1. Introduction
Rotaviruses (RVs) are the most common cause of viral gastroenteritis in infants and animals. RVs are primarily transmitted through the fecal–oral route, and infected individuals typically present with symptoms such as diarrhea, vomiting, and decreased appetite. The infection can spread between humans and animals, posing a serious threat to both human health and animal husbandry [
1,
2,
3,
4]. In 1969, bovine RV was first isolated and cultured in cells and was confirmed to be the primary cause of diarrhea in cattle [
5]. The first human RV was discovered in 1973 [
6]. RVs were subsequently identified in pigs and poultry [
7,
8].
Cultivating RVs in cell cultures was quite challenging until 1984. Subsequently, RVs were successfully cultured in African Green Monkey kidney cells (MA104) after treatment with trypsin, which accelerated the processes of RV isolation and cultivation [
9]. Furthermore, subsequent studies have reported that RVs could replicate in cells such as Vero cells [
10,
11].
Porcine rotaviruses (PoRVs) can damage the cells of the small intestines in pigs, leading to villous atrophy and reduced digestive absorption capacity [
12]. Additionally, RV infection disrupts the balance of Ga
2+, alters cell permeability to Ca
2+, disturbs Ca
2+ homeostasis, and subsequently causes diarrhea [
13].
The genus rotavirus belongs to the family
Reoviridae and order Reovirales. Their genome consists of 11 double-stranded RNA segments, encoding 11 proteins, including 6 structural proteins (virion protein [VP]1–VP4, VP6, and VP7) and 5 nonstructural proteins (NSP1–NSP5/6) [
12]. RVs are classified into 10 subgroups (A–J) based on the VP6 antigen [
14]. The outer shell proteins VP7 and VP4 induce neutralizing antibodies and form the basis for the G and P dual classification system [
15]. The most common groups infecting humans and animals are Group A RV (RVA), Group B RV (RVB), and Group C RV (RVC), with RVA having the highest prevalence and causing the most significant harm [
12]. For RVA strains with high genetic diversity, the dual (G/P) typing system was expanded in 2008 to a complete genome sequence-based classification system. This system established the nucleotide homology cutoffs for all 11 gene segments, labeling the genes as Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Ex-Hx. Subsequently, the Rotavirus Classification Working Group (RCWG) was established to develop classification guidelines and maintain the proposed classification system of RVs [
16]. To date, 42 “G” and 58 “P” genotypes have been reported in the RCWG database (
https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg). RVA strains with G3, G5, G9, and G11 genotypes in combination with P[5], P[6], P[13], P[23], P[27], and P[28] genotypes are considered the most common worldwide [
17].
In this study, a G5P[23] genotype PoRV strain was isolated from the diarrheal samples of piglets in a farm in Jiangsu Province, China, and named RVA/Pig/China/JS/2023/G5P[23](JS). The whole genome of the JS strain was sequenced, and its genetic evolution was determined. The JS strain was also used to infect 15-day-old piglets to investigate its pathogenicity.
2. Materials and Methods
2.1. Clinical Samples, Cells, and Antibodies
On January 6, 2023, 28 diarrheal samples were collected from piglets in a pig farm infected by PoRV in Jiangsu Province, China. The virus isolated from these samples was identified as PoRV through RT–PCR and qRT–PCR using
VP6 (
Supplementary Table S1) [
18,
19,
20]. Samples of the intestinal contents were homogenized in serum-free Dulbecco’s modified Eagle medium [DMEM] (Invitrogen, CA, USA) containing 1% penicillin–streptomycin [10000 units/mL penicillin and 10000 μg/mL streptomycin] (Gibco™, MT, USA) and 0.3% trypsin phosphate broth (Sigma-Aldrich, Germany). The samples were then centrifuged for 30 min at 3500 rpm and 4°C. The supernatant was filtered through a 0.22-μm pore filter (Merck Millipore, Germany) to remove the bacteria and was stored at −80°C until use as an inoculum for virus isolation. MA104 cells - Supplier: American Type Culture Collection (ATCC) - Catalog Number: CRL-2378.MA104 cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum [FBS] (Invitrogen, Australia) and 1% antibiotics [10000 units/mL penicillin, 10000 μg/mL streptomycin, and 25 μg/mL Fungizone
®] (Gibco™, MT, USA). These cells were maintained at 37°C in a humidified incubator under 5% CO
2. Mouse monoclonal antibodies (McAbs) against PoRV VP6 protein (Zoonogen
®, Beijing, China) were purchased and stored in the laboratory.
2.2. Virus Isolation Assay
PoRV isolation was performed using MA104 cells via a previously described method with modifications [
21]. Specifically, prior to inoculation, cells were rinsed thrice with sterile phosphate-buffered saline (PBS; pH 7.2) to remove FBS completely. Simultaneously, the stored inoculum was briefly vortexed and used for cultivation. In total, 300 μL of the inoculum was added to 6-well plates. After incubation at 37°C for 1 h, the inoculum was removed, and the maintenance medium containing trypsin at a final concentration of 4 μg/mL was added. The inoculated cells were maintained at 37°C in a humidified incubator under 5% CO
2, and the CPEs were monitored daily. When CPEs were observed in 90% of the cells, the flask was subjected to three rounds of freezing and thawing. The cells and the supernatant were mixed with a pipette, aliquoted, and stored at −80°C. The harvested cell culture was used as the seed stock for the next generation. For successive passages, the scale of the culture was gradually increased until the PoRV strains were properly propagated and continuously passaged using T-25 flasks.
2.3. Titration and Growth Curve Determination
The growth curve of the 10
th generation of the virus was constructed. Initially, the virus was inoculated onto a 96-well plate at a multiplicity of infection (MOI) of 0.01. Cell culture supernatants were collected at 6, 12, 18, 24, 30, 36, 42, and 48 hpi. Subsequently, based on the Reed–Muench method [
22], the viral titers of these samples were determined using the TCID50 assay. After washing thrice with PBS, 100 μL of the experimental supernatant was mixed with 900 μL of DMEM to obtain serial dilutions ranging from 10
−1 to 10
−10. Each dilution was added to a monolayer of MA104 cells in 10 vertical wells of a 96-well plate. The plate was then placed in a cell culture incubator at 37°C for 5 days, and the viral titers were determined by observing the CPEs under a microscope. Finally, the growth curves for each virus were constructed based on the viral titers at different time points postinfection.
2.4. Immunofluorescence Assay
MA104 cells cultured in 6-well plates were either mock-infected or infected with PoRV at a MOI of 0.01. At 0, 12, and 24 hpi, the cells were fixed with 4% paraformaldehyde at 4°C for 30 min, followed by permeabilization with 0.25% Triton X-100 (Solarbio, Beijing, China) for 10 min at room temperature (RT). Blocking was performed using 5% bovine serum albumin (Solarbio, China) in PBS at RT for 1 h. McAbs against the PoRV VP6 protein and Alexa Fluor® 488-conjugated goat anti-mouse IgG (Abcam, UK) were used as primary and secondary antibodies, respectively. Nuclei were stained with DAPI (VectorLabs, CA, USA) for 20 min at RT. After washing with PBS, the stained cells were observed under a fluorescence microscope (Olympus, Japan).
2.5. Transmission Electron Microscopy
To visualize PoRV particles, MA104 cells were harvested when CPEs were observed in 90% of the cells. The cell culture was subjected to three freeze–thaw cycles, followed by centrifugation for 30 min at 10,000 rpm and 4°C. The supernatant was filtered through a 0.22-mm filter to remove cell debris and was then mixed overnight with polyethylene glycol 8000 [PEG-8000] (Solarbio, China) at a final concentration of 10%. The mixture was subsequently ultracentrifuged for 2 h at 12,000 rpm and 4°C to obtain the PoRV particles. These particles were then resuspended in Tris-buffered saline and negatively stained with 2% phosphotungstic acid. The viruses within the infected MA104 cells were examined using a transmission electron microscope (TEM)(Leica, Germany), and further fixation and imaging were performed based on the methods described in previous studies.
2.6. Sequence Analysis
The complete genome of the RVA strain JS was sequenced using the MiSeq high-throughput sequencing platform (Illumina, CA, USA). The total RNA from the cell culture supernatant was extracted using TRIzol reagent (Invitrogen, CA, USA) following the manufacturer’s instructions. rRNA was removed using RiBo-Zero Magnetic Gold kit (Epicenter Biotechnologies, WI, USA), and the remaining RNA was sequenced using NEBNext® Ultra™ Directional RNA Library Prep Kit (New England Biolabs, MA, USA) and the Illumina MiSeq platform (GENEWIZ, Guangzhou, China).
The obtained raw reads were trimmed using Trimmomatic v0.39 software [
16] and then aligned with the Sscrofa 11.1 reference genome using Bowtie2 v2.4.1 software [
17] to eliminate the reads corresponding to the sequences of the pig genome. The remaining reads were reassembled into contigs using MEGAHIT v1.2.9 software [
18]. Viral sequences were identified using BLASTn and BLASTx searches against a custom-built virus nucleotide reference database from GenBank (Taxonomy ID 10239) or UniProt virus classification database.
2.7. Recombination Analysis
The sequences involved in recombination were analyzed using RDP4 software [
23]. DP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 1, vev003 (
https://doi.org/10.1093/ve/vev003). The RDP, GENECONV, Chimaera, MaxChi, Bootscan, SiScan, and 3Seq methods were employed with their default parameters.
2.8. Animal Experiments
Sixteen 15-day-old conventional piglets were purchased from a commercial pig farm with no history of PoRV outbreak or vaccination. Additionally, these piglets tested negative for various viruses, including porcine epidemic diarrhea virus, transmissible gastroenteritis virus, porcine respiratory and reproductive syndrome virus, porcine circovirus, classical swine fever virus, and pseudorabies virus.
All animal experiments were approved by the Ethics Committee of the South China Agricultural University, Guangzhou, China (approval ID: SYXK-2019-0136). The 18 piglets were randomly divided into 2 groups, with 8 piglets in each group: PoRV-infection and mock-infection groups. The piglets of the PoRV-infection group were orally inoculated with 1 mL of 1.0 × 106 TCID50 PoRV, whereas those of the mock-infection group were fed with an equivalent amount of DMEM. The animals were manually fed with milk replacer every 4 h during the experiment. In piglets showing signs of anorexia, 60–100 mL of milk replacer was administered by gavage every 4 h. Clinical symptoms such as diarrhea, vomiting, anorexia, and depression were recorded daily. Fecal consistency was assessed daily using a scoring system based on solid (0 points), paste-like (1 point), semiliquid (2 points, mild diarrhea), and liquid (3 points, severe diarrhea) standards.
Histopathological and immunohistochemical examinations were performed on the small intestines. Intestinal tissue samples from a randomly selected piglet were examined and collected on day 9 postinfection. After fixation in 4% paraformaldehyde at RT for 48 h, the processed tissue samples were embedded in paraffin, sectioned using a microtome (Leica, Germany), deparaffinized with xylene, and washed with reducing concentrations of ethanol. Subsequently, conventional Hematoxylin and Eosin (BaSo, Zhuhai, China) [H&E] staining was performed for HE, and PoRV-VP6-specific monoclonal antibody was used IHC [
24].
Rectal swabs were collected daily for the qRT–PCR-based determination of viral copy numbers. The animals were euthanized on day 9 postinfection, and the segments of the lungs, duodenum, jejunum, ileum, etc., were collected for HE, IHC, and qRT–PCR.
2.9. Statistical Analysis
All statistical analyses were performed using the GraphPad Prism software version 8.0 (graphpad.com). The statistical significance of the differences between the experimental groups was ascertained using the Student’s t-test. The differences were considered statistically significant at P < 0.05.
3. Results
3.1. Virus Isolation
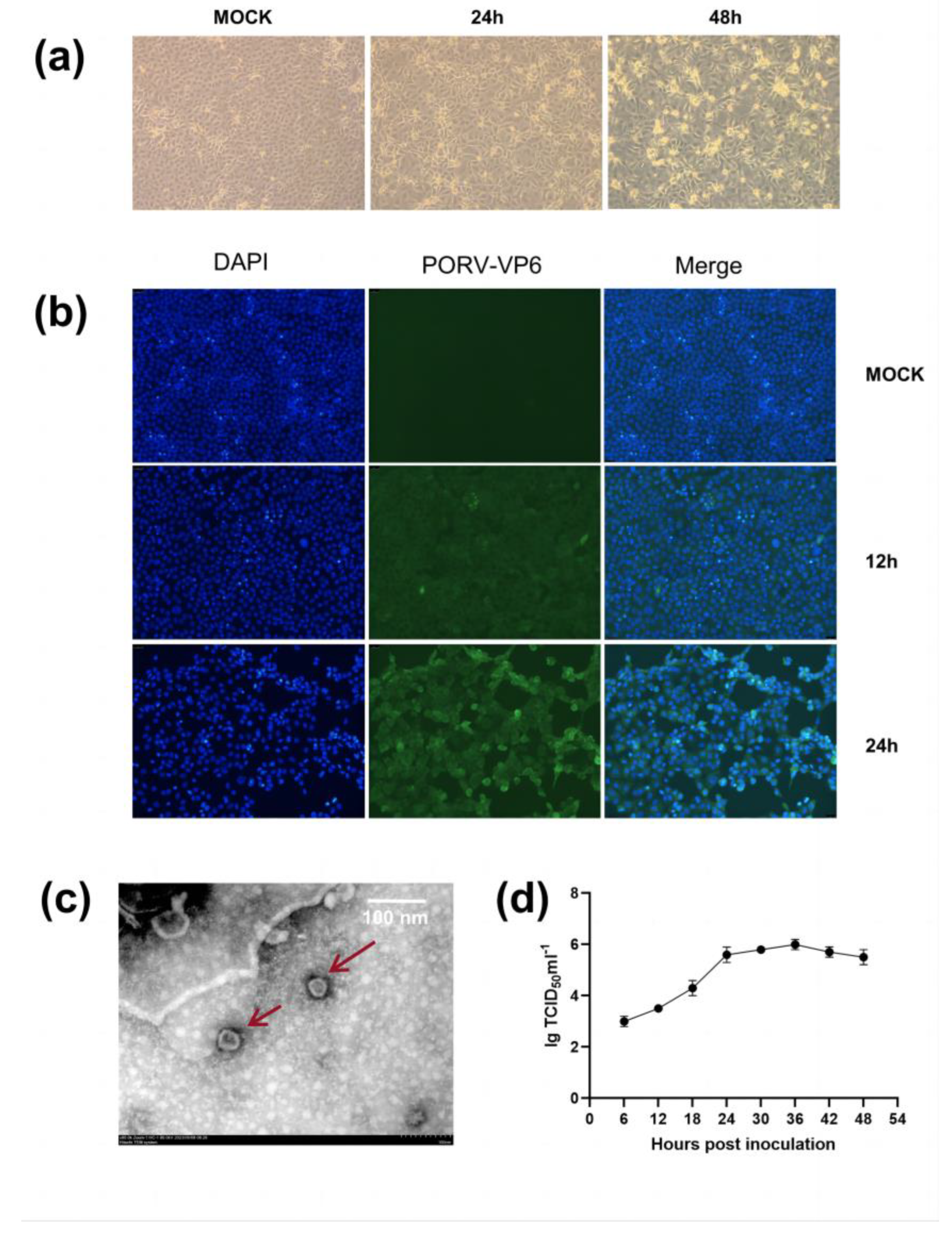
The PoRV-positive samples at a 10-fold dilution were inoculated into MA104 cells. Visible cytopathic effects (CPEs) were observed in the second generation, 24 hours postinfection [hpi]. Complete CPE was observed at 48 hpi (
Figure 1a). Compared with noninoculated cells, the inoculated cells were initially characterized by syncytium and bubble formation, followed by elongation, detachment, and loss of motility in later stages (
Figure 1a).
Furthermore, the presence of PoRV antigen was confirmed via immunofluorescence assay (IFA) using PoRV-VP6-specific monoclonal antibodies and 4′,6-diamidino-2-phenylindole (DAPI) staining. Replication of the JS strain was confirmed in the nuclei of cells in the 10
th generation. From 12 h onward, pronounced green signals were observed in the infected MA104 cells but not in the uninfected cells. These signals tended to markedly intensify with time (0, 12, and 24 hpi) (
Figure 1b).
We conducted transmission electron microscopy (TEM) of PoRV particles obtained from the MA104 cell culture medium and MA104 cells infected with the JS strain. The typical RV particles were observed in the cell culture medium (
Figure 1c). The viral particles appeared wheel-shaped, with a diameter of 80–100 nm, exhibiting characteristic surface projections unique to RVs. TEM confirmed the successful reproduction of PoRV in MA104 cells. The growth curves of the JS strain at different time points postinfection were plotted based on the 50% tissue culture infective dose (TCID50). The results indicated that the viral replication titer peaked at ~1 × 10
6/mL TCID50, and the titer enhanced rapidly from 6 to 36 hpi (
Figure 1d).
3.2. Sequence Analysis
Based on the latest classification and nomenclature system established by the RCWG and the sequences of the 11 gene segments, the genotype of this RV was identified as G5-[P23]-I5-R1-C1-M1-A8-N1-T1-E1-H1 and named RVA/Pig/China/JS/2023/G5P[23]. The sequences of all 11 gene segments were uploaded to GenBank with accession numbers OR644644 to OR644654.
Homology and phylogenetic analyses were conducted between the closely related reference RVA strains of the same genotype. Several gene segments were highly homologous with RV strains of porcine origin, including
VP1,
VP2,
VP4,
VP6,
NSP1,
NSP3, and
NSP5. Additionally, some segments such as
VP3,
VP7,
NSP2, and
NSP4 exhibited an even higher homology with RV strains of human origin, suggesting that the JS-G5P[23] strain was likely a recombinant human–porcine virus (
Supplementary Table S2).
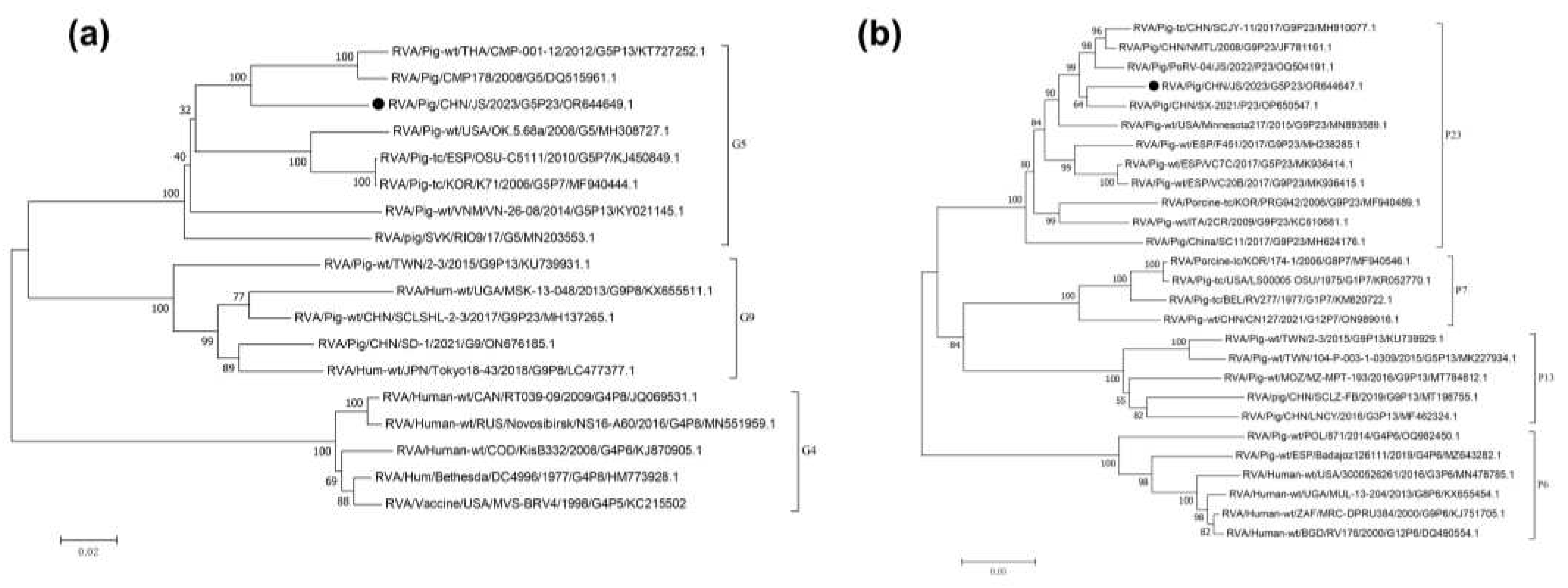
Sequence analysis of
VP7 revealed that it comprised 981 nucleotides (nts), encoding 326 amino acids (aa). A phylogenetic tree was constructed using the full-length
VP7 sequence (981 nts) and selected G genotype sequences obtained from GenBank. The
VP7 of JS strain is grouped within the G5 genotype (glycolated), primarily consisting of strains of porcine origin. However, the segment from the JS strain formed a smaller clade within this genotype, containing sequences derived from both pig and human sources. The most closely related sequence to the JS strain fragment was identified to be derived from a human RVA strain, RVA/Pig-wt/THA/CMP-001-12/2012/G5P[13] (KT727252.1), which was isolated from a patient in Thailand in 2014 (
Figure 2a) [
25]. Furthermore, using ViPR automatic genotyping tool, we revealed that RVA/Pig/China/JS/2023/G5P[23] was closely related to multiple RVA G5 strains [
26]; this sequence belongs to the G5 genotype (glycolated), with the most similar query sequence being RVA/Human-tc/BRA/IAL28/1992/G5P[8] (EF672588) (
Supplementary Table S2) [
27].
Sequence analysis of
VP4 revealed that it comprised 2331 nts, encoding 776 aa. A phylogenetic tree was constructed using the full-length
VP4 sequence from the JS strain and selected P genotype sequences obtained from GenBank. JS was most closely related to a porcine-origin RVA strain, RVA/Pig/CHN/SX-2021/P23 (OP650547.1), isolated from China (
Figure 2b). Additionally, the ViPR automatic genotyping tool revealed that RVA/Pig/China/JS/2023/G5P[23] was closely related to RVA P[23] strains [
21]; this sequence belongs to the P[23] genotype, with the highest similarity to the PoRV strain P23-RVA/Pig-tc/VEN/A34/1985/G5P[23] (AY174094) (
Supplementary Table S2) [
26,
28].
3.3. Recombination Analysis
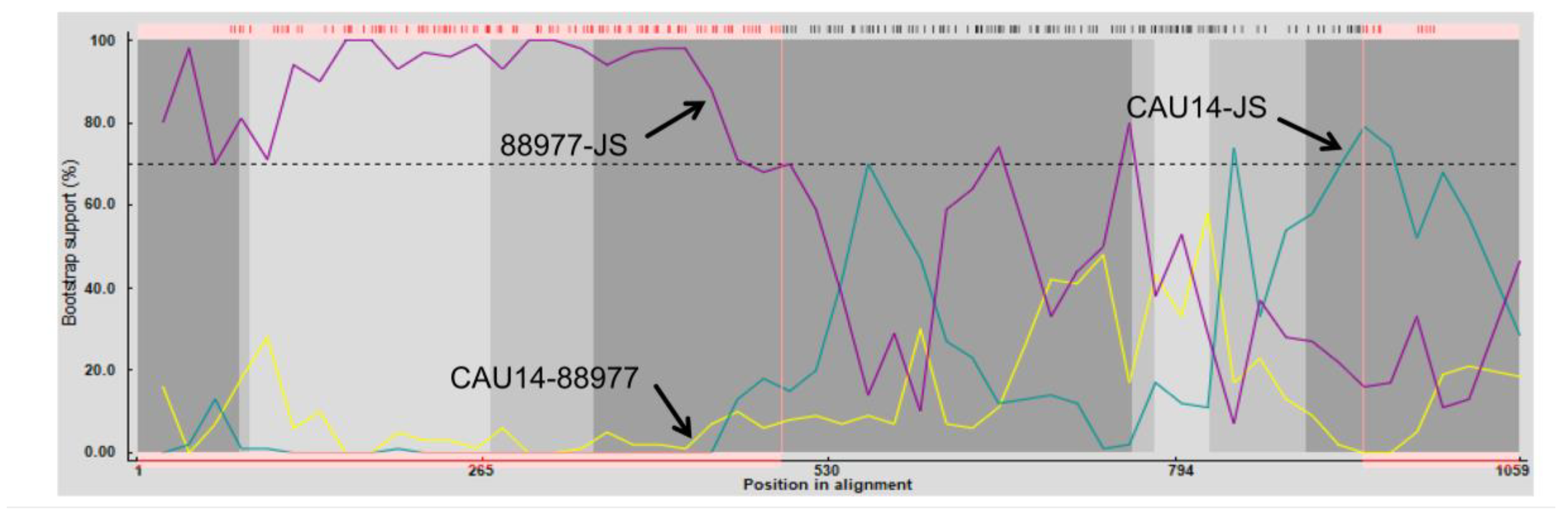
To further analyze the association between
NSP2 from JS and the existing isolates, a genetic analysis was conducted between them using RDP4 software [
29]. The sequence of
NSP2 from JS is indicated in
Figure 3. Crossover points for a potential recombination zone were located at nucleotides 1–494 and 939–994 of
NSP2 (
Figure 3), but no recombination was detected in other genes encoding viral proteins. The major parental strain for the recombination was RVA/Human-tc/KOR/CAU14-1-262/2014/G3P[9] (KR262156.1), and the minor strain was RVA/Dog-wt/GER/88977/2013/G8P1/(KJ940158.1), indicating that C1 of JS was the gene recombination product of C3 and C2 RVAs [
30,
31].
3.4. Clinical Signs and Histological Changes
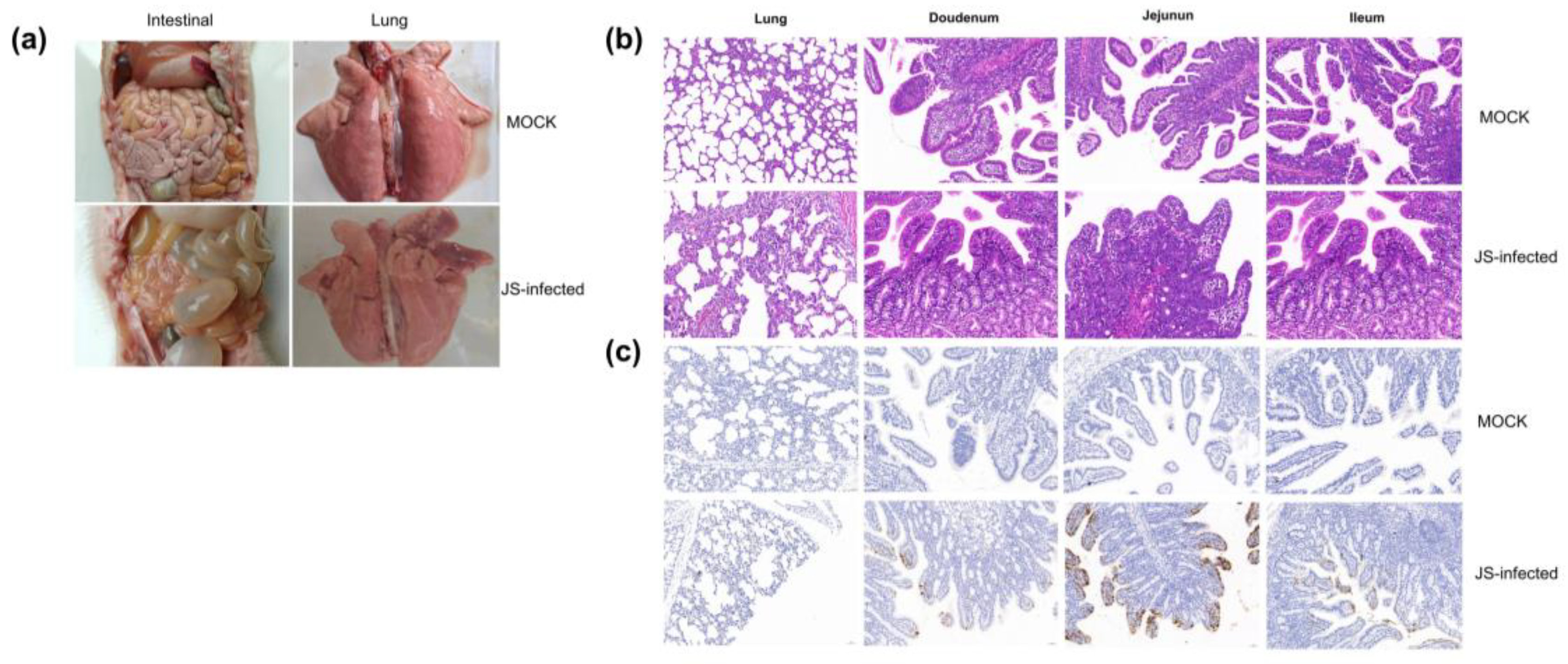
The piglets of the JS strain infection group showed diarrhea symptoms, manifested as dark green, watery feces (
Figure 4a). One piglet succumbed on day 3 postinfection, and two piglets on day 6. On autopsy, the lungs showed congestion and swelling, the intestinal wall became thin and transparent, and the intestines bulged and were filled with yellow water-like liquid (
Figure 4a). On the contrary, in the simulated infection group, except for a few piglets whose feces briefly appeared pasty, none experienced diarrhea, and none succumbed at the end of the experimental cycle. No pathological changes were found in the lungs and intestinal tissues of these piglets during autopsy (
Figure 4a).
HE of the tissues showed that all piglets with diarrhea exhibited viral enteritis, with gastric and intestinal distension; vacuolization of small intestinal cells; and shortened, fused, and sloughed villi in the small intestine (
Figure 4b). The JS strain caused a slight thickening of the alveolar wall and pulmonary inflammation. A few mucosal epithelial cells underwent pyknosis and necrosis. The number of goblet cells in the mucosal layer of the tissue was conspicuously reduced (
Figure 4b). The intestinal tissue pathology of piglets in the control group was normal (
Figure 4b). In addition, IHC examination revealed that in specific segments of the small intestine with villous atrophy, cytoplasmic staining of the PoRV antigen was dominant, with the highest antigen content in the jejunum (
Figure 4c). No PoRV antigen was detected in the small intestines of piglets in the negative control group (
Figure 4c).
3.5. Viral Load in Stool Samples and Tissues
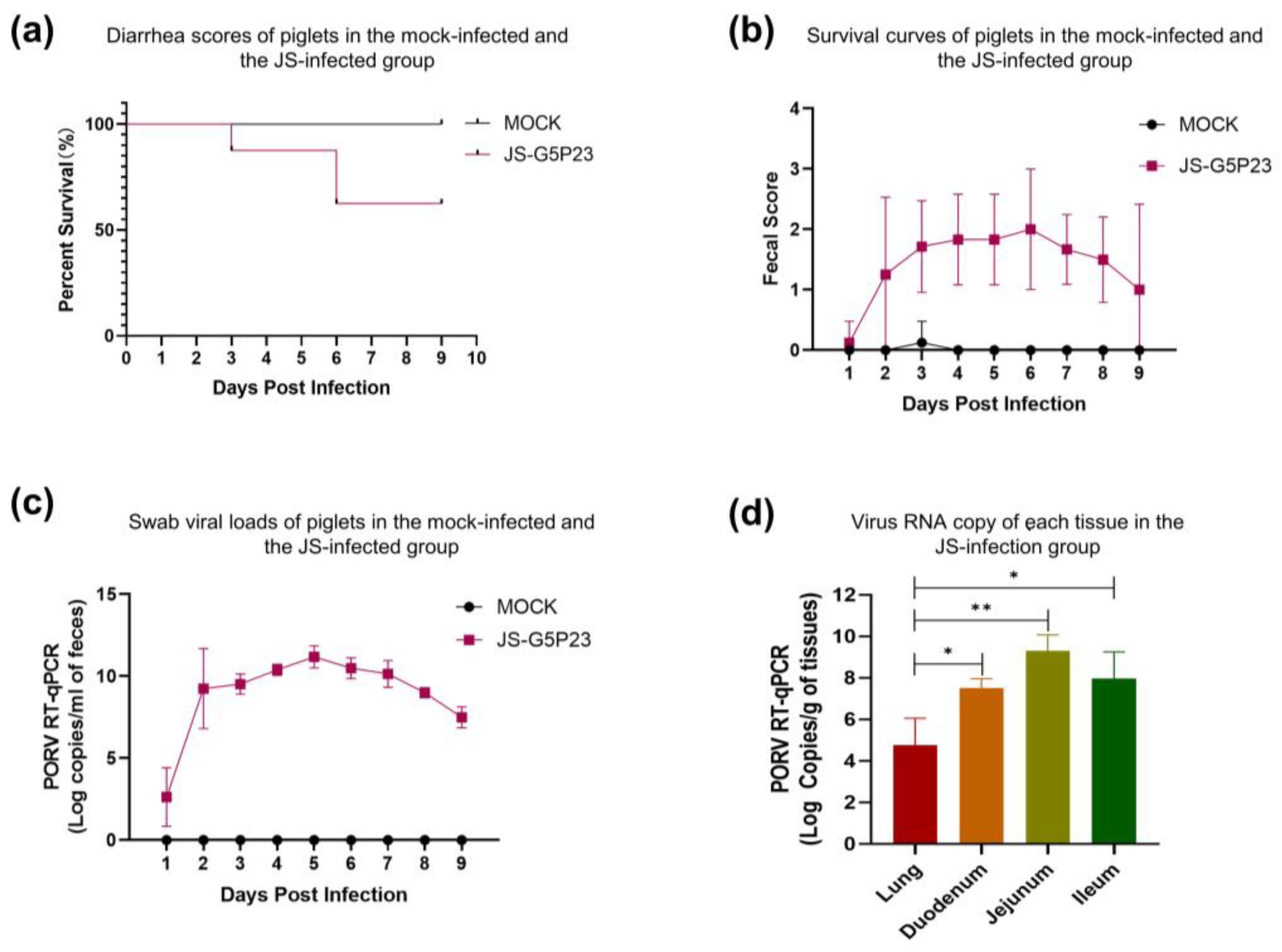
One piglet developed diarrhea on day 2 post-oral inoculation with the JS strain (
Figure 5a). By day 3, all eight piglets exhibited diarrhea, one of them succumbed to the infection on day 3, and two of them on day 6 (
Figure 5b). From day 7 onward, most piglets began to eat, which continued until day 9, when the symptoms of diarrhea began to subside. The piglets infected with JS had a diarrheal rate of 100% (8 of 8;
Figure 5a) and mortality rate of 37.5% (3 of 8;
Figure 5b). In contrast, the control group, apart from a brief period of loose stools in a few piglets, did not show any significant signs of diarrhea, and no deaths occurred (
Figure 5a-b).
Quantitative reverse transcription polymerase chain reaction (qRT–PCR) revealed that the viral RNA was detectable in the feces of infected piglets as early as 24 h postinfection. Viral shedding in rectal swabs of piglets continued to increase from day 1 to day 5 postinfection, reaching its peak on day 5 (
Figure 5c). From day 7 onward, viral shedding began to decline and decreased until the end of the experiment. Additionally, the viral load in the lungs was significantly lower than in various parts of the small intestine (P < 0.05), with the highest viral load detected in the ileum (
Figure 5d). No viral shedding was detected in the feces of the control group, and no viral load was identified in the intestinal tissues (
Figure 5c-d).
4. Discussion
Approximately 50 years ago, RVs were considered the primary cause of diarrhea in infants and young animals [
6]. According to the World Health Organization, RVs cause ~450,000 deaths annually, with 90% of them occurring in developing countries in Asia and Africa [
4,
32,
33]. PoRVs are prevalent worldwide and result in significant economic losses to the swine industry [
12]. Isolation and cell-based cultivation of RVs can help understand their pathogenicity in piglets [
34]. In this study, PoRV-positive samples were treated with 10 μg/mL trypsin for 1 h, and the optimal conditions for the isolation of PoRV were established by adding trypsin to a final concentration of 4 μg/mL in the maintenance medium. A PoRV strain was successfully isolated under these conditions and named JS. After blind passaging of MA104 cells for three generations, stable CPEs including cell elongation and detachment were observed (
Figure 1a), similar to previous studies [
21,
35]. IFA confirmed the replication of the 10th generation of the JS strain in MA104 cells (
Figure 1b). TEM revealed typical RV particles of 80–100 nm in diameter in the cell culture medium (
Figure 1c). The viral titer at the 10
th passage was ~1 × 10
6/mL TCID50 (
Figure 1d).
The complete genome sequence of JS was obtained using high-throughput sequencing. Analysis of genetic evolution revealed that the JS strain was a G5P[23] genotype RV. The genotype G5 PoRV is more common in pigs than in horses, cows, or other animals. It was identified in children in Brazil in the 1980s [
36,
37]. Although the G5 genotype RV has been reported in multiple countries, research on its pathogenicity is limited [
38]. ViPR-based analysis revealed that the genotype of JS strain is G5-P[23]-I5-R1-C1-M1-A8-N1-T1-E1-H1. The structural proteins VP3 and VP7 along with the nonstructural proteins NSP2 and NSP4 of the RVA/Pig/China/JS/2023/G5P[23] were closely related to human RVAs. However, the structural proteins VP1, VP2, VP4, and VP6 along with the nonstructural proteins NSP1, NSP3, and NSP5 were closely related to porcine RVAs. This suggests that JS strain is a recombinant strain of human and porcine RVAs (
Supplementary Table S2). The
NSP2 of the JS strain may be a product of recombination between human and dog strains (
Figure 3).
A previous study reported severe watery diarrhea in 3-day-old piglets within 24 h of infection with the HN03 (G9P[23]) strain and recovery after 72 h [
35]. Similarly, diarrhea was observed in 4-day-old piglets at 16–24 h after infection with the JS-01-20149 (G9P[7]) strain, followed by skin redness and death [
39]. In a previous study, diarrhea was reported in piglets infected with PRG942 (G9P[23]) and PRG9121 (G9P[7]) strains at 1–8 dpi [
40]. Furthermore, Miao et al. reported diarrhea in 1-day-old piglets infected with the CN127 strain after 6–24 h, which continued until euthanization at 48 h, but none of them died due to the infection [
34].
In this study, the pathogenicity of the JS strain was investigated by infecting 15-day-old piglets. Diarrhea and dark green, foamy feces were observed on day 2 postinfection. On day 3, one piglet succumbed to the infection (
Figure 4a). A postmortem examination of the piglets in the JS-infected group showed transparent, distended intestines filled with yellowish fluid and the accumulation of gastric and intestinal gases. RVA infections are not restricted to the intestines and further induce the formation of extraintestinal lesions in humans and animals [
41]. After dissection, the lungs of infected piglets also showed significant lesions, which manifested as congestion and swelling (
Figure 4a). HE revealed viral enteritis in all piglets with diarrhea, characterized by the accumulation of gastric and intestinal gases; vacuolization of small intestinal cells; and presence of shortened, fused, and sloughed villi in the small intestine (
Figure 4b). In contrast, the intestinal tissue pathology of piglets in the control group was normal (
Figure 4b). Additionally, IHC examination revealed dominant cytoplasmic staining of the PoRV antigen in certain segments of the small intestine with villous atrophy (
Figure 4c). However, no PoRV antigen was detected in the small intestines of piglets in the negative control group (
Figure 4c).
At the end of the experimental period, the diarrheal and mortality rates of infected piglets were 100% (8 of 8) and 38.5% (3 of 8), respectively. In contrast, piglets in the control group did not exhibit any apparent symptoms of diarrhea, and no deaths occurred (
Figure 5a-b). TaqMan real-time RT–PCR detected viral RNA shedding in the feces of infected piglets as early as 24 hpi. Viral shedding in the rectal swabs of piglets continued to increase from day 1 to day 5 postinfection but decreased from day 6 onward (
Figure 5c). The viral load in the lungs was significantly lower than in various parts of the small intestine, such as the duodenum, jejunum, and ileum (P < 0.05). The highest viral load was detected in the ileum, consistent with the IHC results (
Figure 5d). Throughout the entire experiment, no viral shedding was detected in the feces, and no viral load was identified in the intestinal tissues of the control group animals (
Figure 5c-d).
5. Conclusion
In summary, we successfully isolated and identified a strain of RVA with a G5P[23] genotype. This strain exhibited excellent adaptability to MA104 cells and demonstrated high virulence in piglets. These findings hold significant importance for understanding the characteristics of PoRV in China and developing novel and effective PoRV vaccines.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1, Table S1. Sequences of primers and probes used in this study; Table S2. Comparison of nucleotide and amino acid characteristics of RVA/Pig/China/JS/2023/G5P[23] strain with the closest strains in the GenBank database.
Author Contributions
Liguo Gao:Writing-review draft and validation. Hanqin Shen: Methodology and resources. Sucan Zhao: Writing-review, and editing. Sheng Chen: Methodology. Puduo Zhu: Writing-review, and editing. Wencheng Lin: Writing, review, and editing. Feng Chen: Methodology and supervision.
Funding
This study was supported by the Science and Technology Program of Yunfu City, Guangdong Province, China (2022020202).
Ethics approval and consent to participate
This study was approved by the Animal Care Committee of the South China Agricultural University (approval ID: SYXK-2019-0136). All study procedures and animal care activities were conducted per the recommendations of the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We thank Qingfeng Zhou for their assistance in the animal study.
Conflicts of Interest
The authors have no competing interests to declare.
Availability of data
GenBank accession numbers for the sample 14 RV sequences are OR644644–OR644654.
List of Abbreviations
BLASTn: Basic local alignment search tool for nucleotides; CPE: cytopathic effect; NCBI: National Center for Biotechnology Information; RCWG: Rotavirus Classification Working Group; NSP: nonstructural protein; PCR: polymerase chain reaction; qRT–PCR: quantitative reverse transcription polymerase chain reaction; RNA: ribonucleic acid; RVA: Group A rotavirus; VP: virion protein
References
- Anderson, E. J.; Weber, S. G. Rotavirus infection in adults. Lancet Infectious Diseases 2004, 4, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Rheingans, R. D.; Antil, L.; Dreibelbis, R.; Podewils, L. J.; Bresee, J. S.; Parashar, U. D. Economic Costs of Rotavirus Gastroenteritis and Cost-Effectiveness of Vaccination in Developing Countries. Journal of Infectious Diseases 2009, 200, S16–S27. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.; Bridger, J.; Kendall, K.; Gomara, M. I.; El-Attar, L.; Gray, J. The zoonotic potential of rotavirus. Journal of Infection 2004, 48, 289–302. [Google Scholar] [CrossRef]
- Martella, V.; Bányai, K.; Matthijnssens, J.; Buonavoglia, C.; Ciarlet, M. Zoonotic aspects of rotaviruses. Veterinary Microbiology 2010, 140, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Mebus, C. A.; Nr, U.; Rhodes, M. B.; Twiehaus, M. J. In Calf Diarrhea (Scours): Reproduced with a Virus from a Field Outbreak, 1969; 1969.
- Bishop, R. F.; Davidson, G. P.; Holmes, I. H.; Ruck, B. J. Virus particles in epithelial cells of duodenal mucosa from children with acute non-bacterial gastroenteritis. Lancet (London, England) 1973, 2, 1281–1283. [Google Scholar] [CrossRef]
- Chasey, D.; Bridger, J. C.; McCrae, M. A. A new type of atypical rotavirus in pigs. Archives of virology 1986, 89, 235–243. [Google Scholar] [CrossRef]
- McNulty, M. S.; Allan, G. M.; McFerran, J. B. Prevalence of antibody to conventional and atypical rotaviruses in chickens. The Veterinary record 1984, 114, 219. [Google Scholar] [CrossRef] [PubMed]
- Bohl, E. H.; Theil, K. W.; Saif, L. J. Isolation and serotyping of porcine rotaviruses and antigenic comparison with other rotaviruses. Journal of clinical microbiology 1984, 19, 105–111. [Google Scholar] [CrossRef]
- Welter, M. W.; Welter, C. J.; Chambers, D. M.; Svensson, L. Adaptation and serial passage of porcine group C rotavirus in ST-cells, an established diploid swine testicular cell line. Archives of virology 1991, 120, 297–304. [Google Scholar] [CrossRef]
- Esona, M. D.; Foytich, K.; Wang, Y. H.; Shin, G.; Wei, G.; Gentsch, J. R.; Glass, R. I.; Jiang, B. M. Molecular characterization of human rotavirus vaccine strain CDC-9 during sequential passages in Vero cells. Human Vaccines 2010, 6, 247–253. [Google Scholar] [CrossRef]
- Vlasova, A. N.; Amimo, J. O.; Saif, L. J. Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses-Basel 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Díaz, Y.; Chemello, M. E.; Peña, F.; Aristimuño, O. C.; Zambrano, J. L.; Rojas, H.; Bartoli, F.; Salazar, L.; Chwetzoff, S.; Sapin, C.; Trugnan, G.; Michelangeli, F.; Ruiz, M. C. Expression of Nonstructural Rotavirus Protein NSP4 Mimics Ca2+ Homeostasis Changes Induced by Rotavirus Infection in Cultured Cells. Journal of Virology 2008, 82, 11331–11343. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Otto, P. H.; Ciarlet, M.; Desselberger, U.; Van Ranst, M.; Johne, R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Archives of Virology 2012, 157, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, H. B.; Estes, M. K. Rotaviruses: From Pathogenesis to Vaccination. Gastroenterology 2009, 136, 1939–1951. [Google Scholar] [CrossRef]
- Maes, P.; Matthijnssens, J.; Rahman, M.; Van Ranst, M. RotaC: A web-based tool for the complete genome classification of group A rotaviruses. Bmc Microbiology 2009, 9. [Google Scholar] [CrossRef] [PubMed]
- Ren, X. L.; Saleem, W.; Haes, R.; Xie, J. X.; Theuns, S.; Nauwynck, H. J. Milk lactose protects against porcine group A rotavirus infection. Frontiers in Microbiology 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, J. N.; Yao, G.; Guo, Q. Y.; Wang, J. Q.; Liu, G. L. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Applied Microbiology and Biotechnology 2019, 103, 4943–4952. [Google Scholar] [CrossRef]
- Marthaler, D.; Homwong, N.; Rossow, K.; Culhane, M.; Goyal, S.; Collins, J.; Matthijnssens, J.; Ciarlet, M. Rapid detection and high occurrence of porcine rotavirus A, B, and C by RT-qPCR in diagnostic samples. Journal of Virological Methods 2014, 209, 30–34. [Google Scholar] [CrossRef]
- Zhu, J. H.; Rawal, G.; Aljets, E.; Yim-Im, W.; Yang, Y. L.; Huang, Y. W.; Krueger, K.; Gauger, P.; Main, R.; Zhang, J. Q. Development and Clinical Applications of a 5-Plex Real-Time RT-PCR for Swine Enteric Coronaviruses. Viruses-Basel, 2022; 14. [Google Scholar]
- Park, G. N.; Kim, D. I.; Choe, S.; Shin, J.; An, B. H.; Kim, K. S.; Hyun, B. H.; Lee, J. S.; An, D. J. Genetic Diversity of Porcine Group A Rotavirus Strains from Pigs in South Korea. Viruses-Basel, 2022; 14. [Google Scholar]
- REED, L. J.; MUENCH, H. A SIMPLE METHOD OF ESTIMATING FIFTY PER CENT ENDPOINTS12. American Journal of Epidemiology 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Martin, D. P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution 2015, 1. [Google Scholar] [CrossRef]
- Liu, X. S.; Zhang, Q. L.; Zhang, L. P.; Zhou, P.; Yang, J.; Fang, Y. Z.; Dong, Z. L.; Zhao, D. H.; Li, W. Y.; Feng, J. X.; Cui, B. F.; Zhang, Y. G.; Wang, Y. L. A newly isolated Chinese virulent genotype GIIb porcine epidemic diarrhea virus strain: Biological characteristics, pathogenicity and immune protective effects as an inactivated vaccine candidate. Virus Research 2019, 259, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Yodmeeklin, A.; Khamrin, P.; Chuchaona, W.; Saikruang, W.; Kongkaew, A.; Vachirachewin, R.; Kumthip, K.; Okitsu, S.; Ushijima, H.; Maneekarn, N. Great genetic diversity of rotaviruses detected in piglets with diarrhea in Thailand. Archives of Virology 2016, 161, 2843–2849. [Google Scholar] [CrossRef] [PubMed]
- Pickett, B. E.; Sadat, E. L.; Zhang, Y.; Noronha, J. M.; Squires, R. B.; Hunt, V.; Liu, M. Y.; Kumar, S.; Zaremba, S.; Gu, Z. P.; Zhou, L. W.; Larson, C. N.; Dietrich, J.; Klem, E. B.; Scheuermann, R. H. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Research 2012, 40, D593–D598. [Google Scholar] [CrossRef] [PubMed]
- Heiman, E. M.; McDonald, S. M.; Barro, M.; Taraporewala, Z. F.; Bar-Magen, T.; Patton, J. T. Group A Human Rotavirus Genomics: Evidence that Gene Constellations Are Influenced by Viral Protein Interactions. Journal of Virology 2008, 82, 11106–11116. [Google Scholar] [CrossRef] [PubMed]
- Liprandi, F.; Gerder, M.; Bastidas, Z.; López, J. A.; Pujol, F. H.; Ludert, J. E.; Joelsson, D. B.; Ciarlet, M. A novel type of VP4 carried by a porcine rotavirus strain. Virology 2003, 315, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Martin, D. P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus evolution 2015, 1, vev003. [Google Scholar] [CrossRef]
- Jeong, S.; Than, V. T.; Lim, I.; Kim, W. Whole-Genome Analysis of a Rare Human Korean G3P 9 Rotavirus Strain Suggests a Complex Evolutionary Origin Potentially Involving Reassortment Events between Feline and Bovine Rotaviruses (vol 9, e97127, 2014). Plos One 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Sieg, M.; Rückner, A.; Köhler, C.; Burgener, I.; Vahlenkamp, T. W. A bovine G8P 1 group A rotavirus isolated from an asymptomatically infected dog. Journal of General Virology 2015, 96, 106–114. [Google Scholar] [CrossRef]
- Tate, J. E.; Burton, A. H.; Boschi-Pinto, C.; Steele, A. D.; Duque, J.; Parashar, U. D.; S, W. H. O. C. G. R. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infectious Diseases 2012, 12, 136–141. [Google Scholar] [CrossRef]
- Parashar, U. D.; Nelson, E. A. S.; Kang, G. Diagnosis, management, and prevention of rotavirus gastroenteritis in children. Bmj-British Medical Journal 2013, 347. [Google Scholar] [CrossRef]
- Miao, Q.; Pan, Y. D.; Gong, L.; Guo, L. J.; Wu, L.; Jing, Z. Y.; Zhang, G. H.; Tian, J.; Feng, L. Full genome characterization of a human-porcine reassortment G12P 7 rotavirus and its pathogenicity in piglets. Transboundary and Emerging Diseases 2022, 69, 3506–3517. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. Y.; Lv, C. C.; Xu, X.; Li, X. D.; Yao, Y. L.; Gao, X. J.; Sun, Z.; Wang, Y. Z.; Sun, Y. J.; Xiao, Y.; Tian, K. G. The dynamics of a Chinese porcine G9P 23 rotavirus production in MA-104 cells and intestines of 3-day-old piglets. Journal of Veterinary Medical Science 2018, 80, 790–797. [Google Scholar] [CrossRef] [PubMed]
- da Silva, M. F. M.; Tort, L. F. L.; Goméz, M. M.; Assis, R. M. S.; Volotao, E. D.; de Mendonça, M. C. L.; Bello, G.; Leite, J. P. G. VP7 Gene of Human Rotavirus A Genotype G5: Phylogenetic Analysis Reveals the Existence of Three Different Lineages Worldwide. Journal of Medical Virology 2011, 83, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Gouvea, V.; de Castro, L.; Timenetsky, M. C.; Greenberg, H.; Santos, N. Rotavirus serotype G5 associated with diarrhea in Brazilian children. Journal of clinical microbiology 1994, 32, 1408–1409. [Google Scholar] [CrossRef] [PubMed]
- Papp, H.; László, B.; Jakab, F.; Ganesh, B.; De Grazia, S.; Matthijnssens, J.; Ciarlet, M.; Martella, V.; Bányai, K. Review of group A rotavirus strains reported in swine and cattle. Veterinary Microbiology 2013, 165, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. W.; Zhang, Z.; Wang, Y. F.; Wang, X.; Xia, M. Q.; Wu, H. Isolation, molecular characterization and evaluation of the pathogenicity of a porcine rotavirus isolated from Jiangsu Province, China. Archives of Virology 2015, 160, 1333–1338. [Google Scholar] [CrossRef] [PubMed]
- Kim, H. H.; Park, J. G.; Matthijnssens, J.; Kim, H. J.; Kwon, H. J.; Son, K. Y.; Ryu, E. H.; Kim, D. S.; Lee, W. S.; Kang, M. I.; Yang, D. K.; Lee, J. H.; Park, S. J.; Cho, K. O. Pathogenicity of porcine G9P 23 and G9P 7 rotaviruses in piglets. Veterinary Microbiology 2013, 166, 123–137. [Google Scholar] [CrossRef]
- Ciarlet, M.; Conner, M. E.; Finegold, M. J.; Estes, M. K. Group A rotavirus infection and age-dependent diarrheal disease in rats: A new animal model to study the pathophysiology of rotavirus infection. Journal of Virology 2002, 76, 41–57. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).