Submitted:
03 November 2023
Posted:
06 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
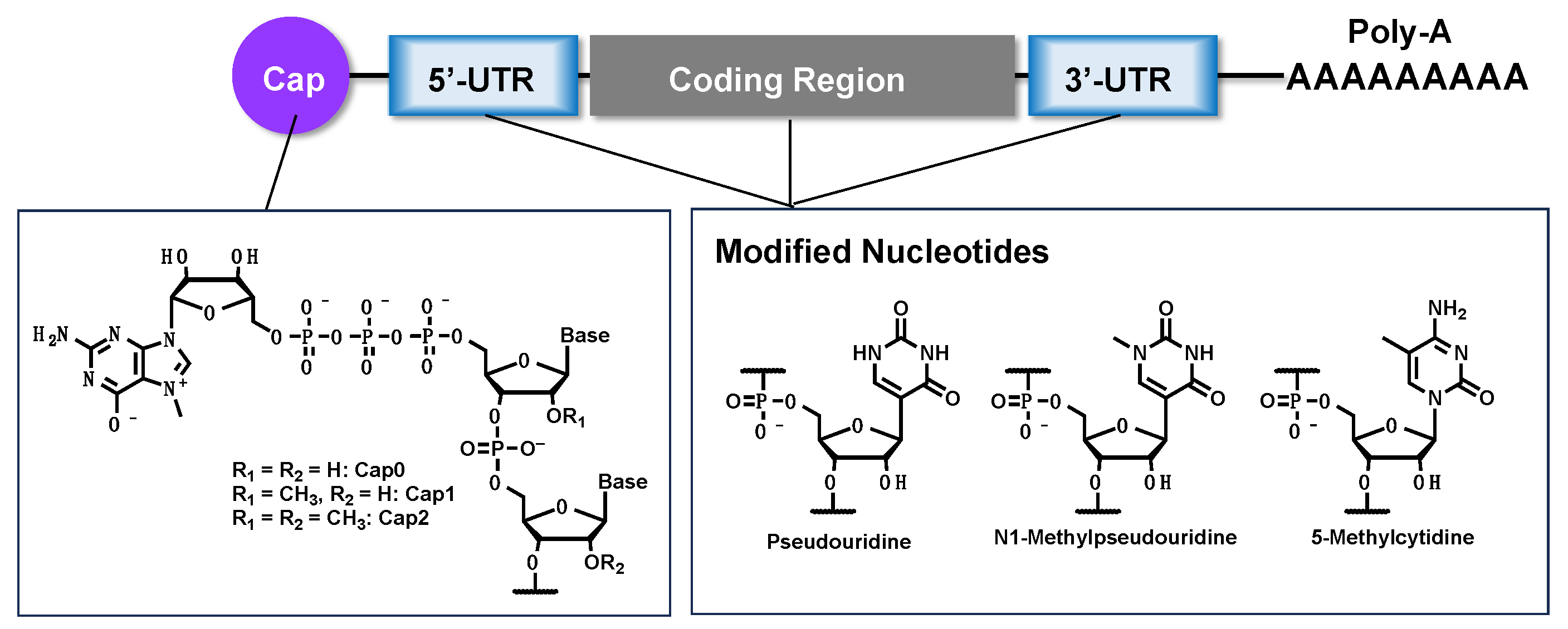
2. Synthesis of mRNA
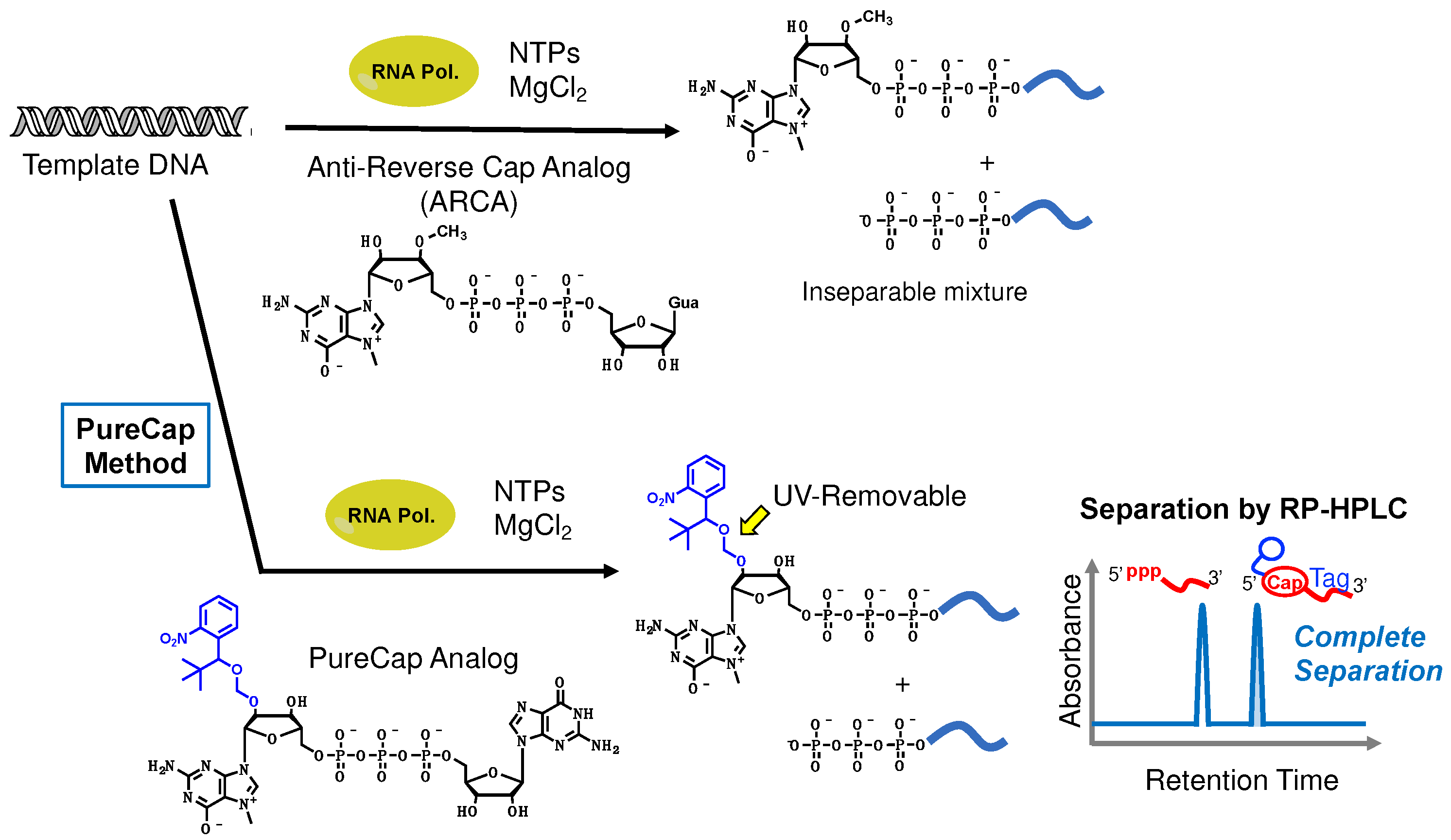
3. Key Technologies for mRNA Therapeutics
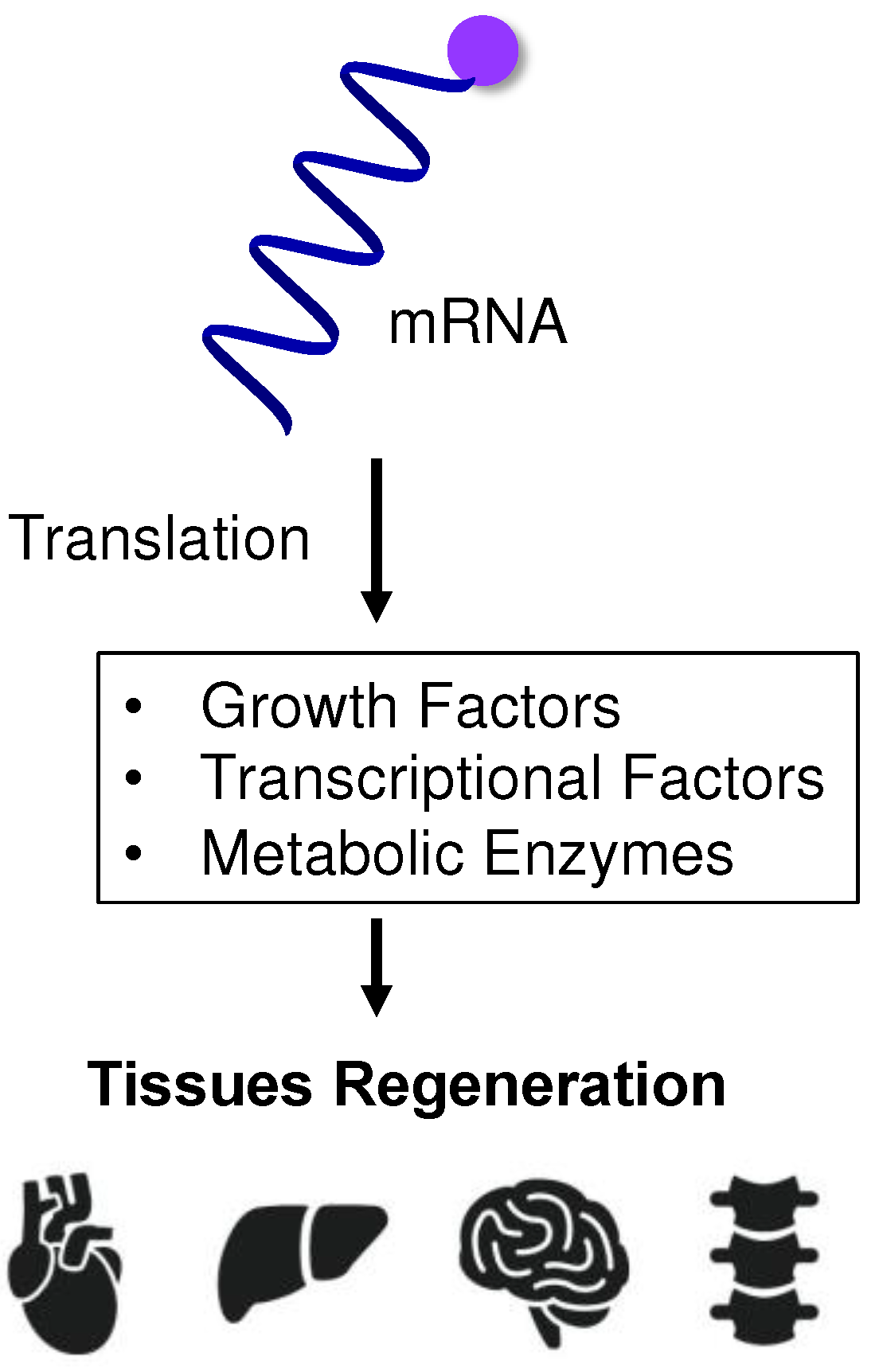
4. mRNA-Based Protein Supplementation for Regenerative Medicine
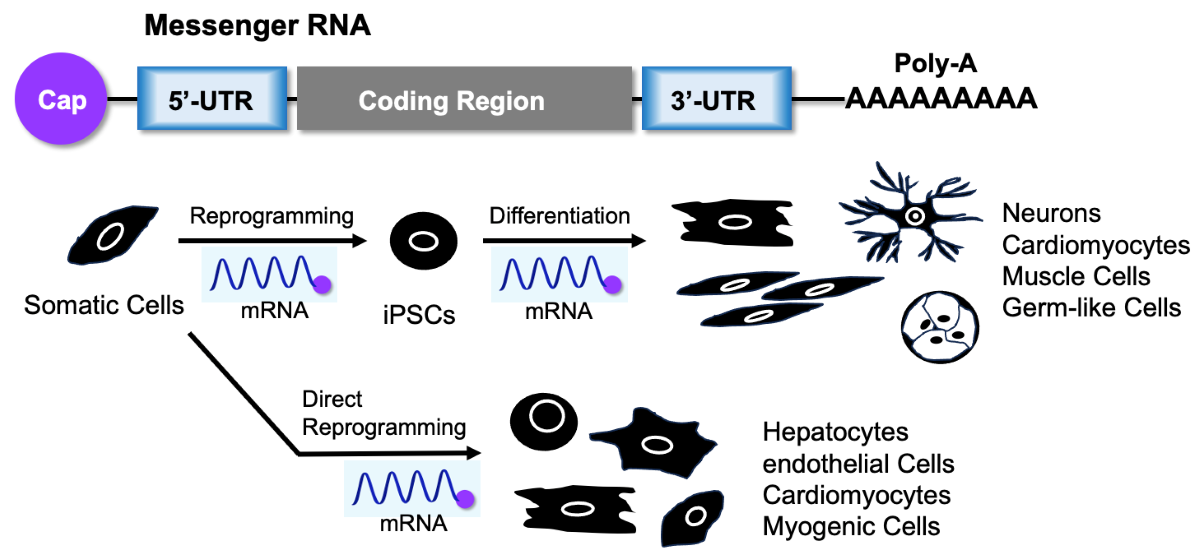
5. mRNA for Cell Reprogramming
6. mRNA-Induced Cell Differentiation from iPSCs
7. mRNA for Both Cell Reprogramming and Differentiation Induction
8. mRNA-Induced Direct Reprogramming without Passage through Pluripotent Stem Cells
9. mRNA-Based Purification Method of iPSCs and iPSC-Derived Cells
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ye, Q.; Wu, M.; Zhou, C.; Lu, X.; Huang, B.; Zhang, N.; Zhao, H.; Chi, H.; Zhang, X.; Ling, D.; et al. Rational development of a combined mRNA vaccine against COVID-19 and influenza. NPJ Vaccines 2022, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Ladak, R. J.; He, A. J.; Huang, Y. H.; Ding, Y. The Current Landscape of mRNA Vaccines Against Viruses and Cancer-A Mini Review. Front. Immunol. 2022, 13, 885371. [Google Scholar] [CrossRef]
- Sahu, I.; Haque, A.; Weidensee, B.; Weinmann, P.; Kormann, M. S. D. Recent Developments in mRNA-Based Protein Supplementation Therapy to Target Lung Diseases. Mol. Ther. 2019, 27, 803–823. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Tang, X.; Chen, Y.; Chen, K.; Fan, N.; Xiao, W.; Zheng, Q.; Li, G.; Teng, Y.; Wu, M.; Song, X. mRNA-based therapeutics: powerful and versatile tools to combat diseases. Signal Transduct. Target. Ther. 2022, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Li, C.; Yang, T.; Hu, B.; Zhang, M.; Guo, S.; Xiao, H.; Liang, X. J.; Huang, Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020, 40, 107534. [Google Scholar] [CrossRef] [PubMed]
- Muttach, F.; Muthmann, N.; Rentmeister, A. Synthetic mRNA capping. Beilstein J. Org. Chem. 2017, 13, 2819–2832. [Google Scholar] [CrossRef]
- Furuichi, Y.; Morgan, M.; Muthukrishnan, S.; Shatkin, A. J. Reovirus messenger RNA contains a methylated, blocked 5'-terminal structure: m-7G(5')ppp(5')G-MpCp. Proc. Natl. Acad. Sci. USA. 1975, 72, 362–366. [Google Scholar] [CrossRef]
- Furuichi, Y.; Miura, K. A blocked structure at the 5' terminus of mRNA from cytoplasmic polyhedrosis virus. Nature 1975, 253, 374–375. [Google Scholar] [CrossRef]
- Hinnebusch, A. G.; Ivanov, I. P.; Sonenberg, N. Translational control by 5'-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416. [Google Scholar] [CrossRef]
- Chatterjee, S.; Pal, J. K. Role of 5'- and 3'-untranslated regions of mRNAs in human diseases. Biol. Cell. 2009, 101, 251–262. [Google Scholar] [CrossRef]
- Schuster, S. L.; Hsieh, A. C. The Untranslated Regions of mRNAs in Cancer. Trends Cancer 2019, 5, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Nobuta, R.; Machida, K.; Sato, M.; Hashimoto, S.; Toriumi, Y.; Nakajima, S.; Suto, D.; Imataka, H.; Inada, T. eIF4G-driven translation initiation of downstream ORFs in mammalian cells. Nucleic Acids Res. 2020, 48, 10441–10455. [Google Scholar] [CrossRef]
- Passmore, L. A.; Coller, J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell Biol. 2022, 23, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Meet the authors: Katalin Kariko and Drew Weissman. Immunity 2021, 54, 2673–2675. [CrossRef] [PubMed]
- Nance, K. D.; Meier, J. L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Grudzien-Nogalska, E.; Stepinski, J.; Jemielity, J.; Zuberek, J.; Stolarski, R.; Rhoads, R. E.; Darzynkiewicz, E. Synthesis of anti-reverse cap analogs (ARCAs) and their applications in mRNA translation and stability. Methods Enzymo.l 2007, 431, 203–227. [Google Scholar]
- Henderson, J. M.; Ujita, A.; Hill, E.; Yousif-Rosales, S.; Smith, C.; Ko, N.; McReynolds, T.; Cabral, C. R.; Escamilla-Powers, J. R.; Houston, M. E. Cap 1 Messenger RNA Synthesis with Co-transcriptional CleanCap((R)) Analog by In Vitro Transcription. Curr. Protoc. 2021, 1, e39. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, M.; Abe, N.; Li, Z.; Nakashima, Y.; Acharyya, S.; Ogawa, K.; Kawaguchi, D.; Hiraoka, H.; Banno, A.; Meng, Z.; et al. Cap analogs with a hydrophobic photocleavable tag enable facile purification of fully capped mRNA with various cap structures. Nat. Commun. 2023, 14, 2657. [Google Scholar] [CrossRef] [PubMed]
- Roth, M. J.; Hurwitz, J. RNA capping by the vaccinia virus guanylyltransferase. Structure of enzyme-guanylate intermediate. J. Biol. Chem. 1984, 259, 13488–13494. [Google Scholar] [CrossRef]
- Ohno, H.; Akamine, S.; Mochizuki, M.; Hayashi, K.; Akichika, S.; Suzuki, T.; Saito, H. Versatile strategy using vaccinia virus-capping enzyme to synthesize functional 5' cap-modified mRNAs. Nucleic Acids Res. 2023, 51, e34. [Google Scholar] [CrossRef]
- Melton, D. A.; Krieg, P. A.; Rebagliati, M. R.; Maniatis, T.; Zinn, K.; Green, M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984, 12, 7035–7056. [Google Scholar] [CrossRef]
- Malone, R. W.; Felgner, P. L.; Verma, I. M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA. 1989, 86, 6077–6081. [Google Scholar] [CrossRef]
- Yeh, T. F.; Lin, C.; Sung, H. C. A review of technological developments in lipid nanoparticle application for mRNA vaccination. Hum. Vaccin. Immunother. 2023, 19, 2256040. [Google Scholar] [CrossRef]
- Bailey, A. L.; Cullis, P. R. Modulation of membrane fusion by asymmetric transbilayer distributions of amino lipids. Biochemistry 1994, 33, 12573–12580. [Google Scholar] [CrossRef]
- Egli, M.; Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [Google Scholar] [CrossRef]
- Geall, A. J.; Verma, A.; Otten, G. R.; Shaw, C. A.; Hekele, A.; Banerjee, K.; Cu, Y.; Beard, C. W.; Brito, L. A.; Krucker, T.; et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012, 109, 14604–14609. [Google Scholar] [CrossRef]
- Gilleron, J.; Querbes, W.; Zeigerer, A.; Borodovsky, A.; Marsico, G.; Schubert, U.; Manygoats, K.; Seifert, S.; Andree, C.; Stoter, M.; et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013, 31, 638–646. [Google Scholar] [CrossRef]
- Xu, X.; Xia, T. Recent Advances in Site-Specific Lipid Nanoparticles for mRNA Delivery. ACS Nanosci. Au 2023, 3, 192–203. [Google Scholar] [CrossRef]
- Kariko, K.; Kuo, A.; Barnathan, E. S.; Langer, D. J. Phosphate-enhanced transfection of cationic lipid-complexed mRNA and plasmid DNA. Biochim. Biophys. Acta. 1998, 1369, 320–334. [Google Scholar] [CrossRef]
- Kariko, K.; Kuo, A.; Barnathan, E. Overexpression of urokinase receptor in mammalian cells following administration of the in vitro transcribed encoding mRNA. Gene. Ther. 1999, 6, 1092–1100. [Google Scholar] [CrossRef]
- Kariko, K.; Ni, H.; Capodici, J.; Lamphier, M.; Weissman, D. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 2004, 279, 12542–12550. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.; Sorensen, E. W.; Mintri, S.; Rabideau, A. E.; Zheng, W.; Besin, G.; Khatwani, N.; Su, S. V.; Miracco, E. J.; Issa, W. J.; et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020, 6, eaaz6893. [Google Scholar] [CrossRef] [PubMed]
- Kariko, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, K. J.; Mir, F. F.; Jhunjhunwala, S.; Kaczmarek, J. C.; Hurtado, J. E.; Yang, J. H.; Webber, M. J.; Kowalski, P. S.; Heartlein, M. W.; DeRosa, F.; Anderson, D. G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 2016, 109, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Ilic, D.; Liovic, M. Industry updates from the field of stem cell research and regenerative medicine in August 2023. Regen. Med. 2023. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Ge, G.; Yang, P.; Wang, L.; Qiao, Y.; Pan, G.; Yang, H.; Bai, J.; Cui, W.; Geng, D. An Update on Adipose-Derived Stem Cells for Regenerative Medicine: Where Challenge Meets Opportunity. Adv. Sci. (Weinh) 2023, 10, e2207334. [Google Scholar] [CrossRef] [PubMed]
- Rizzino, A. A challenge for regenerative medicine: proper genetic programming, not cellular mimicry. Dev. Dyn. 2007, 236, 3199–3207. [Google Scholar] [CrossRef] [PubMed]
- Trommelmans, L. The challenge of regenerative medicine. Hastings Cent. Rep. 2010, 40, 24–26. [Google Scholar] [CrossRef]
- Liu, G.; David, B. T.; Trawczynski, M.; Fessler, R. G. Advances in Pluripotent Stem Cells: History, Mechanisms, Technologies, and Applications. Stem Cell Rev. Rep. 2020, 16, 3–32. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Okano, H.; Yamanaka, S. iPS cell technologies: significance and applications to CNS regeneration and disease. Mol. Brain 2014, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Akiba, R.; Takahashi, M.; Baba, T.; Mandai, M. Progress of iPS cell-based transplantation therapy for retinal diseases. Jpn. J. Ophthalmol. 2023, 67, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Wisitrasameewong, W.; Champaiboon, C.; Surisaeng, T.; Sa-Ard-Iam, N.; Freire, M.; Pardi, N.; Pichyangkul, S.; Mahanonda, R. The Impact of mRNA Technology in Regenerative Therapy: Lessons for Oral Tissue Regeneration. J. Dent. Res. 2022, 101, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Collen, A.; Bergenhem, N.; Carlsson, L.; Chien, K. R.; Hoge, S.; Gan, L. M.; Fritsche-Danielson, R. VEGFA mRNA for regenerative treatment of heart failure. Nat. Rev. Drug. Discov. 2022, 21, 79–80. [Google Scholar] [CrossRef] [PubMed]
- Anttila, V.; Saraste, A.; Knuuti, J.; Hedman, M.; Jaakkola, P.; Laugwitz, K. L.; Krane, M.; Jeppsson, A.; Sillanmaki, S.; Rosenmeier, J.; et al. Direct intramyocardial injection of VEGF mRNA in patients undergoing coronary artery bypass grafting. Mol. Ther. 2023, 31, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Panos, J. A.; Coenen, M. J.; Nagelli, C. V.; McGlinch, E. B.; Atasoy-Zeybek, A.; De Padilla, C. L.; Coghlan, R. F.; Johnstone, B.; Ferreira, E.; Porter, R. M.; et al. IL-1Ra gene transfer potentiates BMP2-mediated bone healing by redirecting osteogenesis toward endochondral ossification. Mol. Ther. 2023, 31, 420–434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Fukushima, Y.; Nozaki, K.; Nakanishi, H.; Deng, J.; Wakabayashi, N.; Itaka, K. Enhancement of bone regeneration by coadministration of angiogenic and osteogenic factors using messenger RNA. Inflamm. Regen. 2023, 43, 32. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Ryals, R. C.; Weller, K. K.; Pennesi, M. E.; Sahay, G. Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J. Control. Release 2019, 303, 91–100. [Google Scholar] [CrossRef]
- Fukushima, Y.; Uchida, S.; Imai, H.; Nakatomi, H.; Kataoka, K.; Saito, N.; Itaka, K. Treatment of ischemic neuronal death by introducing brain-derived neurotrophic factor mRNA using polyplex nanomicelle. Biomaterials 2021, 270, 120681. [Google Scholar] [CrossRef]
- Lin, C. Y.; Crowley, S. T.; Uchida, S.; Komaki, Y.; Kataoka, K.; Itaka, K. Treatment of Intervertebral Disk Disease by the Administration of mRNA Encoding a Cartilage-Anabolic Transcription Factor. Mol. Ther. Nucleic Acids 2019, 16, 162–171. [Google Scholar] [CrossRef]
- Hadas, Y.; Vincek, A. S.; Youssef, E.; Zak, M. M.; Chepurko, E.; Sultana, N.; Sharkar, M. T. K.; Guo, N.; Komargodski, R.; Kurian, A. A.; et al. Altering Sphingolipid Metabolism Attenuates Cell Death and Inflammatory Response After Myocardial Infarction. Circulation 2020, 141, 916–930. [Google Scholar] [CrossRef] [PubMed]
- Magadum, A.; Singh, N.; Kurian, A. A.; Munir, I.; Mehmood, T.; Brown, K.; Sharkar, M. T. K.; Chepurko, E.; Sassi, Y.; Oh, J. G.; et al. Pkm2 Regulates Cardiomyocyte Cell Cycle and Promotes Cardiac Regeneration. Circulation 2020, 141, 1249–1265. [Google Scholar] [CrossRef] [PubMed]
- Zangi, L.; Lui, K. O.; von Gise, A.; Ma, Q.; Ebina, W.; Ptaszek, L. M.; Spater, D.; Xu, H.; Tabebordbar, M.; Gorbatov, R.; et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013, 31, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Yla-Herttuala, S. Cardiovascular gene therapy with vascular endothelial growth factors. Gene 2013, 525, 217–219. [Google Scholar] [CrossRef] [PubMed]
- Anttila, V.; Saraste, A.; Knuuti, J.; Jaakkola, P.; Hedman, M.; Svedlund, S.; Lagerstrom-Fermer, M.; Kjaer, M.; Jeppsson, A.; Gan, L. M. Synthetic mRNA Encoding VEGF-A in Patients Undergoing Coronary Artery Bypass Grafting: Design of a Phase 2a Clinical Trial. Mol. Ther. Methods Clin. Dev. 2020, 18, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Gan, L. M.; Lagerstrom-Fermer, M.; Carlsson, L. G.; Arfvidsson, C.; Egnell, A. C.; Rudvik, A.; Kjaer, M.; Collen, A.; Thompson, J. D.; Joyal, J.; et al. Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun. 2019, 10, 871. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Poenisch, M.; Khanal, R.; Hu, Q.; Dai, Z.; Li, R.; Song, G.; Yuan, Q.; Yao, Q.; Shen, X.; et al. Therapeutic HNF4A mRNA attenuates liver fibrosis in a preclinical model. J. Hepatol. 2021, 75, 1420–1433. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, F.; Everton, E.; Smith, A. R.; Liu, H.; Osota, E.; Beattie, M.; Tam, Y.; Pardi, N.; Weissman, D.; Gouon-Evans, V. Murine liver repair via transient activation of regenerative pathways in hepatocytes using lipid nanoparticle-complexed nucleoside-modified mRNA. Nat. Commun. 2021, 12, 613. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Bonaguidi, M.; Muro, K.; Kessler, J. A. Generation of embryonic stem cells: limitations of and alternatives to inner cell mass harvest. Neurosurg. Focus 2008, 24, E4. [Google Scholar] [CrossRef]
- Fung, R. K.; Kerridge, I. H. Uncertain translation, uncertain benefit and uncertain risk: ethical challenges facing first-in-human trials of induced pluripotent stem (ips) cells. Bioethics 2013, 27, 89–96. [Google Scholar] [CrossRef]
- Tsujimoto, H.; Osafune, K. Current status and future directions of clinical applications using iPS cells-focus on Japan. FEBS J. 2022, 289, 7274–7291. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Wu, W. S. Gene-delivery systems for iPS cell generation. Expert. Opin. Biol. Ther. 2010, 10, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Hong, H.; Takahashi, K.; Yamanaka, S. Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nature Protocols 2010, 5, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Saintcome, C.; Acker, G. R.; Strand, F. L. Development and Regeneration of Motor Systems under the Influence of Acth Peptides. Psychoneuroendocrino. 1985, 10, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Saintcome, C.; Acker, G.; Strand, F. L. Development and Regeneration of Motor Systems under the Influence of Acth Peptides. Neuroendocrinol. Lett. 1983, 5, 136–136. [Google Scholar]
- Yu, M.; Wang, L.; Hu, Y.; Lian, Z. M.; Hua, J. L. ALK Family Inhibitor A83-01 Promotes the Proliferation of Mouse Male Germline Stem Cells (mGSCs) Under Serum- and Feeder-Free Conditions. J. Integr. Agr. 2013, 12, 1839–1846. [Google Scholar] [CrossRef]
- Kim, Y.; Jeong, J.; Choi, D. Small-molecule-mediated reprogramming: a silver lining for regenerative medicine. Exp. Mol. Med. 2020, 52, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, Y. J.; Jung, H. Protein Kinases and Their Inhibitors in Pluripotent Stem Cell Fate Regulation. Stem. Cells Int. 2019, 2019, 1569740. [Google Scholar] [CrossRef]
- Dong, Z. M.; Huo, J. R.; Liang, A.; Chen, J. Z.; Chen, G. W.; Liu, D. Z. Gamma-Secretase Inhibitor (DAPT), a potential therapeutic target drug, caused neurotoxicity in planarian regeneration by inhibiting Notch signaling pathway. Sci. Total. Environ. 2021, 781, 146735. [Google Scholar] [CrossRef]
- Minato, Y.; Nakano-Doi, A.; Maeda, S.; Nakagomi, T.; Yagi, H. A Bone Morphogenetic Protein Signaling Inhibitor, LDN193189, Converts Ischemia-Induced Multipotent Stem Cells into Neural Stem/Progenitor Cell-Like Cells. Stem Cells Dev. 2022, 31, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Suo, N.; Guo, Y. E.; He, B. Q.; Gu, H. F.; Xie, X. Inhibition of MAPK/ERK pathway promotes oligodendrocytes generation and recovery of demyelinating diseases. Glia 2019, 67, 1320–1332. [Google Scholar] [CrossRef]
- Petkov, S.; Hyttel, P.; Niemann, H. The Small Molecule Inhibitors PD0325091 and CHIR99021 Reduce Expression of Pluripotency-Related Genes in Putative Porcine Induced Pluripotent Stem Cells. Cellular Reprogramming 2014, 16, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Gerbeth, L.; Gerling, M.; Rosenthal, R.; Steiger, K.; Weidinger, C.; Keye, J.; Wu, H.; Schmidt, F.; Weicheret, W.; et al. HDAC inhibitors promote intestinal epithelial regeneration via autocrine TGFβ1 signalling in inflammation. Mucosal. Immunol. 2019, 12, 656–667. [Google Scholar] [CrossRef]
- Titan, A. L.; Davitt, M.; Foster, D. Partial Tendon Injury at the Tendon-to-Bone Enthesis Activates Skeletal Stem Cells. Stem Cell Transl. Med. 2022, 11, 889–889. [Google Scholar] [CrossRef] [PubMed]
- Fukui, L.; Henry, J. J. FGF Signaling Is Required for Lens Regeneration in Xenopus laevis. Biol. Bull. 2011, 221, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Mohseni, R.; Shoae-Hassani, A.; Verdi, J. Reprogramming of endometrial adult stromal cells in the presence of a ROCK inhibitor, thiazovivin, could obtain more efficient iPSCs. Cell. Biol. Int. 2015, 39, 515–518. [Google Scholar] [CrossRef]
- Laurencin, C. T.; Ashe, K. M.; Henry, N.; Kan, H. M.; Lo, K. W. H. Delivery of small molecules for bone regenerative engineering: preclinical studies and potential clinical applications. Drug. Discov. Today 2014, 19, 794–800. [Google Scholar] [CrossRef]
- Shah, S.; Solanki, A.; Sasmal, P. K.; Lee, K. B. Single Vehicular Delivery of siRNA and Small Molecules to Control Stem Cell Differentiation. J. Am. Chem. Soc. 2013, 135, 15682–15685. [Google Scholar] [CrossRef]
- Chen, B. Z.; Dodge, M. E.; Tang, W.; Lu, J. M.; Ma, Z. Q.; Fan, C. W.; Wei, S. G.; Hao, W. N.; Kilgore, J.; Williams, N. S.; et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 2009, 5, 100–107. [Google Scholar] [CrossRef]
- Lin, S. L.; Chang, D. C.; Chang-Lin, S.; Lin, C. H.; Wu, D. T. S.; Chen, D. T.; Ying, S. Y. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 2008, 14, 2115–2124. [Google Scholar] [CrossRef] [PubMed]
- Subramanyam, D.; Lamouille, S.; Judson, R. L.; Liu, J. Y.; Bucay, N.; Derynck, R.; Blelloch, R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nature Biotechnology 2011, 29, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Ueyama, T.; Ihara, D.; Harada, Y.; Nakagawa, S.; Saito, K.; Nakao, S.; Kawamura, T. c-Myc/microRNA-17-92 Axis Phase-Dependently Regulates PTEN and p21 Expression via ceRNA during Reprogramming to Mouse Pluripotent Stem Cells. Biomedicines 2023, 11, 1737. [Google Scholar] [CrossRef] [PubMed]
- He, X. P.; Cao, Y.; Wang, L. H.; Han, Y. L.; Zhong, X. Y.; Zhou, G. X.; Cai, Y. P.; Zhang, H. F.; Gao, P. Human Fibroblast Reprogramming to Pluripotent Stem Cells Regulated by the miR19a/b-PTEN Axis. Plos One 2014, 9, e95213. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P. N. N.; Choo, K. B.; Huang, C. J.; Sugii, S.; Cheong, S. K.; Kamarul, T. miR-524-5p of the primate-specific C19MC miRNA cluster targets TP53IPN1-and EMT-associated genes to regulate cellular reprogramming. Stem. Cell Res. Ther. 2017, 8, 214. [Google Scholar] [CrossRef] [PubMed]
- Langroudi, L.; Jamshidi-Adegani, F.; Shafiee, A.; Rad, S. M. A. H.; Keramati, F.; Azadmanesh, K.; Arefian, E.; Soleimani, M. MiR-371-373 cluster acts as a tumor-suppressor-miR and promotes cell cycle arrest in unrestricted somatic stem cells. Tumor Biol. 2015, 36, 7765–7774. [Google Scholar] [CrossRef] [PubMed]
- Lee, M. R.; Mantel, C.; Lee, S. A.; Moon, S. H.; Broxmeyer, H. E. MiR-31/SDHA Axis Regulates Reprogramming Efficiency through Mitochondrial Metabolism. Stem Cell Rep 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Bailly, A.; Milhavet, O.; Lemaitre, J. M. RNA-Based Strategies for Cell Reprogramming toward Pluripotency. Pharmaceutics 2022, 14, 317. [Google Scholar] [CrossRef] [PubMed]
- Winger, Q. A.; Hill, J. R.; Shin, T. Y.; Watson, A. J.; Kraemer, D. C.; Westhusin, M. E. Genetic reprogramming of lactate dehydrogenase, citrate synthase, and phosphofructokinase mRNA in bovine nuclear transfer embryos produced using bovine fibroblast cell nuclei. Mol. Reprod. Dev. 2000, 56, 458–464. [Google Scholar] [CrossRef]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M. Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef]
- Yakubov, E.; Rechavi, G.; Rozenblatt, S.; Givol, D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem. Bioph. Res. Co. 2010, 394, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Krug, C.; Birkholz, K.; Hombach, A.; Reuter, S.; Kershaw, M.; Kämpgen, E.; Schwenkert, M.; Fey, G.; Schule, G.; Abken, H.; et al. Reprogramming T cells with cancer specificity by non-viral transfer of mRNA encoding recombinant immunoreceptors. Hum. Gene Ther. 2009, 20, 1531–1531. [Google Scholar]
- Warren, L.; Manos, P. D.; Ahfeldt, T.; Loh, Y. H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P. K.; Smith, Z. D.; Meissner, A.; et al. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, P. M.; Weissman, S. M. MRNA Mediated T Cell Reprogramming for Adoptive Immunotherapy. J. Immunother. 2012, 35, 727–727. [Google Scholar]
- Pandit, V.; Nesbitt, R. S.; Macione, J.; Kotha, S. P. Reprogramming of cells using modified mRNA. IEEE 37th Annual Northeast Bioengineering Conference (NEBEC) 2011, 01-03 April.
- Mehta, A.; Verma, V.; Nandihalli, M.; Ramachandra, C. J. A.; Sequiera, G. L.; Sudibyo, Y.; Chung, Y. Y.; Sun, W.; Shim, W. A Systemic Evaluation of Cardiac Differentiation from mRNA Reprogrammed Human Induced Pluripotent Stem Cells. Plos One 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, A.; Ohlemacher, S. K.; Langer, K. B.; Meyer, J. S. Robust Differentiation of mRNA-Reprogrammed Human Induced Pluripotent Stem Cells Toward a Retinal Lineage. Stem Cell Transl. Med. 2016, 5, 417–426. [Google Scholar] [CrossRef]
- Lee, K.; Yu, P. Z.; Lingampalli, N.; Kim, Y. J.; Tang, R.; Murthy, N. Peptide-enhanced mRNA transfection in cultured mouse cardiac fibroblasts and direct reprogramming towards cardiomyocyte-like cells. Int. J. Nanomed. 2015, 10, 1841–1854. [Google Scholar]
- Guo, X. R.; Wang, X. L.; Li, M. C.; Yuan, Y. H.; Chen, Y.; Zou, D. D.; Bian, L. J.; Li, D. S. PDX-1 mRNA-induced reprogramming of mouse pancreas-derived mesenchymal stem cells into insulin-producing cells in vitro. Clin. Exp. Med. 2015, 15, 501–509. [Google Scholar] [CrossRef]
- Carlsten, M.; Li, L. H.; Su, S.; Berg, M.; Reger, R.; Peshwa, M. V.; Childs, R. Clinical-Grade mRNA Electroporation of NK Cells: A Novel and Highly Efficient Method to Genetically Reprogram Human NK Cells for Cancer Immunotherapy. Blood 2014, 124, 2153. [Google Scholar] [CrossRef]
- Gaignerie, A.; Lefort, N.; Rousselle, M.; Forest-Choquet, V.; Flippe, L.; Francois-Campion, V.; Girardeau, A.; Caillaud, A.; Chariau, C.; Francheteau, Q.; et al. Urine-derived cells provide a readily accessible cell type for feeder-free mRNA reprogramming. Sci. Rep. 2018, 8, 14363. [Google Scholar] [CrossRef]
- Xu, Y. T.; Huang, L.; Kirschman, J. L.; Vanover, D. A.; Tiwari, P. M.; Santangelo, P. J.; Shen, X. L.; Russell, D. G. Exploitation of Synthetic mRNA To Drive Immune Effector Cell Recruitment and Functional Reprogramming In Vivo. J. Immunol. 2019, 202, 608–617. [Google Scholar] [CrossRef]
- Irene, S.; Ioannis, G.; Ioanna, V.; Anastasios, M.; Angeliki, K.; Marianna, T.; Myrto, P.; Anny, M.; Maria, R. G.; Kalliope, S.; et al. Reprogramming of bone marrow derived mesenchymal stromal cells to human induced pluripotent stem cells from pediatric patients with hematological diseases using a commercial mRNA kit. Blood Cell. Mol. Dis. 2019, 76, 32–39. [Google Scholar]
- Grace, H. E.; Galdun, P.; Lesnefsky, E. J.; West, F. D.; Lyer, S. mRNA Reprogramming of T8993G Leigh's Syndrome Fibroblast Cells to Create Induced Pluripotent Stem Cell Models for Mitochondrial Disorders. Stem Cells Dev. 2019, 28, 846–859. [Google Scholar] [CrossRef]
- Wolfson, D. W.; Zureick, N.; Fernandez, N.; Beyersdorf, J.; Sheng, C.; Park, S. J.; Santangelo, P.; Cho, H. Synthetic TBX18 mRNA Induces Durable Reprogramming of Cardiac Myocytes to Pacemaker Cells. Circulation 2020, 142, A16575. [Google Scholar] [CrossRef]
- Harris, J. K.; Rohde, C. B.; Angel, M. Mesenchymal Stem Cells (MSCs) Generated Using mRNA Reprogramming Show Enhanced Growth Potential, Secretome, and Therapeutic Efficacy in a Demyelinating Disease Model. Molecular Therapy 2020, 28, 366–367. [Google Scholar]
- Wang, A. Y. L. Application of Modified mRNA in Somatic Reprogramming to Pluripotency and Directed Conversion of Cell Fate. Int. J. Mol. Sci. 2021, 22, 8148. [Google Scholar] [CrossRef]
- Chabanovska, O.; Galow, A. M.; David, R.; Lemcke, H. mRNA - A game changer in regenerative medicine, cell-based therapy and reprogramming strategies. Adv. Drug Deliver. Rev. 2021, 179, 114002. [Google Scholar] [CrossRef]
- Warren, L.; Lin, C. mRNA-Based Genetic Reprogramming. Molecular Therapy 2019, 27, 729–734. [Google Scholar] [CrossRef]
- Uchida, S.; Kataoka, K.; Itaka, K. Screening of mRNA Chemical Modification to Maximize Protein Expression with Reduced Immunogenicity. Pharmaceutics 2015, 7, 137–151. [Google Scholar] [CrossRef]
- Connor, B.; Firmin, E.; McCaughey-Chapman, A.; Monk, R.; Lee, K.; Liot, S.; Geiger, J.; Rudolph, C.; Jones, K. Conversion of adult human fibroblasts into neural precursor cells using chemically modified mRNA. Heliyon 2018, 4, e00918. [Google Scholar] [CrossRef]
- Elkhalifa, D.; Rayan, M.; Negmeldin, A. T.; Elhissi, A.; Khalil, A. Chemically modified mRNA beyond COVID-19: Potential preventive and therapeutic applications for targeting chronic diseases. Biomed. Pharmacother. 2022, 145, 112385. [Google Scholar] [CrossRef] [PubMed]
- McGrath, P. S.; McGarvey, S. S.; Kogut, I.; Bilousova, G. Efficient RNA-Based Reprogramming of Disease-Associated Primary Human Fibroblasts into Induced Pluripotent Stem Cells. Methods Mol. Biol. 2020, 2117, 271–284. [Google Scholar] [PubMed]
- Preskey, D.; Allison, T. F.; Jones, M.; Mamchaoui, K.; Unger, C. Synthetically modified mRNA for efficient and fast human iPS cell generation and direct transdifferentiation to myoblasts. Biochem. Biophys. Res. Commun. 2016, 473, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P. K.; Rossi, D. J. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat. Protoc. 2013, 8, 568–582. [Google Scholar] [CrossRef]
- Warren, L.; Ni, Y.; Wang, J.; Guo, X. Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Sci. Rep. 2012, 2, 657. [Google Scholar] [CrossRef]
- Yakubov, E.; Rechavi, G.; Rozenblatt, S.; Givol, D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem. Biophys. Res. Commun. 2010, 394, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Warren, L.; Manos, P. D.; Ahfeldt, T.; Loh, Y. H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P. K.; Smith, Z. D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef]
- Heng, B. C.; Heinimann, K.; Miny, P.; Iezzi, G.; Glatz, K.; Scherberich, A.; Zulewski, H.; Fussenegger, M. mRNA transfection-based, feeder-free, induced pluripotent stem cells derived from adipose tissue of a 50-year-old patient. Metab. Eng. 2013, 18, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Varela, I.; Karagiannidou, A.; Oikonomakis, V.; Tzetis, M.; Tzanoudaki, M.; Siapati, E. K.; Vassilopoulos, G.; Graphakos, S.; Kanavakis, E.; Goussetis, E. Generation of human beta-thalassemia induced pluripotent cell lines by reprogramming of bone marrow-derived mesenchymal stromal cells using modified mRNA. Cell Reprogram. 2014, 16, 447–455. [Google Scholar] [CrossRef]
- Velasquez-Mao, A. J.; Tsao, C. J. M.; Monroe, M. N.; Legras, X.; Bissig-Choisat, B.; Bissig, K. D.; Ruano, R.; Jacot, J. G. Differentiation of spontaneously contracting cardiomyocytes from non-virally reprogrammed human amniotic fluid stem cells. PLoS One 2017, 12, e0177824. [Google Scholar] [CrossRef]
- Liu, C. Y.; Zhang, L. G.; Zhu, W. H.; Guo, R. Q.; Sun, H. M.; Chen, X.; Deng, N. Barriers and Strategies of Cationic Liposomes for Cancer Gene Therapy. Mol. Ther.-Meth. Clin. D. 2020, 18, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Polajzer, T.; Miklavcic, D. Immunogenic Cell Death in Electroporation-Based Therapies Depends on Pulse Waveform Characteristics. Vaccines-Basel 2023, 11, 1036. [Google Scholar] [CrossRef] [PubMed]
- Supakul, S.; Leventoux, N.; Tabuchi, H.; Mimura, M.; Ito, D.; Maeda, S.; Okano, H. Establishment of KEIOi005-A iPSC line from urine-derived cells (UDCs) of a mild Alzheimer's disease (AD) donor with multiple risk SNPs for sporadic Alzheimer's disease (sAD). Stem Cell Res. 2022, 62, 102802. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Benda, C.; Duzinger, S.; Huang, Y. H.; Li, X. Y.; Li, Y. H.; Guo, X. P.; Cao, G. K.; Chen, S.; Hao, L. L.; et al. Generation of Induced Pluripotent Stem Cells from Urine. J. Am. Soc. Nephrol. 2011, 22, 1221–1228. [Google Scholar] [CrossRef] [PubMed]
- Riebeling, C.; Schlechter, K.; Buesen, R.; Spielmann, H.; Luch, A.; Seiler, A. Defined culture medium for stem cell differentiation: Applicability of serum-free conditions in the mouse embryonic stem cell test. Toxicol. in Vitro 2011, 25, 914–921. [Google Scholar] [CrossRef] [PubMed]
- Liang, S. H.; Zhao, J.; Baker, R. K.; Tran, E.; Zhan, L. S.; Kieffer, T. J. Differentiation of stem cell-derived pancreatic progenitors into insulin-secreting islet clusters in a multiwell-based static 3D culture system. Cell. Rep. Methods 2023, 3, 100466. [Google Scholar] [CrossRef] [PubMed]
- Sart, S.; Tsai, A. C.; Li, Y.; Ma, T. Three-Dimensional Aggregates of Mesenchymal Stem Cells: Cellular Mechanisms, Biological Properties, and Applications. Tissue Eng. Part B-Rev. 2014, 20, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M. H.; Kuraishy, A. I.; Deshpande, C.; Hong, J. S.; Cacalano, N. A.; Gatti, R. A.; Manis, J. P.; Damore, M. A.; Pellegrini, M.; Teitell, M. A. AID-Induced Genotoxic Stress Promotes B Cell Differentiation in the Germinal Center via ATM and LKB1 Signaling. Mol. Cell 2010, 39, 873–885. [Google Scholar] [CrossRef]
- Meier-Stiegen, F.; Schwanbeck, R.; Bernoth, K.; Martini, S.; Hieronymus, T.; Ruau, D.; Zenke, M.; Just, U. Activated Notch1 Target Genes during Embryonic Cell Differentiation Depend on the Cellular Context and Include Lineage Determinants and Inhibitors. Plos One 2010, 5, e1148. [Google Scholar] [CrossRef]
- Bories, J. C.; Cayuela, J. M.; Loiseau, P.; Sigaux, F. Expression of Human Recombination Activating Genes (Rag1 and Rag2) in Neoplastic Lymphoid-Cells - Correlation with Cell-Differentiation and Antigen Receptor Expression. Blood 1991, 78, 2053–2061. [Google Scholar] [CrossRef]
- Kim, H. J.; Park, J. M.; Lee, S.; Cho, H. B.; Park, J. I.; Kim, J. H.; Park, J. S.; Park, K. H. Efficient CRISPR-Cas9-based knockdown of RUNX2 to induce chondrogenic differentiation of stem cells. Biomater. Sci. 2022, 10, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Rubio, A.; Luoni, M.; Giannelli, S. G.; Radice, I.; Iannielli, A.; Cancellieri, C.; Di Berardino, C.; Regalia, G.; Lazzari, G.; Menegon, A.; et al. Rapid and efficient CRISPR/Cas9 gene inactivation in human neurons during human pluripotent stem cell differentiation and direct reprogramming. Sci. Rep. 2016, 6, 37540. [Google Scholar] [CrossRef] [PubMed]
- Goparaju, S. K.; Kohda, K.; Ibata, K.; Soma, A.; Nakatake, Y.; Akiyama, T.; Wakabayashi, S.; Matsushita, M.; Sakota, M.; Kimura, H.; et al. Rapid differentiation of human pluripotent stem cells into functional neurons by mRNAs encoding transcription factors. Sci. Rep. 2017, 7, 42367. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, T.; Sato, S.; Ko, S. B. H.; Sano, O.; Sato, S.; Saito, M.; Nagai, H.; Ko, M. S. H.; Iwata, H. Synthetic mRNA-based differentiation method enables early detection of Parkinson's phenotypes in neurons derived from Gaucher disease-induced pluripotent stem cells. Stem Cell Transl. Med. 2021, 10, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Jmoudiak, M.; Futerman, A. H. Gaucher disease: pathological mechanisms and modern management. Brit. J. Haematol. 2005, 129, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Kurolap, M.; del Toro, M.; Spiegel, R.; Gutstein, A.; Shafir, G.; Cohen, I. J.; Barrabés, J. A.; Feldman, H. B. Gaucher disease type 3c: New patients with unique presentations and review of the literature. Mol. Genet. Metab. 2019, 127, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Spitz, M.; Rozenberg, R.; Silveira, P. A. A.; Barbosa, E. R. Parkinsonism in type 1 Gaucher's disease. J. Neurol. Neurosur. Ps 2006, 77, 709–710. [Google Scholar] [CrossRef] [PubMed]
- Musashi Kubiura-Ichimaru, C. P. , Kazuaki Kojima, Constance Dollet, Haruka Yabukami, Katsunori Semi, Yasuhiro Takashima, Thorsten Boroviak, Hideya Kawaji, Knut Woltjen, Aki Minoda, Erika Sasaki, Toshiaki Watanabe. mRNA-based generation of marmoset PGCLCs capable of differentiation into gonocyte-like cells. Stem Cell Rep. 2023, 18, 1987–2002. [Google Scholar]
- Cho, S.; Aakash, P.; Lee, S.; Yoon, Y. S. Endothelial cell direct reprogramming: Past, present, and future. J. Mol. Cell Cardiol. 2023, 180, 22–32. [Google Scholar] [CrossRef]
- Davis, R. L.; Weintraub, H.; Lassar, A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Ieda, M.; Fu, J. D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B. G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef]
- Muraoka, N.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Isomi, M.; Nakashima, H.; Akiyama, M.; Wada, R.; Inagawa, K.; et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014, 33, 1565–1581. [Google Scholar] [CrossRef]
- Zhao, Y.; Londono, P.; Cao, Y.; Sharpe, E. J.; Proenza, C.; O'Rourke, R.; Jones, K. L.; Jeong, M. Y.; Walker, L. A.; Buttrick, P. M.; et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015, 6, 8243. [Google Scholar] [CrossRef]
- Muraoka, N.; Nara, K.; Tamura, F.; Kojima, H.; Yamakawa, H.; Sadahiro, T.; Miyamoto, K.; Isomi, M.; Haginiwa, S.; Tani, H.; et al. Role of cyclooxygenase-2-mediated prostaglandin E2-prostaglandin E receptor 4 signaling in cardiac reprogramming. Nat. Commun. 2019, 10, 674. [Google Scholar] [CrossRef]
- Cao, N.; Huang, Y.; Zheng, J.; Spencer, C. I.; Zhang, Y.; Fu, J. D.; Nie, B.; Xie, M.; Zhang, M.; Wang, H.; et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 2016, 352, 1216–1220. [Google Scholar] [CrossRef]
- Simeonov, K. P.; Uppal, H. Direct reprogramming of human fibroblasts to hepatocyte-like cells by synthetic modified mRNAs. PLoS One 2014, 9, e100134. [Google Scholar] [CrossRef]
- Van Pham, P.; Vu, N. B.; Dao, T. T.; Le, H. T.; Phi, L. T.; Phan, N. K. Production of endothelial progenitor cells from skin fibroblasts by direct reprogramming for clinical usages. In Vitro Cell Dev. Biol. Anim. 2017, 53, 207–216. [Google Scholar] [CrossRef]
- Kaur, K.; Hadas, Y.; Kurian, A. A.; Zak, M. M.; Yoo, J.; Mahmood, A.; Girard, H.; Komargodski, R.; Io, T.; Santini, M. P.; et al. Direct reprogramming induces vascular regeneration post muscle ischemic injury. Mol. Ther. 2021, 29, 3042–3058. [Google Scholar] [CrossRef]
- Qabrati, X.; Kim, I.; Ghosh, A.; Bundschuh, N.; Noe, F.; Palmer, A. S.; Bar-Nur, O. Transgene-free direct conversion of murine fibroblasts into functional muscle stem cells. NPJ Regen. Med. 2023, 8, 43. [Google Scholar] [CrossRef]
- Lee, K.; Yu, P.; Lingampalli, N.; Kim, H. J.; Tang, R.; Murthy, N. Peptide-enhanced mRNA transfection in cultured mouse cardiac fibroblasts and direct reprogramming towards cardiomyocyte-like cells. Int. J. Nanomedicine 2015, 10, 1841–1854. [Google Scholar]
- Pruszak, J.; Sonntag, K. C.; Aung, M. H.; Sanchez-Pernaute, R.; Isacson, O. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell Populations. Stem Cells 2007, 25, 2257–2268. [Google Scholar] [CrossRef] [PubMed]
- Riordon, D. R.; Boheler, K. R. Immunophenotyping of Live Human Pluripotent Stem Cells by Flow Cytometry. Surfaceome: Methods and Protocols 2018, 1722, 127–149. [Google Scholar]
- Fujita, Y.; Hirosawa, M.; Hayashi, K.; Hatani, T.; Yoshida, Y.; Yamamoto, T.; Saito, H. A versatile and robust cell purification system with an RNA-only circuit composed of microRNA-responsive ON and OFF switches. Sci. Adv. 2022, 8, eabj1793. [Google Scholar] [CrossRef] [PubMed]
- Okuzaki, A.; Tsuda, M.; Konagaya, K.; Tabei, Y. A novel strategy for promoting homoplasmic plastid transformant production using the barnase-barstar system. Plant Biotechnol.-Nar. 2020, 37, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xu, Q.; Li, X. W.; Zhang, X. L. MicroRNA-21 regulates the proliferation and apoptosis of cervical cancer cells via tumor necrosis factor-α. Mol. Med. Rep. 2017, 16, 4659–4663. [Google Scholar] [CrossRef]
- Abe, N.; Imaeda, A.; Inagaki, M.; Li, Z. M.; Kawaguchi, D.; Onda, K.; Nakashima, Y.; Uchida, S.; Hashiya, F.; Kimura, Y.; Abe, H. Complete Chemical Synthesis of Minimal Messenger RNA by Efficient Chemical Capping Reaction. Acs. Chem. Biol. 2022, 17, 1308–1314. [Google Scholar] [CrossRef]


| Reprogramming Methods |
Advantages | Disadvantages |
|---|---|---|
| Retroviral Vectors | ● Well-investigated and established | ● Undesired transgene into nuclear genomic DNA |
| ● High cellular introduction efficiency | ● Carcinogenesis and a risk of tumor formation | |
| Plasmids Vectors | ● Low risk of genome insertion | ● Insufficient cellular introduction and reprogramming efficiency |
| Small Molecules | ● Simple handling | ● Required relatively high-dosage |
| ● Low cost | ● Necessary to care for dose-dependent cytotoxicity | |
| ● High cellular introduction efficiency | ● Difficult to cover all applications | |
| microRNA | ● Fast reprogramming | ● Low physiological stability |
| ● No risk of genome insertion | ● Fewer examples relative to other methods | |
| mRNA | ● Fast reprogramming | ● Required more effective intracellular delivery methods |
| ● No risk of genome insertion and tumor development | ● Required multiple injections (every day) | |
| ● High reprogramming efficiency |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




