Submitted:
02 November 2023
Posted:
02 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant material
2.2. Experimental design and cultivation
2.3. Sampling
2.4. Plant morphological traits
2.5. DNA extraction
2.6. Sequencing and bioinformatic analyses
2.7. Statistical analyses
3. Results
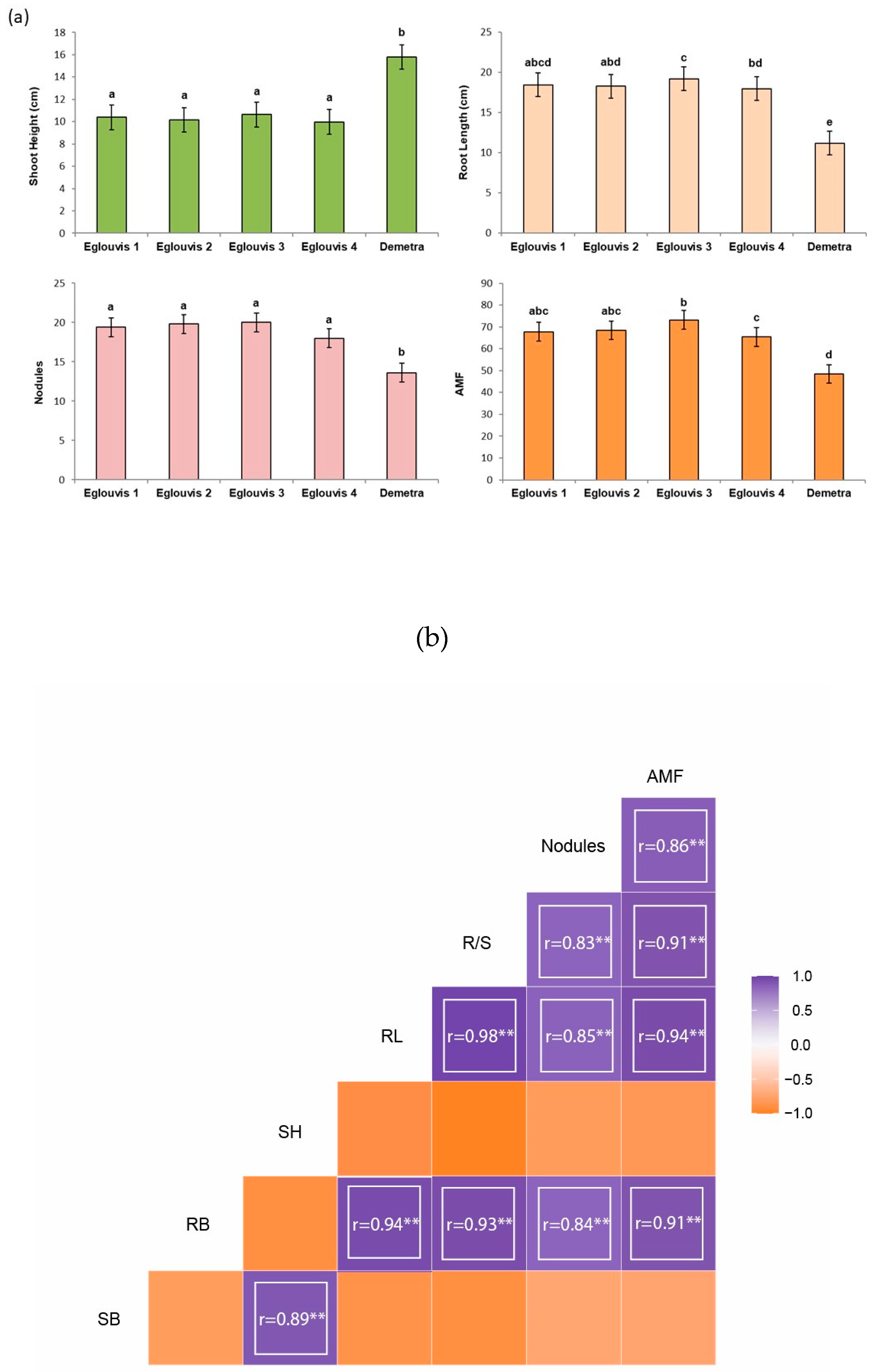
3.1. Assessment of plant morphological characters
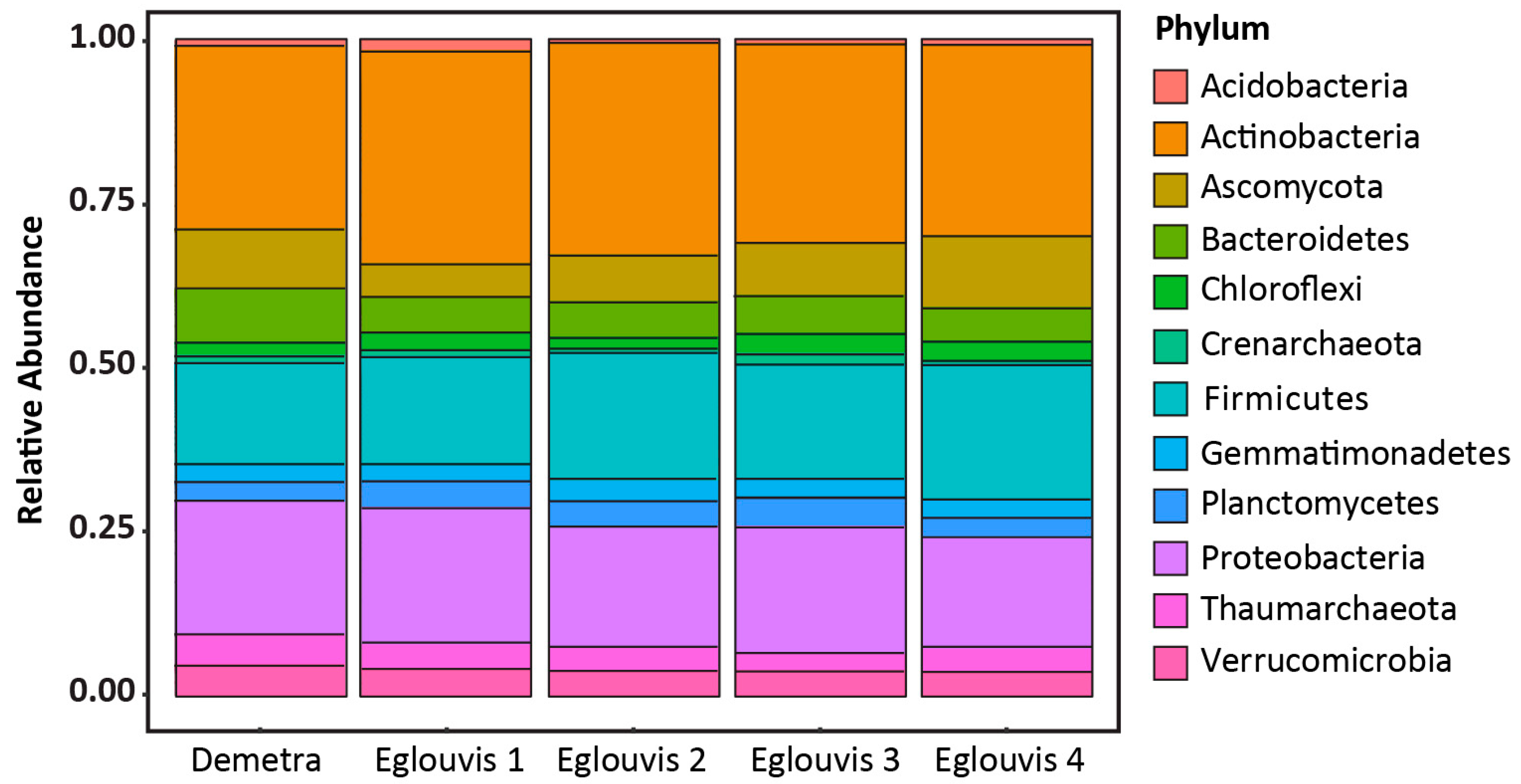
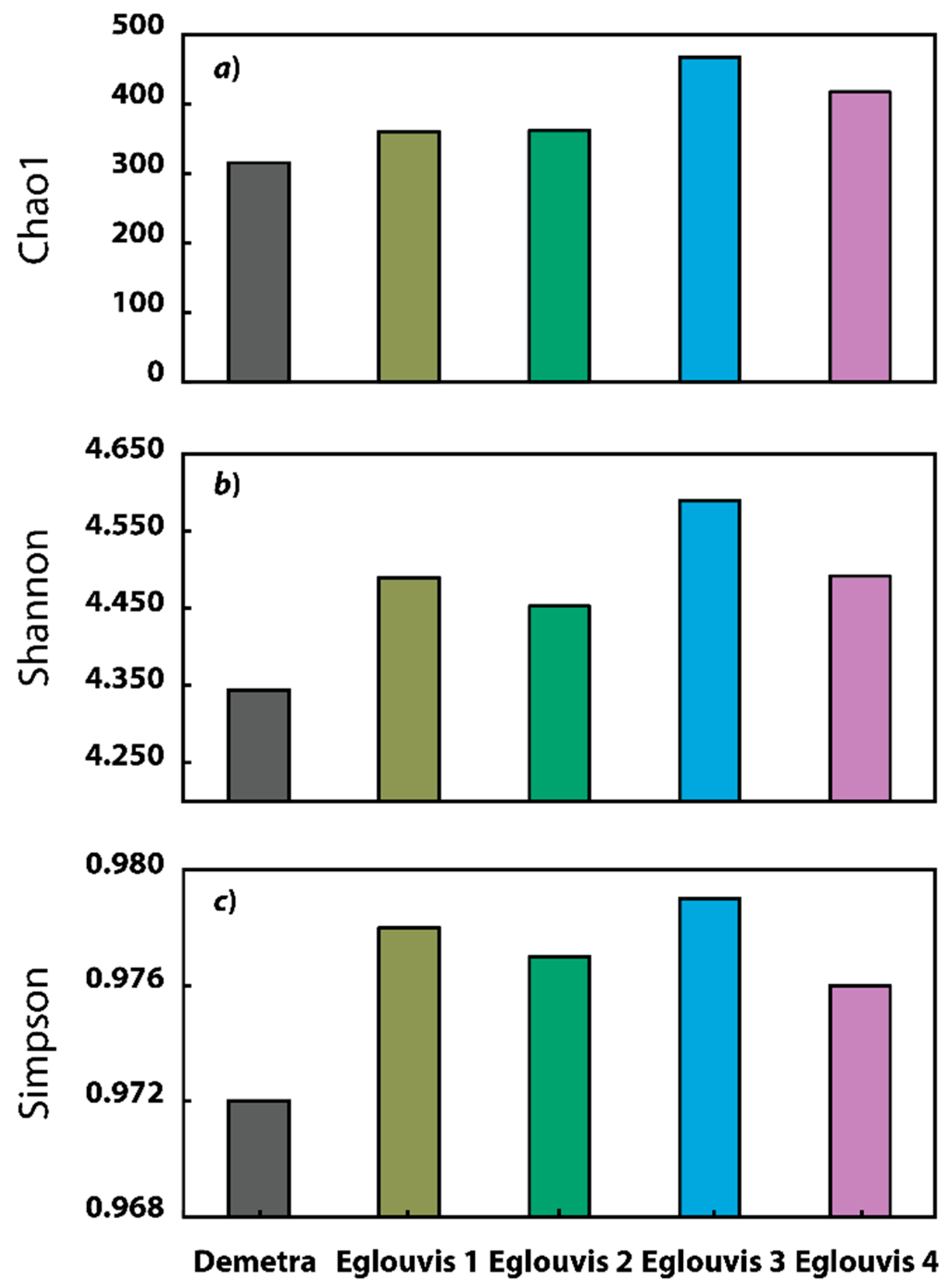
3.2. Sequence analyses
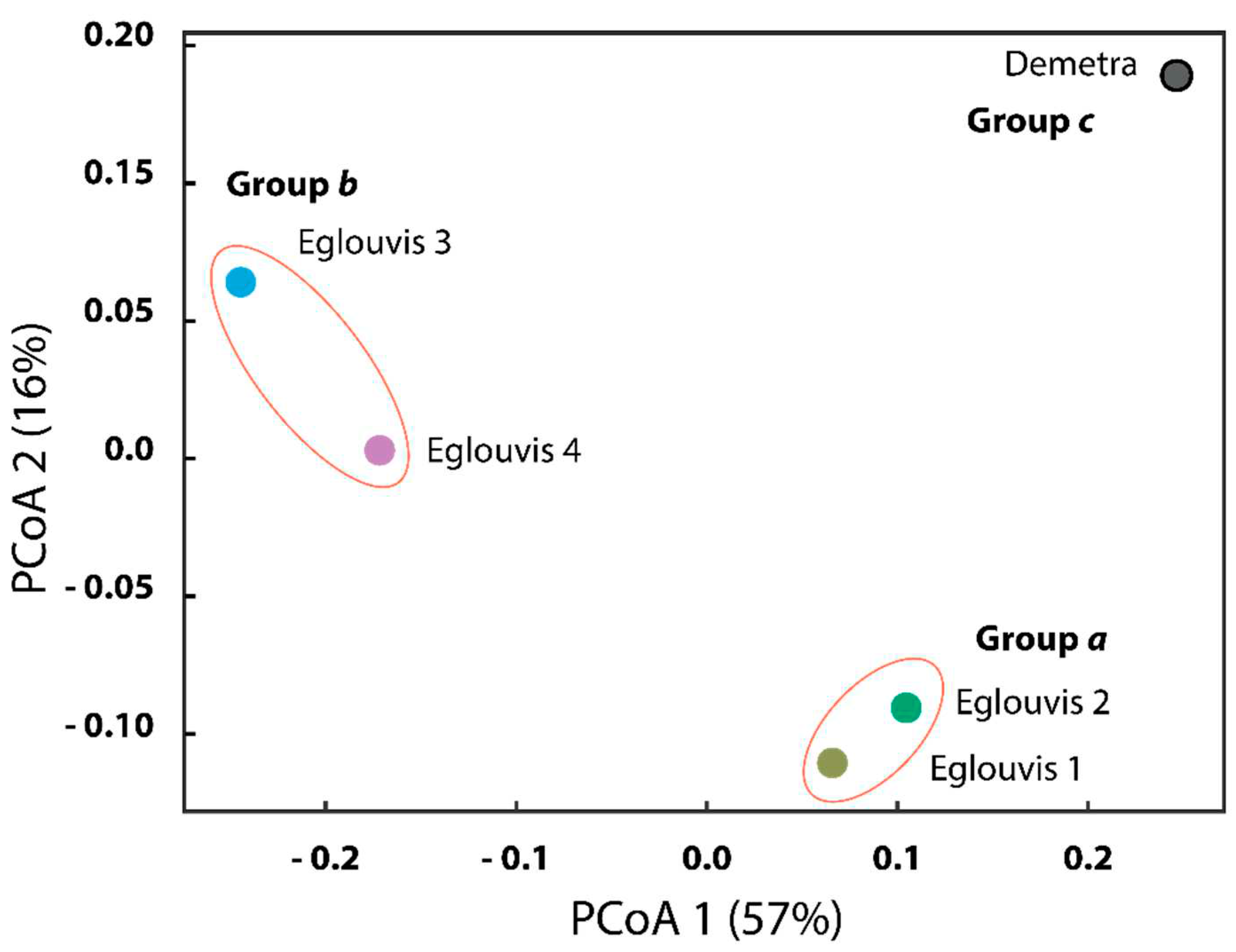
Microbial diversity among genotypes
Unique microbial taxa
4. Discussion
4.1. Plant traits impact microbial diversity
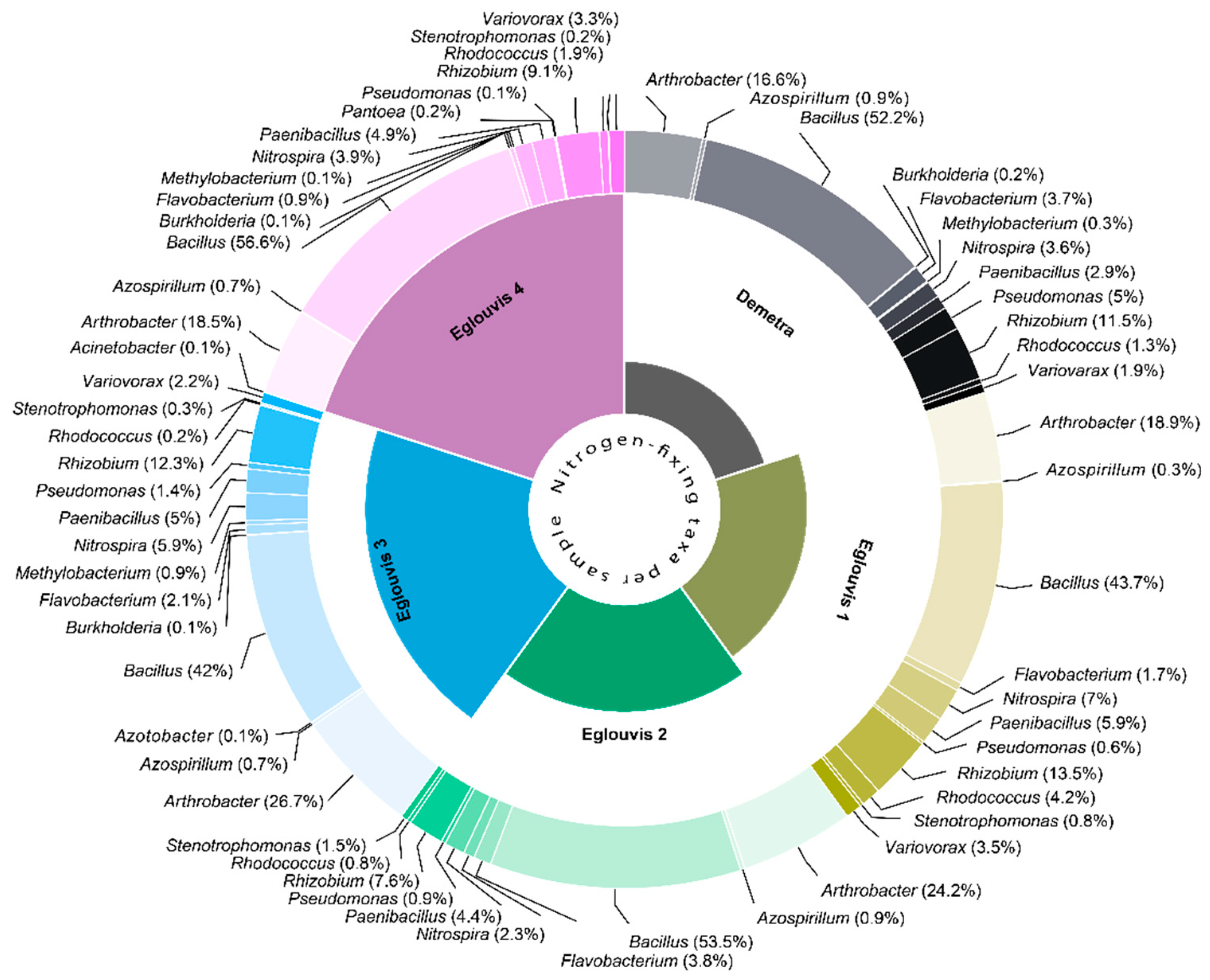
4.2. Microbial Diversity
4.3. Lentil microbial pathogens
4.4. Practical implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Global Economy of Pulses; FAO, 2019. [CrossRef]
- Tsanakas, G.F.; Mylona, P.V.; Koura, K.; Gleridou, A.; Polidoros, A.N. Genetic Diversity Analysis of the Greek Lentil (Lens Culinaris) Landrace ‘Eglouvis’ Using Morphological and Molecular Markers. Plant Genetic Resources 2018, 16, 469–477. [Google Scholar] [CrossRef]
- Polidoros, A.N.; Avdikos, I.D.; Gleridou, A.; Kostoula, S.D.; Koura, E.; Sakellariou, M.A.; Stavridou, E.; Gerasopoulos, D.; Lagopodi, A.; Mavromatis, A.; et al. Lentil Gene Pool for Breeding. In Cash Crops; Priyadarshan, P.M., Jain, S.M., Eds.; Springer International Publishing: Cham, 2022. [Google Scholar] [CrossRef]
- Sakellariou, M.; Psiloglou, B.E.; Giannakopoulos, C.; Mylona, P.V. Integration of Abandoned Lands in Sustainable Agriculture: The Case of Terraced Landscape Re-Cultivation in Mediterranean Island Conditions. Land 2021, 10, 457. [Google Scholar] [CrossRef]
- Cregger, M.A.; Veach, A.M.; Yang, Z.K.; Crouch, M.J.; Vilgalys, R.; Tuskan, G.A.; Schadt, C.W. The Populus Holobiont: Dissecting the Effects of Plant Niches and Genotype on the Microbiome. Microbiome 2018, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Lund, M.; Agerbo Rasmussen, J.; Ramos-Madrigal, J.; Sawers, R.; Gilbert, M.T.P.; Barnes, C.J. Rhizosphere bacterial communities differ among traditional maize landraces. Environmental DNA 2022, 4, 1241–1249. [Google Scholar] [CrossRef]
- Awad, M.; Giannopoulos, G.; Mylona, P.V.; Polidoros, A.N. Comparative Analysis of Grapevine Epiphytic Microbiomes among Different Varieties, Tissues, and Developmental Stages in the Same Terroir. Applied Sciences 2022, 13, 102. [Google Scholar] [CrossRef]
- Morales Moreira, Z.P.; Helgason, B.L.; Germida, J.J. Assembly and Potential Transmission of the Lens culinaris Seed Microbiome. FEMS Microbiology Ecology 2022, 97, fiab166. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Bandara, M.; Hamel, C.; Knight, J.D.; Gan, Y. Intensifying Crop Rotations with Pulse Crops Enhances System Productivity and Soil Organic Carbon in Semi-Arid Environments. Field Crops Research 2020, 248, 107657. [Google Scholar] [CrossRef]
- Pramanik, K.; Das, A.; Banerjee, J.; Das, A.; Chatterjee, S.; Sharma, R.; Kumar, S.; Gupta, S. Metagenomic Insights into Rhizospheric Microbiome Profiling in Lentil Cultivars Unveils Differential Microbial Nitrogen and Phosphorus Metabolism under Rice-Fallow Ecology. IJMS 2020, 21, 8895. [Google Scholar] [CrossRef] [PubMed]
- Gruet, C.; Muller, D.; Moënne-Loccoz, Y. Significance of the Diversification of Wheat Species for the Assembly and Functioning of the Root-Associated Microbiome. Front. Microbiol. 2022, 12, 782135. [Google Scholar] [CrossRef]
- Brescia, F.; Sillo, F.; Franchi, E.; Pietrini, I.; Montesano, V.; Marino, G.; Haworth, M.; Zampieri, E.; Fusini, D.; Schillaci, M. The ‘Microbiome Counterattack’: Insights on the Soil and Root-associated Microbiome in Diverse Chickpea and Lentil Genotypes after an Erratic Rainfall Event. Environ Microbiol Rep 2023, 1758–2229.13167. [Google Scholar] [CrossRef]
- Kuppe, C.W.; Schnepf, A.; Von Lieres, E.; Watt, M.; Postma, J.A. Rhizosphere Models: Their Concepts and Application to Plant-Soil Ecosystems. Plant Soil 2022, 474, 17–55. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends in Plant Science 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Bazghaleh, N.; Prashar, P.; Woo, S.; Vandenberg, A. Effects of Lentil Genotype on the Colonization of Beneficial Trichoderma Species and Biocontrol of Aphanomyces Root Rot. Microorganisms 2020, 8, 1290. [Google Scholar] [CrossRef] [PubMed]
- Schaedel, M.; Hidrobo, G.; Grossman, J. From Microns to Meters: Exploring Advances in Legume Microbiome Diversity for Agroecosystem Benefits. Front. Sustain. Food Syst. 2021, 5, 668195. [Google Scholar] [CrossRef]
- Quiza, L.; Tremblay, J.; Pagé, A.P.; Greer, C.W.; Pozniak, C.J.; Li, R.; Haug, B.; Hemmingsen, S.M.; St-Arnaud, M.; Yergeau, E. The Effect of Wheat Genotype on the Microbiome Is More Evident in Roots and Varies through Time. ISME COMMUN. 2023, 3, 32. [Google Scholar] [CrossRef]
- Cordero, J.; De Freitas, J.R.; Germida, J.J. Bacterial Microbiome Associated with the Rhizosphere and Root Interior of Crops in Saskatchewan, Canada. Can. J. Microbiol. 2020, 66, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Vierheilig, H.; Schweiger, P.; Brundrett, M. An Overview of Methods for the Detection and Observation of Arbuscular Mycorrhizal Fungi in Roots †. Physiologia Plantarum 2005, 125, 393–404. [Google Scholar] [CrossRef]
- Keegan, K.P.; Glass, E.M.; Meyer, F. MG-RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function. In Microbial Environmental Genomics (MEG); Martin, F., Uroz, S., Eds.; Methods in Molecular Biology; Springer New York: New York, NY, 2016; Volume 1399, pp. 207–233. [Google Scholar] [CrossRef]
- R Core Team, 2021. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing. 2021. Available online: https://www.R-project.org/ (accessed on 28 August 2023).
- R Core Team. 2022. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.r-project.org/.
- Chen, H.; Boutros, P.C. VennDiagram: A Package for the Generation of Highly-Customizable Venn and Euler Diagrams in R. BMC Bioinformatics 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J. , Blanchet, F.G., Kindt, R., Legendre, P., Minchin, P.R., O’Hara, R.B., Simpson, G.L., Solymos, P., Stevens, M.H.H. and Wagner, H. Vegan: Community Ecology Package. R Package Version 2.2-0., 2014. http://CRAN.Rproject.org/package=vegan.
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2016., 2nd ed.; Use R!; Springer International Publishing : Imprint: Springer: Cham, 2016. [Google Scholar] [CrossRef]
- Oksanen, A.J.; Blanchet, F.G.; Kindt, R.; Legen-, P.; Minchin, P.R.; Hara, R.B.O.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H. Community Ecology Package. … Ecology Package ….
- Casañas, F.; Simó, J.; Casals, J.; Prohens, J. Toward an Evolved Concept of Landrace. Front. Plant Sci. 2017, 08. [Google Scholar] [CrossRef]
- Lombardi, M.; Materne, M.; Cogan, N.O.I.; Rodda, M.; Daetwyler, H.D.; Slater, A.T.; Forster, J.W.; Kaur, S. Assessment of Genetic Variation within a Global Collection of Lentil (Lens culinarisMedik.) Cultivars and Landraces Using SNP Markers. BMC Genet 2014, 15, 150. [Google Scholar] [CrossRef]
- Khazaei, H.; Caron, C.T.; Fedoruk, M.; Diapari, M.; Vandenberg, A.; Coyne, C.J.; McGee, R.; Bett, K.E. Genetic Diversity of Cultivated Lentil (Lens Culinaris Medik.) and Its Relation to the World’s Agro-Ecological Zones. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Idrissi, O.; Piergiovanni, A.R.; Toklu, F.; Houasli, C.; Udupa, S.M.; De Keyser, E.; Van Damme, P.; De Riek, J. Molecular Variance and Population Structure of Lentil ( Lens Culinaris Medik.) Landraces from Mediterranean Countries as Revealed by Simple Sequence Repeat DNA Markers: Implications for Conservation and Use. Plant Genet. Resour. 2018, 16, 249–259. [Google Scholar] [CrossRef]
- Brisson, V.L.; Schmidt, J.E.; Northen, T.R.; Vogel, J.P.; Gaudin, A.C.M. Impacts of Maize Domestication and Breeding on Rhizosphere Microbial Community Recruitment from a Nutrient Depleted Agricultural Soil. Sci Rep 2019, 9, 15611. [Google Scholar] [CrossRef]
- Brown, S.P.; Grillo, M.A.; Podowski, J.C.; Heath, K.D. Soil Origin and Plant Genotype Structure Distinct Microbiome Compartments in the Model Legume Medicago Truncatula. Microbiome 2020, 8, 139. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Liu, K.; Poppy, L.; Mulenga, A.; Gampe, C. Minimizing Lentil Harvest Loss through Improved Agronomic Practices in Sustainable Agro-Systems. Sustainability 2021, 13, 1896. [Google Scholar] [CrossRef]
- Hartman, K.; Van Der Heijden, M.G.; Roussely-Provent, V.; Walser, J.-C.; Schlaeppi, K. Deciphering Composition and Function of the Root Microbiome of a Legume Plant. Microbiome 2017, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Priya, S.; Bansal, R.; Kumar, G.; Dikshit, H.K.; Kumari, J.; Pandey, R.; Singh, A.K.; Tripathi, K.; Singh, N.; Kumari, N.K.P.; Kumar, S.; Kumar, A. Root Trait Variation in Lentil (Lens Culinaris Medikus) Germplasm under Drought Stress. Plants 2021, 10, 2410. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; Bosse, M.; Ferrão, L.F.V.; De Hollander, M.; Garcia, A.A.F.; Ramírez, C.A.; Mendes, R.; Raaijmakers, J.M. Linking Rhizosphere Microbiome Composition of Wild and Domesticated Phaseolus Vulgaris to Genotypic and Root Phenotypic Traits. ISME J 2017, 11, 2244–2257. [Google Scholar] [CrossRef]
- Saleem, M.; Law, A.D.; Sahib, M.R.; Pervaiz, Z.H.; Zhang, Q. Impact of Root System Architecture on Rhizosphere and Root Microbiome. Rhizosphere 2018, 6, 47–51. [Google Scholar] [CrossRef]
- Kavamura, V.N.; Robinson, R.J.; Hughes, D.; Clark, I.; Rossmann, M.; Melo, I.S.D.; Hirsch, P.R.; Mendes, R.; Mauchline, T.H. Wheat Dwarfing Influences Selection of the Rhizosphere Microbiome. Sci Rep 2020, 10, 1452. [Google Scholar] [CrossRef]
- Lei, S.; Xu, X.; Cheng, Z.; Xiong, J.; Ma, R.; Zhang, L.; Yang, X.; Zhu, Y.; Zhang, B.; Tian, B. Analysis of the Community Composition and Bacterial Diversity of the Rhizosphere Microbiome across Different Plant Taxa. MicrobiologyOpen 2019, 8, e00762. [Google Scholar] [CrossRef] [PubMed]
- Ellouze, W.; Hamel, C.; Vujanovic, V.; Gan, Y.; Bouzid, S.; St-Arnaud, M. Chickpea Genotypes Shape the Soil Microbiome and Affect the Establishment of the Subsequent Durum Wheat Crop in the Semiarid North American Great Plains. Soil Biology and Biochemistry 2013, 63, 129–141. [Google Scholar] [CrossRef]
- Hamel, C.; Gan, Y.; Sokolski, S.; Bainard, L.D. High Frequency Cropping of Pulses Modifies Soil Nitrogen Level and the Rhizosphere Bacterial Microbiome in 4-Year Rotation Systems of the Semiarid Prairie. Applied Soil Ecology 2018, 126, 47–56. [Google Scholar] [CrossRef]
- Wattenburger, C.J.; Gutknecht, J.; Zhang, Q.; Brutnell, T.; Hofmockel, K.; Halverson, L. The Rhizosphere and Cropping System, but Not Arbuscular Mycorrhizae, Affect Ammonia Oxidizing Archaea and Bacteria Abundances in Two Agricultural Soils. Applied Soil Ecology 2020, 151, 103540. [Google Scholar] [CrossRef]
- Morales Moreira, Z.P.; Helgason, B.L.; Germida, J.J. Environment Has a Stronger Effect than Host Plant Genotype in Shaping Spring Brassica Napus Seed Microbiomes. Phytobiomes Journal 2021, 5, 220–230. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Serna-Chavez, H.M.; Fierer, N.; Van Bodegom, P.M. Global Drivers and Patterns of Microbial Abundance in Soil. Global Ecology and Biogeography 2013, 22, 1162–1172. [Google Scholar] [CrossRef]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere Bacteriome Structure and Functions. Nat Commun 2022, 13, 836. [Google Scholar] [CrossRef] [PubMed]
- Gqozo, M.P.; Bill, M.; Siyoum, N.; Labuschagne, N.; Korsten, L. Fungal Diversity and Community Composition of Wheat Rhizosphere and Non-Rhizosphere Soils from Three Different Agricultural Production Regions of South Africa. Applied Soil Ecology 2020, 151, 103543. [Google Scholar] [CrossRef]
- Lu, J.; Yang, F.; Wang, S.; Ma, H.; Liang, J.; Chen, Y. Co-Existence of Rhizobia and Diverse Non-Rhizobial Bacteria in the Rhizosphere and Nodules of Dalbergia Odorifera Seedlings Inoculated with Bradyrhizobium Elkanii, Rhizobium Multihospitium–Like and Burkholderia Pyrrocinia–Like Strains. Front. Microbiol. 2017, 8, 2255. [Google Scholar] [CrossRef]
- Martínez-Hidalgo, P.; Hirsch, A.M. The Nodule Microbiome: N 2 -Fixing Rhizobia Do Not Live Alone. Phytobiomes Journal 2017, 1, 70–82. [Google Scholar] [CrossRef]
- Alexander, A.; Singh, V.K.; Mishra, A.; Jha, B. Plant Growth Promoting Rhizobacterium Stenotrophomonas maltophilia BJ01 Augments Endurance against N2 Starvation by Modulating Physiology and Biochemical Activities of Arachis Hypogea. PLoS ONE 2019, 14, e0222405. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.N. The PGPR Stenotrophomonas Maltophilia SBP-9 Augments Resistance against Biotic and Abiotic Stress in Wheat Plants. Front. Microbiol. 2017, 8, 1945. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-L.; Yang, W.-L.; Fang, W.-W.; Zhao, Y.-X.; Guo, L.; Dai, Y.-J. The Plant Growth-Promoting Rhizobacterium Variovorax Boronicumulans CGMCC 4969 Regulates the Level of Indole-3-Acetic Acid Synthesized from Indole-3-Acetonitrile. Appl Environ Microbiol 2018, 84, e00298-18. [Google Scholar] [CrossRef] [PubMed]
- Sumbul, A.; Ansari, R.A.; Rizvi, R.; Mahmood, I. Azotobacter: A Potential Bio-Fertilizer for Soil and Plant Health Management. Saudi Journal of Biological Sciences 2020, 27, 3634–3640. [Google Scholar] [CrossRef] [PubMed]
- Mylona, P.V.; Premakumar, R.; Pau, R.N.; Bishop, P.E. Characteristics of Orf1 and Orf2 in the anfHDGK Genomic Region Encoding Nitrogenase 3 of Azotobacter Vinelandii. J Bacteriol 1996, 178, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Taffner, J.; Erlacher, A.; Bragina, A.; Berg, C.; Moissl-Eichinger, C.; Berg, G. What Is the Role of Archaea in Plants? New Insights from the Vegetation of Alpine Bogs. mSphere 2018, 3, e00122–18. [Google Scholar] [CrossRef] [PubMed]
- Akinola, S.A.; Babalola, O.O. The Fungal and Archaeal Community within Plant Rhizosphere: A Review on Their Contribution to Crop Safety. Journal of Plant Nutrition 2021, 44, 600–618. [Google Scholar] [CrossRef]
- Mittelstrass, J.; Sperone, F.G.; Horton, M.W. Using Transects to Disentangle the Environmental Drivers of Plant-microbiome Assembly. Plant Cell & Environment 2021, 44, 3745–3755. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and Heritability of the Maize Rhizosphere Microbiome under Field Conditions. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Majdura, J.; Jankiewicz, U.; Gałązka, A.; Orzechowski, S. The Role of Quorum Sensing Molecules in Bacterial–Plant Interactions. Metabolites 2023, 13, 114. [Google Scholar] [CrossRef] [PubMed]
- Zafeiriou, I.; Ntoanidou, S.; Baira, E.; Kasiotis, K.M.; Barmpouni, T.; Machera, K.; Mylona, P.V. Ingenious Characterization and Assessment of Lentil Germplasm Collection to Aphid Acyrthosiphon Pisum Stress Unveils Distinct Responses. Front. Plant Sci. 2022, 13, 1011026. [Google Scholar] [CrossRef]
- Berlanga-Clavero, M.V.; Molina-Santiago, C.; De Vicente, A.; Romero, D. More than Words: The Chemistry behind the Interactions in the Plant Holobiont. Environmental Microbiology 2020, 22, 4532–4544. [Google Scholar] [CrossRef] [PubMed]
- Panth, M.; Hassler, S.C.; Baysal-Gurel, F. Methods for Management of Soilborne Diseases in Crop Production. Agriculture 2020, 10, 16. [Google Scholar] [CrossRef]
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, P.S.; Staton, M.E. Soil Indigenous Microbiome and Plant Genotypes Cooperatively Modify Soybean Rhizosphere Microbiome Assembly. BMC Microbiol 2019, 19, 201. [Google Scholar] [CrossRef]
- Barret, M.; Guimbaud, J.; Darrasse, A.; Jacques, M. Plant Microbiota Affects Seed Transmission of Phytopathogenic Microorganisms. Molecular Plant Pathology 2016, 17, 791–795. [Google Scholar] [CrossRef]
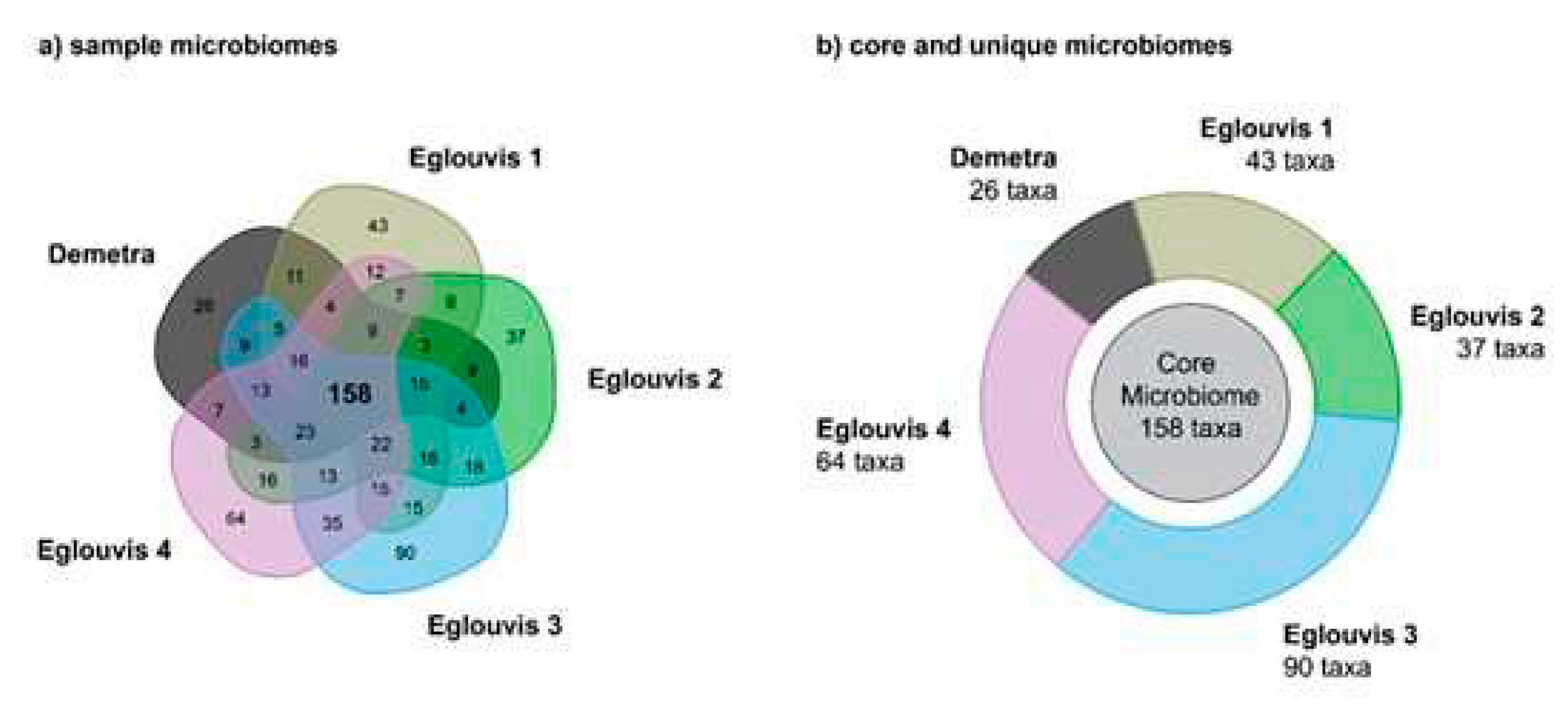
| Accession | Genotype | Collection site /origin | Cropping system |
|---|---|---|---|
| Eglouvis 1 | Landrace | Eglouvis region, Lefkada Island, Greece | Conventional |
| Eglouvis 2 | “ | “ | Conventional |
| Eglouvis 3 | “ | “ | Organic |
| Eglouvis 4 | ” | GRC1015, IPB&GR, Greece | Conventional |
| Demetra | Commercial cultivar | IPB&GR, Greece | Conventional |
| Soil Property (units) | Value |
|---|---|
| Soil type | Loam |
| Silt (%) | 42 |
| Clay (%) | 20 |
| Sand (%) | 36 |
| pH | 7.8 |
| EC (mS/cm) | 0.6 |
| SOM (%) | 1.6 |
| NO3-N (kg ha-1) | 63.2 |
| P (kg ha-1) | 17.2 |
| K (kg ha-1) | 538.5 |
| Sample | Bacterial | Fungal | Total |
|---|---|---|---|
| Eglouvis 1 | 41 | 2 | 43 |
| Eglouvis 2 | 37 | 0 | 37 |
| Eglouvis 3 | 88 | 2 | 90 |
| Eglouvis 4 | 62 | 2 | 64 |
| Demetra | 25 | 1 | 26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





