Submitted:
30 October 2023
Posted:
31 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Parameters Characterizing Inbreeding Load and Purging
3. Softwares
4. Correlation among Inbreeding Coefficients
5. Estimates of Lethal Equivalents in Populations of Different Species
6. Studies Signalling Purging Based on Ancestral Inbreeding or Inbreeding-Purging Model
7. Application Possibilities of Purging, Future Perspective
Author Contributions
Conflicts of Interest
References
- Allendorf, F.W.; Funk, W.C.; Aitken, S.N; Byrne, M.; Luikart, G. Conservation and the Genomics and Populations, 3rd ed.; Publisher: Oxford University Press, Oxford, United Kingdom, 2022; pp. 1–672.
- Henderson, C.R. Best Linear Unbiased Estimation and Prediction under a selection model. Biometrics 1975, 31, 423-447. [CrossRef]
- Mehrabani-Yeganeh, H.; Gibson, J.P.; Schaeffer, L.R. Including coefficients of inbreeding in BLUP evaluation and its effect on response to selection. J. Anim. Breed. Genet. 2000, 117, 145–151. [CrossRef]
- Curik, I.; Ferencakovic, M.; Sölkner, J. Genomic dissection of inbreeding depression: A gate to new opportunities. Revista Brasileira de Zootecnia, 2017, 46, 773–782. [CrossRef]
- Schäler, J.; Krüger, B.; Thaller, G.; Hinrichs, D. Comparison of ancestral, partial, and genomic inbreeding in a local pig breed to achieve genetic diversity. Conserv. Genet. Resour., 2018, 12, 77-86. [CrossRef]
- Haig, S.; M., Ballou, J. D.; Derrickson, S. R. Management options for preserving genetic diversity: reintroduction of Guam rails to the wild. Conserv. Biol., 1990, 4, 290-300. [CrossRef]
- Zhang, Q.; Calus, M.P.; Guldbrandtsen, B.; Lund, M. S.; Sahana, G. Estimation of inbreeding using pedigree, 50k SNP chip genotypes and full sequence data in three cattle breeds. BMC Genet., 2015, 16(1), 1-11. [CrossRef]
- Frankham, R. Conservation genetics. Annu. Rev. Genetics, 1995, 29, 305-327.
- Kardos, M.; Luikart, G.; Allendorf, F.W. Measuring individual inbreeding in the age of genomics: marker-based measures are better than pedigrees. Heredity, 2015, 115(1), 63-72. [CrossRef]
- Malecot, G. (1970): Mathematics of Heredity. Publisher: Freeman & Company, Ltd. New York, US, pp. 1-88.
- Galla, S.J.; Brown, L.; Couch-Lewis (Ngāi Tahu: Te Hapū o Ngāti Wheke, Ngāti Waewae), Y., Cubrinovska, I.; Eason, D.; Gooley, R.M.; Hamilton, J.A.; Heath, J.A.; Hauser, S.S.; Latch, E.K., Matocq, M.D., Richardson, A.; Wold, J.R., Hogg, C.J., Santure, A.W.; Steeves, T.E. The relevance of pedigrees in the conservation genomics era. Mol. Ecol., 2022, 31, 41– 54.
- Hedrick, P.W.; García-Dorado, A. Understanding Inbreeding Depression, Purging, and Genetic Rescue. Trends Ecol. Evol., 2016, 31, 940-952. [CrossRef]
- Frankham, R.; Gilligan, D.M.; Morris, D.; Briscoe, D.A. Inbreeding and extinction: effects of purging. Conserv. Genet., 2001, 2, 279-284. [CrossRef]
- Crnokrak, P.; Roff; D.A, Inbreeding depression in the wild. Heredity, 1999, 83, 260-270. [CrossRef]
- Keller, L.F.; Waller, D.M. Inbreeding effects in wild populations. Trends Ecol. Evol.2002, 17, 230–241. [CrossRef]
- Leroy, G. Inbreeding depression in livestock species: review and meta-analysis. Anim. Genet., 2014, 45, 618–628. [CrossRef]
- Kardos, M.; Husby, A.; McFarlane, S. E.; Qvarnstrom, A.; Ellegren, H. Whole-genome resequencing of extreme phenotypes in collared flycatchers highlights the difficulty of detecting quantitative trait loci in natural populations. Mol. Ecol. Resour., 2016, 16, 727–741. [CrossRef]
- Doekes, H.P., Bijma, P., Windig, J.J. How depressing is inbreeding? A meta-analysis of 30 years of research on the effects of inbreeding in livestock. Genes, 2021, 12, 926. [CrossRef]
- Gutiérrez-Reinoso M.A.; Aponte P.M.; García-Herreros M.A. A review of inbreeding depression in dairy cattle: current status, emerging control strategies, and future prospects. J. Dairy Res., 2022, 28:1-10. [CrossRef]
- Vega-Trejo, R., de Boer, R.A., Fitzpatrick, J.L., Kotrschal, A. Sex-specific inbreeding depression: a meta-analysis. Ecol. Lett. 2022, 25, 1009–1026. [CrossRef]
- Sonesson, A.K.; Meuwissen, T.H.E. Mating schemes for optimum contribution selection with constrained rates of inbreeding. Genet. Sel. Evol., 2000, 32:231. [CrossRef]
- Sonesson, A.K.; Meuwissen, T.H.E. Minimization of rate of inbreeding for small populations with overlapping generations. Genet. Res. 2001, 77: 285-292. [CrossRef]
- Weigel, K. Controlling inbreeding in modern breeding programs. J. Dairy Sci. 2001, 84:E177–84. [CrossRef]
- Gebregiwergis, G.T.; Sørensen, A.C.; Henryon, M.; Meuwissen, T.H.E. Controlling coancestry and thereby future inbreeding by optimum-contribution selection using alternative genomic-relationship matrices. Front. Genet., 2020, 11, 345. [CrossRef]
- Kristensen, T.N.; Sørensen, A.C. Inbreeding–lessons from animal breeding, evolutionary biology and conservation genetics. Anim. Sci. 2005, 80, 121–33. [CrossRef]
- Templeton, A.R.; Read, B. The elimination of inbreeding depression in a captive herd of Speke’s Gazelle. In: C.M. Schonewald-Cox, S.M., MacBryde, B. and I. Thomas, editors, Genetics and conservation. Benjamin/Cummings, Menlo Park, California. 1983, p. 241-261.
- Read, B.; Frueh, R.J. Management and breeding of Speke’s gazelle, Gazella spekei, at the St. Louis Zoo, with a note on artificial insemination. — Int. Zoo Yb., 1980, 20: 99-106.
- Templeton, A.R.; Read, B. Factors eliminating inbreeding depression in a captive herd of Speke's gazelle (Gazella spekei). Zoo Biol., 1984, 3, 177-199. [CrossRef]
- Willis, K.; Wiese, R.J. Elimination of inbreeding depression from captive populations: Speke’s gazelle revisited. Zoo Biol., 1997, 16, 9–16.
- Templeton, A.R; Read, B. Elimination of inbreeding depression from a captive population of Speke’s gazelle: validity of the original statistical analysis and confirmation by permutation testing. Zoo Biol, 1998, 17:77–94. [CrossRef]
- Kalinowski, S.T.; Hedrick, P.W., Miller, P.S. Inbreeding depression in the Speke’s gazelle captive breeding program. Conserv. Biol. 2000, 14, 1375–84. [CrossRef]
- Templeton, A.R. The Speke’s gazelle breeding program as an illustration of the importance of multilocus genetic diversity in conservation biology: Response to Kalinowski et al. Conserv. Biol. 2002, 16: 1151-1155. [CrossRef]
- Bijlsma,B.; V. Putten, V. Environmental dependence of inbreeding depression and purging in Drosophila melanogaster J. Evolution. Biol.,1999, 12, 1125-1137. [CrossRef]
- Miller, P.S.; Hedrick, P.W.; Purging of inbreeding depression and fitness decline in bottlenecked populations of Drosophila melanogaster. J. Evol. Biol., 2001, 14, 595–601. [CrossRef]
- Pérez-Pereira, N., Pouso, R.; Rus, A., Vilas, A.; López-Cortegano, E., García-Dorado A., Quesada, H., Caballero, A. Long-term exhaustion of the inbreeding load in Drosophila melanogaster. Heredity, 2021, 127, 373–383. [CrossRef]
- Moreno, E.; Pérez-González, J.; Carranza, J.; Moya-Laraño, J. Better fitness in captive Cuvier’s gazelle despite inbreeding increase: evidence of purging? PloS ONE, 2015, 10:e0145111. [CrossRef]
- Hinrichs, D.; Bennewitz, J.; Wellmann, R.; Thaller, G. Estimation of ancestral inbreeding effects on stillbirth, calving ease and birthweight in German Holstein dairy cattle. J. Anim. Breed. Genet., 2015, 132, 59-67. [CrossRef]
- Mc Parland, S.; Kearney, F.; Berry, D. P. Purging of inbreeding depression within the Irish Holstein-Friesian population. Genet. Sel. Evol., 2009, 41, 1-8. [CrossRef]
- Wright, S. Coefficients of inbreeding and relationship. Am. Nat., 1922, 56, 330-338. [CrossRef]
- Ballou, J.D. Ancestral inbreeding only minimally affects inbreeding depression in mammalian populations. J. Hered., 1997, 88(3), 169-178. [CrossRef]
- Baumung, R.; Farkas, J.; Boichard, D.; Mészáros, G.; Sölkner, J.; Curik, I. GRAIN: a computer program to calculate ancestral and partial inbreeding coefficients using a gene dropping approach. J. Anim. Breed. Genet., 2015, 132, 100-108. [CrossRef]
- Slatis, H.M. An analysis of inbreeding in the European bison. Genetics, 1960, 45, 275-287. [CrossRef]
- Kennedy, E.S;, Grueber, C. E.; Duncan, R.P.; Jamieson, I.G. Severe inbreeding depression and no evidence of purging in an extremely inbred wild species—the Chatham Island black robin. Evolution, 2014, 68(4), 987-995. [CrossRef]
- Hinrichs, D.; Meuwissen, T.H.E.; Ødegard, J.; Holt, M.; Vangen, O.; Woolliams, J.A.; Analysis of inbreeding depression in the first litter size of mice in a long-term selection experiment with respect to the age of the inbreeding. Heredity, 2007, 99, 81–88. [CrossRef]
- López-Cortegano, E.; Moreno, E.; García-Dorado, A. Genetic purging in captive endangered ungulates with extremely low effective population sizes. Heredity, 2021, 127, 433-442. [CrossRef]
- Morton, N.E.; Crow, J.F.; Muller, H.J. An estimate of the mutational damage in man from data on consanguineous marriages. Proc. Natl. Acad. Sci. 1956, 42, 855–863. [CrossRef]
- Hoeck, P.E. A.; Wolak, M.E.; Switzer, R.A., Kuehler, C. M.; Lieberman, A.A. Effects of inbreeding and parental incubation on captive breeding success in Hawaiian crows. Biol. Conserv. 2015, 184, 357–364. [CrossRef]
- Boichard D. PEDIG: a fortran package for pedigree analysis suited for large populations. In Proceedings of the 7th world congress on genetics applied to livestock production: 19–23 August 2002; Montpellier. 2002, 525–8.
- Gutiérrez, J.P.; Goyache, F. A note on ENDOG: a computer program for analysing pedigree information. J. Anim. Breed. Genet., 2005, 122: 172-176. [CrossRef]
- Coster, A.; Coster, M.A. Package ‘pedigree’. R Package Version 1.4. 2010. Available online: https: //cran.r-project.org/web/packages/pedigree/index.html (accessed on 21 October 2023).
- Meuwissen, T.H.E.; Luo, Z. Computing inbreeding coefficients in large populations. Genet. Sel. Evol. 1992, 15, 24, 305–13. [CrossRef]
- Doekes, H.P.; Curik, I.; Nagy, I.; Farkas, J.; Kövér, G.; Windig, J.J. Revised calculation of Kalinowski’s ancestral and new inbreeding coefficients. Diversity, 2020, 12, 155. [CrossRef]
- MacCluer, J.W.; VandeBerg, J.L.; Read, B.; Ryder, O.A. Pedigree analysis by computer simulation. Zoo Biol., 1986, 5, 147–160. [CrossRef]
- Suwanlee, S.; Baumung, R.; Sölkner, J.; Curik, I. Evaluation of ancestral inbreeding coefficients: ballou’s formula versus gene dropping. Conserv Genet. 2007, 8, 489–95. [CrossRef]
- García-Dorado, A.; Wang, J.; López-Cortegano, E. Predictive model and software for inbreeding-purging analysis of pedigreed populations. G3, 2016, 6, 3593–3601. [CrossRef]
- López-Cortegano, E. PurgeR: Inbreeding and purging in pedigreed populations. Bioinformatics,2022, 38, 564–565. [CrossRef]
- Gilmour, A.R.; Gogel, B.J.; Cullis, B.R.; Thompson, R. ASReml User Guide Release 3.0. 2009, VSN International Ltd, Hemel Hempstead, HP1 ES, UK.
- Bates, D.; Machler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48.
- Bates, D.; Vazquez, A.I. Fit pedigree based mixed-effect models; 2014. http://pedigreemm.R-Forge.R-project.org. Accessed 22 Mar 2020.
- Curik, I.; Kover, G.; Farkas, J.; Szendrő, Z.; Romvari, R.; Sölkner, J.; Nagy, I. Inbreeding depression for kit survival at birth in a rabbit population under long-term selection. Genet. Sel. Evol., 2020, 52, 39. [CrossRef]
- Piles, M.; Sánchez, J.P.; Pascual, M.; Rodríguez-Ramilo, S.T. Inbreeding depression on growth and prolificacy traits in two lines of rabbit. J. Anim. Breed. Genet. 2023, 140, 39-48. [CrossRef]
- Ceballos, F.C.; Álvarez, G. Royal dynasties as human inbreeding laboratories: the Habsburgs. Heredity, 2013, 111, 114–21. [CrossRef]
- Jamieson, I.G.; Tracy, L.N., Fletcher, D.; Armstrong, D.P. Moderate inbreeding depression in a reintroduced population of North Island robins. Anim. Conserv. 2007, 10, 95–102. [CrossRef]
- Szulkin, M.; Garant, D.; McCleery, H.; Sheldon, B.C. Inbreeding depression along a life-history continuum in the great tit. J. Evol. Biol. 2007, 20, 1531–1543. [CrossRef]
- Laws, R.J.; Jamieson, I.G. Is lack of evidence of inbreeding depression in a threatened New Zealand robin indicative of reduced genetic load? Anim. Conserv, 2011, 14, 47-55. [CrossRef]
- Grueber, C. E.; Laws, R. J.; Nakagawa, S.; Jamieson, I.G. Inbreeding depression accumulation across life-history stages of the endangered takahe. Conserv. Biol. 2010, 24, 1617–1625. [CrossRef]
- Kennedy, E. S.; Grueber, C.E.; Duncan, R.P.; Jamieson, I.G. Severe inbreeding depression and no evidence of purging in an extremely inbred wild species - the Chatham Island black robin. Evolution 2014, 68, 987–995. [CrossRef]
- Hoeck, P.E.; Wolak, M.E.; Switzer, R.A.; Kuehler, C.M.; Lieberman, A.A. Effects of inbreeding and parental incubation on captive breeding success in Hawaiian crows. Biol. Conserv. 2015, 184, 357–364. [CrossRef]
- Stoffel, M.A.; Johnston, S.E.; Pilkington, J.G.; Pemberton, J.M.; Genetic architecture and lifetime dynamics of inbreeding depression in a wild mammal. Nat. Commun, 2021, 12, 2972. [CrossRef]
- Frankham, R. Effects of genomic homozygosity on total fitness in an invertebrate: lethal equivalent estimates for Drosophila melanogaster. Conserv. Genet. 2023, 24, 193-201. [CrossRef]
- Kardos, M.; Zhang, Y.; Parsons, K.M.; Kang, H.; Xu, X.; Liu, X.; Matkin, C.O.; Zhang, P.; Ward, E.J.; Hanson, M.B. Inbreeding depression explains killer whale population dynamics. Nat Ecol Evol. 2023, 7, 675–686. Sutter, J.;
- Tabah, L. Effets de la consanguinité et de l'endogamie. Une enquête en Morbihan et Loir-et-Cher. Population, 1952, 7, 249-266. [CrossRef]
- Tabah, L. Structure de la mortalité dans les familles consanguines. Population, 1953, 8, 511-526. [CrossRef]
- Falconer D.S. 1960. Genetics of the litter size in mice. J. Cell. Comp. Physiol. 56 (Suppl. 1) 153-167.
- Armbruster, P.; Reed, D.H. Inbreeding depression in benign and stressful environments. Heredity, 2005, 95, 235–242. [CrossRef]
- Grueber, C.E.; Nakagawa, S.; Laws, R.J.; Jamieson, I.G. Multimodel inference in ecology and evolution: challenges and solutions. J. Evol. Biol., 2011, 24, 699-711.
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, 2nd ed. New York: Springer-Verlag; 2002.
- Nietlisbach, P.; Muff, S.; Reid, J.M.; Whitlock, M.C.; Keller, L.F. Nonequivalent lethal equivalents: Models and inbreeding metrics for unbiased estimation of inbreeding load. Evol. Appl, 2019, 12, 266–279. [CrossRef]
- Armstrong, D.P.; Cassey, P. Estimating the effect of inbreeding on survival. Anim Conserv. 2007, 10:487-92. [CrossRef]
- Kalinowski, S.T.; Hedrick, P.W. An improved method for estimating inbreeding depression in pedigrees. Zoo Biol., 1998, 17, 481-97. [CrossRef]
- Boakes, E.H.; Wang, J.; Amos, W. An investigation of inbreeding depression and purging in captive pedigreed populations. Heredity, 2007, 98, 172-182. [CrossRef]
- Lacy, R.C.; Ballou, J.D. Effectiveness of selection in reducing the genetic load in populations of Perommyscus polinotus during generations of inbreeding. Evolution, 1998, 52, 900-909. [CrossRef]
- Ács, V.; Kövér, G.; Farkas, J.; Bokor, Á.; Nagy, I. Effects of Long-Term Selection in the Border Collie Dog Breed: Inbreeding Purge of Canine Hip and Elbow Dysplasia. Animals, 2020, 10, 1743. [CrossRef]
- Bersabé, D.; García-Dorado, A. On the genetic parameter determining the efficiency of purging: An estimate for Drosophila egg-to-pupae viability.J. Evol. Biol., 2013, 26, 375–385. [CrossRef]
- López-Cortegano, E.; Vilas, A.; Caballero, A.; García-Dorado, A. Estimation of genetic purging under competitive conditions. Evolution,2016, 70, 1856–1870. [CrossRef]
- López-Cortegano, E.; Bersabé, D.; Wang, J.; García-Dorado, A. Detection of genetic purging and predictive value of purging pa- rameters estimated in pedigreed populations. Heredity,2018, 121, 38–51. [CrossRef]
- Kövér, Gy.; Curik, I.; Vostry, L.; Farkas, J.; Mezőszentgyörgyi, D.; Nagy, I. Analysis of Inbreeding Effects on Survival at Birth of Pannon White Rabbits Using the Inbreeding-Purging Model. Diversity, 2023, 15, 71. [CrossRef]
- Khan, A.; Patel. K.; Shukla, H.; Viswanathan, A.; van der Valk, T.; Borthakur, U.; Nigam, P.; Zachariah, A.; Jhala, Y.V.; Kardos, M.; Ramakrishnan, U. Genomic evidence for inbreeding depression and purging of deleterious genetic variation in Indian tigers. PNAS, 2021, 118, e2023018118. [CrossRef]
- Dussex, N.; van der Valk, T.; Morales, H.E.; Wheat, C.W.; Díez-del-Molino, D.; von Seth, J.; Foster, Y.; Kutschera, V.E.; Guschanski, K.; Rhie, A.; et al. Population genomics of the critically endangered Kākāpō. Cell Genom, 2021, 1, 100002.
- Kleinman-Ruiz, D.; Lucena-Perez, M.; Villanueva, B.; Fernández, J.; Saveljev, A.P.; Ratkiewicz, M.; Schmidt, K.; Galtier, N.; García-Dorado, A.; Godoy, J.A. Purging of deleterious burden in the endangered Iberian lynx. PNAS, 2022, 119, e2110614119. [CrossRef]
- Boakes, E.; Wang, J. A simulation study on detecting purging of inbreeding depression in captive populations. Genet Res. 2005, 86, 139–48. [CrossRef]
- Hedrick, P.W. Purging inbreeding depression and the probability of extinction: full-sib mating. Heredity, 1994, 73, 363–72. [CrossRef]
- Nagy, I.; Curik, I.; Radnai, I.; Cervantes, I.; Gyovai, P.; Baumung, R.; Farkas, J.; Szendrő, Z. Genetic diversity and population structure of the synthetic Pannon White rabbit revealed by pedigree analysis. J. Anim. Sci. 2010, 88, 1267–1275. [CrossRef]
- García-Dorado, A. Understanding and predicting the fitness decline of shrunk populations: inbreeding, purging, mutation, and standard selection. Genetics, 2012, 190, 1461–76. [CrossRef]
- Theodorou, K.; Couvet, D. The efficiency of close inbreeding to reduce genetic adaptation to captivity. Heredity, 2015, 114: 38–47. [CrossRef]
- Pérez-Pereira, N.; Caballero, A.; García-Dorado, A. Reviewing the consequences of genetic purging on the success of rescue programs. Conserv. Genet, 2022a, 23, 1–17. [CrossRef]
- Pérez-Pereira, N.; Caballero, A.; García-Dorado, A. Reviewing the consequences of genetic purging on the success of rescue programs. Conserv. Genet. 2022b, 23, 1–17. [CrossRef]
- De Cara, M.A.R.; Villanueva, B.; Toro, M.A.; Fernández, J. Purging deleterious mutations in conservation programs: Combining optimal contributions with inbred mattings. Heredity,2013, 110, 530–537. [CrossRef]
- Caballero, A.; Bravo, I.; Wang, J. The risk of forcing inbreed- ing in conservation programmes: A reply to Theodorou and Couvet. Heredity, 2017, 19, 51–53. [CrossRef]
- Ralls, K.; Sunnucks, P.; Lacy, R.C.; Frankham, R. Genetic Rescue: A Critique of the Evidence Supports Maximizing Genetic Diversity Rather than Minimizing the Introduction of Putatively Harmful Genetic Variation. Biol. Conserv., 2020, 251, 108784. [CrossRef]
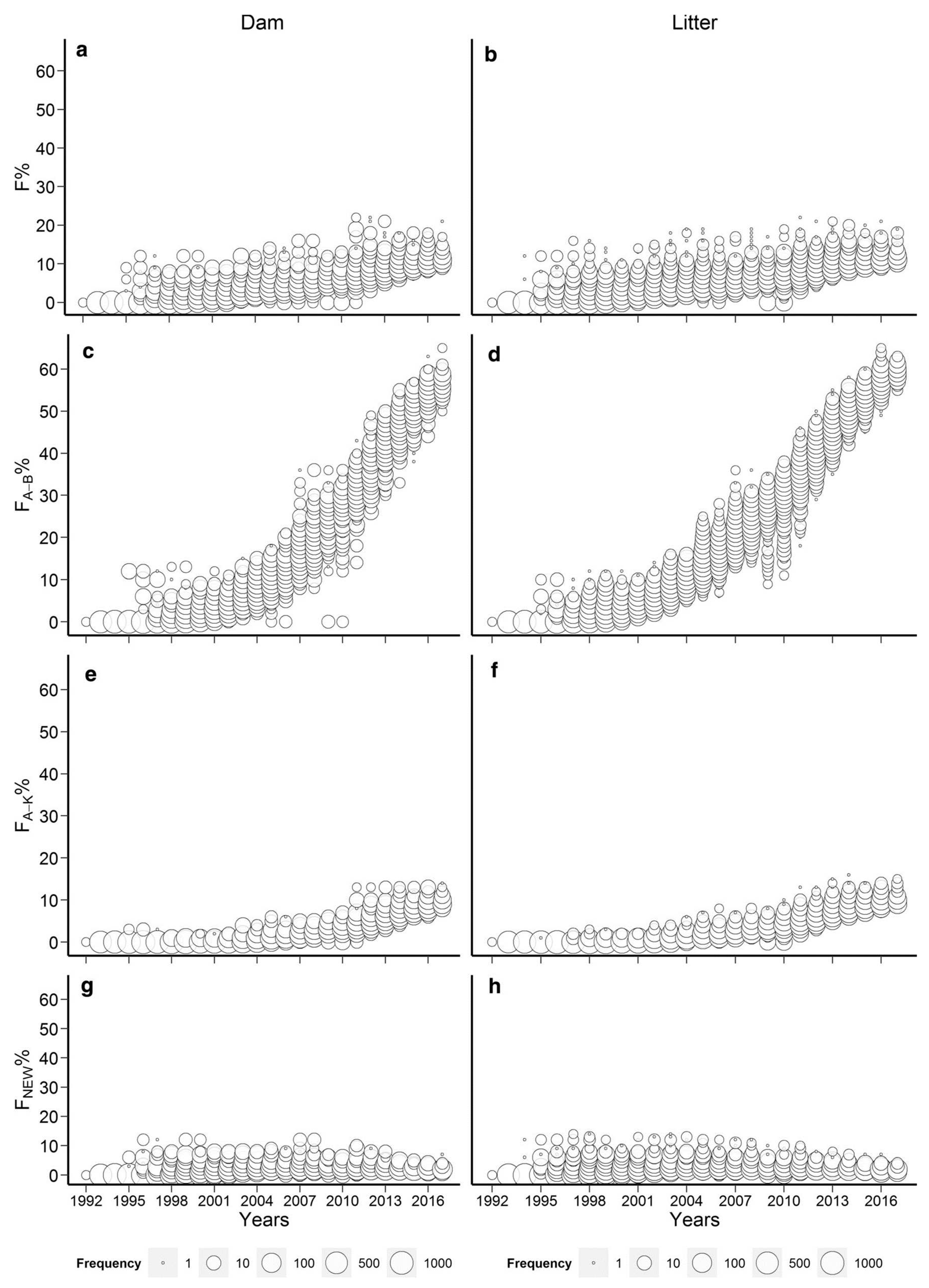
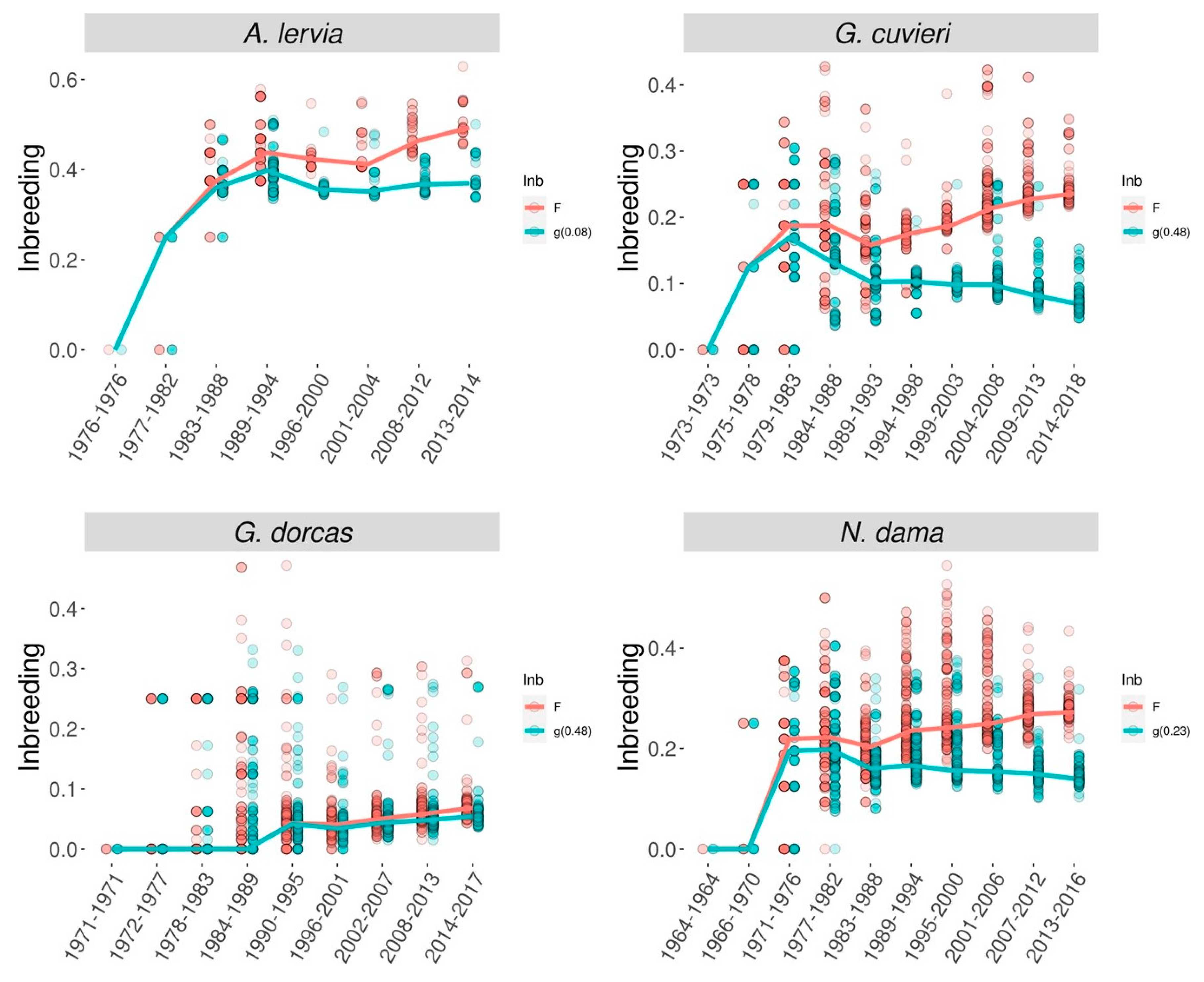
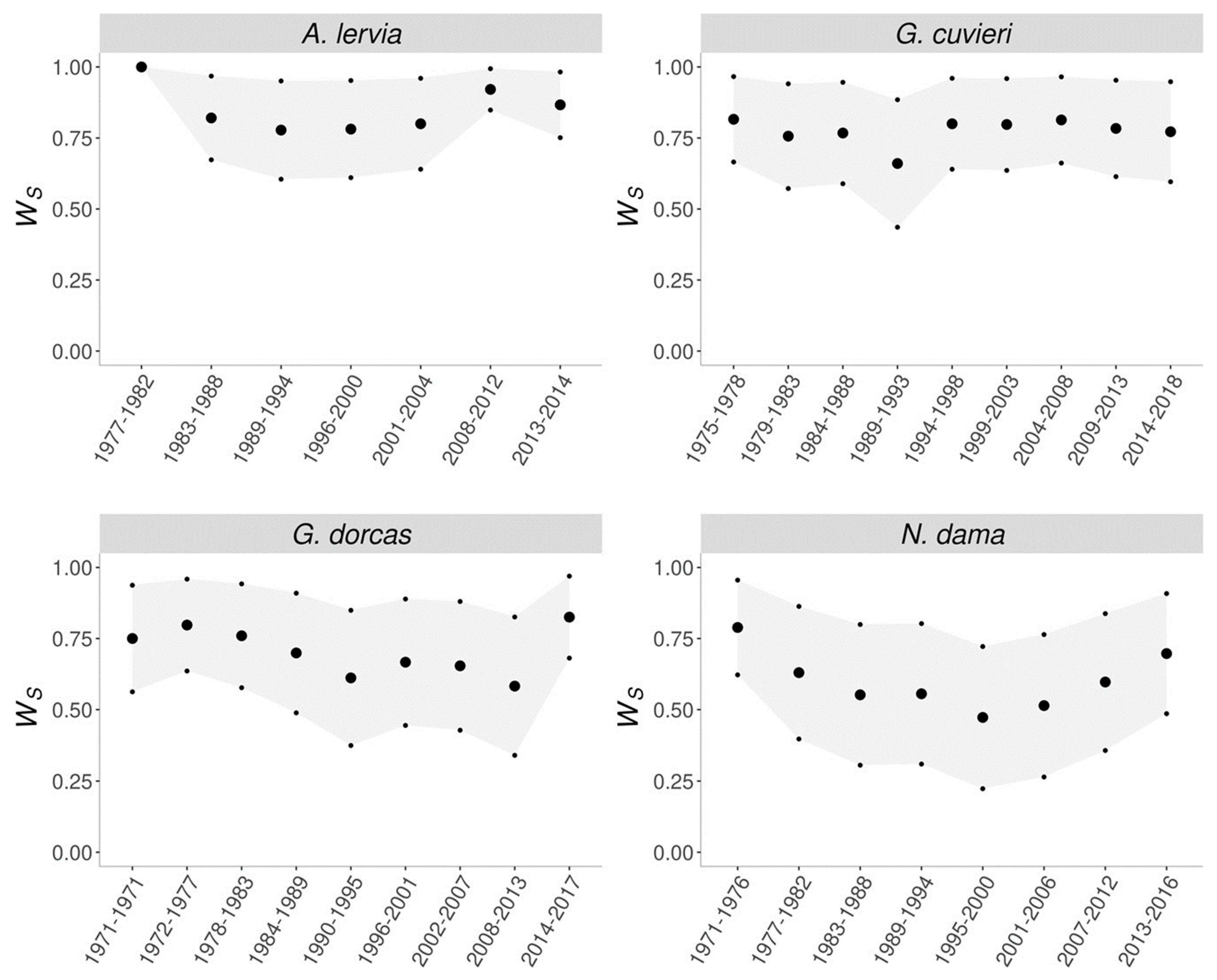
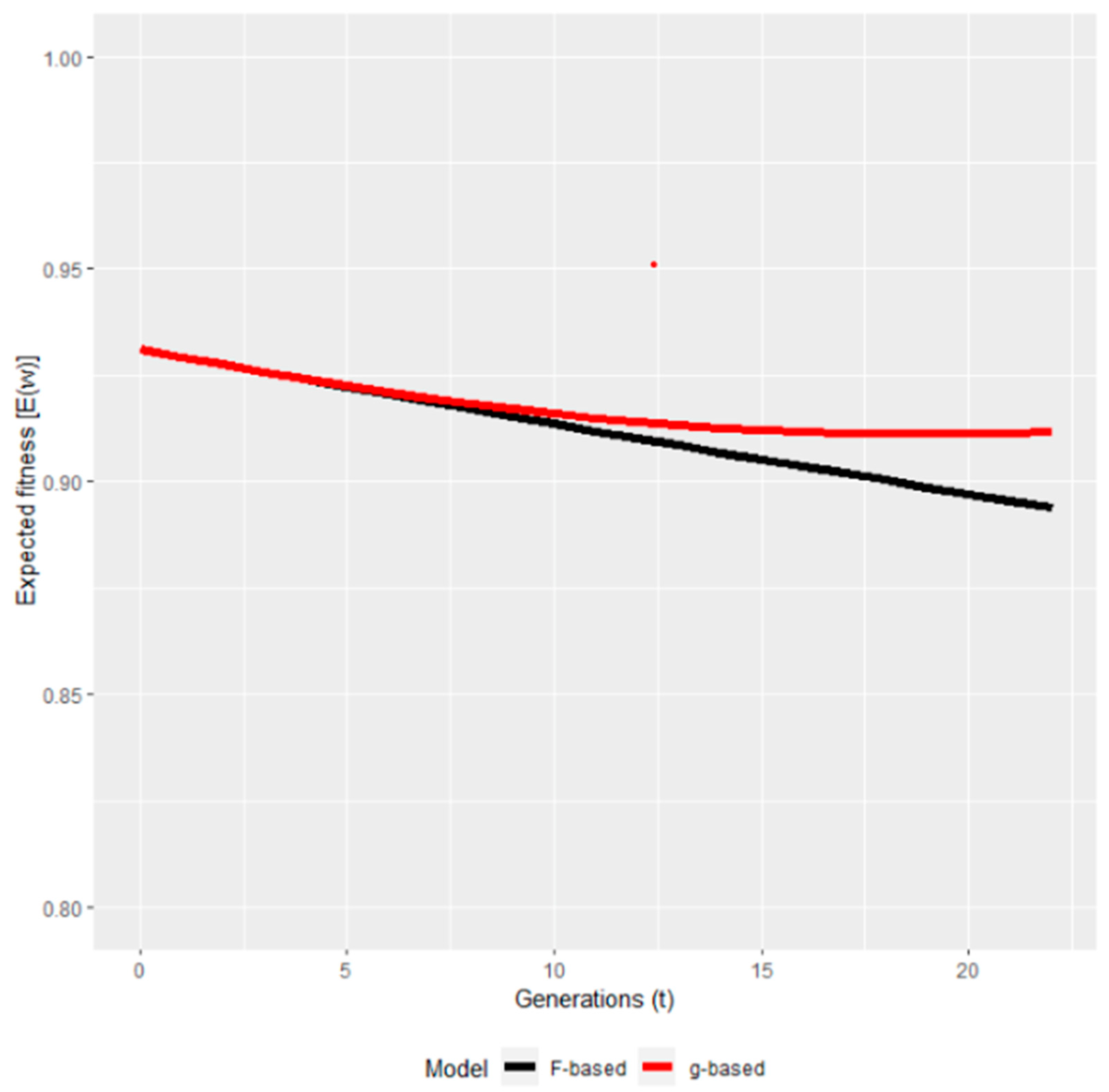
| Species | Life history trait | Estimated lethal equivalent | Reference |
|---|---|---|---|
| Homo sapiens | Stillbirth and neonatal birth | 1.124a 0.574b |
[46] |
| Infant and juvenile death | 1.431a | ||
| 0.908b | |||
| Homo sapiens | Infant survival | 0.396c | [62] |
| 2.745d | |||
| Survival to 10 years | 4.373c | ||
| 3.674d | |||
| Speke’s Gazelle | 30-day viability | 2.97e | [26] |
| 1.59f | |||
| North Island robin | Juvenile survival | 4.14 | [63] |
| Great tit | Survival to hatching | 0.4 | [64] |
| Survival to fledging | 0.4 | ||
| Survival to recruitment | 1.3 | ||
| Survival to adulthood | 2.12 | ||
| New Zealand robin | Juvenile survival | 0.24 | [65] |
| Takahe | Hatching rate | 0.691 | [66] |
| Fledging rate | 3.339 | ||
| 2-year survival | 0.952 | ||
| Offspring recruitment | 3.383 | ||
| The Catham Island Black robin | Juvenile survival | 3.42 | [67] |
| Hawaiian crow | 2-year survival | 6.9 | [68] |
| Soay sheep | 1-year survival | 2.285 | [69] |
| Drosophila melanogaster | Total fitness | 5.04 | [70] |
| Killer whale | 1-year survival | 0.10g | [71] |
| 0.14h | |||
| 40-year survival | 2.74g | ||
| 3.74h |
| Species/Breeds | Analyzed trait | Used methodology | Reference |
|---|---|---|---|
| German Holstein-Frisean | Birthweight | Ancestral inbreeding | [37] |
| Irish Holstein-Frisean | Milk yield | Ancestral inbreeding | [38] |
| Protein yield | Ancestral inbreeding | [38] | |
| Sumatran tiger | Neonatal survival rate | Ancestral inbreeding | [40] |
| Gazella cuvieri | Early survival | Inbreeding-Purging Model | [45] |
| Nanger dama | Early survival | Inbreeding-Purging Model | [45] |
| Pannon white rabbit | Survival at birth | Ancestral inbreeding | [60] |
| Prat rabbit line | Weaning weight | Ancestral inbreeding | [61] |
| Prat rabbit line | Slaughter weight | Ancestral inbreeding | [61] |
| Amur tiger | Survival to 7 days | Ancestral inbreeding | [81] |
| Black-footed ferret | Survival to 7 days | Ancestral inbreeding | [81] |
| Lesser kudu | Survival to 7 days | Ancestral inbreeding | [81] |
| Grey dorcopsis wallaby | Survival to 30 days | Ancestral inbreeding | |
| Hippopotamus | Survival to 30 days | Ancestral inbreeding | |
| Congo peafowl | Survival to 30 days | Ancestral inbreeding | [81] |
| Black-footed ferret | Survival to 30 days | Ancestral inbreeding | [81] |
| Bontebok | Survival to 30 days | Ancestral inbreeding | [81] |
| Goeldi's marmoset | Survival to 30 days | Ancestral inbreeding | [81] |
| Wied's Black-tufted-ear marmoset | Survival to 30 days | Ancestral inbreeding | [81] |
| Wyoming toad | Survival to 30 days | Ancestral inbreeding | [81] |
| Golden lion tamarin | Survival to 30 days | Ancestral inbreeding | [81] |
| Reindeer | Survival to 30 days | Ancestral inbreeding | [81] |
| Gunther's dik-dik | Survival to 30 days | Ancestral inbreeding | [81] |
| Peromyscus polinatus rhoadsi | Litter size | Ancestral inbreeding | [82] |
| Peromyscus polinatus rhoadsi | Litter weight and weaning | Ancestral inbreeding | [82] |
| Border collie dog | Hip dysplasia | Ancestral inbreeding | [83] |
| Drosophila melanogaster | Egg to pupae viability | Inbreeding-Purging Model | [84 |
| Drosophila melanogaster | Noncompetitive pupae productivity | Inbreeding-Purging Model | [85] |
| Drosophila melanogaster | Competitive productivity | Inbreeding-Purging Model | [86] |
| Pannon white rabbit | Survival at birth | Inbreeding-Purging Model | [87] |
| Indian tiger | NA | Whole genome analysis | [88] |
| Kākāpō | NA | Whole genome analysis | [89] |
| Iberian lynx | NA | Whole genome analysis | [90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





