1. Introduction
Lentinula edodes (Berk.) Pegler, known as “Xianggu” in China and “shiitake” in Japan, is the second most cultivated and the most popular edible fungus in the world [
1,
2]. In China, the
L. edodes is of the greatest importance in the industry of edible fungi, since its annual yield has account for over one-third of the total mushroom production according to the latest production statistics [
3].
L. edodes can be consumed either for therapeutic purposes or as nutritional food. It harbors abundant bioactive molecules with proven pharmacological properties comprising polysaccharide, lentinan, eritadenine, ergosterol, etc. [
4]. These bioactive compounds have been shown confer it multiple therapeutic capacities to prevent and treat diseases involving cancer, depressed immune function, hyperlipidemia, obesity, etc.[
4,
5].
L. edodes is the first medicinal macrofungus to enter the realm of modern biotechnology. As food, it is rich in various of minerals, vitamins, unsaturated fatty acid, proteins and dietary fiber [
4,
6]. It has been cultivated and consumed around the globe, especially in ester Asia [
7].
L. edodes possess characteristic aroma. The aroma of fresh
L. edodes fruiting body is largely eight-carbon (C8) compounds comprising 1-Octen-3-ol, 2-Octen-1-ol, 1-Octen-3-one, 3-Octanone, 3-Octenol etc. [
8,
9,
10]. The content of C8 compounds can account for more than 70% of the total volatile emission [
10]. Among those C8 compounds, the so called “mushroom alcohol” 1-Octen-3-ol is the main contributor of the aroma of fresh
L. edodes [
11]. C8 compounds are synthesized from the oxidation and cleavage of fatty acid linoleic acid through lipoxygenase (LOX) pathway[
12,
13]. The C8 compounds, however, can describe many of the fresh mushrooms [
14,
15,
16]. Thus, it cannot be treated as the key characteristic distinguishing it with other mushrooms. The typical aroma of dry
L. edodes can be characterized by a blend of sulfur-containing cyclic and straight chain compounds, including Lethionine, 1,2,4-Trithiolane, 1,2,4,5-Tetrathiane, Dimethyl disulfide and Dimethyl trisulfide, etc. [
8,
9,
17,
18]. The content of sulfur flavor compounds can account for 22.91% to 74.74% to the total content of volatiles emission in dried shiitake [
19] and 19.41%– 21.53% in fresh shiitake [
20,
21]. The synthesis of sulfur flavor compounds involves both enzymatic and nonenzymatic reactions. The biosynthesis of volatile sulfur compounds starts from the precursor Lentinic acid followed by a two-step enzyme-catalyzed reaction: The γ-Glutamyl peptide bond of Lentinic acid is hydrolyzed by γ-Glutamyl transpeptidase (GTT) and releases γ-Glutamylpeptides. Then, γ-Glutamylpeptides are hydrolyzed by Cysteine sulfoxide lyase (C-S lyase) and produce Thiosulfate. The Thiosulfate was thereafter subjected to complex nonenzymatic reaction through which various of sulfur-containing flavor compounds, including Lenthionine, were formed [
17,
18,
22]. The characteristic aroma, which conferred by the sulfur-containing compounds, is one of the key factors determining the quality of
L. edodes [
23,
24,
25].
Members in
Agaricomycetes, including
L. edodes, develop distinct specialized structures including the pileus skin, context, gill and stipe of the fruiting body. It was found that in
Agaricus bisporus, the cap and gills emitted more 1-Octen-3-ol than in the stipe and the cap produced more desirable fresh and cooked aroma [
26]. However, in
Pleurotus ostreatus, the highest content of 1-octen-3-ol was found in the stipe, followed by the cap and base of fruiting body [
27]. In
L. edodes, the flavor profile in different parts of fruiting body can also be different. Chen et al.[
28] reported that the concentration of most of the C8 compounds and sulfur compounds are markedly higher in the pileus than in the stipe of
L. edodes. However, the two predominating compounds 1-Octen-3-ol and Lenthionine showed comparable content in the pileus and stipe [
28]. While another study showed that the content of 1-Octen-3-ol in pileus was significantly lower than in stipe of
L. edodes [
29]. Taken together, the volatile flavor profile can be different depending on different parts of the fruiting body.
Overall, the tissue-dependent variations of flavor profile in L. edodes are still not well understood. To date, only few studies compared the differences of flavor profiles between cap and stipe of fruiting body. However, the flavor profile of skin and gills remains to be a gap. In this study, we investigated the volatile flavor profile of pileus skin, context, gill and stipe of the L. edodes fruiting body using gas chromatography–mass spectrometry (GC–MS) technique combined with multivariate analysis, providing comprehensive understanding on the tissue-specific variation of flavor biosynthesis.
2. Materials and Methods
2.1. Sample preparation
The mature fruiting body of fresh L. edodes (strain 0912 and T2) was selected for experiment use. The fruiting body of L. edodes was taken from the base of North Wild Edible Fungus Development co. LTD located in Guangling county, Shaanxi province, China. Different tissues including the skin of cap, context, gill and stipe of fruiting body were carefully split up and then cut into thin slices for following volatile flavor sampling.
2.2. Volatile compounds extraction
Fresh homogenized samples (ca.16 g) of different tissues were transferred to glass headspace amber vials (15ml, Anpel Laboratory Technologies, Shanghai, China). The vials containing samples were then incubated in water bath at 55 ℃ for 1 h to boost volatile emission. Volatile compounds were extracted by solid phase microextraction (SPME) method for 1h using a fused silica fiber (75-μm length) coated with a 50/30-μm layer of divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS) (Supelco® Analytical, Merck KGaA, Darmstadt, Germany). Delta-2-Carene were used as internal standard.
2.3. Volatile compounds measurements with GC-MS
The extracted volatile compounds were analyzed by GC-MS (GC type 7890A, MS type 7000C; Agilent Technologies, Palo Alto, CA, USA) using a 5% phenyl-methylpolysiloxane phase capillary column (30 m × 250 μm × 0.25 μm J&W HP-5MS; Agilent Technologies, CA, United States). The volatile compounds were desorbed from the extraction fiber in the GC injection port at 250 ℃ for 10 min [
11]. GC–MS conditions were configured as follows: carrier gas, 99.999% helium gas; flow rate, 1.0 mL/min; oven temperature program, initial temperature at 40 ℃, maintained for 1 min, heating rate at 10 ℃/min to 250 ℃, which was maintained for 1 min; ionization mode, EI; ion source temperature, 230 ℃; quadrupole temperature, 150 °C; mass scan range, 35–550 u. Non-isothermal Kovats retention indices were calculated according to generally accepted standards [
30] , based on chromatography retention times of a saturated alkane mixture (C7-C30; Anpel Laboratory Technologies Inc., Shanghai, China). The annotation of peaks was performed by comparison of the mass spectra against libraries of reference spectra (NIST 11, Wiley 275) and non-isothermal Kovats retention indices found in literature followed [
31] and [
32]. The quantification of volatile compounds was performed following the well-established method [
11,
31,
33]. The GC-MS data were normalized to the fresh weight of fruiting body used for the volatile sampling.
2.4. Statistics
The tissue-dependent volatile profile patterns were analyzed using orthogonal projections to latent structures discriminant analysis (OPLS-DA) using SIMCA-P (SIMCA 14.1, Umetrics, Umeå, Sweden). Otherwise, all data analysis and visualization were performed with the R Studio version 2023.6.1.524 integrated development environment (IDE) [
34] using R version 4.3.1 (R Core Team, 2018) [
35]. The pairwise spearman correlations and corresponding significance (p-values) between compounds were calculated using “Hmisc” package. The significant correlations (p < 0.05) were selected for further network analysis. The correlation matrix were constructed and visualized using “corrplot” package [
36]. The highly interconnected communities were computed using the “ggraph”package[
37] with Fruchterman-Reingold layout algorithm [
38] from ”igraph” package [
39]. Random Forest-Recursive Feature Elimination (RF-RFE) algorithm [
40,
41,
42] was performed to identify the volatile biomarkers of different strains and tissues of
L. edodes. Number of variables randomly sampled as candidates at each split (mtry) and number of trees to grow (ntree). mtry and ntree were tuned (grid search) using caret package to get the optimal predictive ability and accuracy [
43]. The size of volatile biomarkers was determined with a prediction accuracy over 80% with the Cohen’s kappa value over 0.7, which guaranteed a good prediction performance [
44].
3. Results
3.1. Overall volatile emission from different tissues of L. edodes fruiting body
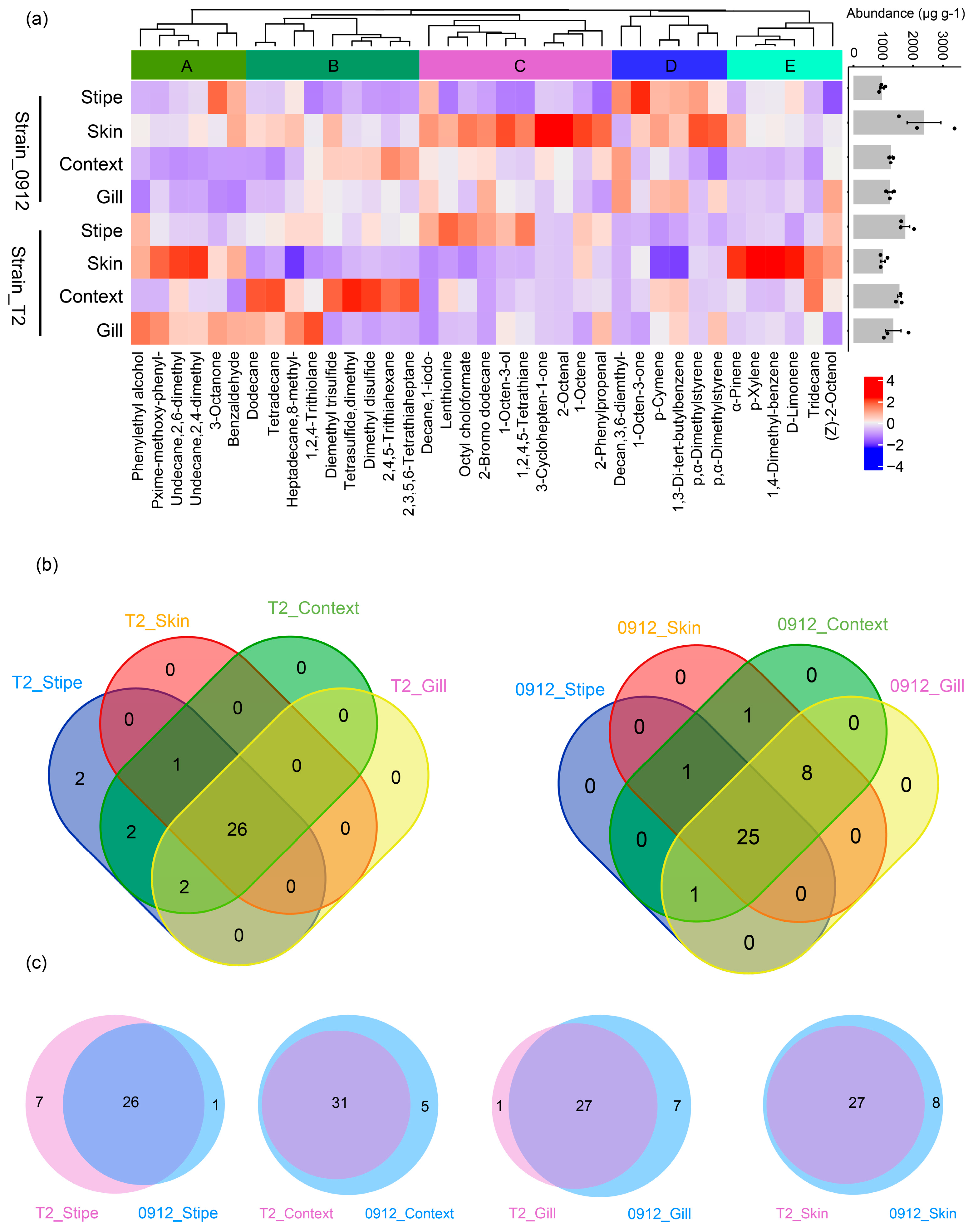
Overall, 33 and 36 volatile compounds can be detected from different tissues of strain T2 and strain 0912 fruiting body, respectively (
Table 1,
Table S1). For strain T2, the total emission level was highest in stipe and lowest in the skin, whereas a contrary trend was observed for the strain 0912. For both strains, total emission levels were comparable in the context and gill (
Table 1,
Figure 1). With the hierarchical cluster, we showed that the detected compounds could be gathered to 5 groups (A, B, C, D, E). Those subgroups of compounds demonstrated some of the clear strain or tissue chemotypes, for example, the group A was generally rich for the strain T2, whereas the group C and D were abundant for the strain 0912. The group B, comprising most of the sulfur compounds, was abundant in the context, in particular for the strain T2. The group E, which containing largely terpenes and benzenoids, was characteristic for the skin of the strain T2. Venn analysis showed that most of the compounds were shared with different tissues for both strains, through their abundances showed huge variations. For the strain T2, two compounds i.e.,Decane,1-iodo- and 2-Bromo dodecane were found unique for the stipe. Two typical acyclic sulfur flavor compound Dimethyl trisulfide and Tetrasulfide, dimethyl were only detected in the stipe and context (
Figure 1b). The same tissue of different strains shared most of the compounds. For the stipes, 1 compound (Decan,3,6-diemthyl-) was found unique for the strain 0912, and 7 compounds were unique for the strain T2 (Dimethyl trisulfide, Tetrasulfide, dimethyl, Lenthionine, 2,3,5,6-Tetrathiaheptane, 2,4,5-Trithiahexane, 1-Octene, 2-Bromo dodecane). There were no compounds unique for the skin of the strain T2. For the context, 5 compounds (Decane,1-iodo-, Cyclohepten-1-one, Decan,3,6-diemthyl-, 2-Octenal, 2-Bromo dodecane) were only detected in the strain 0912, whereas no compounds were unique for context of the strain T2. The gill of the strain 0912 had 7 unique compounds (Dimethyl trisulfide, Decane,1-iodo-, Tetrasulfide, dimethyl, Decan,3,6-diemthyl-, 2-Octenal, 1-Octene, 2-Bromo dodecane), while only one compounds (Phenylethyl alcohol) was unique for the strain T2 (
Figure 1C).
3.2. Chemical diversity of the volatile compounds in different tissues of L. edodes fruiting body
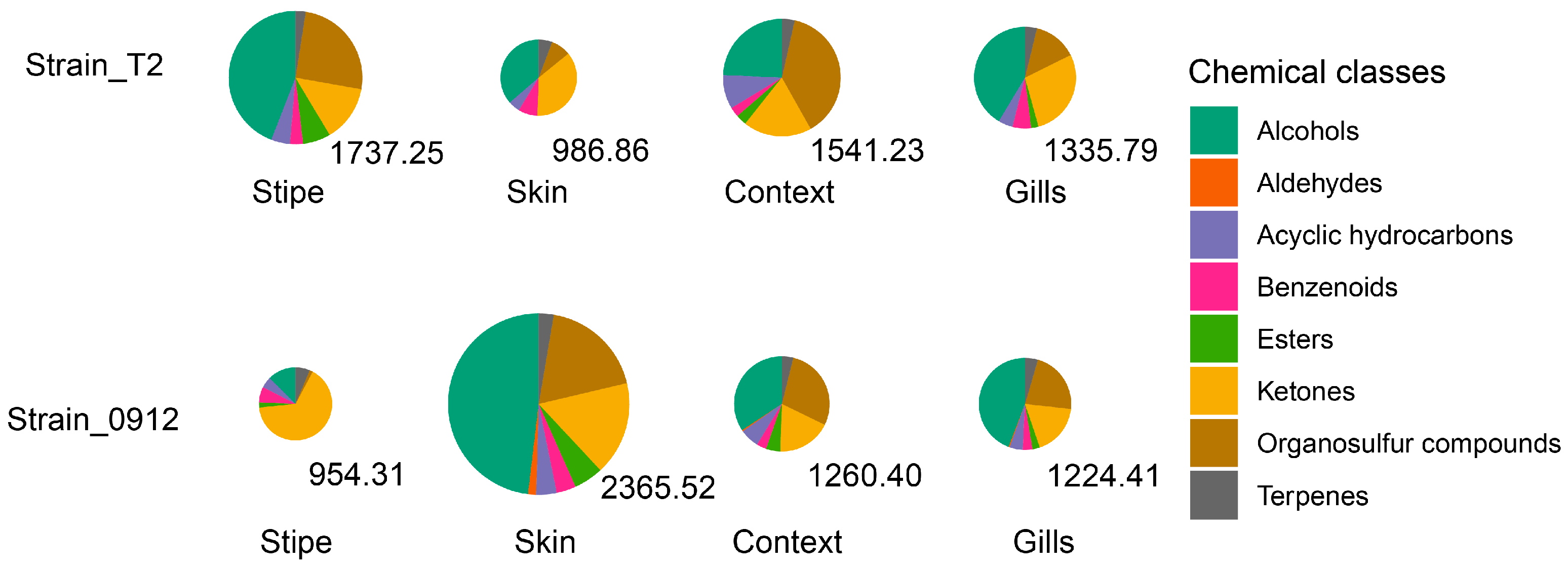
To illustrate the chemical diversity of the volatile profile, we clustered the volatile compounds into 8 chemical classes (alcohols, aldehydes, acyclic hydrocarbon, benzenoids, esters, ketones, sulfur compounds and terpenes). The chemical diversities were similar in different tissues. For both strains, the volatile profiles were dominating by alcohols, ketones and sulfur compounds, which account for more than 80% to the total volatile emission. However, in the stipe of strain 0912, only small amount of sulfur compounds could be detected (1.43%). For both strains, most proportion of the sulfur compounds, which sketching the typical aroma of dried fruiting body, were found in the context. For the strain T2, the lowest amount of sulfur compounds was detected in the skin, whereas for the strain 0912 it was lowest in the stipe (
Figure 2).
3.3. The profiles of C8 and sulfur compounds differed in different tissues and strains
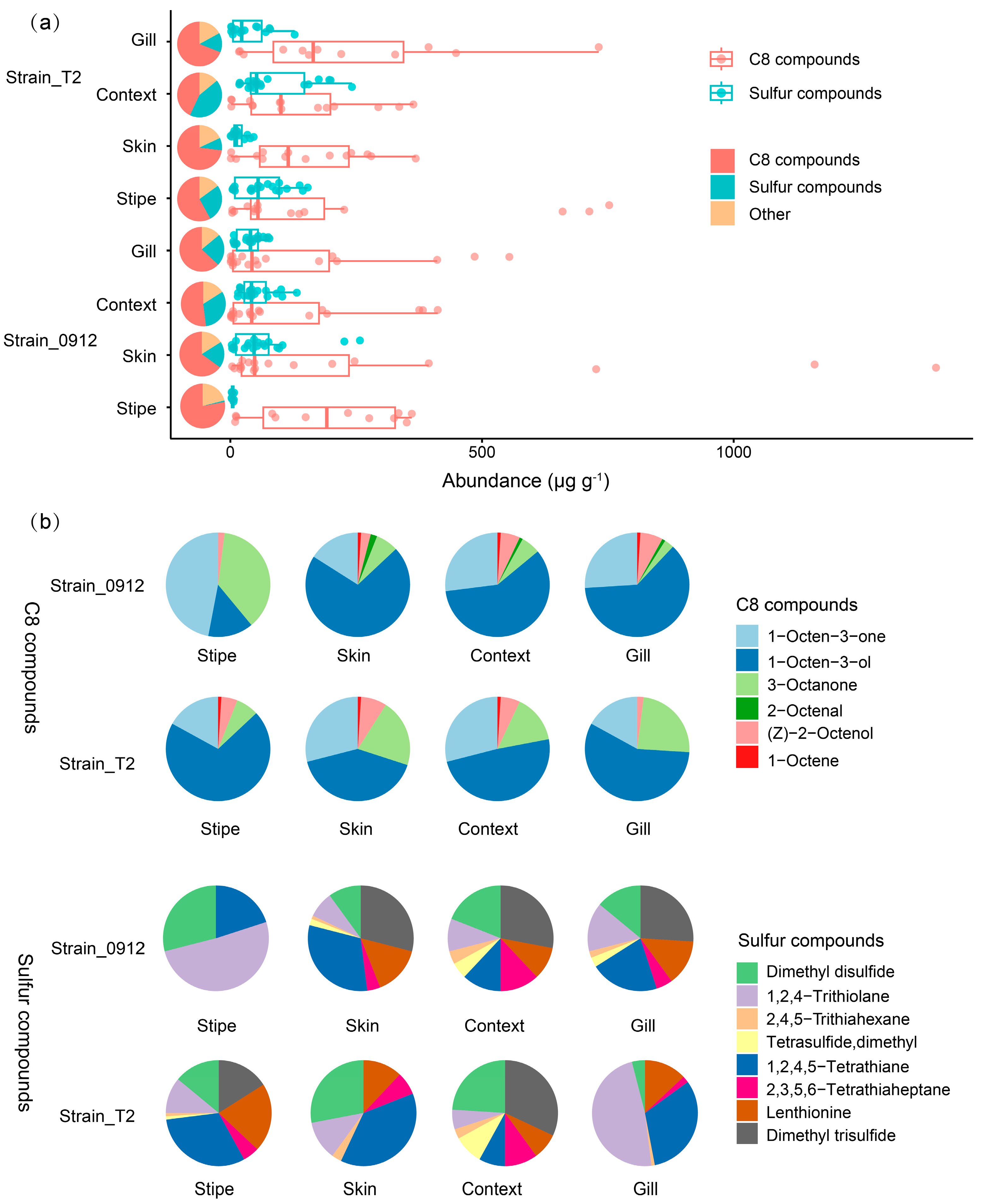
The C8 and sulfur compounds, which characterizing the aroma of fresh and dried L. edodes fruiting body, dominating the volatile profiles of different tissues in the two strains. The C8 compounds could account for 43%-78% to the content of total volatile emissions of different tissues in the two strains, following by sulfur compounds that account for 19%-43%, except for the stipe of strain 0912 which contained only 1.4% of the sulfur compounds (
Figure 3a). For the strain T2, the total content of C8 compounds was highest in the stipe, followed by the gill, skin and context, while for the strain 0912 it was highest in the skin, followed by the gill, stipe and context. The sulfur compounds were highest in the context, followed by the stipe, gill and skin for the strain T2, whereas it was highest in the skin, followed by the context, gill and stipe in the strain 0912 (
Figure 3a).
There are 6 C8 compounds and 8 sulfur compounds were detected in all samples. For the strain 0912, the stipe had the lowest diversity of C8 compounds. The proportion of mushroom alcohol, 1-Octen-3-ol, was predominant in all tissues for both strains, except for the stipe of the strain 0912. Additionally, two ketone compounds 1-Octen-3-one and 3-Octanone also pronouncedly contributed for the C8 profiles of different tissues of the two strains. These 3 C8 compounds accounted for 91%-98% of the total C8 content (
Figure 3b). Similar to the trend of C8 compounds, the stipe of the strain 0912 possessed the lowest diversity of sulfur compounds. The most important sulfur flavor compounds Lenthionine was not detected in the stipe of the strain 0912, whereas it accounted for the largest proportion to the total sulfur compounds in the stipe of the strain T2. Surprisingly, the ratio of Lenthionine was always low in the context in comparison with other tissue. Except for Lenthionine, the two straight chain sulfur compounds Dimethyl disulfide and Dimethyl trisurfide, together with a cyclic sulfur compounds 1,2,4-Trithiolane, dominated the sulfur flavor profiles of different tissues for both strains, through their ratio were changing depending on tissues and strains (
Figure 3b).
3.4. Correlations of C8 and sulfur compounds
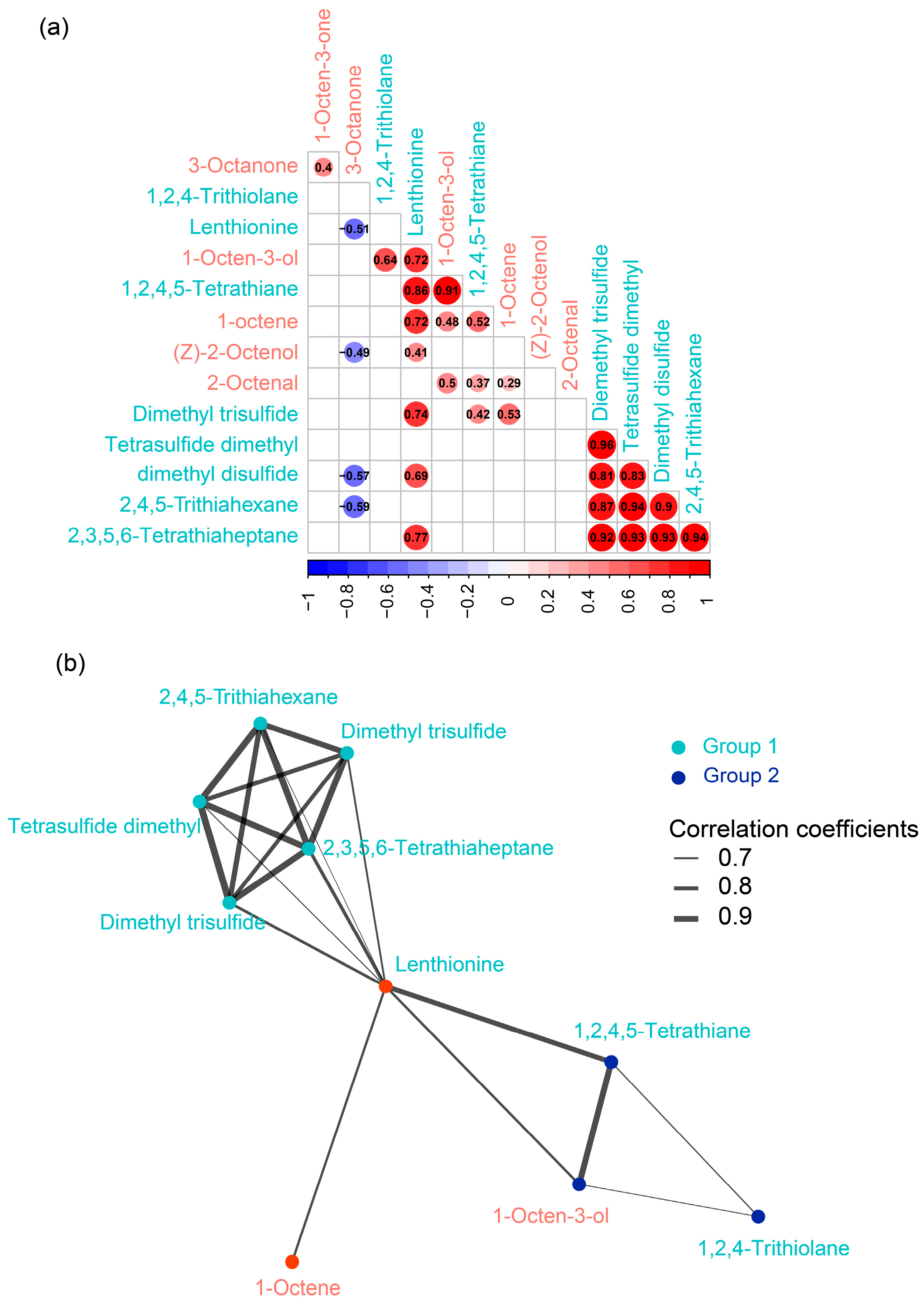
To infer relationships of individual C8 and sulfur compounds, we conducted the spearman’s rank correlation analysis. Results showed that the significant (p < 0.05) paired correlations were largely positive. Only 4 moderate (0.4 < r < 0.6) negative correlations were found between 3-Octanone with Lenthionine, Z-2-Octenol, Dimethyl disulfide and 2,4,5-Trithiahexane, respectively (
Figure 4a). Further, we performed community detection algorithm to identity the module of highly connected compounds. Results showed that most of the sulfur flavor compounds were highly connected except for the 1,2,4-Trithiolane. Surprisingly, none of the C8 compounds were connected with each other (
Figure 4b).
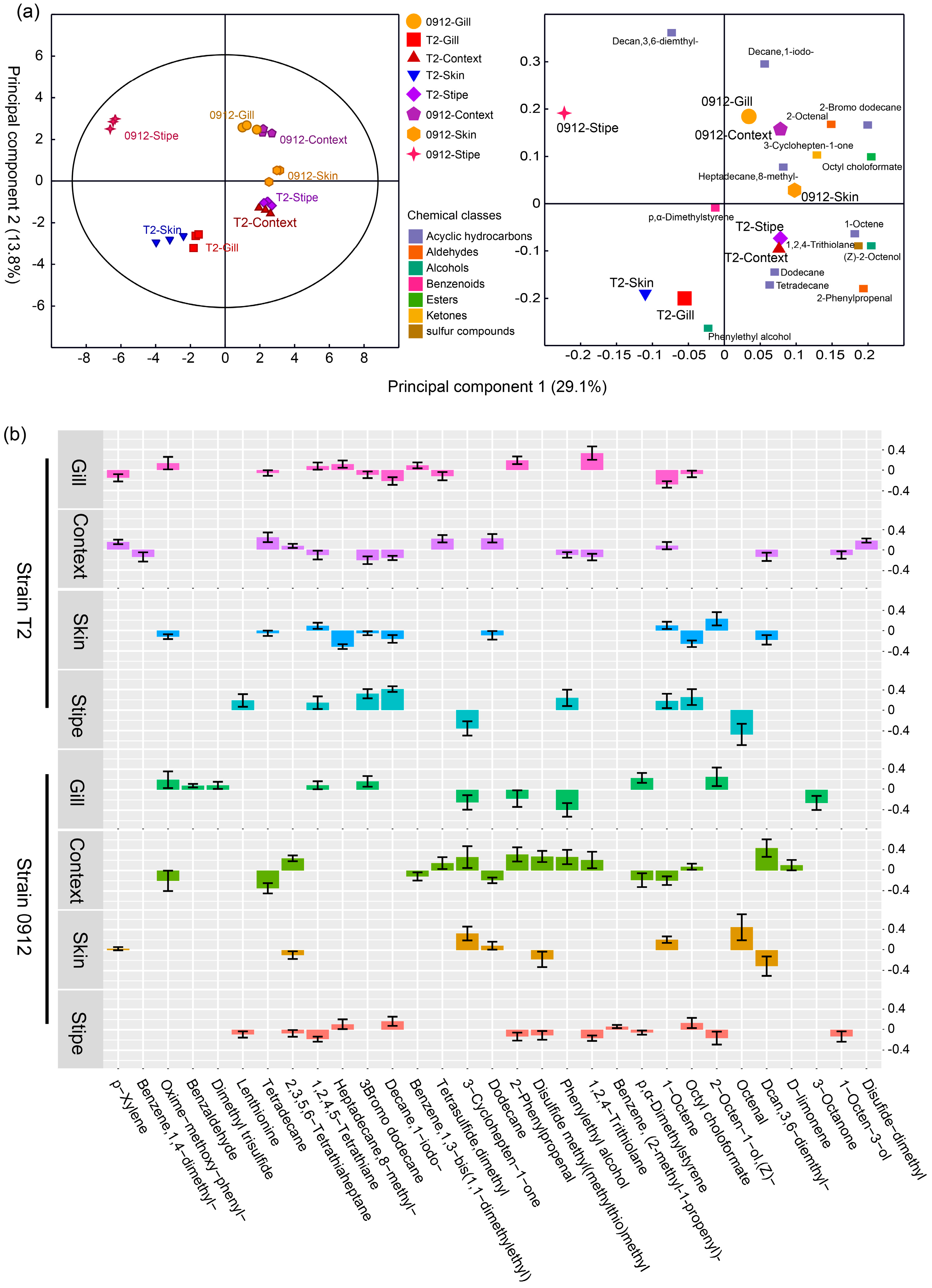
3.5. The tissue-dependent volatile profile patterns predicts different L. edodes strains and tissues
Using OPLS-DA model, the volatile profiles showed significant difference (CV-ANOVA, P < 0.05) in different tissues and strains. The first two principal components could explain 42.9% of the total variation. Bar plots (
Figure 5b) illustrate the significant regression coefficients of each compound to different tissues. The number of compounds with significant correlations varied depending on tissues and strains. The skin of the strain T2 had the lowest number of significantly correlated compounds, otherwise, the number were showed in comparable level. The Lenthionine was showed only positively correlated with the stipe for the strain T2, whereas was negatively correlated with the stipe of the strain 0912. The 1-Octen-3-ol was merely negatively correlated with the context in the strain T2 and the stipe in the strain 0912, respectively. There were some compounds showed only one significant correlation such as Dimethyl disulfide, Dimethyl trisulfide and 3-Octanone (
Figure 5b).
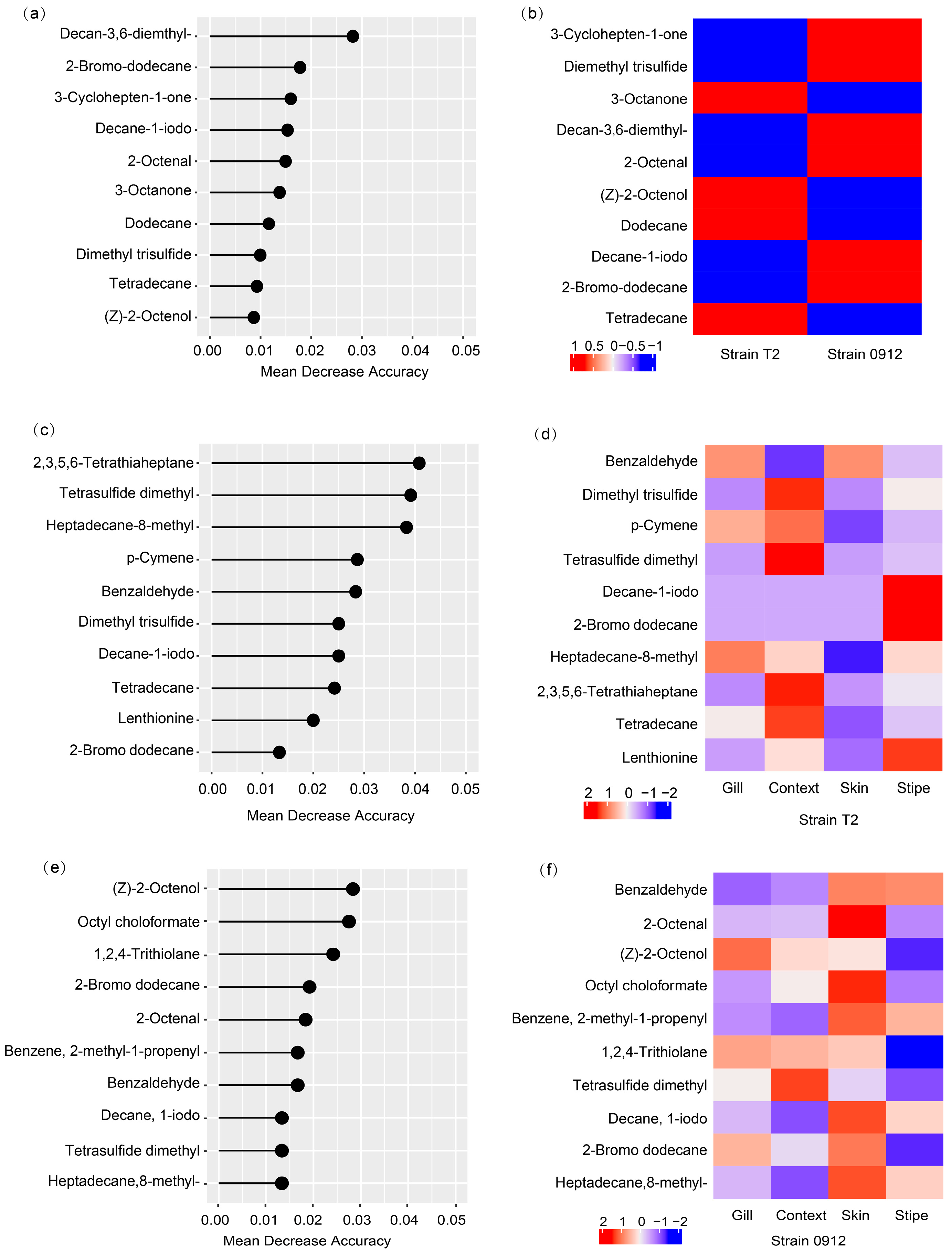
3.6. Volatile biomarkers of L. edodes strains and tissues
As the volatile profiles could characterize different strains and tissues of
L. edodes, we performed random forest algorithm to identify the corresponding volatile biomarkers. For group of the strain T2 and 0912, random forest model could return a prediction accuracy of 100% (p < 0.05,
Table 2). The top 10 compounds (Decan-3,6-diemthyl-, 2-Bromo-dodecane, 3-Cyclohepten-1-one, Decane-1-iodo, 2-Octenal, 3-Octanone, Dodecane, Dimethyl trisulfide, Tetradecane, (Z)-2-Octenol) could give a model accuracy of 88.1% (Kappa = 0.71). These biomarkers could use to distinguish the strain T2 and 0912 (
Figure 6). To predict the different tissues of the strain T2, their volatile profiles could give an overall prediction accuracy of 100% (p < 0.01). The prediction accuracy for the stipe, skin, context and gill was also 100% (sensitivity =1, specificity =1). The top 14 volatile biomarkers (2,3,5,6-Tetrathiaheptane, Tetrasulfide dimethyl, Heptadecane, 8-methyl-, p-Cymene, Benzaldehyde, Dimethyl trisulfide, Decane, 1-iodo,Tetradecane,Lenthionine, 2-Bromo dodecane, (Z)-2-Octenol, 2,4,5-Trithiahexane, 1,2,4,5-Tetrathiane, Octyl choloformate) could give a prediction accuracy of 90% (kappa = 0.78). For the strains 0912, the overall model accuracy was relatively low (75%, p-value = 0.05, Kappa = 0.67), due to the low prediction ability for the skin (balanced accuracy = 0.50, sensitivity = 0). Nevertheless, some of the compounds still had the prediction capacities, for example the Tetrasulfide dimethyl for the context (
Figure 6).
4. Discussion
Mushrooms are popular owing to their medical value, balanced nutrition, texture and aroma [
45]. Of those characteristics, aroma can be the key determinant of consumer preference, in particular for the mushrooms have a relatively strong odor such as
L. edodes [
23,
45]. For
L. edodes, there are considerable studies have investigated the overall volatile profiles of its fruiting body in either different cultivation conditions, different developmental stages or different drying methods [
11,
19,
20,
46,
47,
48,
49,
50]. Those literatures provide an overall perception to its unique odor. However, the volatile flavor variation of different tissues of the fruiting are largely overlooked. To the best of our knowledge, our study reveals for the first time the comprehensive tissue-specific aroma variations of
L. edodes fruiting body.
In our work, we showed that the stipe of the strains T2 and skin of the strain 0912, not the context, had the highest volatile emissions per unit weight. Nevertheless, volatiles from the context may contribute the largest part of the total content of the volatile emission since the context can represent over 85% of the total fruiting body fresh weight [
51]. For both strains, the C8 compounds dominated the total volatiles profiles of the fresh fruiting body, which is consistent with other studies [
10,
11]. More specifically, the few compounds including the 1-Octen-3-ol, 1-Octen-3-one and 3−Octanone governed the odor of fresh
L. edodes fruiting body. These three C8 compounds are commonly predominant for many of other mushrooms, for example,
Pleurotus salmoneostramineus and
Pleurotus sajor-caju [
52],
A. bisporus [
53] and truffles [
54]. The individual C8 compounds showed interesting variations depending on tissues and strains. For example, the mushroom alcohol 1-Octen-3-ol was found to be the most predominant one for the stipe of the strain T2, whereas to be the weakest contributor for the C8 profile of the stipe of the strain 0912. Such tissue-driven variations also present in other mushrooms, for example, in
A. bisporus the cap and gill emitted most abundant of mushroom alcohol than in the stipe, however in
P. ostreatus the stipe was showed to be the strongest emitter [
26,
27] .
The volatile profiles of stipes of different strains showed markedly differences. Notably, the sulfur flavor compounds were rather low (1.4%) in the stipe of the strain 0912. As a results, the utilization of the stipe, which commonly regarded as waste byproducts, are suggested to consider the strain differences, particularly for the strategy which aimed to recycle the flavor ingredient. Conversely, the volatile sulfur compounds represent significant proportion (43%) in the context of the strain T2, suggesting a good sensory quality of this part. Similarly, the proportion of volatile sulfur compounds showed also the largest in context of the strain 0912. The abundance of individual sulfur flavor compounds varied greatly in different tissues. One of the most important sulfur flavor compounds Lenthionine was absent from the stipe of the strain 0912, whereas it represented one of the main contributors for the sulfur flavor profile of the strain T2. Further, in other study, the Lenthionine content could have comparable level in the pileus and stipe [
28]. The proportion of another key sulfur flavor compounds 1,2,4-Trithiolane was also changed remarkably depending on different tissues.
Through some of literatures documented the differences of the volatile profiles in different tissue of
L. edodes fruiting body [
28,
29], our data showed for the first time the clear patterns of the volatile profiles for different tissues and strains. It suggested that the volatile profiles were capable of predict either the tissues or different strains. Using a random forest machine learning algorithm, it was found that the volatile profile could 100% predict the strain 0912 and T2. The prediction accuracy for different tissues of the studied strains could also reach 100%. These results further confirm and highlight the strain- and tissue-specific variations of the volatile emissions of
L. edodes fruiting body.
Supplementary Materials
Table S1. Annotations of all the detected compounds from different tissues of the strain T2 and 0912 fruiting body.
Author Contributions
Conceptualization, Y. G., Y. L. and S. W.; Data curation, Y. G. and Y. F.; Formal analysis, Y. G. and J. Z.; Funding acquisition, Y. G., Y. L. and S. W.; Investigation, J. Z., H. W., Q. G., S. S., Y. F. and D. Y.; Methodology, Y. G.; Project administration, Y. G., Y. L. and S. W.; Resources, Y. L.; Software, Y. G. and J. Z.; Supervision, Y. L. and S. W.; Validation, Y. G., H. W., Q. G. and Y. F.; Visualization, Y. G. and J. Z.; Writing–original draft, Y. G.; Writing–review & editing, Y. L. and S. W. All authors have read and agreed to the published version of the manuscript
Funding
This research was funded by Beijing Academy of Agriculture and Forestry Sciences (BAAFS) Program (KJCX20230419), China Agriculture Research System (CARS-20) and the Beijing Innovation Consortium of Agriculture Research System (BAIC03).
Data Availability Statement
The raw GC-MS data of this study are available at the National Genomics Data Center, China National Center for Bioinformation, under the PRJCA020932.
Acknowledgments
The authors thank ZJ song for the technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yu, H.; Zhang, L.; Shang, X.; Peng, B.; Li, Y.; Xiao, S.; Tan, Q.; Fu, Y. Chromosomal genome and population genetic analyses to reveal genetic architecture, breeding history and genes related to cadmium accumulation in Lentinula edodes. BMC Genom. 2022, 23, 1-14. [CrossRef]
- Li, C.; Gong, W.; Zhang, L.; Yang, Z.; Nong, W.; Bian, Y.; Kwan, H.-S.; Cheung, M.K.; Xiao, Y. Association mapping reveals genetic loci associated with important agronomic traits in Lentinula edodes, shiitake mushroom. Front. microbiol. 2017, 8, 237. [CrossRef]
- [3]China Edible fungi Association. Available online: (accessed on 30 October 2023) https://mp.weixin.qq.com/s/M6ZQN5IYkkMCOmsojfLtUw.
- Ahmad, I.; Arif, M.; Xu, M.; Zhang, J.; Ding, Y.; Lyu, F. Therapeutic values and nutraceutical properties of shiitake mushroom (Lentinula edodes): A review. Trends Food Sci. Technol. 2023, 134, 123-135. [CrossRef]
- Bisen, P.S.; Baghel, R.K.; Sanodiya, B.S.; Thakur, G.S.; Prasad, G.B. Lentinus edodes: a macrofungus with pharmacological activities. Curr. Med. Chem. 2010, 17, 2419-2430. [CrossRef]
- Finimundy, T.C.; Dillon, A.J.P.; Henriques, J.A.P.; Ely, M.R. A review on general nutritional compounds and pharmacological properties of the Lentinula edodes mushroom. Food Sci. Nutr. 2014, 2014. [CrossRef]
- Chung, I.M.; Kim, S.Y.; Han, J.G.; Kong, W.-S.; Jung, M.Y.; Kim, S.H. Fatty acids and stable isotope ratios in shiitake mushrooms (Lentinula edodes) indicate the origin of the cultivation substrate used: A preliminary case study in Korea. Foods (Basel, Switzerland) 2020, 9, 1210. [CrossRef]
- Chen, C.C.; Liu, S.E.; Wu, C.M.; Ho, C.T. Enzymic Formation of Volatile Compounds in Shiitake Mushroom ( Lentinus edodes Sing.) in Biogeneration of Aromas. Parliament, T.H., Croteau, R. ACS Symposium Series, 1986, 317, pp. 176-183.
- [9 Wu, C.M.; Wang, Z. Volatile compounds in fresh and processed shiitake mushrooms (Lentinus edodes Sing.). Food Sci. Technol. Res. 2000, 6, 166-170. [CrossRef]
- Cho, D.B.; Seo, H.Y.; Kim, K.S. Analysis of the volatile flavor compounds produced during the growth stages of the shiitake mushrooms (Lentinus edodes). Prev. Nutr. Food Sci. 2003, 8, 306-314. [CrossRef]
- Li, W.; Wang, J.; Chen, W.; Yang, Y.; Zhang, J.; Feng, J.; Yu, H.; Li, Q. Analysis of volatile compounds of Lentinula edodes grown in different culture substrate formulations. Food Res. Int. 2019, 125, 108517. [CrossRef]
- Sun, L.; Xin, G.; Hou, Z.; Zhao, X.; Xu, H.; Bao, X.; Xia, R.; Li, Y.; Li, L. Biosynthetic mechanism of key volatile biomarkers of harvested Lentinula edodes triggered by spore release. J. Agric. Food Chem. 2021, 69, 9350-9361. [CrossRef]
- Combet, E.; Eastwood, D.C.; Burton, K.S.; Henderson, J. Eight-carbon volatiles in mushrooms and fungi: properties, analysis, and biosynthesis. Mycoscience 2006, 47, 317-326. [CrossRef]
- Venkateshwarlu, G.; Chandravadana, M.; Tewari, R. Volatile flavour components of some edible mushrooms (Basidiomycetes). Flavour Fragr. J. 1999, 14, 191-194.
- Zhu, R.; Wen, Y.; Wu, W.; Zhang, L.; Salman Farid, M.; Shan, S.; Wen, J.; Farag, M.A.; Zhang, Y.; Zhao, C. The flavors of edible mushrooms: A comprehensive review of volatile organic compounds and their analytical methods. Crit. Rev. Food Sci. Nutr. 2022, 1-15. [CrossRef]
- Moliszewska, E. Mushroom flavour. Acta Universitatis Lodziensis. Folia Biol. Oecol. 2014, 10, 80-88.
- Yasumoto, K. Enzyme-catalyzed evolution of lenthionine from lenthinic acid. Agr. Biol. Chem. 1971, 35.
- Yasumoto, K.; Iwami, K.; Mitsuda, H. A new sulfur-containing peptide from Lentinus edodes acting as a precursor for lenthionine. Agric. Biol. Chem. 1971, 35, 2059-2069.
- Li, W.; Li, R.; Chen, W.; Feng, J.; Wu, D.; Zhang, Z.; Zhang, J.; Yang, Y. The anabolism of sulphur aroma volatiles responds to enzymatic and non-enzymatic reactions during the drying process of shiitake mushrooms. Food Chem. 2022, 371, 131123. [CrossRef]
- Li, W.; Chen, W.C.; Wang, J.B.; Feng, J.; Wu, D.; Zhang, Z.; Zhang, J.S.; Yang, Y. Effects of enzymatic reaction on the generation of key aroma volatiles in shiitake mushroom at different cultivation substrates. Food Sci. Nutr. 2021, 9, 2247-2256. [CrossRef]
- Chen, H.; Bao, D.; Kang, Q.; Wu, Y. Research progress of volatiles in Lentinula edodes. Acta Edulis Fungi 2018, 25, 105-114.
- Sneeden, E.Y.; Harris, H.H.; Pickering, I.J.; Prince, R.C.; Johnson, S.; Li, X.; Block, E.; George, G.N. The Sulfur Chemistry of Shiitake Mushroom. J. Am. Chem. Soc. 2004, 126, 458-459. [CrossRef]
- Hiraide, M.; Yokoyama, I. The smell and odorous components of dried shiitake mushroom, Lentinula edodes IV: survey of trends in consumer preferences and changes in sensory evaluation. J. Wood Sci. 2007, 53, 458-461. [CrossRef]
- Hiraide, M.; Yokoyama, I.; Miyazaki, Y. The smell and odorous components of dried shiitake mushroom, Lentinula edodes II: sensory evaluation by ordinary people. J. Wood Sci. 2005, 51, 628-633. [CrossRef]
- Hiraide, M.; Miyazaki, Y.; Shibata, Y. The smell and odorous components of dried shiitake mushroom, Lentinula edodes I: relationship between sensory evaluations and amounts of odorous components. J. Wood Sci. 2004, 50, 358-364. [CrossRef]
- Wurzenberger, M.; Grosch, W. Bestimmung von 1-oeten-3-ol in pilzen und pilzprodukten. Zeitschrift für Lebensmittel-Untersuchung und Forschung 1983, 176, 16-19.
- Tasaki, Y.; Kobayashi, D.; Sato, R.; Hayashi, S.; Joh, T. Variations in 1-octen-3-ol and lipoxygenase gene expression in the oyster mushroom <i>Pleurotus ostreatus</i> according to fruiting body development, tissue specificity, maturity, and postharvest storage. Mycoscience 2019, 60, 170-176.
- Chen, D.; Wang, S.; Li, M.; Hao, T.; Lin, S. The dynamic changes in product attributes of shiitake mushroom pilei and stipes during dehydration by hot air drying. J. Food Process. Preserv. 2021, 45, e15648. [CrossRef]
- Li, S.; Wang, A.; Liu, L.; Tian, G.; Wei, S.; Xu, F. Evaluation of nutritional values of shiitake mushroom (Lentinus edodes) stipes. J. Food Meas. Charact. 2018, 12, 2012-2019. [CrossRef]
- van Den Dool, H.; Kratz, P.D. A generalization of the retention index system including linear temperature programmed gas-liquid partition chromatography. J. Chromatogr. 1963,11, 463-471. [CrossRef]
- Guo, Y.; Ghirardo, A.; Weber, B.; Schnitzler, J.-P.; Benz, J.P.; Rosenkranz, M. Trichoderma species differ in their volatile profiles and in antagonism toward ectomycorrhiza Laccaria bicolor. Front. Microbiol. 2019, 10, 891. [CrossRef]
- Kreuzwieser, J.; Scheerer, U.; Kruse, J.; Burzlaff, T.; Honsel, A.; Alfarraj, S.; Georgiev, P.; Schnitzler, J.-P.; Ghirardo, A.; Kreuzer, I.; et al. The Venus flytrap attracts insects by the release of volatile organic compounds. J. Exp. Bot. 2014, 65, 755-766. [CrossRef]
- Xu, L.; Fang, X.; Wu, W.; Chen, H.; Mu, H.; Gao, H. Effects of high-temperature pre-drying on the quality of air-dried shiitake mushrooms (Lentinula edodes). Food Chem. 2019, 285, 406-413. [CrossRef]
- RStudio, T. RStudio: integrated development environment for R. PBC, Boston. 2023.
- Team, R.D.C. R: R Core Team. R: a language and environment for statistical computing. R Foundation for statistical computing, Vienna, Austria. <https://www.R-project.org/>. 2023.
- Wei, T.; Simko, V. package ‘corrplot’: visualization of a correlation matrix (version 0.92). 2021.
- Pedersen, T. ggraph: An implementation of grammar of graphics for graphs and 502 networks. 2022.
- Fruchterman, T.M.; Reingold, E.M. Graph drawing by force-directed placement. Software: Practice and experience 1991, 21, 1129-1164. [CrossRef]
- Csárdi, G.; Nepusz, T.; Traag, V.; Kirill, S.; Zanini, H.; Noom, F. igraph: network analysis and visualization in R. 2023. URL https://CRAN. R-project. org/package= igraph. R package version 1.
- Breiman, L. Random forests. Machine learning 2001, 45, 5-32.
- Darst, B.F.; Malecki, K.C.; Engelman, C.D. Using recursive feature elimination in random forest to account for correlated variables in high dimensional data. BMC Genet. 2018, 19, 1-6. [CrossRef]
- Guo, Y.; Jud, W.; Ghirardo, A.; Antritter, F.; Benz, J.P.; Schnitzler, J.P.; Rosenkranz, M. Sniffing fungi–phenotyping of volatile chemical diversity in Trichoderma species. New Phyto. 2020, 227, 244-259.
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1-26.
- Deviaene, M.; Testelmans, D.; Borzée, P.; Buyse, B.; Van Huffel, S.; Varon, C. Feature selection algorithm based on random forest applied to sleep apnea detection. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2019; pp. 2580-2583.
- Deng, G.; Li, J.; Liu, H.; Wang, Y. Volatile compounds and aroma characteristics of mushrooms: a review. Crit. Rev. Food Sci. Nutr., 1-18. [CrossRef]
- Luo, D.; Wu, J.; Ma, Z.; Tang, P.; Liao, X.; Lao, F. Production of high sensory quality Shiitake mushroom (Lentinus edodes) by pulsed air-impingement jet drying (AID) technique. Food Chem. 2021, 341, 128290,. [CrossRef]
- Wang, Y.; Yang, Z.; Chen, X.; Han, D.; Han, J.; Wang, L.; Ren, A.; Yu, H.; Zhao, M. Lenthionine, a key flavor substance in Lentinula edodes, is regulated by cysteine under drought Stress. J. Agric. Food Chem. 2021, 69, 12645–12653. [CrossRef]
- Li, W.; Chen, W.-C.; Wang, J.-B.; Feng, J.; Wu, D.; Zhang, Z.; Zhang, J.-S.; Yang, Y. Screening candidate genes related to volatile synthesis in shiitake mushrooms and construction of regulatory networks to effectively improve mushroom aroma. J. Sci. Food Agric. 2021,101, 5618-5626. [CrossRef]
- Xiaokang, W.; Brunton, N.P.; Lyng, J.G.; Harrison, S.M.; Carpes, S.T.; Papoutsis, K. Volatile and non-volatile compounds of shiitake mushrooms treated with pulsed light after twenty-four hour storage at different conditions. Food Biosci. 2020, 36, 100619. [CrossRef]
- Qin, L.; Gao, J.X.; Xue, J.; Chen, D.; Lin, S.Y.; Dong, X.P.; Zhu, B.W. Changes in aroma profile of Shiitake mushroom (Lentinus edodes) during different stages of hot air drying. Foods (Basel, Switzerland) 2020, 9. [CrossRef]
- Xu, S.; Wang, F.; Fu, Y.; Li, D.; Sun, X.; Li, C.; Song, B.; Li, Y. Effects of mixed agro-residues (corn crop waste) on lignin-degrading enzyme activities, growth, and quality of Lentinula edodes. RSC adv. 2020, 10, 9798-9807. [CrossRef]
- Usami, A.; Nakaya, S.; Nakahashi, H.; Miyazawa, M. Chemical composition and aroma evaluation of volatile oils from edible mushrooms (Pleurotus salmoneostramineus and Pleurotus sajor-caju). J. Oleo Sci. 2014, 63, 1323-1332. [CrossRef]
- Feng, T.; Yang, M.; Ma, B.; Zhao, Y.; Zhuang, H.; Zhang, J.; Chen, D. Volatile profiles of two genotype Agaricus bisporus species at different growth stages. Food Res. Int. 2021, 140, 109761. [CrossRef]
- Culleré, L.; Ferreira, V.; Venturini, M.E.; Marco, P.; Blanco, D. Potential aromatic compounds as markers to differentiate between Tuber melanosporum and Tuber indicum truffles. Food Chem. 2013, 141, 105-110. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).