1. Introduction
During infection with RNA viruses (such as SARS-COV-2), pathogen-associated molecular patterns (PAMPs) are recognized by retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) [
1]. Following the recognition of PAMPs, RIG-I undergoes dephosphorylation and subsequent polyubiquitination mediated by the ubiquitin E3 ligase tripartite-motif protein 25 (TRIM25) [
2]. Ubiquitinated RIG-I then interacts with the Mitochondrial Antiviral Signaling protein (MAVS), leading to the activation of TANK-binding kinase 1 (TBK1) and inhibitor of κB kinase-ε (IKKε), resulting in the phosphorylation of interferon regulatory factor 3 (IRF3) and the production of interferon β (IFN-β). Ultimately, IFN-β binds to specific receptors, inducing the expression of interferon-stimulated genes (ISGs) and initiating a host’s antiviral response [
3]. In this process, RIG-I activates MAVS and initiates the first line of defense against viral replication.
Severe and critical COVID-19 patients display a distinct phenotype characterized by a severe impairment of the type I interferon (IFN I) response. This is evident from the absence of IFN-β production, reduced IFN-α production and activity, leading to a persistent viral load in the bloodstream and an exacerbated inflammatory response [
4]. SARS-CoV-2 has been shown to encode multiple proteins that manipulate or evade host’s antiviral response, enabling successful infection [
5,
6,
7]. The nucleocapsid protein (N) protein, which is the most conservative protein among human β coronaviruses, including SARS-CoV-2 responsible for COVID-19, has been implicated in interfering with the host’s innate immune response. The SARS-COV-2 N protein has been previously documented to interact with RIG-I at the DExD/H domain, inhibiting type I interferon (IFN I) signaling by disrupting the binding of immunostimulatory RNAs [
8].
Recent study indicates that the N protein of SARS-CoV-2 may employ additional mechanisms to evade the host's innate immune response. One such mechanism involves the interaction between the N protein and the MAVS, which hinders the IFN-β signaling pathway by suppressing the Lys63-linked poly-ubiquitination of MAVS [
9]. However, the precise underlying mechanism still requires further investigation for a comprehensive understanding.
In this study, we revealed that N protein plays a crucial role in modulating MAVS SUMOylation by promoting an enhanced interaction between MAVS and Ubc9. Furthermore, we demonstrated that this N protein induced MAVS SUMOylation change leads to a reduction in its Lys63-linked ubiquitination, consequently inhibiting phosphorylation events involving IKKα, TBK1, and IRF3. As a result, the IFN-β signaling pathway is disrupted.
These findings elucidate the mechanism by which human β coronaviruses N protein inhibits MAVS ubiquitination, resulting in the suppression of IFN-β expression and evasion of the host's innate immune response. This knowledge can potentially contribute to the development of targeted therapeutic strategies to combat severe COVID-19 and enhance our overall understanding of the virus's pathogenesis.
2. Materials and Methods
2.1. Cells and Reagent
In this study, various cell lines, including HEK293T, VERO, VERO E6, Huh7 and HRT18, were obtained from the American Type Culture Collection (ATCC). HEK293T and Huh7 cells were cultured in DMEM media, VERO and VERO E6 cells were cultured in MEM media, and HRT18 cells were cultured in RPMI media. All media were supplemented with 10% fetal bovine serum and 100 U/ml penicillin/streptomycin. The cells were maintained at 37°C in a humidified atmosphere with 5% CO2. It's noteworthy that all cell lines used in the study were regularly tested and confirmed to be free from mycoplasma contamination. To trigger the innate immune response, the researchers utilized Sendai virus (SeV) stimulation.
2.2. Plasmid Constructs and Transfection
To create the necessary plasmids for the study, the coding sequences of five human beta coronavirus N proteins (or truncated N proteins), UBC9 (or UBC9C93A mutant), MAVS, SUMO3, and Ub-K63 (ubiquitin contains a substitution of arginine for all lysine residues, except the lysine at position 63) were amplified using standard PCR techniques. Subsequently, these amplified sequences were inserted into different vectors, namely pCDNA3.1, pET28a, pGEX-6P-1, pEGFP-N1, and pDsRED-mono-N1, as required for the subsequent experiments. To ensure the accuracy of the plasmid constructs, rigorous verification was conducted through DNA sequencing, which was performed by Wuhan Aoke Botai Biotechnology Co., Ltd. For the transfection assay, the researcher followed the instruction provided by the manufacturer and utilized Lipofectamine™ 3000 Transfection Reagent (Cat No: L3000008) obtained from Thermo Fisher Scientific Inc.
2.3. Immunoblotting
For the immunoblot assay, whole cells were lysed with RIPA buffer (Solarbio, Cat No: R0010) at 4 °C for 10 minutes. The total concentration of protein was quantified using a BCA Protein Assay Kit (Beyotime, Cat No: P0010S) according to the manufacturer’s instruction. Subsequently, the protein was separated by SDS-PAGE and transferred to PVDF membranes (Millipore, Cat No: ISEQ00010). After blocking with 5% skimmed milk, the membranes were incubated overnight at 4 °C with primary antibodies which included: anti-FLAG (Sigma–Aldrich, Cat No: F1804), anti-GFP (Proteintech, Cat No: 66002-1-Ig), anti-GST (Proteintech, Cat No: 66001-2-Ig), anti-His6 (Abcam, Cat No: ab18184), anti-HA tag (Abcam, Cat No: ab13834), anti-UBC9 (Proteintech, Cat No. 60201-1-Ig), anti-K63 (Abcam, Cat No: ab179434), anti-SUMO2/3 (Abcam, Cat No: ab81371), anti-IKKα (Abcam, Cat No: ab32041), anti-p-IKKα at Ser176/180 (Abcam, Cat No: ab17943), anti-TBK1 (Abcam, Cat No: ab40676 ), anti-p-TBK1 at Ser172 (Abcam, Cat No: ab109272), anti-IRF3 (Abcam, Cat No: ab76409), anti-p-IRF3 at Ser396 (Abcam, Cat No: ab81371), anti-GAPDH (Proteintech, Cat No: 60004-1-Ig).
After three washes to remove any unbound primary antibodies, the membranes were probed with horseradish peroxidase-coupled (HRP) secondary antibodies. Subsequently, the membranes were exposed, and signals were recorded using Image Lab Software (Bio-Rad) following incubation with Clarity Western ECL Substrate (Bio-Rad, Cat No: 1705061). If necessary, the relative band intensity was quantified using ImageJ software (Version 1.8).
2.4. Coimmunoprecipitation (Co-IP)
Cells were harvested from 100-mm petri dishes and subjected to lysis using RIPA buffer. This buffer consisted of 50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, and 1 mM protease inhibitor PMSF (Thermo Scientific, Cat No: 36978). The lysis process was carried out for a duration of 30 minutes on ice. Following lysis, immunoprecipitation was performed utilizing either an anti-Flag antibody (Sigma, Cat No: F1084) or an anti-His6 antibody (Abcam, Cat No: ab18184). Pierce™ Protein A/G Magnetic Beads (Thermo Scientific, Cat No: 88802) were employed for the immunoprecipitation. This procedure was conducted over a period of 3 hours at 4°C. The proteins that were precipitated underwent a series of five washes with a washing buffer. Subsequently, these proteins were eluted from the beads using an elution buffer comprised of 50 mM sodium phosphate (pH 8.0), 300 mM sodium chloride, and 300 mM imidazole. The elution process was carried out at a temperature of 98°C. The subsequent detection of the immunocomplexes was achieved through Immunoblotting, employing specific antibodies corresponding to the targeted proteins.
2.5. Confocal Microscope
To ascertain the subcellular localization of UBC9 (or UBC9C93A mutant) and N, a co-expression strategy was employed using pDsRED-mono-N1 (RFP Tag) housing the SARS-COV-2 N protein coding genes, and pEGFP-N1 (GFP Tag) containing the UBC9 or UBC9C93A coding gene. These constructs were simultaneously introduced into HEK293T cells. Subsequently, the transfected cells were exposed to Sev for an additional 8 hours, followed by PBS washing and fixation with 4% paraformaldehyde in PBS for a duration of 20 minutes. Prior to confocal microscopy imaging, the cells were subjected to DAPI staining.
2.6. SUMOylation and Ubiquitination Assay
To explore ubiquitination, cells were collected 24 hours following transfection with the designated recombinant constructs, and they were subsequently exposed to SeV stimulation. Following this, cellular lysis was conducted using a lysis buffer supplemented with 5 μM MG132 (Sigma-Aldrich, Cat No: M7449-200UL), and the lysis process was maintained for 30 minutes on ice. Following immunoprecipitation, the presence of ubiquitination was investigated utilizing the anti-Ubiquitin (linkage-specific K63) antibody (Abcam, Cat No: ab179434). For the purpose of identifying SUMOylation, cells were subjected to lysis using a lysis buffer containing an additional 20 μM N-Ethylmaleimide (Sigma-Aldrich, Cat No: E3876-5G). This lysis process was carried out over a 30-minute period while maintaining the samples on ice. After immunoprecipitation, the assessment of SUMOylation was performed using the anti-SUMO2/3 antibody (Abcam, Cat No: ab81371).
2.7. Statistical Analyses
The mean and standard deviation (x ̅±SD) of three independent experiments were reported. Statistical analyses were conducted using GraphPad Prism 5 (GraphPad Software, 5.0) and SPSS (v19.0). An unpaired Student's t-test was utilized to compare the two groups, while ANOVA was used for multiple group comparisons. Symbols *, **, ***, and "ns" were used to represent p-values of less than 0.05, less than 0.01, less than 0.001, and no significant difference, respectively.
3. Results
3.1. N Protein Interacts with Human SUMO-Conjugating E2 Enzyme UBC9.
A previous study indicated that the N protein of SARS-CoV interacts with the human SUMO-conjugating E2 enzyme UBC9, and that mutations in UBC9 at the active site (specifically the C93A mutation) severely disrupt the interaction with the SARS-CoV N protein [
10]. To investigate if the N protein of SARS-CoV-2 also interacts with UBC9, Co-IP experiments were performed. The results demonstrated that the N protein of SARS-CoV-2 specifically binds to UBC9 (
Figure 1A). To further confirm this interaction, SARS-CoV-2 N protein and UBC9 (or UBC9
C93A) were co-expressed in HEK293T cells and visualized using confocal microscopy. The results revealed that the N protein and UBC9 were co-localized in the cytoplasm. However, the UBC9
C93A mutation seems disrupted the co-localization with N protein (
Figure 1B).
Subsequently, plasmids containing the N protein coding genes with a fused HIS
6 tag (in pET28a vector) and UBC9 or UBC9
C93A coding genes with a fused GST tag (in pGEX-6P-1 vector) were co-expressed in BL21 (DE3)
E. coli cells. After induction with IPTG (1mM) for 6 hours, cell lysates were subjected to Co-IP. The results confirmed that mutations in UBC9 at the C93A site impaired its ability to bind to the N protein (
Figure 1C). We also verified interaction of SARS-COV-2 N protein with endogenous UBC9 in HEK293T cells. Co-IP assays proved that SARS-COV-2 N protein could endogenous UBC9 in HEK293T cells (
Figure 1D).
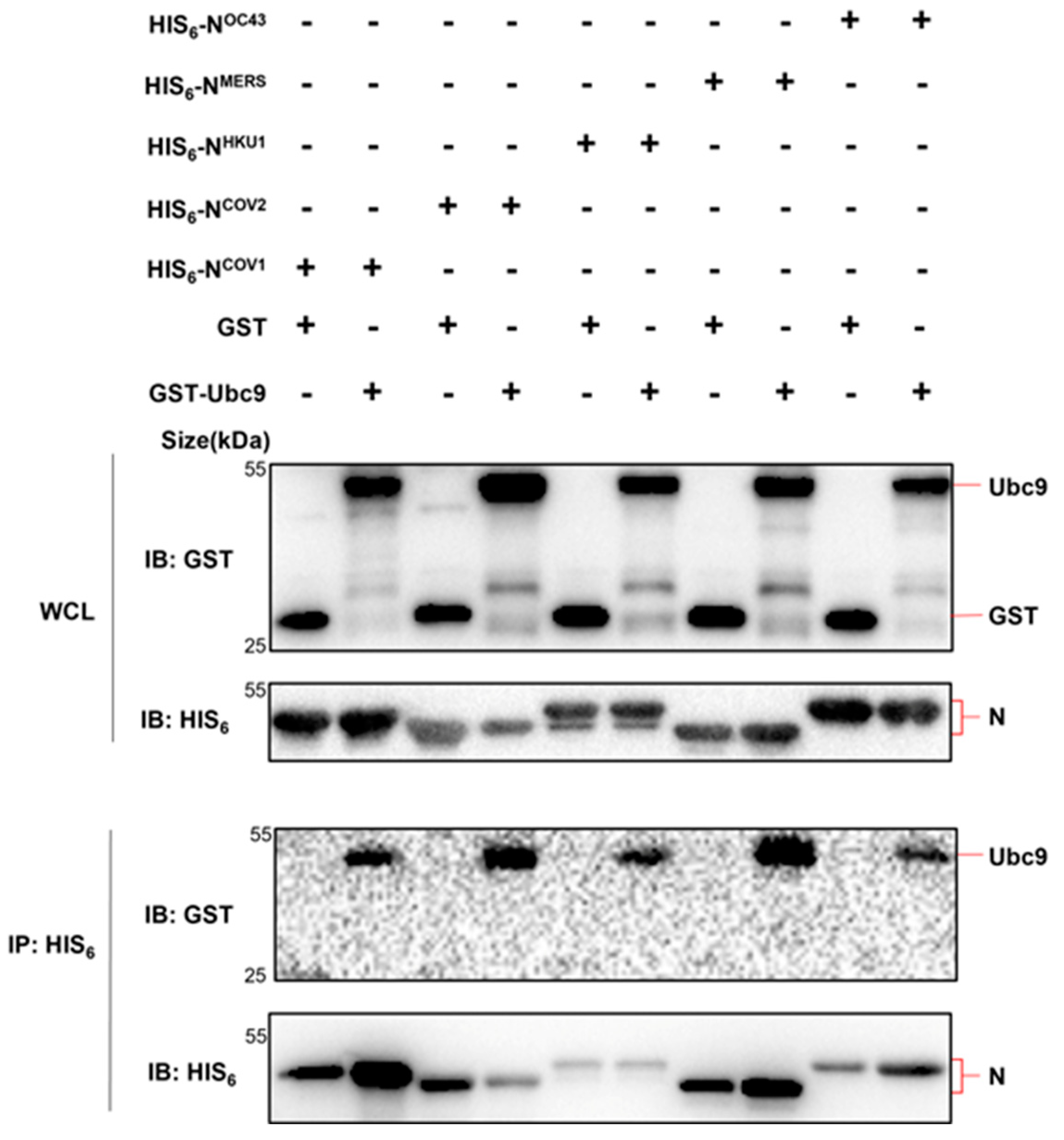
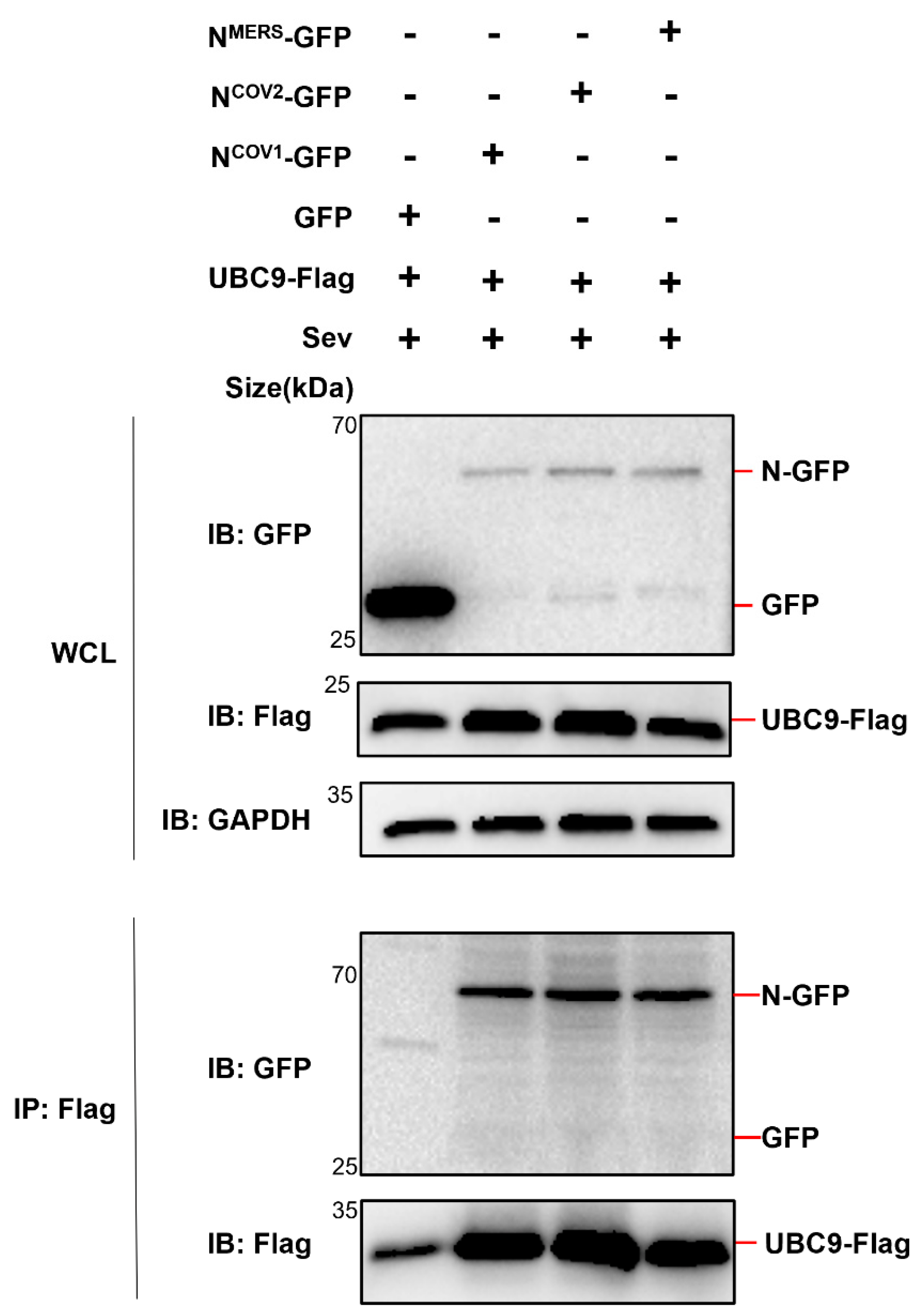
Considering the conservation of the N protein among human infectious β coronaviruses [
11], further investigation was conducted to determine if all five human β coronavirus (SARS-CoV, MERS-CoV, SARS-CoV-2, HCoV-OC43, HCoV-HKU1) N proteins could interact with UBC9. Co-expression of these N proteins in
E. coli or HEK293T cells, followed by Co-IP experiments, demonstrated that all five human β coronavirus N proteins were capable of interacting with the SUMO-conjugating enzyme E2 UBC9 (
Figure 2 and
Figure 3).
Taken together, these results demonstrated that human beta coronavirus N protein interacts with human SUMO-conjugating E2 enzyme UBC9.
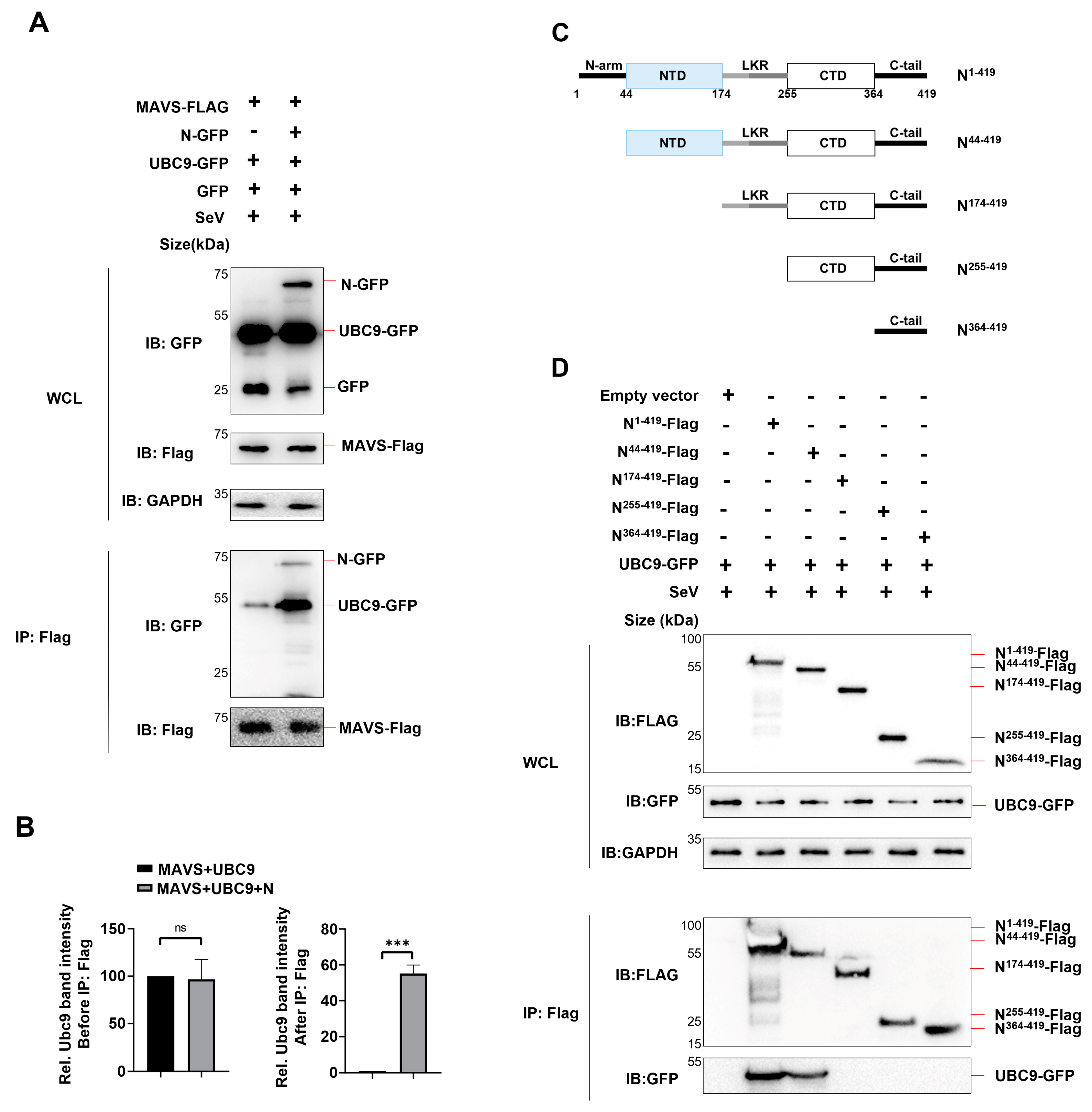
3.2. SARS-COV-2 N Protein Enhanced Molecular Interaction between UBC9 and MAVS
Previous studies have reported that the N protein of SARS-CoV-2 interacts with MAVS and inhibits the Lys63-linked poly-ubiquitination of MAVS [
9]. Based on our findings regarding the interaction between the N protein and UBC9, we further investigated whether the N protein mediates the molecular interaction between UBC9 and MAVS.
To examine this, we co-expressed the N protein, MAVS, and UBC9 in HEK293T cell and performed a Co-IP assay. The results revealed that MAVS could interact with UBC9 even in the absence of the N protein. However, the presence of the N protein enhanced the molecular interaction between UBC9 and MAVS (
Figure 4A,B).
It is known that SARS-CoV-2 N protein consists of two domains, the N-terminal domain (NTD; 44–174) and the C-terminal domain (CTD; 255–364) [
12]. (
Figure 4C). The NTD is responsible for RNA-binding and the CTD for dimerization and oligomerization. Two domains are connected by a flexible linker, which contains Serine/Arginine-rich region and Leucin/Glutamine-rich region (LKR; 174–255). Here, we determined the domain of SARS-CoV-2 N Protein involved in the interaction with UBC9 by constructing four mutants, N
44-419, N
174-419, N
255-419, and N
364-419, which lack N-arm, NTD, LKR, CTD, in sequence respectively. Co-IP results showed that, like full length N Protein, mutants N
44-419 could interact with UBC9, except mutant N
174-419, N
255-419, and N
364-419. This suggests that the NTD domain of N Protein interacts with the UBC9 protein (
Figure 4C,D).
Taken together, these results demonstrate that the N-terminal domain (NTD) of the N Protein interacts with the UBC9 protein, enhancing the molecular interaction between UBC9 and MAVS.
3.3. The Interaction between MAVS and UBC9 Inhibits MAVS Ubiquitination by Enhancing Its SUMOylation
SUMOylation has a narrower range of enzymatic machinery compared to Ubiquitylation. SUMOylation primarily relies on a single E1-activating enzyme complex called SAE1/SAE2, and a sole E2 SUMO-conjugating enzyme known as UBC9 [
12]. Consequently, the increased molecular interaction observed between MAVS and UBC9 could potentially result in an up-regulation of SUMOylation on MAVS, impacting its function.
Previous studies have reported that when the RIG-I/MAVS antiviral pathway is activated, polymeric chains of SUMO3, but not SUMO1 and SUMO2, are observed on MAVS [
13]. Additionally, K63 ubiquitination is known to be essential for MAVS activation and propagation of the antiviral signaling cascade [
14].
MAVS boasts several lysine residues distributed across different domains. For instance, previous research has reported that TRIM31 interacts with MAVS and orchestrates Lys63 (K63)-linked polyubiquitination on specific lysine residues, including Lys10, Lys311, and Lys461 [
15]. Moreover, certain lysine residues, such as Lys461 and Lys500, have been identified as sites for SUMOylation on MAVS. Substituting either of these two sites with arginine, as demonstrated in cases like MAVS-K461R or MAVS-K500R, significantly inhibits poly-SUMOylation of MAVS. A double mutation of Lys461 and Lys500 to arginine not only hampers MAVS poly-SUMOylation but also drastically diminishes K63-linked MAVS polyubiquitination [
16]. These findings strongly suggest that ubiquitination and SUMOylation of MAVS can indeed take place on the same lysine residue, indicating a close interaction between these two post-translational modification processes.
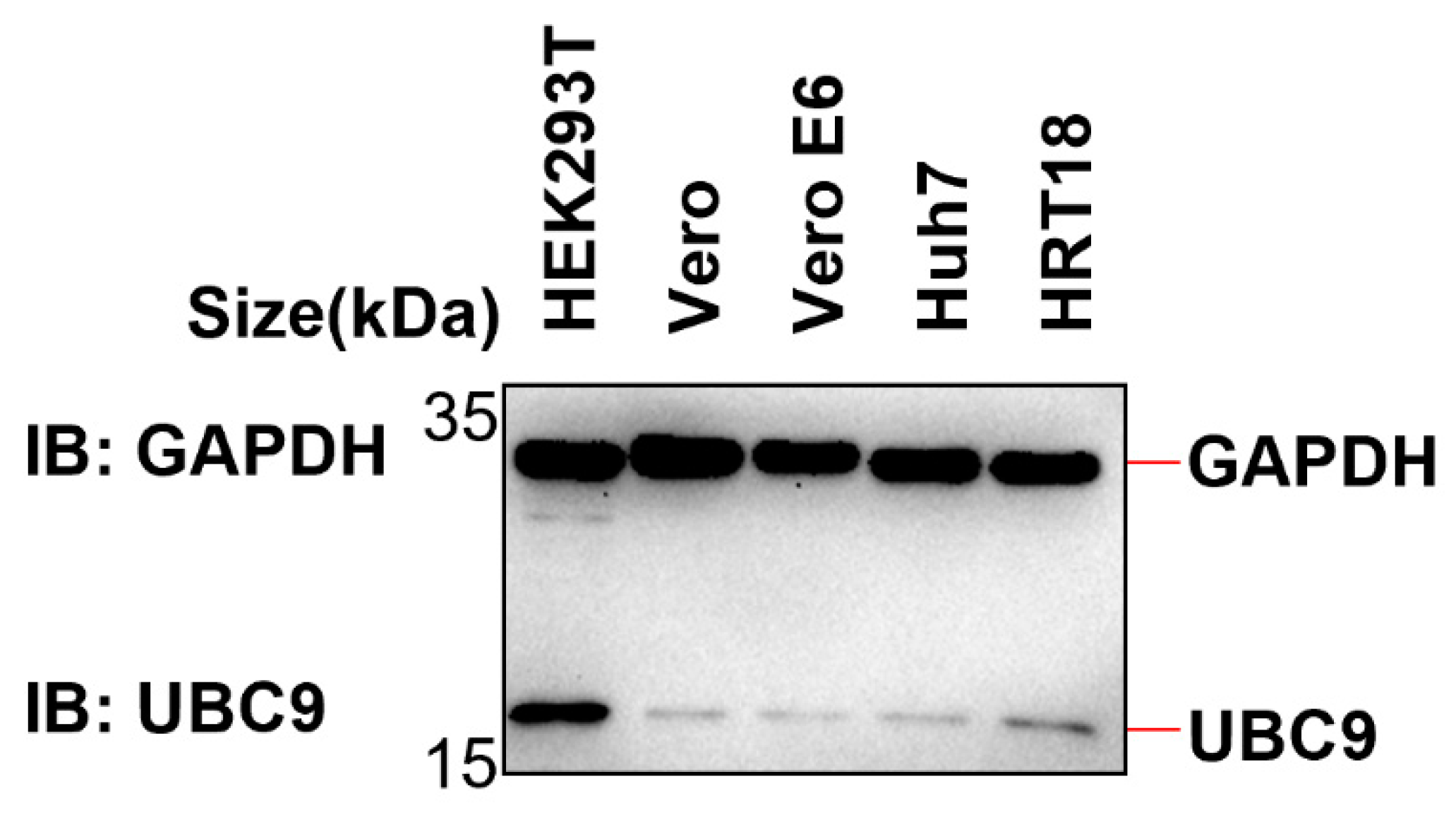
Given this interplay between ubiquitination and SUMOylation on the same lysine residue, we embarked on further investigations to ascertain whether the augmented interaction between MAVS and UBC9 curtails MAVS ubiquitination by intensifying its SUMOylation. First, we examined the endogenous expression level of UBC9 in different cell lines (HEK293T, Vero, Vero E6, Huh7, and HRT18). The results from western blotting showed that UBC9 is highly expressed in HEK293T cells but underexpressed in Vero E6 cells (
Figure 5).
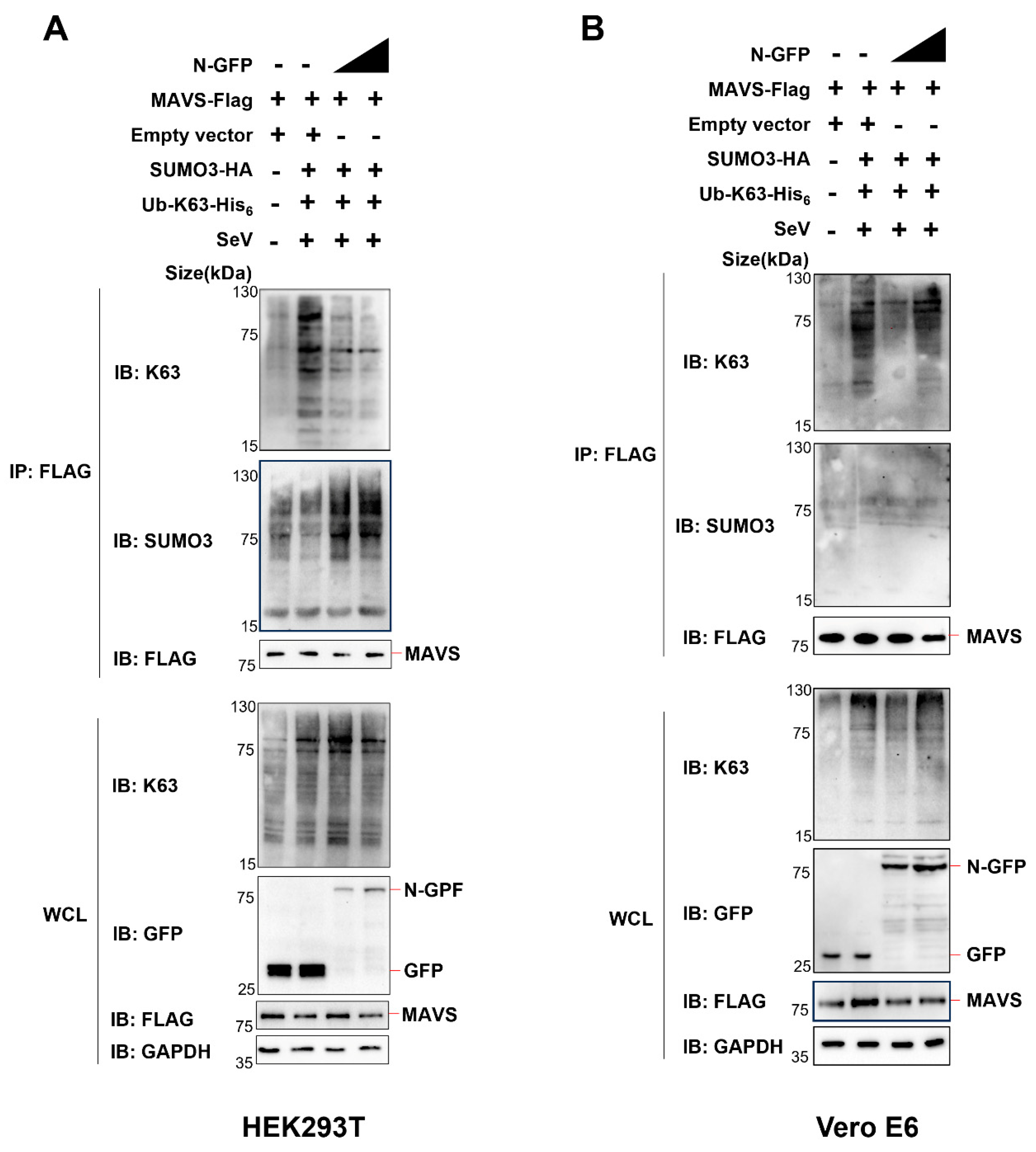
Next, we co-expressed Ub-K63-his6 (ubiquitin containing a substitution of arginine for all lysine residues, except the lysine at position 63), SUMO3-HA, MAVS-Flag, and N-GFP in HEK293T cells or Vero E6 cells. Subsequently, we conducted Co-IP analysis employing an anti-Flag antibody to examine alterations in the ubiquitination and SUMOylation of MAVS.
In HEK293T cells (endogenous UBC9 is highly expressed), we made an interesting observation. As we incrementally expressed the N protein, there was a notable decrease in K63 ubiquitination and a simultaneous increase in SUMOylation of MAVS (
Figure 6A).
However, the pattern differed in Vero E6 cells, where endogenous UBC9 is underexpressed. In this context, we did not observe the same trend. There was no significant decrease in K63 ubiquitination, nor was there an increase in SUMOylation of MAVS when we incrementally expressed the N protein, as shown in
Figure 6B.
Taken together, these results demonstrate that the N Protein interacts with the UBC9 protein, enhancing the molecular interaction between UBC9 and MAVS. This enhanced interaction between MAVS and UBC9 inhibits MAVS ubiquitination by enhancing its SUMOylation.
3.4. The Interaction of N Protein and UBC9 Plays a Crucial Role in Regulating the SUMOylation and Ubiquitination Modifications of MAVS
To further investigate this phenomenon, we conducted experiments in HEK293T cells, where endogenous UBC9 is highly expressed, using a non-conjugatable SUMO3
KR mutant (a SUMO3 mutant with arginine substitutions for all lysine residues). In these cells, we co-expressed K63-his
6, WT SUMO3 or SUMO3
KR mutant, along with MAVS-Flag and N-GFP. Surprisingly, in contrast to the significant decrease in K63 ubiquitination and increase in SUMOylation observed when overexpressing SUMO3, there was no such effect when overexpressing the SUMO3
KR mutant in HEK293 cells (
Figure 7A).
As we demonstrated that the UBC9
C93A mutant impairs its ability to bind to the N protein (
Figure 1C), we further explored its impact. We co-expressed K63-his
6, WT UBC9 or the UBC9
C93A mutant, along with MAVS-Flag and N-GFP in Vero E6 cells, where endogenous UBC9 is underexpressed. As expected, the overexpression of the UBC9
C93A mutant did not lead to a significant decrease in K63 ubiquitination or an increase in SUMOylation of MAVS in Vero E6 cells (
Figure 7B).
Moreover, our results revealed that UBC9 interacts with the NTD domain of the SARS-CoV-2 N Protein (
Figure 4C,D). In order to explore the impact of this interaction, we co-expressed WT N protein, the N
44-419 mutant (containing the NTD domain), or the N
174-419 mutant (lacking the NTD domain) with K63-his
6, SUMO3-HA, and MAVS-Flag in HEK293T cells. The results demonstrated that both the WT N protein and the N
44-419 mutant (with the NTD domain) caused a notable decrease in K63 ubiquitination and a simultaneous increase in SUMOylation of MAVS. However, the N
174-419 mutant, lacking the NTD domain and failing to interact with endogenous UBC9, did not result in a significant decrease in K63 ubiquitination or an increase in SUMOylation of MAVS in HEK293T cells (
Figure 7C).
Taken together, these results suggest that the interaction of N protein and UBC9 plays a crucial role in regulating the SUMOylation and Ubiquitination modifications of MAVS during SARS-COV-2 infection.
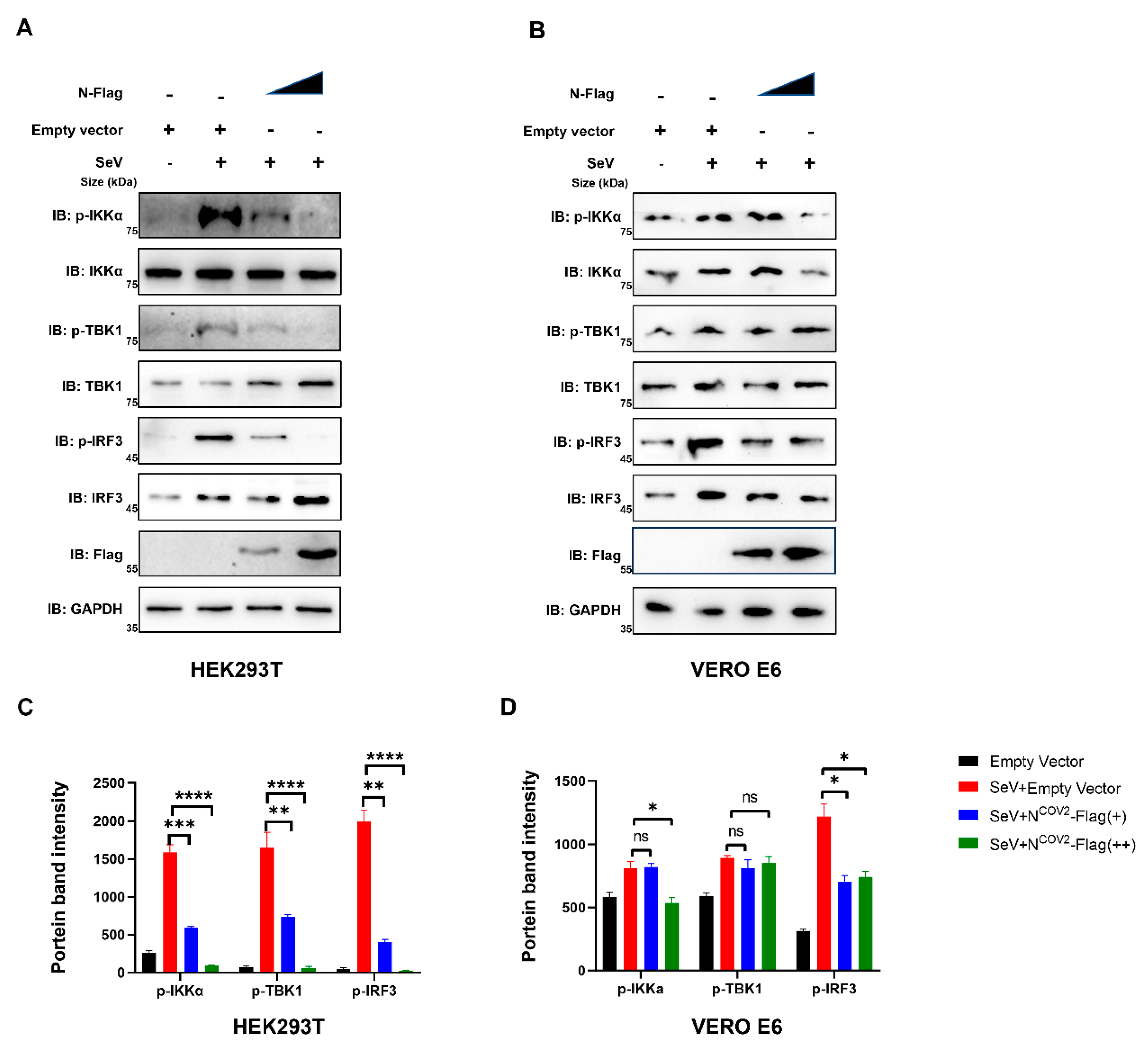
3.5. UBC9 Plays a Critical Role in the Process of Impaired IFN I Response Caused by the In-Teraction between the N Protein and MAVS during SARS-CoV-2 Infection
The decrease in phosphorylation of IKKα, TBK1, and IRF3 indicates impaired IFN I response upon incremental expression of the N protein [
17]. We investigated the impact of incremental N protein expression on the phosphorylation levels of IKKα, TBK1, and IRF3 in HEK293T cells, where UBC9 is highly expressed. Our results demonstrated a significant reduction in phosphorylation levels in HEK293T cells (
Figure 8A,C). Conversely, this phenomenon was less pronounced in UBC9-underexpressed VeroE6 cells (
Figure 8B,D).
These results demonstrate that UBC9 plays a critical role in the process of impaired IFN I response caused by the interaction between the N protein and MAVS during SARS-CoV-2 infection.
4. Discussion
The ongoing global pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has underscored the urgent need for a comprehensive understanding of the virus and its interactions with host cellular machinery. Despite extensive efforts to combat the virus, the molecular mechanisms by which SARS-CoV-2 overcomes host defense systems to cause respiratory symptoms and potentially fatal outcomes remain largely elusive [
18].
Host defense responses are initiated by the recognition of pathogen-related molecular patterns through various receptors [
19,
20]. Upon sensing viral RNA duplexes, key players like RIG-I and Toll-like receptor family members trigger a signaling cascade that induces the transcription of interferons (IFN-β and IFN-α) to mount an antiviral defense [
14,
21,
22,
23,
24]. In this process, Lys63-linked polyubiquitination of MAVS plays a crucial role in facilitating the aggregation of MAVS, thereby initiating IFN-β signaling and downstream antiviral responses [
9,
14,
25]. It has been demonstrated that the SARS-CoV-2 N protein inhibits Lys63-linked polyubiquitination and aggregation of MAVS, subsequently suppressing the innate antiviral immune response [
9]. However, the precise molecular mechanism by which SARS-CoV-2 N protein regulates the polyubiquitination of MAVS remains elusive.
In this study we initial found that there is interaction between the human beta coronavirus N protein and the SUMO-conjugating E2 enzyme UBC9, as illustrated in
Figure 1,
Figure 2 and
Figure 3. This interaction holds substantial significance, particularly in light of previous reports indicating that the SARS-CoV-2 N protein plays a pivotal role in modulating MAVS’s interaction with RIG-I and TRIM31, thereby suppressing innate immune signaling at the MAVS level [
9]. Our subsequent investigations have provided further validation that the interaction between N and UBC9 amplifies the molecular interplay between UBC9 and MAVS (
Figure 4A,B). In addition, we have identified that the N protein’s interaction with UBC9 is contingent on the presence of the N-terminal domain (NTD) (
Figure 4C,D).
In addition to its role in ubiquitination, SUMOylation is a crucial protein modification process with multifaceted involvement in antiviral defense mechanisms, as supported by recent research [
12,
14,
26]. Notably, recent studies have illuminated the ability of the SARS-CoV-2 Nsp5 protein to induce MAVS SUMOylation through its interaction with UBC9 [
27]. Building upon our initial findings, our subsequent study aimed to delve deeper into the potential implications of the intensified molecular interaction between MAVS and UBC9 on both MAVS ubiquitination and SUMOylation. To explore this further, we conducted experiments using cell models with varying UBC9 expression levels and observed that UBC9 is highly expressed in HEK293T cells but underexpressed in Vero E6 cells (
Figure 5). Intriguingly, we discovered that the SARS-CoV-2 N protein inhibits MAVS ubiquitination while enhancing its SUMOylation in HEK293T cells but not in VERO E6 cells (
Figure 6A,B). Notably, this phenomenon indicates that the N protein-induced SUMOylation of MAVS is contingent upon the presence of UBC9.
In the subsequent experiments, we proceeded to overexpress SUMO3
KR, a non-conjugatable SUMO3 mutant that contains arginine substitutions for all lysine residues in SUMO3, and UBC9
C93A, a UBC9 mutant that impairs its ability to bind to the N protein, in HEK293T and VERO E6 cells, respectively. Interestingly, we observed that the lack of SUMO3 or the absence of interaction between N and UBC9 both resulted in the inhibition of N protein-induced MAVS SUMOylation (
Figure 7A,B). Furthermore, our previous findings indicated that in the absence of the NTD domain, the N protein fails to interact with UBC9. This lack of interaction also results in an inability to inhibit MAVS ubiquitination while simultaneously enhancing its SUMOylation (
Figure 7C).
Hence, we have successfully elucidated how the SARS-CoV-2 N protein enhances the molecular interaction between UBC9 and MAVS. This interaction, as we have shown, hinders MAVS ubiquitination by promoting its SUMOylation. As a result, we proceeded to investigate whether this interaction has any impact on the MAVS-TBK1-IKK complex within the MAVS-IRF3 signaling cascade. As anticipated, the gradual increase in N protein expression led to reduced phosphorylation levels of IKKα, TBK1, and IRF3 in UBC9 highly expressed HEK293T cells, while this effect was not observed in UBC9 underexpressed VERO E6 cells (
Figure 8).
In summary, our results highlight the significance of the interaction between the SARS-CoV-2 N protein and UBC9. This interaction appears to hinder the ubiquitination of MAVS by facilitating its SUMOylation, subsequently impairing the host's interferon (IFN) type I response. These findings illuminate the complex interplay between various post-translational modifications and provide insights into the regulatory mechanisms that govern MAVS-mediated antiviral responses.
Author Contributions
Data curation, C.H., Y.Y. and P.P.; funding acquisition, Z.L.; methodology, Y.H., S.C., J.C., J.W., G.X., X.T., X.X., J.L., J.Y., Z.J. and B.L.; project administration, Z.T. and Z.L.; resource Z.L.; supervision, Z.L., L.L. and W.D.; writing—original draft, C.H., Y.Y. and P.P.; writing—review and editing, Z.T., W.D., L.L. and Z.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Hubei Provincial Technology Innovation Project (2023BCB058), Hubei Provincial Natural Science Foundation (2023AFB855, 2022CFB324) and Principal Investigator Program at Hubei University of Medicine (HBMUPI202102), Cultivating Project for Young Scholar at Hubei University of Medicine (2020QDJZR024) and Hubei Provincial Training Program of Innovation and Entrepreneurship for Undergraduates (S202310929005). The funders had no role in the study design, data collection, or manuscript preparation.
Institutional Review Board Statement
Not applicable. This study is not involving humans or animals.
Informed Consent Statement
Not applicable. This study is not involving humans.
Data Availability Statement
All processed data are included in the manuscript or provided as supplementary data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Naslund, T.I.; Liljestrom, P.; Weber, F.; Reis e Sousa, C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Pere, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Shemesh, M.; Aktepe, T.E.; Deerain, J.M.; McAuley, J.L.; Audsley, M.D.; David, C.T.; Purcell, D.F.J.; Urin, V.; Hartmann, R.; Moseley, G.W.; et al. SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLoS Pathog. 2021, 17, e1009800. [Google Scholar]
- Xiangbo, Z.; Zhaofang, Y.; Jinjing, G.; Zhuandi, G.; Suocheng, W. Bovine coronavirus nucleocapsid suppresses IFN-beta production by inhibiting RIG-I-like receptors pathway in host cells. Arch. Microbiol. 2022, 204, 536. [Google Scholar] [CrossRef] [PubMed]
- Yelemali, P.; Hao, L.; Liu, Q. Mechanisms of host type I interferon response modulation by the nucleocapsid proteins of alpha- and betacoronaviruses. Arch. Virol. 2022, 167, 1925–1930. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Xiao, F.; Hu, D.; Ge, W.; Tian, M.; Wang, W.; Pan, P.; Wu, K.; Wu, J. SARS-CoV-2 Nucleocapsid Protein Interacts with RIG-I and Represses RIG-Mediated IFN-beta Production. Viruses 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Dai, T.; Qin, Z.; Pan, T.; Chu, F.; Lou, L.; Zhang, L.; Yang, B.; Huang, H.; Lu, H.; et al. Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell Biol. 2021, 23, 718–732. [Google Scholar] [CrossRef]
- Fan, Z.; Zhuo, Y.; Tan, X.; Zhou, Z.; Yuan, J.; Qiang, B.; Yan, J.; Peng, X.; Gao, G.F. SARS-CoV nucleocapsid protein binds to hUbc9, a ubiquitin conjugating enzyme of the sumoylation system. J. Med. Virol. 2006, 78, 1365–1373. [Google Scholar] [CrossRef]
- Matsuo, T. Viewing SARS-CoV-2 Nucleocapsid Protein in Terms of Molecular Flexibility. Biology 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Li, X.; Zhang, L.; Zong, Z.; Wang, F.; Huang, J.; Zeng, L.; Zhang, C.; Yan, H.; Zhang, L.; et al. SUMOylation in Viral Replication and Antiviral Defense. Adv. Sci. 2022, 9, e2104126. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.W.; Lee, Y.; Yun, M.; Kang, J.; Lee, S.B. Formation of SUMO3-conjugated chains of MAVS induced by poly(dA:dT), a ligand of RIG-I, enhances the aggregation of MAVS that drives the secretion of interferon-beta in human keratinocytes. Biochem. Biophys. Res. Commun. 2020, 522, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Hou, F.; Sun, L.; Zheng, H.; Skaug, B.; Jiang, Q.X.; Chen, Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 2011, 146, 448–461. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, M.; Chu, H.; Zhang, H.; Wu, H.; Song, G.; Wang, P.; Zhao, K.; Hou, J.; Wang, X.; et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat. Immunol. 2017, 18, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Zhang, L.; Ran, Y.; Zhang, M.; Yang, B.; Lu, H.; Lin, S.; Zhang, L.; Zhou, F. MAVS deSUMOylation by SENP1 inhibits its aggregation and antagonizes IRF3 activation. Nat. Struct. Mol. Biol. 2023, 30, 785–799. [Google Scholar] [CrossRef]
- Stapleford, K.; Sa Ribero, M.; Jouvenet, N.; Dreux, M.; Nisole, S. Interplay between SARS-CoV-2 and the type I interferon response. PLOS Pathog. 2020, 16, e1008737. [Google Scholar]
- Diamond, M.S.; Kanneganti, T.D. Innate immunity: The first line of defense against SARS-CoV-2. Nat. Immunol. 2022, 23, 165–176. [Google Scholar] [CrossRef]
- Hu, M.M.; Shu, H.B. Cytoplasmic Mechanisms of Recognition and Defense of Microbial Nucleic Acids. Annu. Rev. Cell Dev. Biol. 2018, 34, 357–379. [Google Scholar] [CrossRef]
- Li, S.; Kuang, M.; Chen, L.; Li, Y.; Liu, S.; Du, H.; Cao, L.; You, F. The mitochondrial protein ERAL1 suppresses RNA virus infection by facilitating RIG-I-like receptor signaling. Cell Rep. 2021, 34, 108631. [Google Scholar] [CrossRef]
- Lozhkov, A.A.; Plotnikova, M.A.; Egorova, M.A.; Baranovskaya, I.L.; Elpaeva, E.A.; Klotchenko, S.A.; Vasin, A.V. Simultaneous Detection of RIG-1, MDA5, and IFIT-1 Expression Is a Convenient Tool for Evaluation of the Interferon-Mediated Response. Viruses 2022, 14. [Google Scholar] [CrossRef]
- Wu, B.; Hur, S. How RIG-I like receptors activate MAVS. Curr. Opin. Virol. 2015, 12, 91–98. [Google Scholar] [CrossRef]
- Gori Savellini, G.; Anichini, G.; Gandolfo, C.; Cusi, M.G. SARS-CoV-2 N Protein Targets TRIM25-Mediated RIG-I Activation to Suppress Innate Immunity. Viruses 2021, 13. [Google Scholar] [CrossRef]
- Thoresen, D.; Wang, W.; Galls, D.; Guo, R.; Xu, L.; Pyle, A.M. The molecular mechanism of RIG-I activation and signaling. Immunol. Rev. 2021, 304, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Chen, Y.; Liu, X.; Yuan, F.; Tan, E.Y.J.; Lei, Y.; Song, N.; Han, Y.; Pascal, B.D.; Griffin, P.R.; et al. Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains. Immunity 2021, 54, 2218–2230. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; de Thé, H.; Lallemand-Breitenbach, V. Sumoylation in Physiology, Pathology and Therapy. Cells 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qiao, J.; You, Q.; Zong, S.; Peng, Q.; Liu, Y.; Hu, S.; Liu, W.; Li, S.; Shu, X.; et al. SARS-CoV-2 Nsp5 Activates NF-κB Pathway by Upregulating SUMOylation of MAVS. Front. Immunol. 2021, 12, 750969. [Google Scholar] [CrossRef]
Figure 1.
SARS-COV-2 N protein interacts with UBC9. (A) pCDNA3.1 (3XFlag Tag) with N protein coding genes and pEGFP-N1 (GFP Tag) with UBC9 coding gene were co-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays. (B) pDsRED-mono-N1 (RFP Tag) with SARS-COV-2 N protein coding genes and pEGFP-N1 (GFP Tag) with UBC9 or UBC9C93A coding gene were co-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell was stained with DAPI before confocal microscopy imaging. (C) pET28a (HIS6 tag, Kan+) with SARS-COV-2 N protein coding genes and pGEX-6P-1 (GST tag, Amp+) with UBC9 or UBC9C93A coding gene were co-expressed in BL21 (DE3) E. coli. cell lysates were subjected to Co-IP after IPTG (1 mM) inducing for 6 hours. (D) pCDNA3.1 (3XFlag Tag) with SARS-COV-2 N protein coding genes were over-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays.
Figure 1.
SARS-COV-2 N protein interacts with UBC9. (A) pCDNA3.1 (3XFlag Tag) with N protein coding genes and pEGFP-N1 (GFP Tag) with UBC9 coding gene were co-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays. (B) pDsRED-mono-N1 (RFP Tag) with SARS-COV-2 N protein coding genes and pEGFP-N1 (GFP Tag) with UBC9 or UBC9C93A coding gene were co-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell was stained with DAPI before confocal microscopy imaging. (C) pET28a (HIS6 tag, Kan+) with SARS-COV-2 N protein coding genes and pGEX-6P-1 (GST tag, Amp+) with UBC9 or UBC9C93A coding gene were co-expressed in BL21 (DE3) E. coli. cell lysates were subjected to Co-IP after IPTG (1 mM) inducing for 6 hours. (D) pCDNA3.1 (3XFlag Tag) with SARS-COV-2 N protein coding genes were over-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays.
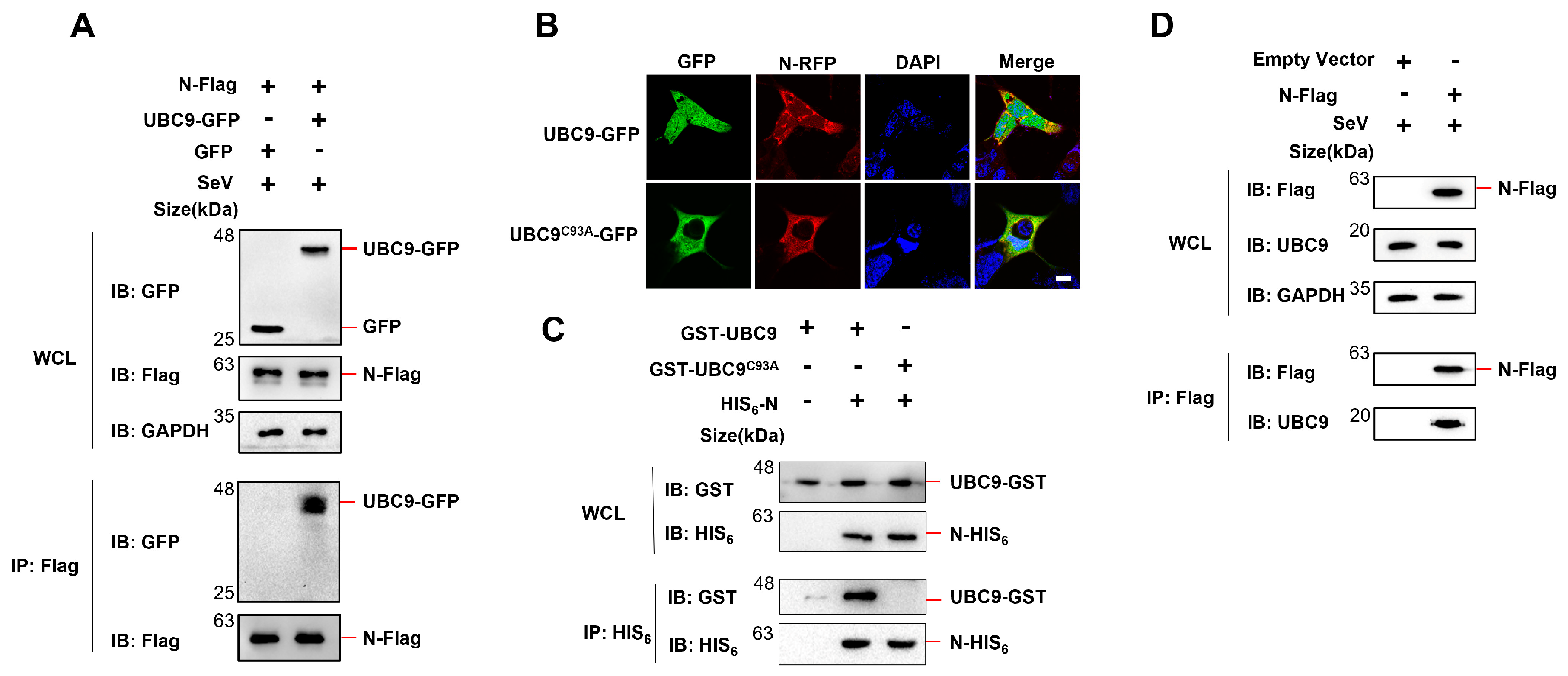
Figure 2.
Five human beta coronavirus N protein interacts with UBC9. pET28a-N (HIS6 tag, Kan+) containing five human beta coronavirus N protein coding genes and pGEX-UBC9 (GST tag, Amp+) harboring UBC9 coding gene were co-expressed in BL21(DE3)E.coli. Cell lysates were prepared and subjected to Co-IP following IPTG induction (1mM) for 6 hours.
Figure 2.
Five human beta coronavirus N protein interacts with UBC9. pET28a-N (HIS6 tag, Kan+) containing five human beta coronavirus N protein coding genes and pGEX-UBC9 (GST tag, Amp+) harboring UBC9 coding gene were co-expressed in BL21(DE3)E.coli. Cell lysates were prepared and subjected to Co-IP following IPTG induction (1mM) for 6 hours.
Figure 3.
Three human beta coronavirus N protein interacts with UBC9. pEGFP-N1 (GFP Tag) with three human beta coronavirus N protein coding genes and pCDNA3.1 (3XFlag Tag) with UBC9 coding gene were co-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays.
Figure 3.
Three human beta coronavirus N protein interacts with UBC9. pEGFP-N1 (GFP Tag) with three human beta coronavirus N protein coding genes and pCDNA3.1 (3XFlag Tag) with UBC9 coding gene were co-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays.
Figure 4.
SARS-COV-2 N protein enhanced the molecular interaction between UBC9 and MAVS. (A) pEGFP-N1 (GFP Tag) with SARS-COV-2 N protein or UBC9 coding genes and pCDNA3.1 (3XFlag Tag) with MAVS coding gene were co-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays. (B) Relative Ubc9 band intensity before or after IP: Flag in (A) were calculated by using ImageJ. ns: no significant difference, *** p < 0.001. (C) Schematic diagram of wild-type SARS-CoV-2 N protein and truncated mutants. (D) pEGFP-N1 (GFP Tag) with UBC9 coding genes and pCDNA3.1 (3XFlag Tag) with SARS-COV-2 N protein or truncation coding gene were co-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays.
Figure 4.
SARS-COV-2 N protein enhanced the molecular interaction between UBC9 and MAVS. (A) pEGFP-N1 (GFP Tag) with SARS-COV-2 N protein or UBC9 coding genes and pCDNA3.1 (3XFlag Tag) with MAVS coding gene were co-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays. (B) Relative Ubc9 band intensity before or after IP: Flag in (A) were calculated by using ImageJ. ns: no significant difference, *** p < 0.001. (C) Schematic diagram of wild-type SARS-CoV-2 N protein and truncated mutants. (D) pEGFP-N1 (GFP Tag) with UBC9 coding genes and pCDNA3.1 (3XFlag Tag) with SARS-COV-2 N protein or truncation coding gene were co-expressed in HEK293T cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays.
Figure 5.
The expression of UBC9 in HEK293T, Vero, Vero E6, Huh7 and HRT18 cells were detected by Western blotting.
Figure 5.
The expression of UBC9 in HEK293T, Vero, Vero E6, Huh7 and HRT18 cells were detected by Western blotting.
Figure 6.
The Interaction between SARS-CoV-2 N protein and UBC9 inhibits MAVS ubiquitination by enhancing its SUMOylation. (A,B) pEGFP-N1 (GFP Tag) with SARS-COV-2 N coding genes or pCDNA3.1 with MAVS-Flag, SUMO3-HA or K63-His6 coding genes were co-expressed in HEK293T cells (endogenous UBC9 highly expressed) or Vero E6 cells (endogenous UBC9 underexpressed). After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays.
Figure 6.
The Interaction between SARS-CoV-2 N protein and UBC9 inhibits MAVS ubiquitination by enhancing its SUMOylation. (A,B) pEGFP-N1 (GFP Tag) with SARS-COV-2 N coding genes or pCDNA3.1 with MAVS-Flag, SUMO3-HA or K63-His6 coding genes were co-expressed in HEK293T cells (endogenous UBC9 highly expressed) or Vero E6 cells (endogenous UBC9 underexpressed). After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays.
Figure 7.
The interaction of N protein and UBC9 plays a crucial role in regulating the SUMOylation and ubiquitination modifications of MAVS. (A) pEGFP-N1 (GFP Tag) with SARS-COV-2 N coding genes or pCDNA3.1 with MAVS-Flag, SUMO3-HA or SUMO3KR-HA, K63-His6 coding genes were co-expressed in HEK293T cells (endogenous UBC9 highly expressed). After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays. (B) pEGFP-N1 (GFP Tag) with SARS-COV-2 N coding genes or pCDNA3.1 with MAVS-Flag, SUMO3-HA, UBC9 or UBC9C93A, K63-His6 coding genes were co-expressed in Vero E6 cells (endogenous UBC9 underexpressed). After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays. (C) pEGFP-N1 (GFP Tag) with SARS-COV-2 N, N44-419 or N174-419 coding genes or pCDNA3.1 with MAVS-Flag, SUMO3-HA, K63-His6 coding genes were co-expressed in Vero E6 cells (endogenous UBC9 underexpressed). After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays.
Figure 7.
The interaction of N protein and UBC9 plays a crucial role in regulating the SUMOylation and ubiquitination modifications of MAVS. (A) pEGFP-N1 (GFP Tag) with SARS-COV-2 N coding genes or pCDNA3.1 with MAVS-Flag, SUMO3-HA or SUMO3KR-HA, K63-His6 coding genes were co-expressed in HEK293T cells (endogenous UBC9 highly expressed). After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays. (B) pEGFP-N1 (GFP Tag) with SARS-COV-2 N coding genes or pCDNA3.1 with MAVS-Flag, SUMO3-HA, UBC9 or UBC9C93A, K63-His6 coding genes were co-expressed in Vero E6 cells (endogenous UBC9 underexpressed). After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays. (C) pEGFP-N1 (GFP Tag) with SARS-COV-2 N, N44-419 or N174-419 coding genes or pCDNA3.1 with MAVS-Flag, SUMO3-HA, K63-His6 coding genes were co-expressed in Vero E6 cells (endogenous UBC9 underexpressed). After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for Co-IP assays.
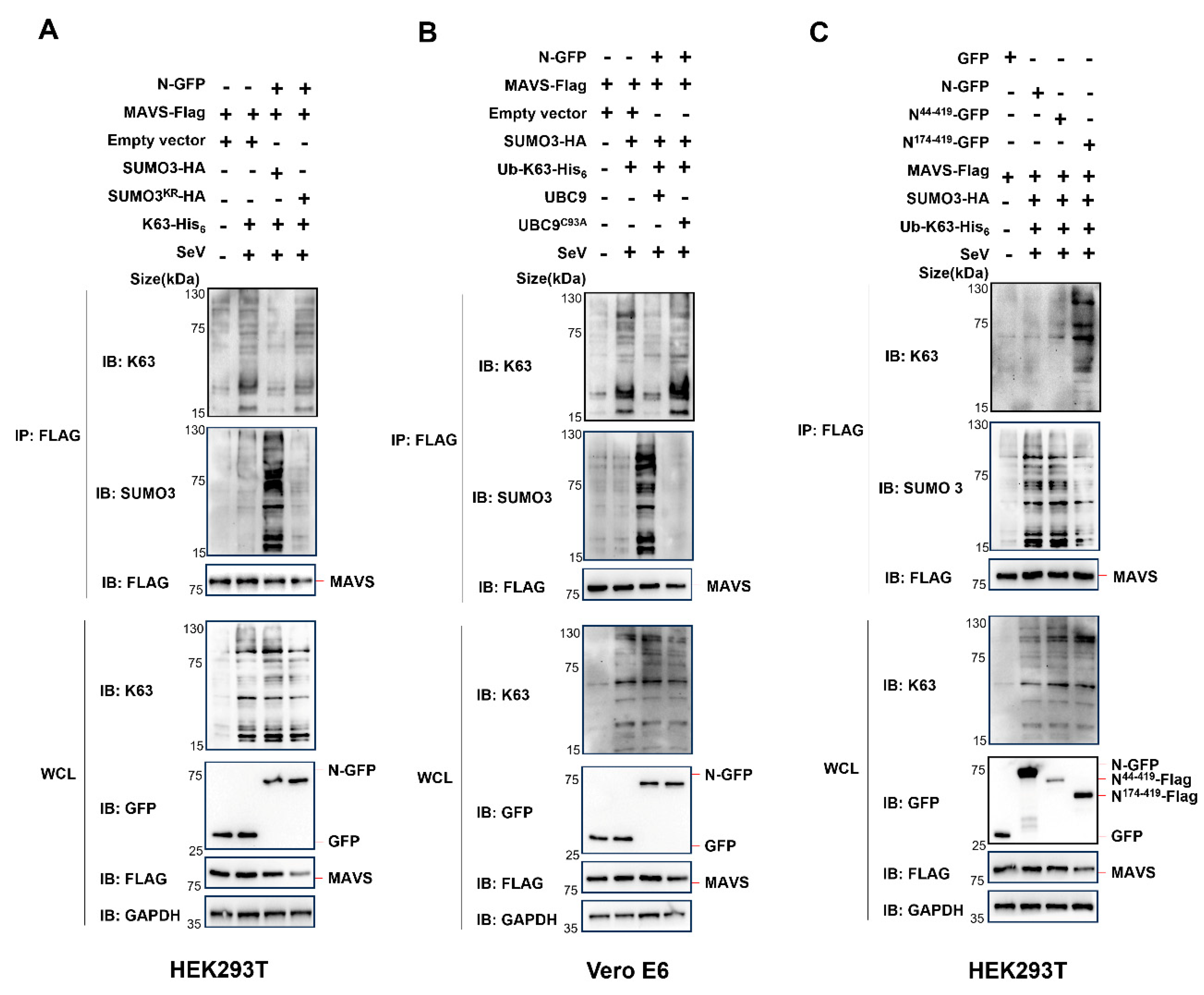
Figure 8.
UBC9 plays a critical role in the process of impaired IFN I response caused by the in-teraction between the N protein and MAVS during SARS-CoV-2 infection. (
A,
B) pCDNA3.1 with SARS-CoV-2 N protein coding genes were gradual increased expressed in HEK293T cells or VERO E6 cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for western blotting. (
C,
D) Relative band intensity of western blotting results in
Figure 7A,B were calculated by using ImageJ. ns: no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001.
Figure 8.
UBC9 plays a critical role in the process of impaired IFN I response caused by the in-teraction between the N protein and MAVS during SARS-CoV-2 infection. (
A,
B) pCDNA3.1 with SARS-CoV-2 N protein coding genes were gradual increased expressed in HEK293T cells or VERO E6 cells. After a 24-hour lipofectamine transfection followed by 8 hours of SeV stimulation, cell lysates were collected for western blotting. (
C,
D) Relative band intensity of western blotting results in
Figure 7A,B were calculated by using ImageJ. ns: no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).