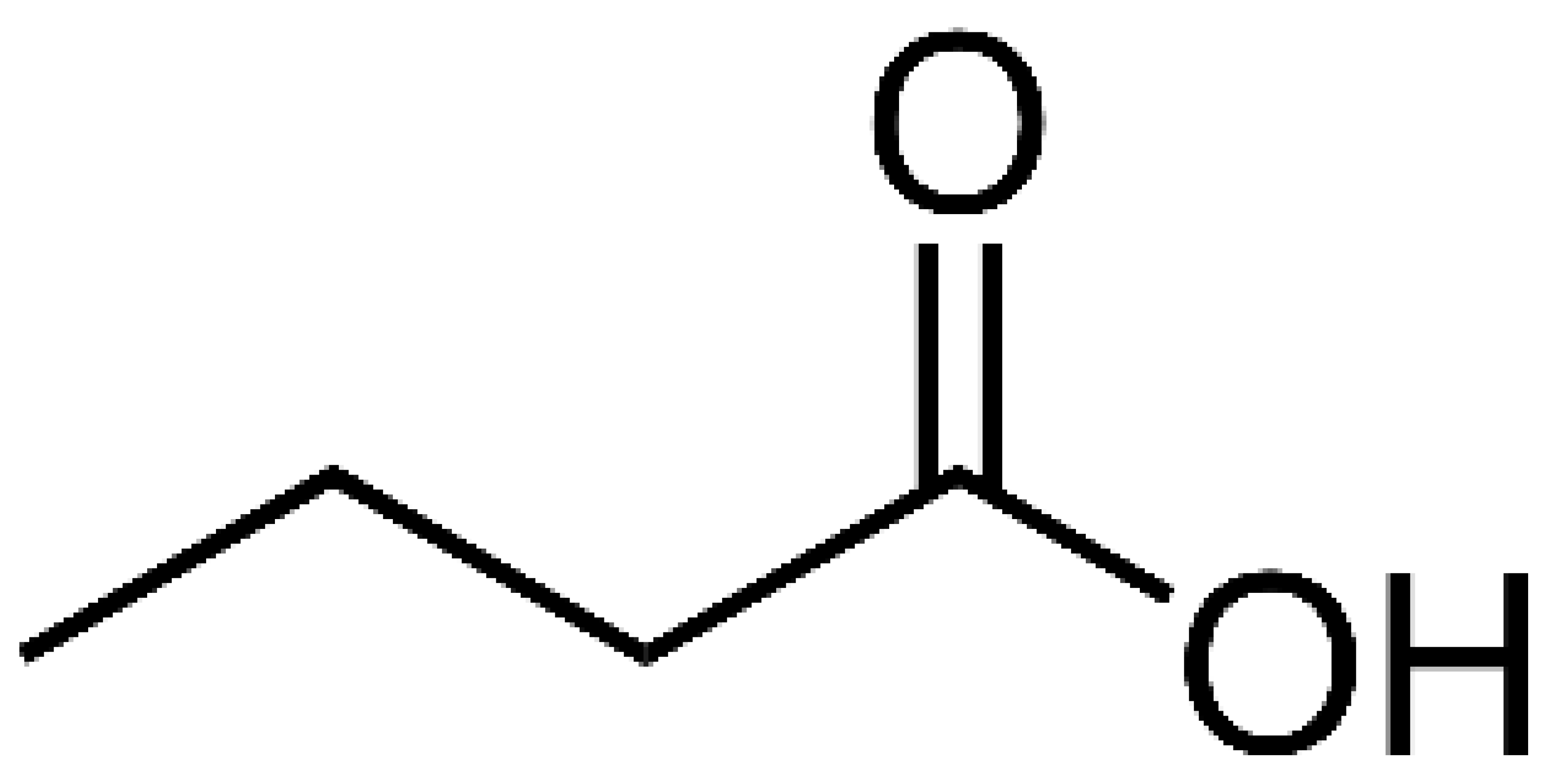 |
Inhibits the activity of DNMT 1, DNMT 3a, DNMT 3b [244] and HDACs [245] and downregulates the expression of HDAC1, HDAC2, and HDAC3 expression [246]. Inhibits HAT activity [247]. Decreases the levels of let-7e-5p, miR-103a-3p, miR-151a-5p, miR-195-5p, miR-222-3p, miR-23a-3p, miR-23b-3p, miR-26a-5p, miR-27a-3p, miR-29b-3p, miR-3195, miR-3651, miR-4281, miR-4459, miR-4516, miR-762, and miR-125b-5p [248]. Induces the expression of miR-3663-3p, miR-1181, miR-3613-3p, miR1281, miR-1539, miR-221-5p, miR-374b, miR-4306, miR-500a-5p, miR590-5p miR-140-3p and miR-221 [249] [250,251]. Inhibits the replication of IAV, HBV, HCV, HSV-1 and HSV-2, HPV, ZIKV and SARS-COV2 [252,253,254,255,256,257,258]. Upregulates miR-548m and inhibit miR-122 expression which modulate HCV infectivity [255]. Upregulates let-7 to increased interferon expression and inhibit IAV infection [197]. |
| Galangin | Inhibits HDAC activity [259] and upregulates miR-455-5p [260]. Exhibits antiviral activity against HSV-1 and CoxB1 [261]. |
|
| Garcinol | 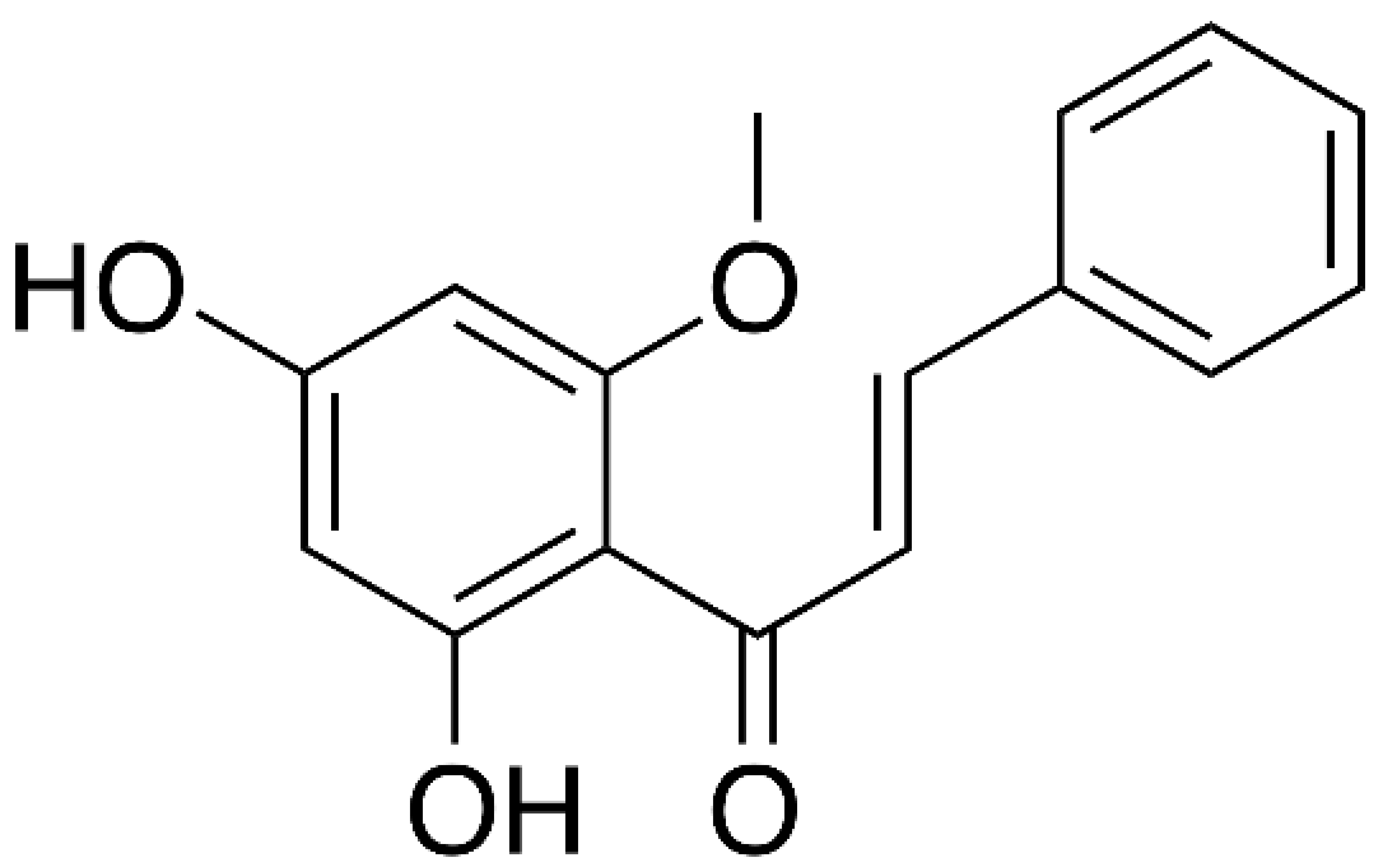 |
Decreases HAT activity of p300 and pCAF [262]. Downregulates miR-21, miR-494, miR-495, and miR-1977 [263]. Upregulates miR-453, miR-128, miR-1280 and miR-720, let-7a, let-7e, let-7f, miR-200b, and miR-200c [264]. Inhibits HIV-1 reverse transcriptase-associated ribonuclease H [265]. |
| Genistein | Reduces HDAC while increasing HAT activity [266]. Inhibits miR-223 and miR-223 expression [267] whom involved in regulation of immune response and viral infections [117,118] [119]. |
|
| Ginkgolic acid | 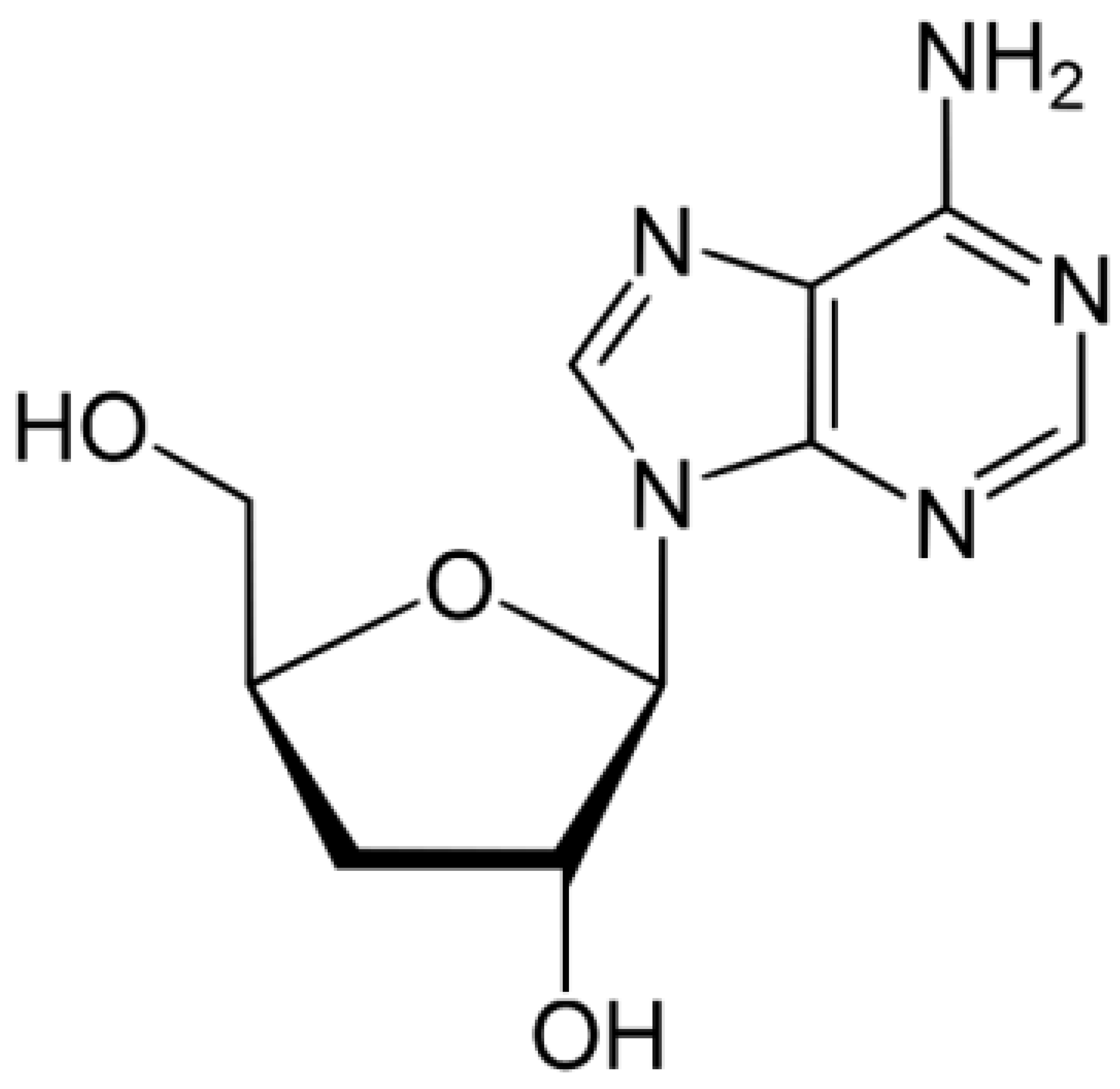 |
Impairs protein SUMOylation [268]. Inhibits HSV-1, HSV-2, VZV, HCMV, ZIKV, IAV, EBV, HIV, EBOV and Coronavirus COVID-19 [269]. |
| Glycyrrhizic acid | Inhibits replications of various viruses including HBV, HCV, IAV H1N1, HIV [270], NDV [271], SARS-COV2 [272,273,274], RSV, VACV, HSV [275] and VSV [275,276]. Exhibits anti-inflammatory effects decreasing IL-6 release [277] by regulating NF-κB, PI3K signaling pathways [278]. Inhibits viral replication of various viruses including HBV, HCV, IAV H1N1 and HIV [270]. |
|
| Grifolin |  |
Reduces Elk1 transcription as well as its binding to the DNMT1 promoter region [279]. Modulates of ERK1/2-Elk1-DNMT1 signaling [280]. |
| Oleacein | down-regulates several class I/II HDACs [281,282]. Exhibits antiviral effect against HIV-1 [283]. |
|
| Plitidepsin |  |
Targets the eukaryotic translation elongation factor 1A (eEF1A) [284]. Exhibits anti-viral activity against RSV and gastroenteritis coronavirus [285] and SARS-CoV-2 [286] [287]. |
| Pterostilbene | Modulates HDAC activity and inhibited SIRT1 [288] [289]. Inhibits SARS-CoV-2 Replication [290] |
|
| Quercetin |  |
Enhances histone H3 acetylation, activates HAT, and inhibits HDAC activities [291]. Inhibits HMT [292]. Inhibits miR-146a expression [293], a regulator of HIV replication [294], and miR-16, miR-217, and miR-145 [295,296,297]. Inhibits replication of IAV H1N1, IVA H3N2, HBV, HCV, DENV, poliovirus, rhinovirus, CHIKV, MERS-CoV, HSV 1/2, EBV, RSV , Arbovirus, EBOV, HIV, Japanese encephalitis virus , hAdV, enterovirus, ZIKV, NDV, Mayaro virus (MAYV) and SARS-COV2 [192,257,298,299,300]. Activates SIRT1 which resulted in inhibition of HCV [301] Upregulates let-7 which restore anti-viral immune response and thus exhibits anti-influenza activity [197]. |
| Resveratrol | Inhibits HDAC [302,303] and activates SIRT1 [304]. Decreases the levels of miR-17, miR-21, miR-25, miR-92a-2, miR-103-1, miR-103-2 [305] and upregulates miR-200c [306]. In humans study increases miR-21, miR-181b, miR-663, and miR-30c, while reduces inflammatory cytokines like IL-6, CCL3, IL-1β, and TNF-α [307]. Inhibits HSV infection [308], beta-corona viruses such as MERS-COV and SARS-COV2 [309], Varicella-zoster virus (VZV) wild-type and DNA polymerase mutants with acyclovir-resistant VZV [310,311], VEEV [312], EBV [313], CV [314] and RSV. SIRT proteins regulate HBV replication and thus SIRTs modulators such as resveratrol are suitable as anti-HBV and anti-RSV infections [315] [316]. |
|
| Silibinin |  |
Inhibits the expression of HDAC1, HDAC2, HDAC3, HDAC6, SET domain proteins (SETD1A, D4, D6), and lysine-specific demethylases (KDM 5B, 5C, 4A) [317]. Inhibits DNMTs [318] [319,320]. Downregulates expression of miR-21 and miR-155 [321]. Exhibits anti-viral activity againsdt HBV, DENV, CHIKV, MAYV, IVA, HIV, and HBV [322]. |
| Silvestrol | Targets the eukaryotic initiation factor-4A (eIF4A) [323]. Exhibits activity against EBOV, ZIKV, CHIKV and coronaviruses, MERS-CoV, HCoV-229E and SARS-CoV-2 [324,325,326] [327,328,329]. |
|
| Sulforaphane |  |
Reduces HDAC activity [330] while increases acetylated histones H3 and H4 expression [330]. Upregulates let-7 expression and exhibits anti-IVA activity [197] and diminishes viral-induced immune cell activation in the lungs [331]. inhibits replications of HCV and DENV [332] |
| Tanshinone IIA | Decreases the expression and activity of HDACs [333]. By inhibiting MAPK p38 compromised the replications of number of viruses including DENV [42], coronavirus [43], VEEV [44], and EV71 [45], SFTSV, HSV-1 and SARS-CoV-2 [46,334]. |
|
| Ursolic acid |  |
Reduces the expression of HDAC1, HDAC2, HDAC3, HDAC8 (Class I), HDAC6 and HDAC7 (Class II) [335]. Exhibits anti-cytomegalovirus activity [336]. |
| Withaferin A | Downregulates HDAC1 [337] [338]. Decreases HMT activity, but enhanced HAT activity [338]. Inhibits Mpro main protease of SARS-COV2 [339]. Attenuates H1N1 IVA [340]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





