Submitted:
19 October 2023
Posted:
23 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Construction and Sequencing of Small Ribonucleic acid Library
2.3. Differentially Expressed miRNA and mRNA Analysis
2.4. Target Gene Prediction of Differentially Expressed miRNAs and KEGG and GO Analysis
2.6. Real-Time qPCR Validation of Differentially Expressed mRNA and miRNAs
3. Results
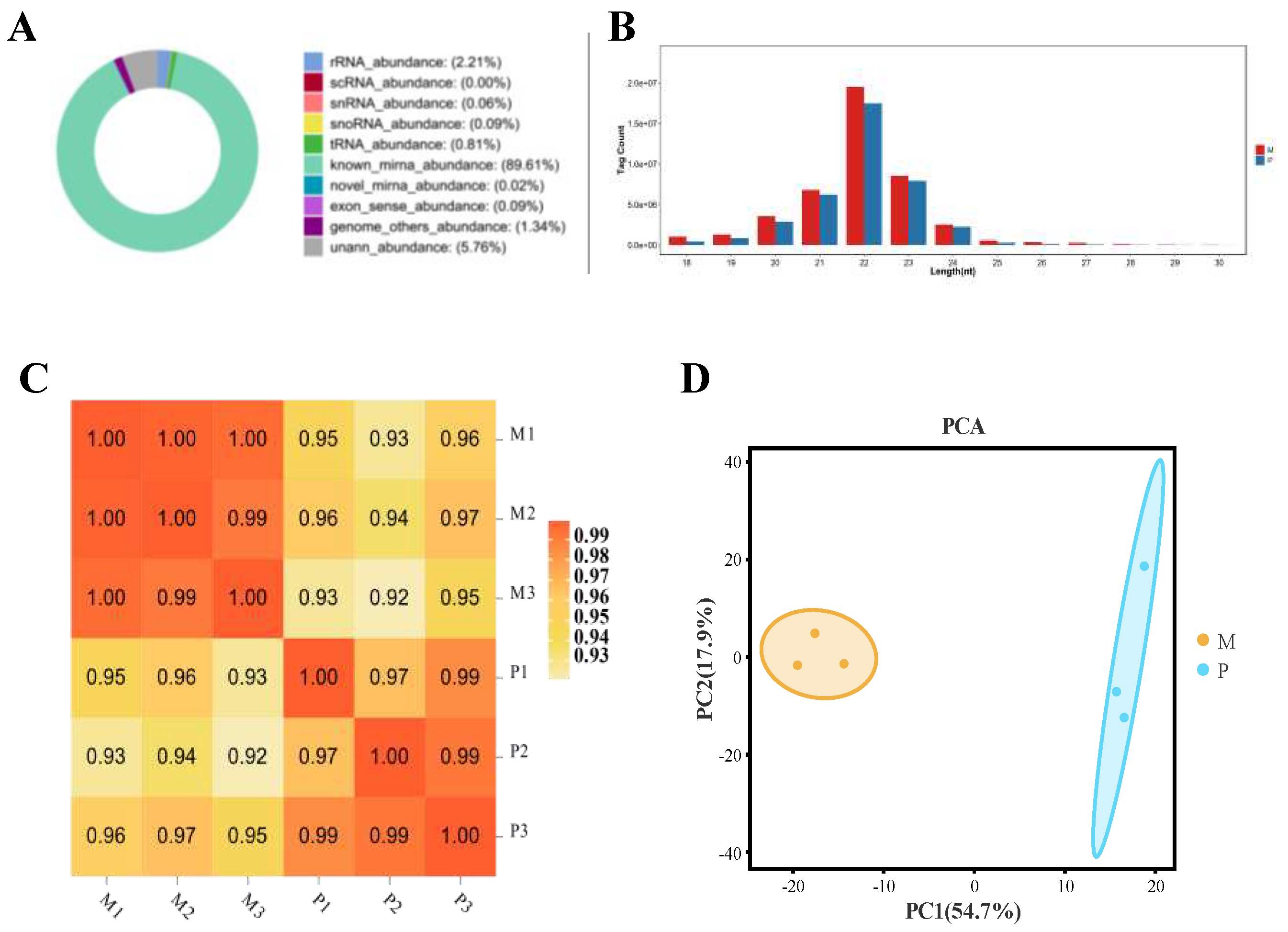
3.1. Summary of Sequencing Small RNA
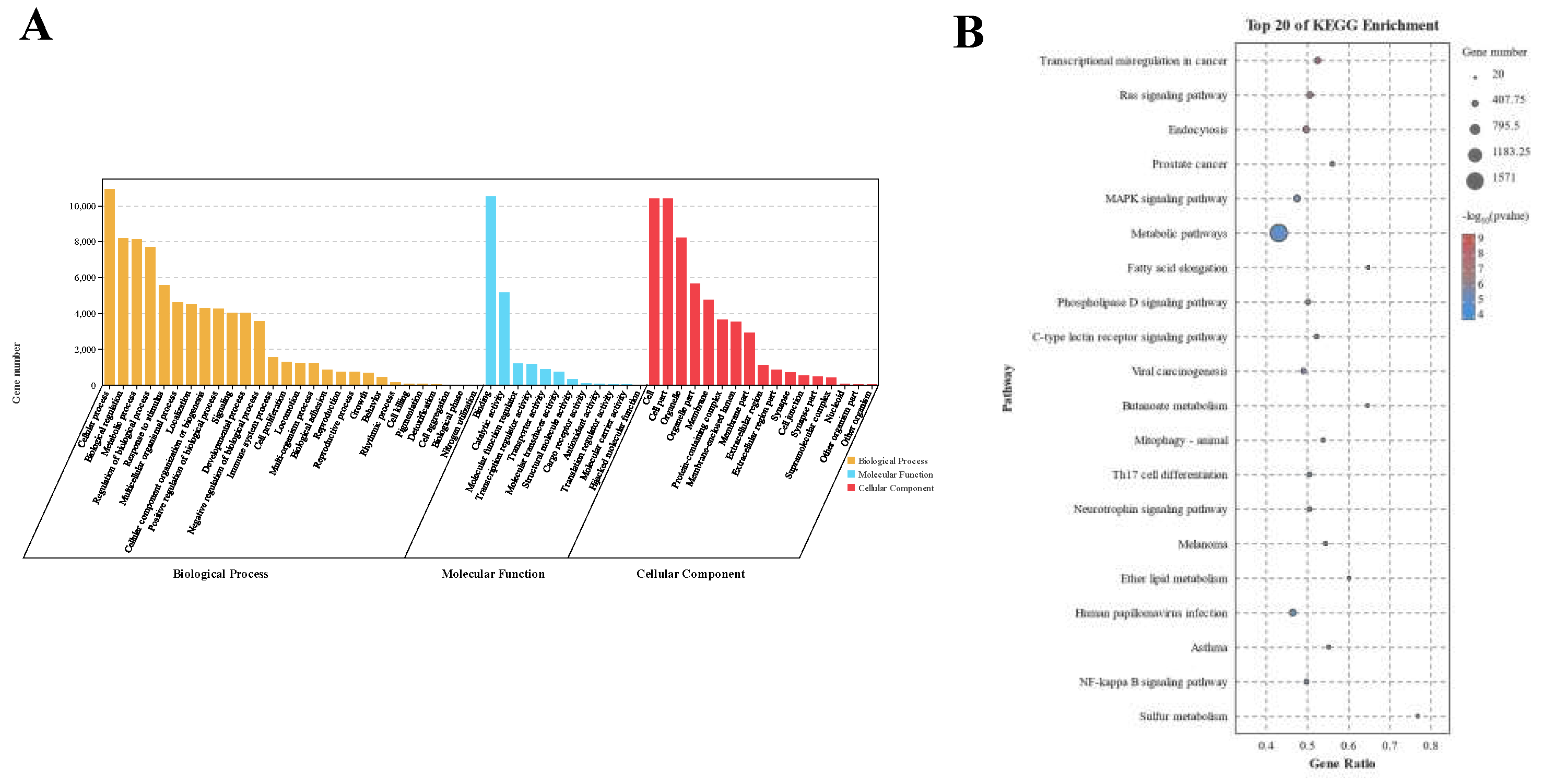
3.2. Differential Expression Analysis of mRNAs
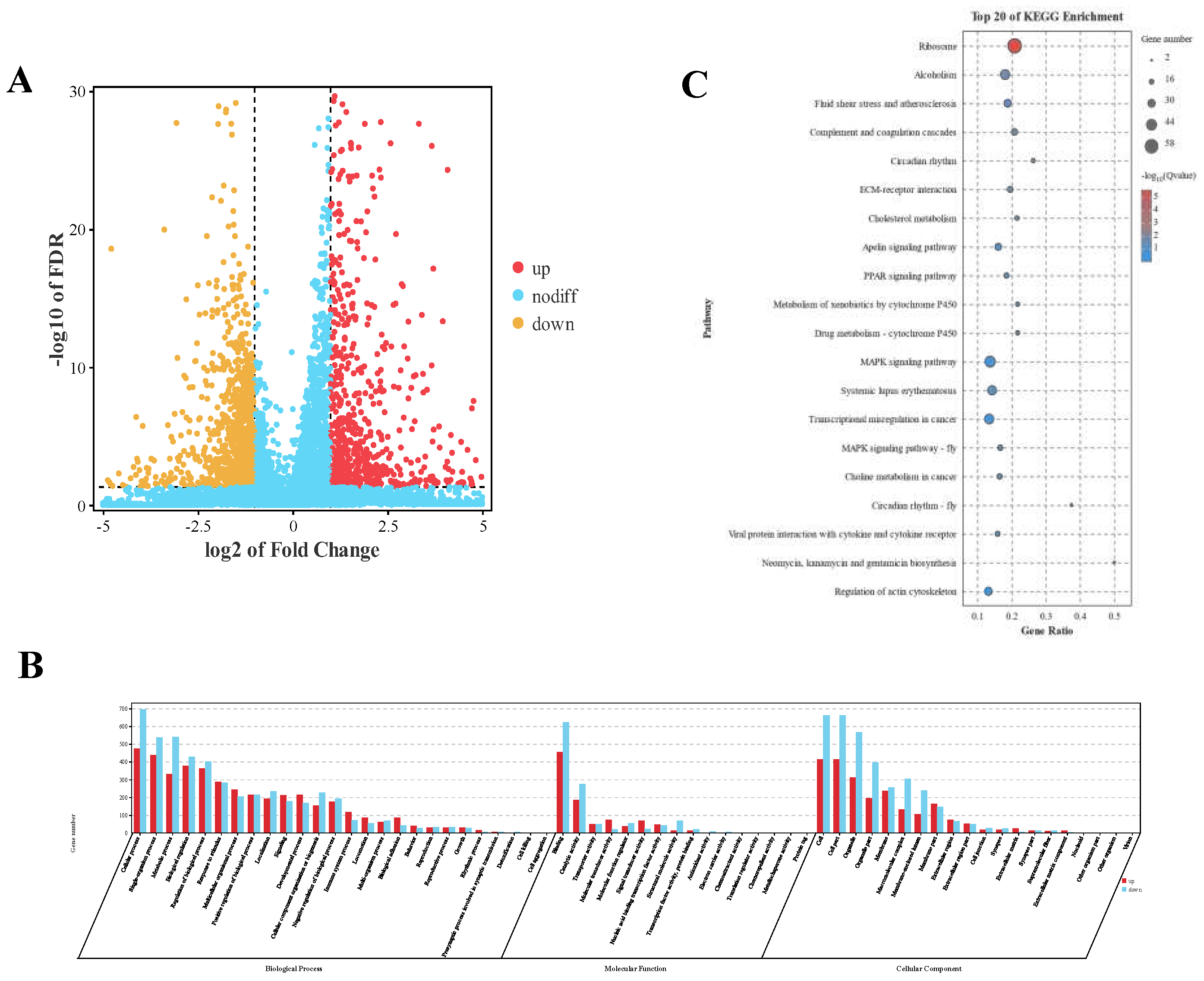
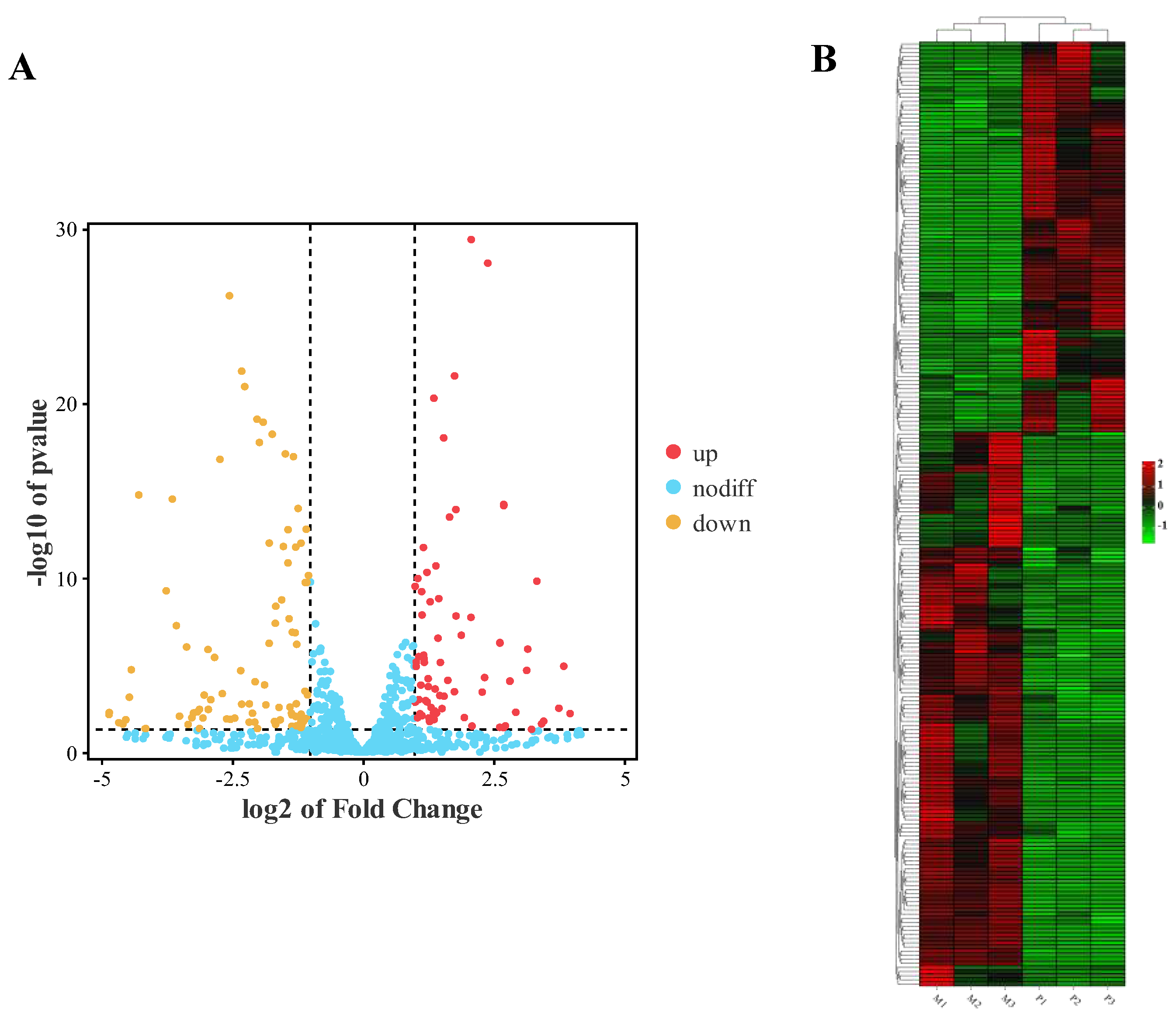
3.3. Differential Expression Analysis of miRNAs
3.4. Target Gene Prediction of Differentially Expressed miRNAs and KEGG and GO Analysis
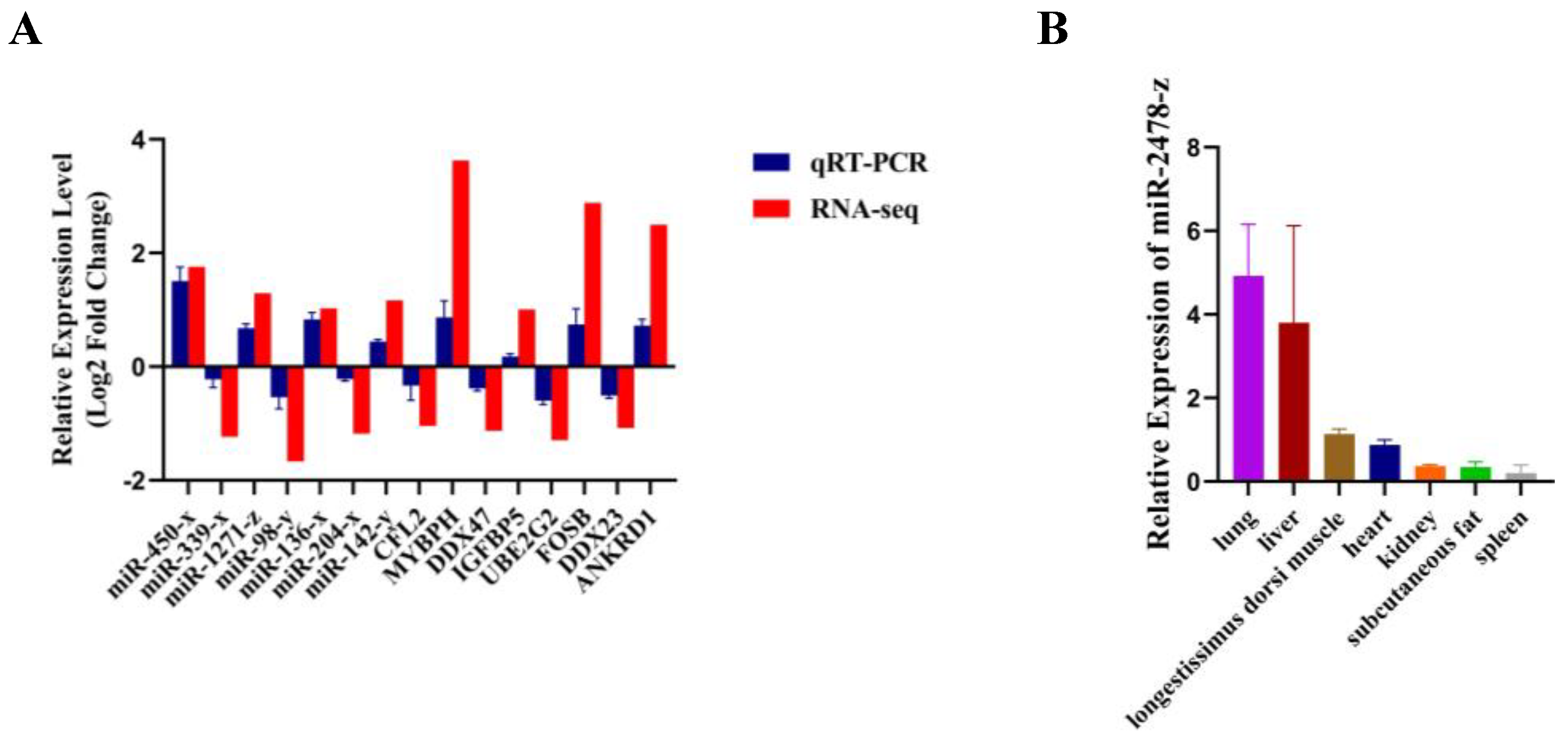
3.5. Validation of differentially expressed miRNAs and mRNAs by qRT-PCR
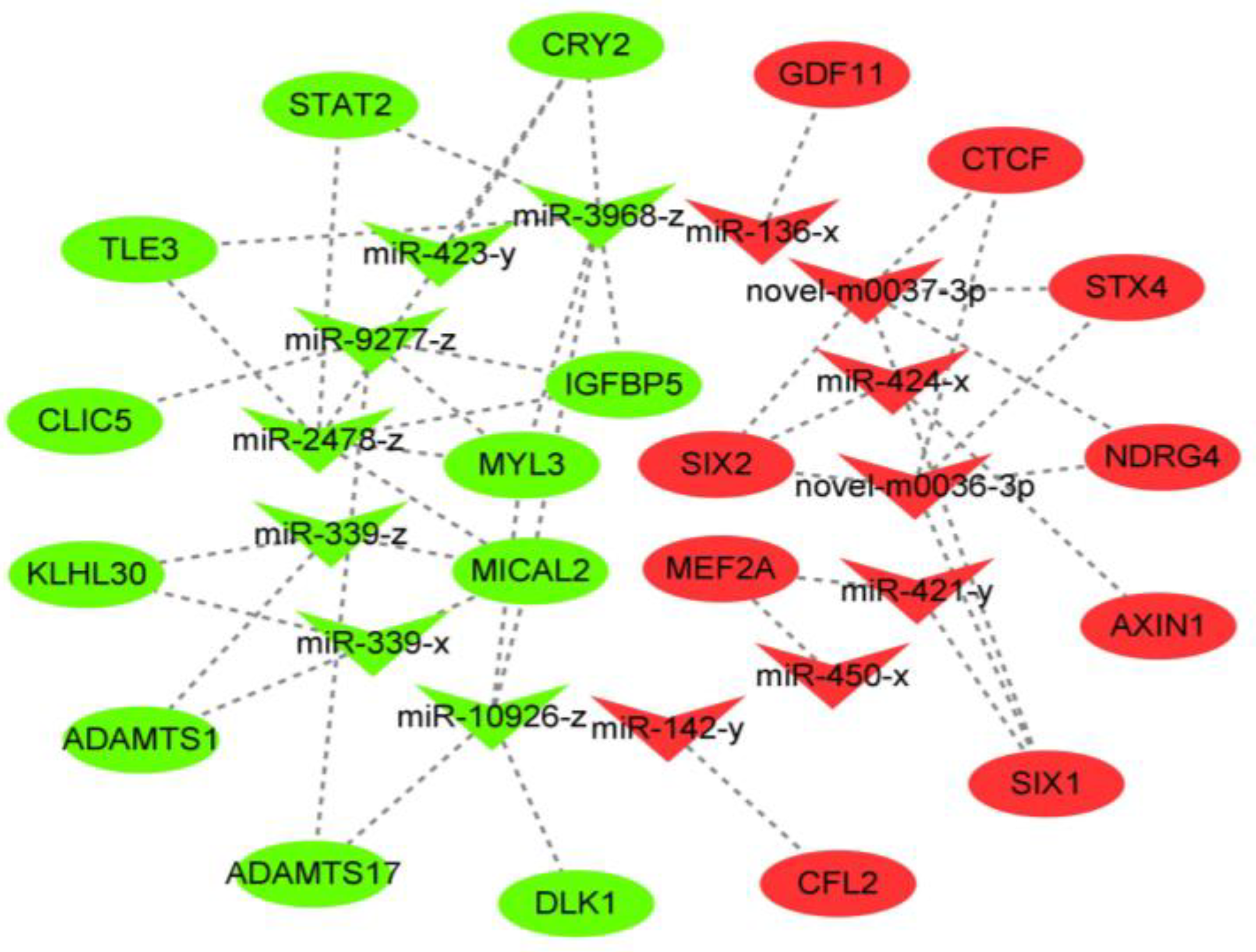
3.6. Building the Network for miRNA-mRNA Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, S.Z. , et al., Determination of growth and development indicators of Jeryak in Gannan alpine pasture area. Chinese Herbivore Science. 2019, 39, 73–75. [Google Scholar]
- Wei, Y. P and Xu J.T, Current status of cattle-yak production and research in Qinghai Province. Chinese Bovine Science. 2010, 36, 60–62. [Google Scholar]
- Yin, H.; He, H.; Cao, X.; Shen, X.; Han, S.; Cui, C.; Zhao, J.; Wei, Y.; Chen, Y.; Xia, L.; Wang, Y.; Li, D.; Zhu, Q. MiR-148a-3p Regulates Skeletal Muscle Satellite Cell Differentiation and Apoptosis via the PI3K/AKT Signaling Pathway by Targeting Meox2. FRONT GENET 2020, 11, 512. [Google Scholar] [CrossRef]
- Liu, J.; Li, F.; Hu, X.; Cao, D.; Liu, W.; Han, H.; Zhou, Y.; Lei, Q. Deciphering the miRNA transcriptome of breast muscle from the embryonic to post-hatching periods in chickens. BMC GENOMICS 2021, 22, 64. [Google Scholar] [CrossRef]
- LUO, W.; ABDALLA, B.A.; NIE, Q.; ZHANG, X. The genetic regulation of skeletal muscle development:insights from chicken studies. FRONT AGRIC SCI ENG 2017, 4, 295–304. [Google Scholar] [CrossRef]
- Hernandez-Hernandez, J.M.; Garcia-Gonzalez, E.G.; Brun, C.E.; Rudnicki, M.A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. SEMIN CELL DEV BIOL 2017, 72, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.V.; Hughes, S.M. Mef2 and the skeletal muscle differentiation program. SEMIN CELL DEV BIOL 2017, 72, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.M.; Pan, Y.; Zou, C.X.; An, Q.; Cheng, J.R.; Li, P.J.; Zheng, Z.H.; Pan, Y.; Feng, W.Y.; Yang, S.F.; Shi, D.S.; Wei, Y.M.; Deng, Y.F. CircUBE2Q2 promotes differentiation of cattle muscle stem cells and is a potential regulatory molecule of skeletal muscle development. BMC GENOMICS 2022, 23, 267. [Google Scholar] [CrossRef]
- Buckingham, M.; Relaix, F. PAX3 and PAX7 as upstream regulators of myogenesis. SEMIN CELL DEV BIOL 2015, 44, 115–125. [Google Scholar] [CrossRef]
- Gao, L.; Yang, M.; Wei, Z.; Gu, M.; Yang, L.; Bai, C.; Wu, Y.; Li, G. MSTN Mutant Promotes Myogenic Differentiation by Increasing Demethylase TET1 Expression via the SMAD2/SMAD3 Pathway. INT J BIOL SCI 2020, 16, 1324–1334. [Google Scholar] [CrossRef]
- Liu, J.; Pan, M.; Huang, D.; Guo, Y.; Yang, M.; Zhang, W.; Mai, K. Myostatin-1 Inhibits Cell Proliferation by Inhibiting the mTOR Signal Pathway and MRFs, and Activating the Ubiquitin-Proteasomal System in Skeletal Muscle Cells of Japanese Flounder Paralichthys olivaceus. CELLS-BASEL, 2020; 9. [Google Scholar]
- Tajbakhsh, S.; Borello, U.; Vivarelli, E.; Kelly, R.; Papkoff, J.; Duprez, D.; Buckingham, M.; Cossu, G. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. DEVELOPMENT 1998, 125, 4155–4162. [Google Scholar] [CrossRef] [PubMed]
- Elia, D.; Madhala, D.; Ardon, E.; Reshef, R.; Halevy, O. Sonic hedgehog promotes proliferation and differentiation of adult muscle cells: Involvement of MAPK/ERK and PI3K/Akt pathways. Biochim Biophys Acta 2007, 1773, 1438–1446. [Google Scholar] [CrossRef] [PubMed]
- Otto, A.; Schmidt, C.; Luke, G.; Allen, S.; Valasek, P.; Muntoni, F.; Lawrence-Watt, D.; Patel, K. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J CELL SCI 2008, 121, 2939–2950. [Google Scholar] [CrossRef] [PubMed]
- Abu-Elmagd, M.; Robson, L.; Sweetman, D.; Hadley, J.; Francis-West, P.; Munsterberg, A. Wnt/Lef1 signaling acts via Pitx2 to regulate somite myogenesis. DEV BIOL 2010, 337, 211–219. [Google Scholar] [CrossRef]
- Liu, S.; Gao, F.; Wen, L.; Ouyang, M.; Wang, Y.; Wang, Q.; Luo, L.; Jian, Z. Osteocalcin Induces Proliferation via Positive Activation of the PI3K/Akt, P38 MAPK Pathways and Promotes Differentiation Through Activation of the GPRC6A-ERK1/2 Pathway in C2C12 Myoblast Cells. Cell Physiol Biochem 2017, 43, 1100–1112. [Google Scholar] [CrossRef] [PubMed]
- Kornasio, R.; Riederer, I.; Butler-Browne, G.; Mouly, V.; Uni, Z.; Halevy, O. Beta-hydroxy-beta-methylbutyrate (HMB) stimulates myogenic cell proliferation, differentiation and survival via the MAPK/ERK and PI3K/Akt pathways. Biochim Biophys Acta 2009, 1793, 755–763. [Google Scholar] [CrossRef]
- Ghini, F.; Rubolino, C.; Climent, M.; Simeone, I.; Marzi, M.J.; Nicassio, F. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. NAT COMMUN 2018, 9, 3119. [Google Scholar] [CrossRef]
- Kooshapur, H.; Choudhury, N.R.; Simon, B.; Muhlbauer, M.; Jussupow, A.; Fernandez, N.; Jones, A.N.; Dallmann, A.; Gabel, F.; Camilloni, C.; Michlewski, G.; Caceres, J.F.; Sattler, M. Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1. NAT COMMUN 2018, 9, 2479. [Google Scholar] [CrossRef]
- Khatri, B.; Seo, D.; Shouse, S.; Pan, J.H.; Hudson, N.J.; Kim, J.K.; Bottje, W.; Kong, B.C. MicroRNA profiling associated with muscle growth in modern broilers compared to an unselected chicken breed. BMC GENOMICS 2018, 19, 683. [Google Scholar] [CrossRef]
- Li, Z.; Abdalla, B.A.; Zheng, M.; He, X.; Cai, B.; Han, P.; Ouyang, H.; Chen, B.; Nie, Q.; Zhang, X. Systematic transcriptome-wide analysis of mRNA-miRNA interactions reveals the involvement of miR-142-5p and its target (FOXO3) in skeletal muscle growth in chickens. MOL GENET GENOMICS 2018, 293, 69–80. [Google Scholar] [CrossRef]
- Xu, S.; Chang, Y.; Wu, G.; Zhang, W.; Man, C. Potential role of miR-155-5p in fat deposition and skeletal muscle development of chicken. BIOSCIENCE REP 2020, 40. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Shan, Y.; Ji, G.; Ju, X.; Tu, Y.; Sheng, Z.; Xie, J.; Zou, J.; Shu, J. miRNA-mRNA network regulation in the skeletal muscle fiber phenotype of chickens revealed by integrated analysis of miRNAome and transcriptome. SCI REP-UK 2020, 10, 10619. [Google Scholar] [CrossRef]
- Liu, J.; Li, F.; Hu, X.; Cao, D.; Liu, W.; Han, H.; Zhou, Y.; Lei, Q. Deciphering the miRNA transcriptome of breast muscle from the embryonic to post-hatching periods in chickens. BMC GENOMICS 2021, 22, 64. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, S.; Xu, Z.; Gao, J.; Mishra, S.K.; Zhu, Q.; Zhao, X.; Wang, Y.; Yin, H.; Fan, X.; Zeng, B.; Yang, M.; Yang, D.; Ni, Q.; Li, Y.; Zhang, M.; Li, D. MiRNA Profiling in Pectoral Muscle Throughout Pre- to Post-Natal Stages of Chicken Development. FRONT GENET 2020, 11, 570. [Google Scholar] [CrossRef]
- Chen, J.F.; Tao, Y.; Li, J.; Deng, Z.; Yan, Z.; Xiao, X.; Wang, D.Z. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J CELL BIOL 2010, 190, 867–879. [Google Scholar] [CrossRef]
- Nakasa, T.; Ishikawa, M.; Shi, M.; Shibuya, H.; Adachi, N.; Ochi, M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J CELL MOL MED 2010, 14, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Ran, L.; Lang, H.; Zhou, M.; Yu, L.; Yi, L.; Zhu, J.; Liu, L.; Mi, M. Myricetin improves endurance capacity by inducing muscle fiber type conversion via miR-499. NUTR METAB 2019, 16, 27. [Google Scholar] [CrossRef]
- Wang, S.; Cao, X.; Ge, L.; Gu, Y.; Lv, X.; Getachew, T.; Mwacharo, J.M.; Haile, A.; Sun, W. MiR-22-3p Inhibits Proliferation and Promotes Differentiation of Skeletal Muscle Cells by Targeting IGFBP3 in Hu Sheep. ANIMALS-BASEL 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Lu, S.; Bai, M.; Xiang, L.; Li, J.; Jia, C.; Jiang, H. Integrative microRNA-mRNA Analysis of Muscle Tissues in Qianhua Mutton Merino and Small Tail Han Sheep Reveals Key Roles for oar-miR-655-3p and oar-miR-381-5p. DNA CELL BIOL 2019, 38, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. NUCLEIC ACIDS RES 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. BIOINFORMATICS 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. GENOME BIOL 2010, 11, R90. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. ELIFE 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. CELL 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Ge, F.; Ma, X.; Dai, R.; Dingkao, R.; Zhaxi, Z.; Burenchao, G.; Bao, P.; Wu, X.; Guo, X.; Chu, M.; Yan, P.; Liang, C. Comprehensive Analysis of mRNA, lncRNA, circRNA, and miRNA Expression Profiles and Their ceRNA Networks in the Longissimus Dorsi Muscle of Cattle-Yak and Yak. FRONT GENET 2021, 12, 772557. [Google Scholar] [CrossRef]
- Campos, C.F.; Costa, T.C.; Rodrigues, R.; Guimaraes, S.; Moura, F.H.; Silva, W.; Chizzotti, M.L.; Paulino, P.; Benedeti, P.; Silva, F.F.; Duarte, M.S. Proteomic analysis reveals changes in energy metabolism of skeletal muscle in beef cattle supplemented with vitamin A. J SCI FOOD AGR 2020, 100, 3536–3543. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Pei, J.; Xiong, L.; Guo, S.; Wang, X.; Kang, Y.; Guo, X. Analysis of Chromatin Openness in Testicle Tissue of Yak and Cattle-Yak. INT J MOL SCI 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.Z. , et al., Determination of slaughter performance and meat quality of Jeryak in alpine pastures. Chinese Herbivore Science. 2019, 39, 4. [Google Scholar]
- Ayuso, M.; Fernandez, A.; Nunez, Y.; Benitez, R.; Isabel, B.; Barragan, C.; Fernandez, A.I.; Rey, A.I.; Medrano, J.F.; Canovas, A.; Gonzalez-Bulnes, A.; Lopez-Bote, C.; Ovilo, C. Comparative Analysis of Muscle Transcriptome between Pig Genotypes Identifies Genes and Regulatory Mechanisms Associated to Growth, Fatness and Metabolism. PLOS ONE 2015, 10, e145162. [Google Scholar] [CrossRef]
- Myers, S.A.; Wang, S.C.; Muscat, G.E. The chicken ovalbumin upstream promoter-transcription factors modulate genes and pathways involved in skeletal muscle cell metabolism. J BIOL CHEM 2006, 281, 24149–24160. [Google Scholar] [CrossRef]
- Estrella, N.L.; Desjardins, C.A.; Nocco, S.E.; Clark, A.L.; Maksimenko, Y.; Naya, F.J. MEF2 transcription factors regulate distinct gene programs in mammalian skeletal muscle differentiation. J BIOL CHEM 2015, 290, 1256–1268. [Google Scholar] [CrossRef]
- Liu, N.; Nelson, B.R.; Bezprozvannaya, S.; Shelton, J.M.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Requirement of MEF2A, C, and D for skeletal muscle regeneration. P NATL ACAD SCI USA 2014, 111, 4109–4114. [Google Scholar] [CrossRef]
- Yang, X.; Ning, Y.; Abbas, R.S.; Mei, C.; Zan, L. MEF2C Expression Is Regulated by the Post-transcriptional Activation of the METTL3-m(6)A-YTHDF1 Axis in Myoblast Differentiation. FRONT VET SCI 2022, 9, 900924. [Google Scholar] [CrossRef]
- Wu, W.; Huang, R.; Wu, Q.; Li, P.; Chen, J.; Li, B.; Liu, H. The role of Six1 in the genesis of muscle cell and skeletal muscle development. INT J BIOL SCI 2014, 10, 983–989. [Google Scholar] [CrossRef]
- Spitz, F.; Demignon, J.; Porteu, A.; Kahn, A.; Concordet, J.P.; Daegelen, D.; Maire, P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. P NATL ACAD SCI USA 1998, 95, 14220–14225. [Google Scholar] [CrossRef]
- Giordani, J.; Bajard, L.; Demignon, J.; Daubas, P.; Buckingham, M.; Maire, P. Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. P NATL ACAD SCI USA 2007, 104, 11310–11315. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhao, T.; Ma, Y.; Wu, X.; Mao, Y.; Yang, Z.; Chen, H. New Insight into Muscle-Type Cofilin (CFL2) as an Essential Mediator in Promoting Myogenic Differentiation in Cattle. BIOENGINEERING-BASEL 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Floss, T.; Arnold, H.H.; Braun, T. A role for FGF-6 in skeletal muscle regeneration. GENE DEV 1997, 11, 2040–2051. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Sun, W.; Chen, S.Y.; Li, Y.; Wang, J.; Lai, S.; Jia, X. The exploration of miRNAs and mRNA profiles revealed the molecular mechanisms of cattle-yak male infertility. FRONT VET SCI 2022, 9, 974703. [Google Scholar] [CrossRef] [PubMed]
- Mitin, N.; Kudla, A.J.; Konieczny, S.F.; Taparowsky, E.J. Differential effects of Ras signaling through NFkappaB on skeletal myogenesis. ONCOGENE 2001, 20, 1276–1286. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Lopez, J.M. Understanding MAPK Signaling Pathways in Apoptosis. INT J MOL SCI 2020, 21. [Google Scholar] [CrossRef]
- Bengal, E.; Aviram, S.; Hayek, T. p38 MAPK in Glucose Metabolism of Skeletal Muscle: Beneficial or Harmful? INT J MOL SCI 2020, 21. [Google Scholar] [CrossRef]
- Galpin, A.J.; Raue, U.; Jemiolo, B.; Trappe, T.A.; Harber, M.P.; Minchev, K.; Trappe, S. Human skeletal muscle fiber type specific protein content. ANAL BIOCHEM 2012, 425, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, S.; Borello, U.; Vivarelli, E.; Kelly, R.; Papkoff, J.; Duprez, D.; Buckingham, M.; Cossu, G. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. DEVELOPMENT 1998, 125, 4155–4162. [Google Scholar] [CrossRef]
- Chen, K.; Gao, P.; Li, Z.; Dai, A.; Yang, M.; Chen, S.; Su, J.; Deng, Z.; Li, L. Forkhead Box O Signaling Pathway in Skeletal Muscle Atrophy. AM J PATHOL 2022, 192, 1648–1657. [Google Scholar] [CrossRef]
- Sandri, M.; Sandri, C.; Gilbert, A.; Skurk, C.; Calabria, E.; Picard, A.; Walsh, K.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. CELL 2004, 117, 399–412. [Google Scholar] [CrossRef]
- Zhang, H.; Stavnezer, E. Ski regulates muscle terminal differentiation by transcriptional activation of Myog in a complex with Six1 and Eya3. J BIOL CHEM 2009, 284, 2867–2879. [Google Scholar] [CrossRef]
- Cui, J.X.; Gong, Z.A.; Zhang, W.T.; Liu, K.; Li, T.; Shao, S.L.; Zhang, W.W. [Effects of transcription factor SIX2 gene on the proliferation of bovine skeletal muscle satellite cells]. Zhongguo Ying Yong Sheng Li Xue Za Zhi 2022, 38, 622–627. [Google Scholar] [PubMed]
- Zhu, M.; Zheng, R.; Guo, Y.; Zhang, Y.; Zuo, B. NDRG4 promotes myogenesis via Akt/CREB activation. Oncotarget 2017, 8, 101720–101734. [Google Scholar] [CrossRef]
- Fedoriw, A.M.; Stein, P.; Svoboda, P.; Schultz, R.M.; Bartolomei, M.S. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. SCIENCE 2004, 303, 238–240. [Google Scholar] [CrossRef]
- Delgado-Olguin, P.; Brand-Arzamendi, K.; Scott, I.C.; Jungblut, B.; Stainier, D.Y.; Bruneau, B.G.; Recillas-Targa, F. CTCF promotes muscle differentiation by modulating the activity of myogenic regulatory factors. J BIOL CHEM 2011, 286, 12483–12494. [Google Scholar] [CrossRef] [PubMed]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; Da, P.I.; Gunsalus, K.C.; Stoffel, M.; Rajewsky, N. Combinatorial microRNA target predictions. NAT GENET 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
| mRNAs | Forward (5′→ 3′) | Reverse (5′→ 3′) |
|---|---|---|
| MYBPH | ATGTGAGTGACAGCTCGGTG | GTCACCTACAGCCAGGTTCC |
| DDX23 | CAATGACAGCACACTGCACC | TCCCTGTCTCGGTCCTTCTT |
| FOSB | CCTCATCTCTTCCATGGCCC | CCACTGCTGTAGCCACTCAT |
| DDX47 | TCTGCCCATTCTCAACGCAT | CAATGACAGCACACTGCACC |
| IGFBP5 | GGCAGAGGAGACCTACTCAC | GGCAGAGGAGACCTACTCAC |
| CFL2 | TGCCATCCTGAGTTTCCCAC | TGCCATCCTGAGTTTCCCAC |
| ANKRD1 | CAGAACCTGTGGATGTGCCT | TGCCAAATGTCCTTCCAAGC |
| UBE2G2 | TGCCATCCTGAGTTTCCCAC | TGCCATCCTGAGTTTCCCAC |
| GAPDH | AGTTCAACGGCACAGTCAAGG | ACCACATACTCAGCACCAGCA |
| mRNAs | Forward (5′→ 3′) | Reverse (5′→ 3′) |
|---|---|---|
| miR-450-x | TTTTGCAATATGTTCCTGAAT | |
| miR-136-x | ACTCCATTTGTTTTGATGATGG | |
| miR-1271-z | CTTGGCACCTAGTAAGTACTCAA | |
| miR-142-y | TGTAGTGTTTCCTACTTTATGG | |
| miR-204-x | TTCCCTTTGTCATCCTATGCCT | |
| miR-98-y | CTATACAACTTACTACTTTCCT | |
| miR-339-x | TCCCTGTCCTCCAGGAGCTCACT | |
| U6 | ACGGACAGGATTGACAGATT | TCGCTCCACCAACTAAGA |
| Samples | Clean_ reads | High_ quality | 3’adapter_null | insert_ null | 5’adapter_contaminants | PolyA (%) | clean_ tags |
|---|---|---|---|---|---|---|---|
| M1 | 17408914 (100%) |
17260011 (99.1447%) | 9612 (0.0557%) |
108519 (0.6287%) | 31981 (0.1853%) |
321 (0.0019%) | 16255077 (93.3721%) |
| M2 | 14015050 (100%) |
13899769 (99.1774%) | 6900 (0.0496%) |
54327 (0.3908%) | 13958 (0.1004%) |
162 (0.0012%) | 13362108 (95.3411%) |
| M3 | 16597591 (100%) |
16448657 (99.1027%) | 10968 (0.0667%) |
83221 (0.5059%) | 20095 (0.1222%) |
247 (0.0015%) | 15172291 (91.4126%) |
| P1 | 14318748 (100%) |
14151985 (98.8354%) | 9475 (0.0670%) |
59453 (0.4201%) | 7319 (0.0517%) |
126 (0.0009%) | 13782038 (96.2517%) |
| P2 | 9195586 (100%) |
9071110 (98.6464%) | 76171 (0.8397%) |
46305 (0.5105%) | 3837 (0.0423%) |
76 (0.0008%) |
8756687 (95.2271%) |
| P3 | 16740804 (100%) |
16610388 (99.2210%) | 66607 (0.4010%) |
74568 (0.4489%) | 6918 (0.0416%) |
134 (0.0008%) | 16180410 (96.6525%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





